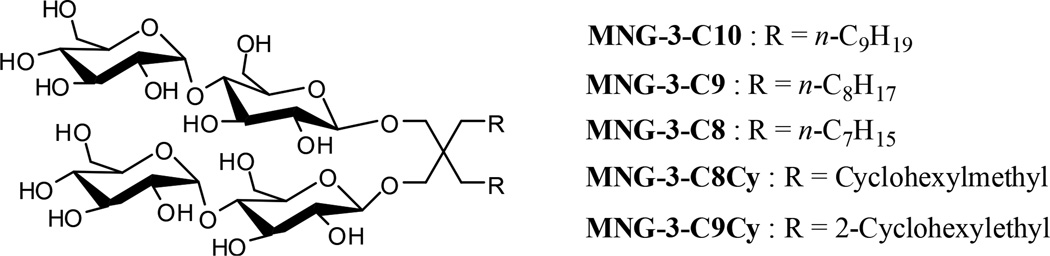Abstract
Detergents are typically used to both extract membrane proteins (MPs) from the lipid bilayer and maintain them in solution. However, MPs encapsulated in detergent micelles are often prone to denaturation and aggregation. Thus, development of novel agents with enhanced stabilization characteristics is necessary to advance MP research. Maltose neopentyl glycol-3 (MNG-3) has contributed to >10 crystal structures including G-protein coupled receptors. Here we prepared MNG-3 analogues and characterised their properties using selected MPs. Most MNGs behaved superior to a conventional detergent, n–dodecyl–β–D–maltopyranoside (DDM), in terms of membrane protein stabilization efficacy. Interestingly, optimal stabilization was achieved with different MNG-3 analogues depending on the target MP. The origin for such detergent specificity could be explained by a novel concept: compatibility between detergent hydrophobicity and MP tendency to denature and aggregate. This set of MNGs represents viable alternatives to currently available detergents for handling MPs, and can be also used as tools to estimate MP sensitivity to denaturation and aggregation.
Introduction
Membrane proteins account for 20–30% of the human proteome1 and are the targets for 50% of current drug molecules.2 Since the first crystal structure of the bacterial photosynthetic reaction center was solved almost 30 years ago,3 hundreds of membrane protein (MP) structures have become available.4 These structures have made an invaluable contribution to the understanding of the mechanism of action of these important molecules and provided templates for rational drug design. However, the available MP structures correspond to only ~ 1% of those available for soluble proteins, illustrating the difficulty of MP structure determination which is mainly attributed to the low stability of the MPs in aqueous solutions.5 MPs are inserted into the lipid bilayers surrounding cells and organelles and the lipid environment has an essential role in maintaining the structural and functional integrity of MPs. Some lipid molecules form specific interactions with the MPs, thereby stabilizing the proteins.6 In addition, due to their cylindrical molecular geometry, the lipids self-assemble into a bilayer which provides a lateral pressure to the MPs.7 However, these large membrane assemblies are not compatible with the current methods used in MP structure determination such as X–ray crystallography and nuclear magnetic resonance (NMR) spectroscopy. Therefore, an essential requirement for structural studies is that MPs are extracted from the membranes and maintained structurally and functionally intact in an aqueous solution.
Amphipathic molecules termed detergents are typically used to extract and solubilize membrane proteins from the native membranes.8 Above a certain concentration, these molecules self-assemble into micelles with a globular or oval shape. These nano-assemblies have the ability to encapsulate the hydrophobic segment of membrane proteins effectively replacing the lipid bilayer and producing protein-detergent complexes (PDCs) ideally containing structurally and functionally intact MP. MPs tend to be significantly less stable in detergent micelles than in the native membrane since encapsulation into the micelles can cause loss of associated lipids and result in temporal exposure of hydrophobic regions of the protein normally buried in the lipidic environment. Together with the limited strength of lateral pressure associated with the micelle compared to the membrane, these factors make the solubilized MPs much more prone to unfolding and non-specific aggregation.9 Conventional detergents such as n–octyl–β–D–glucopyranoside (OG), n–dodecyl–β–D–maltopyranoside (DDM) and lauryldimethylamine–N–oxide (LDAO) are commonly used for membrane protein study. However, membrane protein stability even in these popular agents is often unsatisfactory. Thus it is of great importance to develop novel agents which combine enhanced membrane protein stabilization with the ability to effectively extract membrane proteins from the membrane environment.
