SUMMARY
A new era has begun for single particle cryo-electron microscopy (cryoEM) which can now compete with X-ray crystallography for determination of protein structures. The development of direct detectors constitutes a revolution that has led to a wave of near-atomic resolution cryoEM reconstructions. However, regardless of the sample studied, virtually all high-resolution reconstructions reported to date have been achieved using high-end microscopes. We demonstrate that the new generation of direct detectors coupled to a widely used mid-range electron microscope also enables obtaining cryoEM maps of sufficient quality for de novo modeling of protein structures of different sizes and symmetries. We provide an outline of the strategy used to achieve a 3.7 Å resolution reconstruction of Nudaurelia capensis ω virus and a 4.2 Å resolution reconstruction of the Thermoplasma acidophilum T20S proteasome.
Single-particle cryo-electron microscopy (cryoEM) is currently undergoing a revolution due to the recent development of a new generation of detectors using the complementary metal-oxide semiconductors technology (Kuhlbrandt, 2014). These cameras directly detect incoming electrons, without the need for a scintillator converting the electrons into photons, and are characterized by improved detective quantum efficiencies at all spatial frequencies compared to traditional charge-coupled device cameras or photographic films (Ruskin et al., 2013). The fast read-out rate of these devices also allows for recording movies composed of multiple frames during typical image exposure times enabling correction for beam-induced sample motion and stage drift to reduce image blurring (Brilot et al., 2012; Shigematsu and Sigworth, 2013).
Several algorithms have been developed for tracking beam-induced motion and stage drift in movies recorded with direct detectors (Bai et al., 2013; Campbell et al., 2012; Li et al., 2013b). These data processing strategies have subsequently been used to produce a myriad of high-resolution reconstructions of samples in the MDa mass range (Amunts et al., 2014), (Fernandez et al., 2014; Voorhees et al., 2014) (Wong et al., 2014) (Allegretti et al., 2014) as well as of protein complexes once considered too small for single-particle cryoEM (≤300 kDa) (Cao et al., 2013; Liao et al., 2013; Lu et al., 2014). These achievements constitute a tremendous leap forward for single-particle cryoEM. This method can now compete with X-ray crystallography for determination of protein structures at near-atomic resolution while also offering the unique advantage of enabling the characterization of heterogeneous or flexible protein complexes.
Virtually all high-resolution cryoEM reconstructions reported to date have been obtained using high-end microscopes operated at 300 kV, such as the FEI Titan Krios (Amunts et al., 2014; Fernandez et al., 2014; Lu et al., 2014; Voorhees et al., 2014; Wong et al., 2014; Zhang et al., 2013a; Zhang et al., 2013b), the FEI F30 Polara (Allegretti et al., 2014; Bai et al., 2013; Cao et al., 2013; Li et al., 2013a; Li et al., 2013b; Liao et al., 2013; Wolf et al., 2010; Zhang et al., 2008) or the JEOL JEM3200FSC (Baker et al., 2013; Cong et al., 2010). Pairing one of these microscopes with a direct detector clearly represents an ideal setup to study biological specimens at near-atomic resolution but the costs associated with the purchase and maintenance of this high-end equipment is a major undertaking for many universities and institutes worldwide. To follow-up on our previous studies aiming at characterizing the potential of mid-range electron microscopes when coupled to direct detectors (Campbell et al., 2012; Milazzo et al., 2011; Veesler et al., 2013), this contribution provides an assessment of the achievable resolution limits by single-particle cryoEM when using an FEI TF20 Twin electron microscope coupled to a Gatan K2 Summit camera. We demonstrate that it is possible to obtain cryoEM maps of sufficient quality for de novo modeling of protein structures of different sizes and symmetries when a widely used mid-range electron microscope is combined with a direct detector. This is illustrated by reconstructions at 3.7 Å and 4.2 Å resolution for Nudaurelia capensis ω virus (NωV, icosahedral symmetry) and the Thermoplasma acidophilum 20S proteasome (T20S, D7 symmetry), respectively.