A number of novel amphiphiles have been invented to overcome the limitations of conventional detergents.10 Most of these novel agents are small amphipathic molecules.11 Good examples include tripod amphiphiles (TPAs) with three hydrophobic groups,11a–d hemifluorinated surfactants (HFSs) containing a fluorinated alkyl chain,11e facial amphiphiles (FAs) derived from cholic/deoxycholic acid,11f,g rigid hydrophobic group-bearing amphiphiles (chobimalt and glyco-diosgenin (GDN)) with a multi-fused ring in the lipophilic portion,11h,i glucose or maltose-neopentyl glycols (GNGs and MNGs) with branched diglucoside or maltoside head group,11j–m and calixarene-based ionic surfactants.11n The secondary peptide structures have proved popular as scaffolds for novel amphiphile development.12 The α-helix forming peptides such as peptitergent12a and lipopeptide detergents (LPDs)12b are the most well-known designs. However, the utility of these agents is limited because they are challenging to synthesize. Short peptides composed of several amino acid residues were shown to possess high stabilization efficacy using different MP systems.12c In addition, very recently, a β sheet-forming peptide exhibited promising results for MP stabilization.12d More complex systems than small amphipathic molecules and peptides were invented by innovative approaches, including amphipols with a polyacrylic acid backbone13a,b and nano-assemblies such as nanodiscs (NDs)13c and nanolipodisq.13d Although these membrane mimetic systems were observed to be very effective at membrane protein stabilization, these agents, similar to peptide-based amphiphiles, have as yet not contributed to high resolution MP structure determination. In contrast, some small amphiphilic molecules have garnered significant attention as they have been shown to both confer markedly greater stabilization on a range of MPs and have a proven track record for crystallization. The most outstanding examples in this regard are the GNGs and MNGs. GNG-3 (commercial name: OGNG) was used in the determination of the Na+–pumping pyrophosphatase and human aquaporin 2 (AQZ 2).14a,b MNG-3 (commercial name: LMNG)11j has facilitated the high resolution structure determination of more than 10 G–protein coupled receptors such as the β2 adrenergic receptor (β2AR), opioid receptors, muscarinic acetylcholine receptors and the neurotensin receptor15a–j in addition to the Twin Arginine Translocator, N–methyl–D–aspartate (NMDA) receptor ion channel and Claudin–15 tight junction.15k–m Furthermore, this agent is shown to confer stability on membrane protein complexes such as the β2ARβGs complex and β2ARβarrestin–1 complex.15n,o The reports of these structures clearly demonstrate the superior properties of the GNGs and MNGs with respect to MP stabilization and crystallization. It is expected that MNG-3 and GNG-3 will continue to make a valuable contribution to MP structural studies. However, there is no single amphiphile suitable for all MPs. Thus, the aim of this study was to generate a range of derivatives of MNG-3 and assess their properties with different MP systems. We found that all the MNG-3 derivatives conferred similar or improved MP stability compared to DDM, but the MNG with the best stabilization properties was dependent on the target protein. The origin for this protein dependent detergent efficacy will be discussed in terms of compatibility between detergent hydrophobicity and MP characteristics.
Results and discussion
The new MNGs share the branched dimaltoside headgroup used for the original compound, MNG-3-C10, but have variations in the hydrophobic group. As indicated by the nomenclatures, MNG-3-C10 has C10 alkyl chains while C9 and C8 alkyl chains were introduced to create MNG-3-C9 and MNG-3-C8, respectively (Scheme 1). In contrast, MNG-3-C8Cy and MNG-3-C9Cy contain a cyclohexyl ring at each alkyl chain tip like the commercially available cyclohexyl group-bearing glucosides (Cyglus) and maltosides (Cymals). These cyclohexyl ring-bearing MNGs (MNG-3-C8Cy and MNG-3-C9Cy) are the same as MNG-3-C8 and MNG-3-C9, respectively, in terms of the number of carbon units in their hydrophobic chains. These new agents were devised based on the fact that similar conventional counterparts are available and popularly used for membrane protein research. For instance, for maltoside class detergents, n–decyl–β–D–maltopyranoside (DM), n–undecyl–β–D–maltopyranoside (UDM), DDM and Cymals with C10, C11, C12 and cyclohexyl groups, respectively, are all known to be useful for MP structural determination. All of these new agents were prepared via a convenient synthetic protocol, giving overall synthetic yields of more than 80% (see supporting information for details).