Data collection and processing strategy
We operated our TF20 microscope using an extraction voltage of 4150 V, a gun lens setting of 3 and a spotsize of 6. Careful alignment of the microscope was performed before each data collection to ensure Thon rings were visible up to ~3 Å resolution in the power spectrum of micrographs collected over amorphous carbon using a dose inferior or equal to the one used for data acquisition. Coma-free alignment was carried out before and during each run to align the beam to the column optical axis with the assistance of a Zemlin tableau, as implemented in Leginon (Glaeser et al., 2011). Moreover, we used a C2 aperture size of 50 µm in combination with a beam width of ~1.2 µm at the specimen level to minimize off-axis coma within the imaging area (Glaeser et al., 2011). Fully automated data collection was carried out using Leginon (Suloway et al., 2005) to control both the FEI TF20 Twin microscope and the Gatan K2 Summit camera operated in counting mode at a dose rate of ~11 e−/pixel/s (Li et al., 2013b). Each movie was acquired over 5 sec and comprised 25 frames at a pixel size of 1.25 Å (29,000× magnification).
Whole frame alignment was carried out using the software developed by Li et al (Li et al., 2013b), which is integrated into the Appion pipeline (Lander et al., 2009), to account for stage drift and beam-induced motion. We used a frame offset of 2 or 7 along with a B factor of 150 or 1000 pixels2 for aligning the movie frames of the NωV and T20S datasets, respectively. Projection-matching refinements were performed with the Relion software (Scheres, 2012a; Scheres, 2012b) for the two datasets described in this manuscript. Reported resolutions are based on the gold-standard FSC=0.143 criterion (Scheres and Chen, 2012) and Fourier shell correction curves were corrected for the effects of soft masking by high-resolution noise substitution (Chen et al., 2013).
Structure of NωV at 3.7 Å resolution
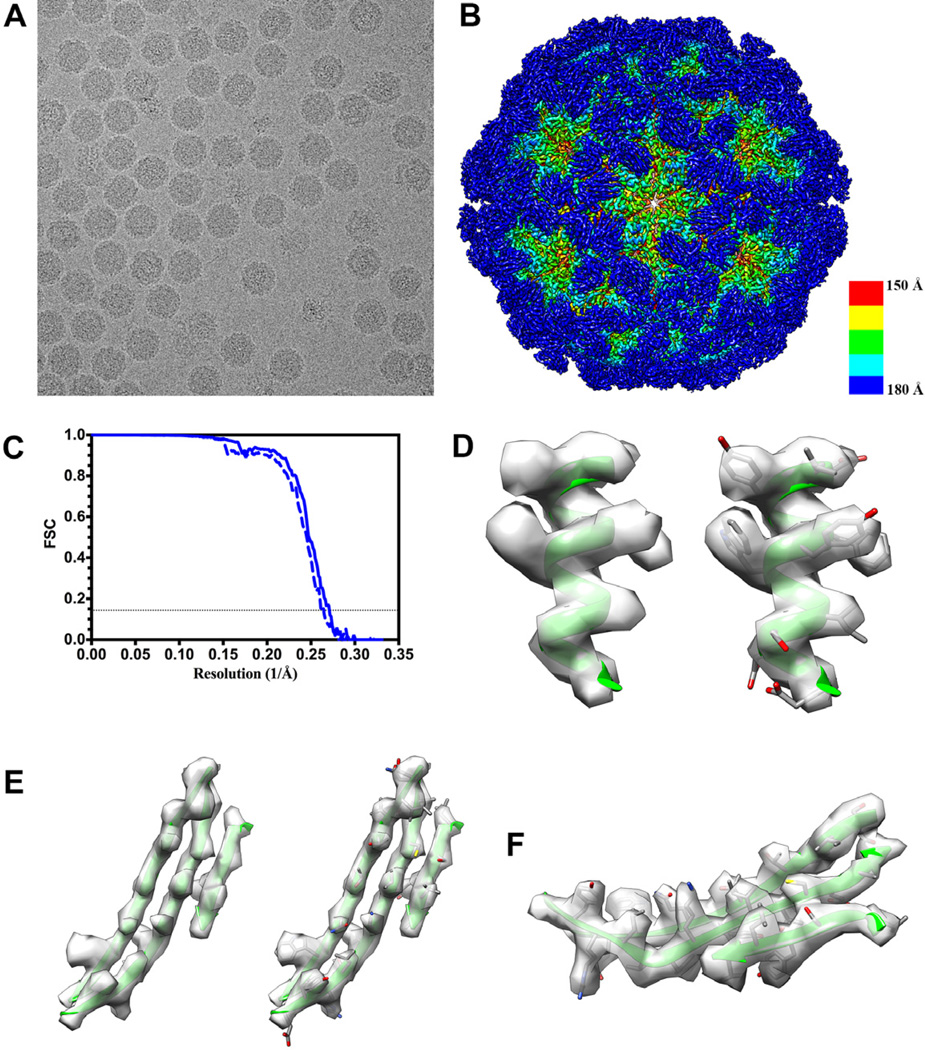
We collected 625 movies of frozen-hydrated mature NωV particles (incubated at pH5 for 24h to induce maturation, followed by an incubation at pH8 for 3.5 minutes immediately prior freezing) with a defocus in the range 1–3 µm and a total exposure of 38 e−/Å2 (Fig. 1 A), which corresponds to 1.5 e−/Å2/frame. We initially computed a 3D reconstruction using 14,884 particle images extracted from the motion-corrected 25-frame averages and the crystal structure of the NωV capsid low-pass filtered to 60 Å resolution as initial model (PDB 1OHF) (Helgstrand et al., 2004; Munshi et al., 1996). The resolution of the resulting map is 3.8 Å, as attested by the FSC at a cutoff of 0.143 and the map resolvability (Fig. 1 B and C). We then applied the statistical refinement procedure implemented in Relion to correct individual particle motions (rotations and translations) (Bai et al., 2013). We used a running average window of 3 frames along with standard deviations of the priors on the rotations and translations of 1° and 2 pixels, respectively. This procedure improved the resolution of the reconstruction to 3.7 Å resolution (Fig. 1 C) which is in agreement with the features observed in the map showing well-defined α-helices, fully resolved β-strands and many amino-acid side chains (Fig. 1 D–F).
Figure 1. NωV reconstruction at 3.7 Å resolution.
(A) A micrograph of ice-embedded NωV virions after movie frame alignment (defocus: 1.5 µm). (B) NωV reconstruction colored by radius. The color key is indicated at the bottom right of the map. (C) Gold-standard Fourier shell correlation curves indicate a resolution of 3.8 Å before (dashed line) and 3.7 Å after (solid line) accounting for beam-induced motion rotations and translations for each particle, respectively. (D) An α-helical segment from one NωV subunit is shown in ribbon representation, without (left) and with (right) side chain atoms, along with the corresponding region of the reconstruction. Most side chains are visible. (E) A β-sheet segment from one NωV subunit is shown in ribbon representation, without (left) and with (right) side chain atoms, along with the corresponding region of the reconstruction. (F) Lateral view of the β-sheet depicted in (D) showing that most side chains are resolved.
Structure of the T20S proteasome at 4.2 Å resolution
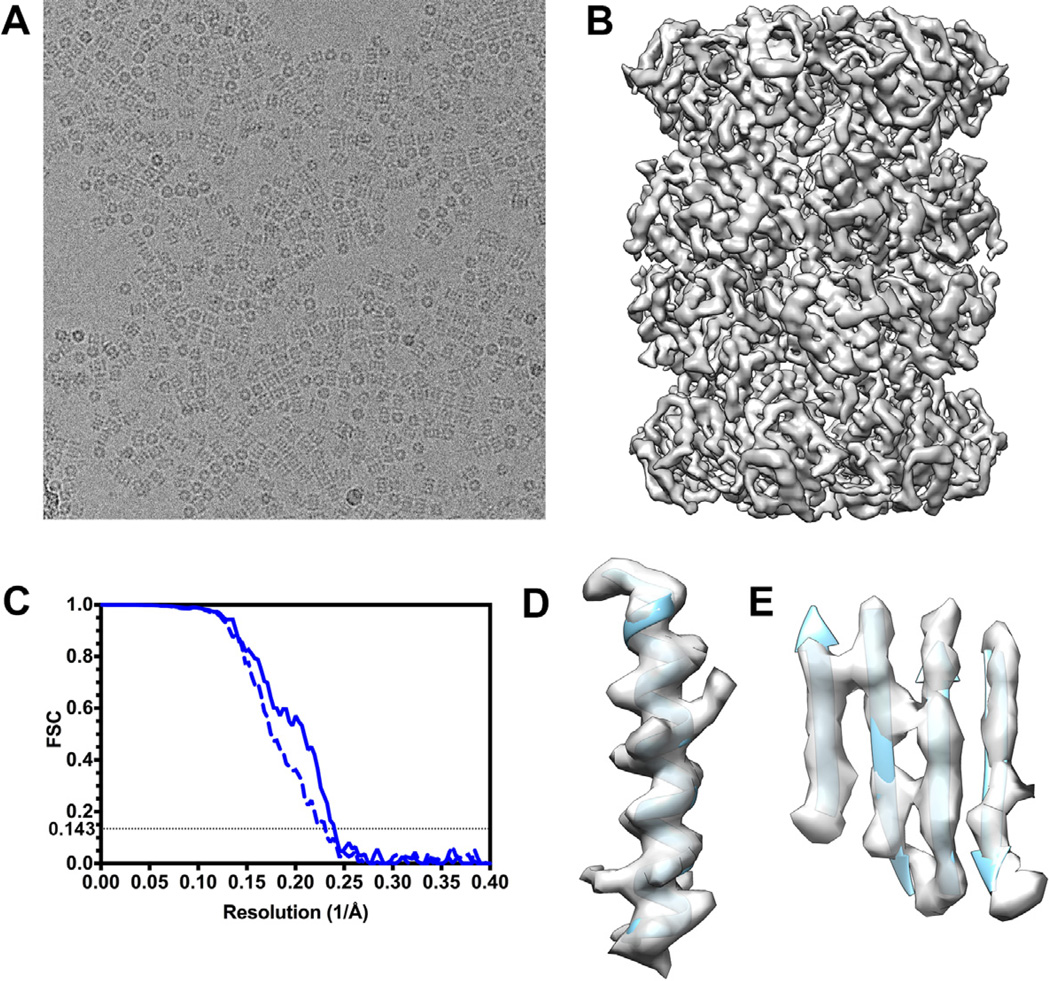