Scheme 1.
Chemical structures of a previously reported MNG-3, designated MNG-3-C10, and new MNG-3 analogues (MNG-3-C9, MNG-3-C8, MNG-3-C8Cy and MNG-3-C9Cy). The new compounds share a branched dimaltoside headgroup with MNG-3-C10, but have variations in the hydrophobic groups.
All the new MNG analogues were highly water-soluble (> 10 wt %). We also prepared the C11 chain version of MNG-3 (MNG-3-C11), which turned out to have limited solubility in water (< 2 wt%) and thus this compound was not further studied. The self-aggregation tendencies of MNG-3-C10 and its hydrophobic analogues were investigated by measuring the critical micelle concentration (CMC). A CMC value for each agent was obtained using a fluorescent probe, diphenylhexatriene (DPH).16 Micelles formed by MNG-3s were characterized in terms of their sizes by determining the hydrodynamic radii (Rh) via dynamic light scattering (DLS). The data for the different detergents are summarized in Table 1. As expected, the CMC values of the new agents decreased with increasing alkyl chain length of the hydrophobic groups; each additional -CH2- unit at the end of both alkyl chains reduced the CMC value approximately by half. Conversely, introduction of a cyclohexyl ring into the lipophilic groups increased CMC values by ~ three times. For example, the CMC value of MNG-3-C9Cy is three times higher than for MNG-3-C9 although both have C9 alkyl chains. A similar trend was found in the CMC values of MNG-3-C8Cy and MNG-3-C8 with C8 alkyl chains. These comparatively high CMC values for the cyclohexane-bearing MNGs are likely due to the bulkiness of the cyclohexane ring relative to the straight alkyl chain.17 A detergent micelle has a very congested interior because many hydrophobic groups pack together in a small space. The inclusion of a large group such as the cyclohexane ring into the micelle interior is sterically unfavorable, thereby decreasing the tendency for micelle formation. Note that all MNG agents tend to form micelles at lower concentration than DDM, indicative of a comparatively strong tendency for self-association. Micelles formed by the MNGs were significantly varied in terms of size, depending on the chain length of the hydrophobic groups. Thus, MNG-3-C8Cy with the shortest alkyl chains formed the smallest micelles while MNG-3-C10 with the longest alkyl chains generated the largest micelles. Interestingly, MNG-3-C10 micelles appeared to be substantially larger than those of the other MNG analogues (Figure S1); the Rh of micelles formed by this agent was more than twice that of micelles formed by DDM. Thus, in terms of micellar volume, MNG-3-C10 is eight times larger than DDM. In contrast, MNG-3-C9 with the alkyl chains shorter than MNG-3-C10 by one carbon unit formed even smaller micelles compared to DDM. The large difference in Rh between MNG-3-C10 and MNG-3-C9, despite a small difference in their alkyl chain lengths, is somewhat surprising. However, a similar trend was found in a previous GNG study.11m Because of the presence of two alkyl chains, a small increase in the alkyl chain length of GNGs or MNGs could induce a substantial change in the molecular geometry from a cone to a cylindrical shape, thus resulting in a significant increase in the micelle size.
Table 1.
Critical micelle concentrations (CMCs) and hydrodynamic radii (Rh) for MNG agents (MNG-3-C10, MNG-3-C9, MNG-3-C8, MNG-3-C8Cy and MNG-3-C9Cy) and a conventional detergent (DDM).
| Amphiphile | MWa | CMC (mM) | CMC (wt %) | Rh (nm)b |
|---|---|---|---|---|
| MNG-3-C10 | 1005.2 | ~0.010 | ~0.0010 | 7.2 ± 0.01 |
| MNG-3-C9 | 977.1 | ~0.018 | ~0.0018 | 3.1 ± 0.01 |
| MNG-3-C8 | 949.1 | ~0.036 | ~0.0034 | 2.7 ± 0.05 |
| MNG-3-C8Cy | 945.1 | ~0.15 | ~0.014 | 2.5 ± 0.06 |
| MNG-3-C9Cy | 973.1 | ~0.058 | ~0.0056 | 2.8 ± 0.04 |
| DDM | 510.1 | 0.17 | 0.0087 | 3.5 ± 0.04 |
Molecular weight of detergents.