We collected 166 movies of frozen-hydrated T20S with a defocus in the range 0.75–3.3 µm and a total exposure of 38 e−/Å2 (Fig. 2 A and Table 1), which corresponds to 1.5 e−/Å2/frame. Particle images were sorted and selected using Xmipp Image sort by statistics (Scheres et al., 2008) and CL2D (Sorzano et al., 2010) retaining both side views and top views. We initially computed a 3D reconstruction with 21,818 particle images extracted from the motion-corrected 25-frame averages and a previous reconstruction of the same specimen low-pass filtered to 50 Å resolution as initial model (EMD-5623) (Li et al., 2013b). The resolution of the resulting reconstruction is 4.4 Å, as indicated by the FSC at a cutoff of 0.143 and the map quality (Fig. 2 B and C). We subsequently used the particle polishing procedure implemented in the version 1.3 of the Relion software to account for individual beam-induced particle translations and to calculate a frequency-dependent weight for the contribution of individual movie frames to the reconstruction (Scheres SHW, 2014). This processing scheme improved the resolution of the reconstruction to 4.2 Å, as attested by a significant enhancement of map quality. The final reconstruction features well-resolved α-helices and β-strands as well as visible density for some aromatic and bulky amino acid side chains in the T20S β subunits. The resolution of the reconstruction is more moderate in the α subunits especially in the most peripheral regions.
Figure 2. T20S proteasome reconstruction at 4.2 Å resolution.
(A) A micrograph of ice-embedded T20S after movie frame alignment (defocus: 2.1 µm). (B) T20S reconstruction. (C) Gold-standard Fourier shell correlation curves indicate a resolution of 4.36 Å before (dashed line) and 4.18 Å after (solid line) accounting for beam-induced translations for each particle, respectively. (C) An α-helical segment from one β subunit is shown in ribbon representation with the corresponding region of the reconstruction. Bulky side chains are visible. (D) A β-sheet segment from one β subunit is depicted in ribbon representation with the corresponding region of the reconstruction showing that individual β-strands are resolved.
Table 1.