Hydrodynamic radius of micelles was determined at 0.5 wt % by dynamic light scattering.
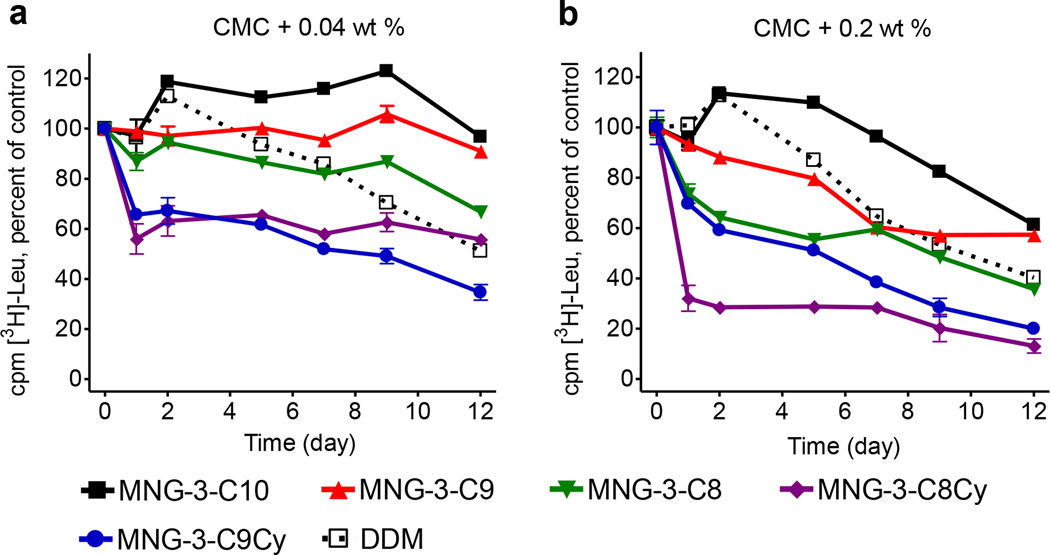
The properties of the new MNG analogues were first evaluated using bacterial wild type leucine transporter (LeuT) from Aquifex aeolicus.18 This transporter was initially extracted from the E. coli membranes with 1.0 wt% DDM and purified in 0.05 wt % of the same detergent. DDM-solubilized LeuT was subsequently diluted into solutions containing individual MNGs or DDM. The final concentration of each test detergent (MNG or DDM) was CMC+0.04 wt% or CMC+0.2 wt%. We monitored protein activity assessing radiolabeled leucine binding by scintillation proximity assay (SPA)19a,b at regular intervals during a 12-day incubation at room temperature. Consistent with our previous study,11j MNG-3 was superior to DDM at both tested detergent concentrations (Figure 1). Out of four new MNG agents, only MNG-3-C9 was found to be better than DDM for the long-term stabilization of the LeuT. MNG-3-C8 was superior to DDM only at the lower concentration, CMC + 0.04 wt%. Both cyclohexane–bearing MNG agents (MNG-3-C9Cy and MNG-3-C8Cy) were inferior to both DDM and the other straight chain MNGs (i.e., MNG-3-C9 and MNG-3-C8) in preserving protein activity. Interestingly, detergent efficacy order for the LeuT stabilization is inversely proportional to the CMC values of the test MNGs. Thus, MNG-3-C10, detergent with the lowest CMC value, was found to be best while MNG-3-C8Cy, detergent with the highest CMC value, was least effective at preserving the activity of this protein. Given that detergent CMC values generally decrease with detergent alkyl chain length (i.e., detergent hydrophobicity), the correlation between detergent CMC value and detergent stabilization efficacy observed for the LeuT indicates that the native structure of this transporter could be effectively maintained by a detergent with high hydrophobicity.
Figure 1.
Long-term activity of LeuT solubilized in test MNG amphiphiles (MNG-3-C10, MNG-3-C9, MNG-3-C8, MNG-3-C8Cy and MNG-3-C9Cy) or DDM at (a) CMC + 0.04 wt% and (b) CMC + 0.2 wt%. DDM-solublized LeuT was incubated with individual compounds at room temperature and the protein activity was estimated based on [3H]-Leu binding using scintillation proximity assay (SPA) over the course of 12 days. Results are expressed as % activity relative to protein activity at day 0 (mean ± s.e.m., n = 2).