Comparisons of the data acquisition conditions between the T20S dataset described in this manuscript and the one reported by Li et al (Li et al., 2013b).
| TF20 | Polara | |
|---|---|---|
| C2 aperture size (µm) | 50 | 30 |
| Size of area illuminated (µm) | ~1.2 | ~3 |
| Beam intensity (counts/pixel/sec) | 11.2 | 8 |
| Frame rate (frame per second) | 5 | 5 |
| Number of frames per movie | 25 | 25 |
| Total exposure (e−/Å2) | 38 | 35 |
| Number of micrographs | 166 | 553 |
| Total number of particles used in the final map | 21,818 | 126,729 |
| Defocus range (µm) | 0.725–3.3 (200kV) | 0.8–1.9 (300kV) |
| Final resolution (Å) | 4.2 | 3.3 |
Prospects for improving the resolution achievable using a TF20 microscope
We describe here the strategy used to achieve near-atomic resolution cryoEM reconstructions of two samples with different sizes and symmetries using an FEI Tecnai TF20 Twin microscope coupled with a Gatan K2 Summit camera operated in counting mode. The outcome of this study demonstrates that this setup enables obtaining maps of sufficient quality for de novo tracing of the protein backbone and of many amino acid side-chains. The possibility to determine protein structures at better than 4 Å resolution using the mid-range electron microscopes most frequently found in research laboratories world-wide expands the potential of single-particle cryoEM and structural biology in general.
We envision that several experimental parameters can be modified to further improve the resolution of the reconstructions obtained using the conditions described in this manuscript. Using a wider beam (e.g. 2–3 µm) in combination with a smaller C2 aperture (e.g. 30 µm) should provide improved illumination conditions and limit phase shifts resulting from off-axis coma (Glaeser et al., 2011). It has also been shown that data acquired with dose rates above 5 e-/pixel/s suffer from significant coincidence loss, resulting in deviation from the quantum efficiency of the camera (Li et al., 2013b). Our datasets were collected with a dose rate of ~11 e−/pixel/s which is estimated to result in undercounting ~25% of the incoming electrons. Hence, reducing the dose rate during data collection should enhance image quality provided that stage drift and beam-induced motion can still be corrected on images acquired with longer acquisition times. Finally, the two datasets used in this study were collected using the counting mode of the Gatan K2 Summit camera; acquiring data in super-resolution mode could further improve micrograph quality (Ruskin et al., 2013).
Data deposition
The reconstructions have been deposited to the Electron Microscopy Data Bank with ID EMD-2791 and EMD-2792 (T20S).
Acknowledgments
This work was supported by a FP7 Marie Curie IOF fellowship (273427) to D.V, an American Hearth Association predoctoral fellowship to M.G.C (14PRE18870036) and a NIH grant (R01GM054076) to J.E.J. Part of this research was conducted at the National Resource for Automated Molecular Microscopy which is supported by the NIH and the NIGMS (GM103310). We are grateful to Yifan Cheng and Kiyoshi Egami for kindly providing the T20S sample used in this study. We are also thankful to Tatiana Domitrovic for assistance in sample preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allegretti M, Mills DJ, McMullan G, Kuhlbrandt W, Vonck J. Atomic model of the F420-reducing [NiFe] hydrogenase by electron cryo-microscopy using a direct electron detector. Elife (Cambridge) 2014;3:e01963. doi: 10.7554/eLife.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A, Brown A, Bai XC, Llacer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SH, Ramakrishnan V. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Hryc CF, Zhang Q, Wu W, Jakana J, Haase-Pettingell C, Afonine PV, Adams PD, King JA, Jiang W, Chiu W. Validated near-atomic resolution structure of bacteriophage epsilon15 derived from cryo-EM and modeling. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:12301–12306. doi: 10.1073/pnas.1309947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilot AF, Chen JZ, Cheng A, Pan J, Harrison SC, Potter CS, Carragher B, Henderson R, Grigorieff N. Beam-induced motion of vitrified specimen on holey carbon film. Journal of structural biology. 2012;177:630–637. doi: 10.1016/j.jsb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MG, Cheng A, Brilot AF, Moeller A, Lyumkis D, Veesler D, Pan J, Harrison SC, Potter CS, Carragher B, Grigorieff N. Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure. 2012;20:1823–1828. doi: 10.1016/j.str.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504:113–118. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, McMullan G, Faruqi AR, Murshudov GN, Short JM, Scheres SH, Henderson R. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, Reissmann S, Kumar RN, Redding-Johanson AM, Batth TS, Mukhopadhyay A, Ludtke SJ, Frydman J, Chiu W. 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4967–4972. doi: 10.1073/pnas.0913774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez IS, Bai XC, Murshudov G, Scheres SH, Ramakrishnan V. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell. 2014;157:823–831. doi: 10.1016/j.cell.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaeser RM, Typke D, Tiemeijer PC, Pulokas J, Cheng A. Precise beam-tilt alignment and collimation are required to minimize the phase error associated with coma in high-resolution cryo-EM. Journal of structural biology. 2011;174:1–10. doi: 10.1016/j.jsb.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgstrand C, Munshi S, Johnson JE, Liljas L. The refined structure of Nudaurelia capensis omega virus reveals control elements for a T = 4 capsid maturation. Virology. 2004;318:192–203. doi: 10.1016/j.virol.2003.08.045. [DOI] [PubMed] [Google Scholar]
- Kuhlbrandt W. Biochemistry. The resolution revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, Lyumkis D, Potter CS, Carragher B. Appion: an integrated, database-driven pipeline to facilitate EM image processing. Journal of structural biology. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zheng SQ, Egami K, Agard DA, Cheng Y. Influence of electron dose rate on electron counting images recorded with the K2 camera. Journal of structural biology. 2013a;184:251–260. doi: 10.1016/j.jsb.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature methods. 2013b;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Bai XC, Ma D, Xie T, Yan C, Sun L, Yang G, Zhao Y, Zhou R, Scheres SH, Shi Y. Three-dimensional structure of human gamma-secretase. Nature. 2014 doi: 10.1038/nature13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milazzo AC, Cheng A, Moeller A, Lyumkis D, Jacovetty E, Polukas J, Ellisman MH, Xuong NH, Carragher B, Potter CS. Initial evaluation of a direct detection device detector for single particle cryo-electron microscopy. Journal of structural biology. 2011;176:404–408. doi: 10.1016/j.jsb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi S, Liljas L, Cavarelli J, Bomu W, McKinney B, Reddy V, Johnson JE. The 2.8 A structure of a T = 4 animal virus and its implications for membrane translocation of RNA. Journal of molecular biology. 1996;261:1–10. doi: 10.1006/jmbi.1996.0437. [DOI] [PubMed] [Google Scholar]
- Ruskin RS, Yu Z, Grigorieff N. Quantitative characterization of electron detectors for transmission electron microscopy. Journal of structural biology. 2013;184:385–393. doi: 10.1016/j.jsb.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. A Bayesian view on cryo-EM structure determination. Journal of molecular biology. 2012a;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology. 2012b;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nature methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Nunez-Ramirez R, Sorzano CO, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nature protocols. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. Elife. 2014 Aug 13;3:e03665. doi: 10.7554/eLife.03665. PMID: 25122622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu H, Sigworth FJ. Noise models and cryo-EM drift correction with a direct-electron camera. Ultramicroscopy. 2013;131:61–69. doi: 10.1016/j.ultramic.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorzano CO, Bilbao-Castro JR, Shkolnisky Y, Alcorlo M, Melero R, Caffarena-Fernandez G, Li M, Xu G, Marabini R, Carazo JM. A clustering approach to multireference alignment of single-particle projections in electron microscopy. Journal of structural biology. 2010;171:197–206. doi: 10.1016/j.jsb.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. Journal of structural biology. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Veesler D, Campbell MG, Cheng A, Fu CY, Murez Z, Johnson JE, Potter CS, Carragher B. Maximizing the potential of electron cryomicroscopy data collected using direct detectors. Journal of structural biology. 2013;184:193–202. doi: 10.1016/j.jsb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Fernandez IS, Scheres SH, Hegde RS. Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution. Cell. 2014;157:1632–1643. doi: 10.1016/j.cell.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M, Garcea RL, Grigorieff N, Harrison SC. Subunit interactions in bovine papillomavirus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:6298–6303. doi: 10.1073/pnas.0914604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W, Bai XC, Brown A, Fernandez IS, Hanssen E, Condron M, Tan YH, Baum J, Scheres SH. Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. Elife (Cambridge) 2014:e03080. doi: 10.7554/eLife.03080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ge P, Yu X, Brannan JM, Bi G, Zhang Q, Schein S, Zhou ZH. Cryo-EM structure of the mature dengue virus at 3.5-A resolution. Nature structural & molecular biology. 2013a;20:105–110. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guo H, Jin L, Czornyj E, Hodes A, Hui WH, Nieh AW, Miller JF, Zhou ZH. A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 A resolution. Elife (Cambridge) 2013b;2:e01299. doi: 10.7554/eLife.01299. [DOI] [PMC free article] [PubMed] [Google Scholar]