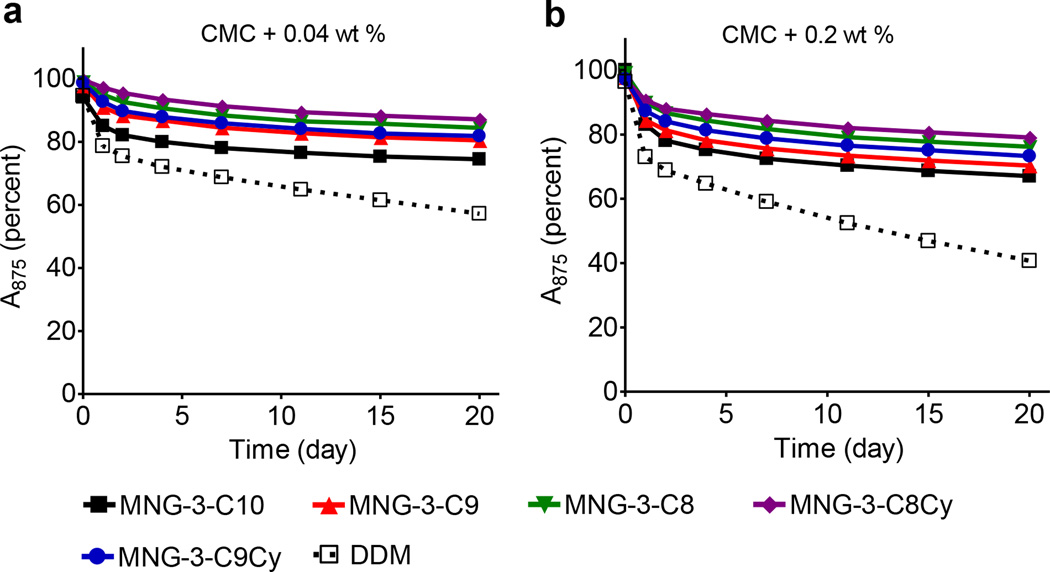
Next, we evaluated the effects of MNG agents on R. capsulatus superassembly stability. This complex is comprised of light harvesting complex I (LHI) together with the reaction center (RC), which contains dozens of individual protein subunits.20 It is known that maintaining the native structure of this complex is challenging. Even the use of a mild detergent such as DDM and DM destroys this complex over time.11b Due to the presence of multiple cofactors, intact superassembly exhibits strong absorbance at 875 nm, a feature utilized to assess protein integrity. For the evaluation of test MNG-3 analogues with this complex, we initially solubilized LHI-RC complex from the membrane with 1.0 wt% DDM and purified in 0.0087 wt% of the same detergent. The DDM-purified LHI-RC complex was subsequently diluted into individual MNG-containing solutions to give the final detergent concentrations of CMC+0.04 wt% or CMC+0.2 wt%. We monitored protein integrity by measuring the absorbance of protein samples at 875 nm during a 20-day incubation at room temperature (Figure 2). All MNG agents were better than DDM. The detergent stabilization efficacy order for this complex was dependent on detergent alkyl chain length/hydrophobicity; MNG-3-C8Cy and MNG-3with the smallest carbon unit (C8) in the alkyl chains was found to be most effective whereas MNG-3-C10 with the longest carbon unit (C10) displayed the worst effect.
Figure 2.
Time course stability of R. capsulatus superassembly encapsulated in MNGs (MNG-3-C10, MNG-3-C9, MNG-3-C8, MNG-3-C8Cy and MNG-3-C9Cy) or DDM at (a) CMC+0.04 wt% and (b) CMC+0.2 wt%. The superassembly was initially purified in DDM at 1xCMC and then diluted with solutions including individual test detergents. Protein integrity was monitored by measuring absorbance at 875 nm over the course of 20 days at room temperature. All spectra were measured between 650 nm and 950 nm.
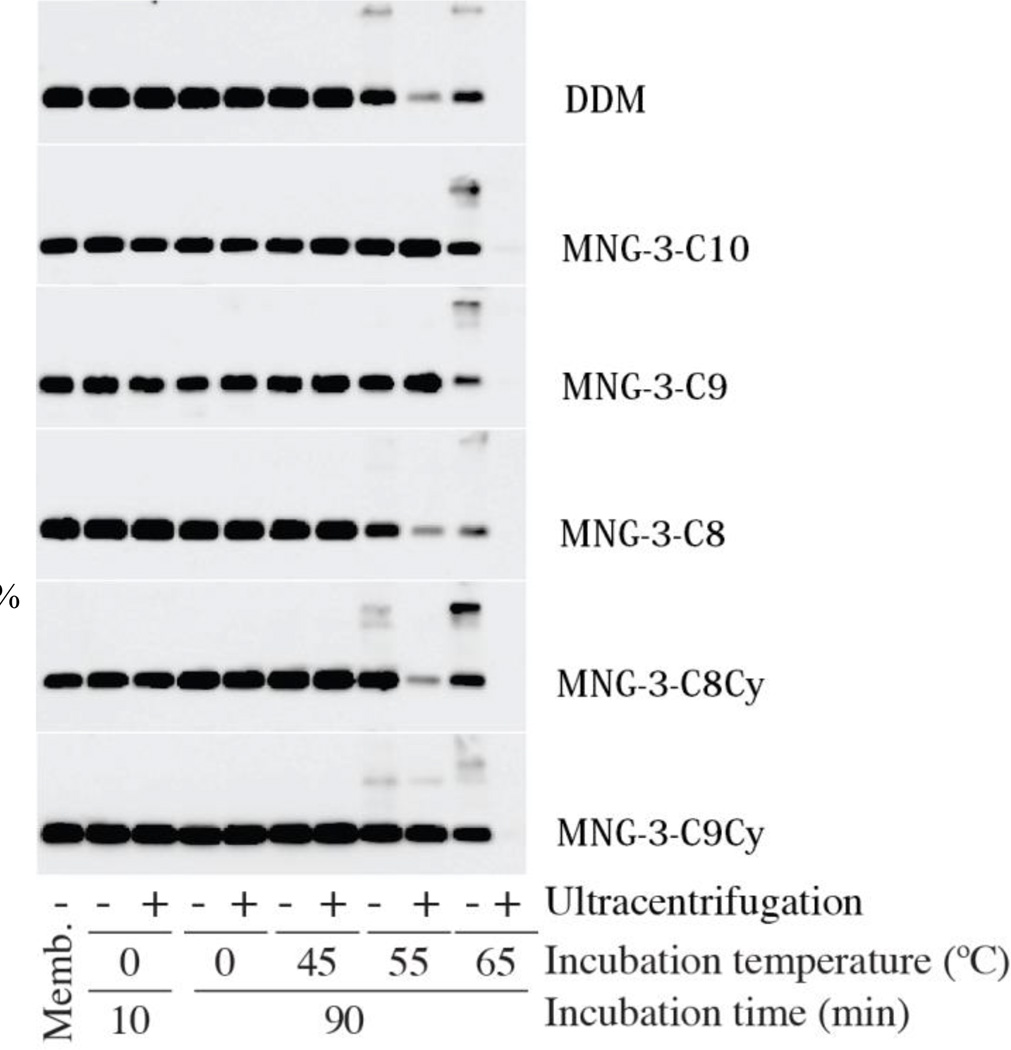
The last MP we selected for the MNG-3 analogues characterization was the Na+-coupled sugar symporter, the melibiose permease from Salmonella typhimurium (MelBSt).21a,b For extraction of this protein from the E. coli membranes, 1.5 wt% DDM or MNGs were used at 0 °C for 10 min. The amounts of MelBSt solubilized by each detergent treatment were analyzed using Western blotting after ultracentrifugation. As can be seen in Figure 3, similar to DDM, all test MNGs showed quantitative protein solubilization under experimental condition. For assessment of protein stabilization efficacy, individual MNG/DDM-solubilized proteins were subsequently incubated at elevated temperatures (45 °C, 55 °C or 65 °C) for 90 min. In this thermostability experiment, DDM gave a high yield of soluble MelB only at 45 °C, but at 55 °C or higher, little soluble protein was obtained, indicating that majority of the proteins was pelleted during ultracentrifugation due to denaturation/aggregation. Similar results were obtained for the MNG-3-C8 and MNG-3-C8Cy with the lowest carbon unit (C8) in the alkyl chains. In contrast, the original MNG (MNG-3-C10) and the other MNGs (i.e., MNG-3-C9 and MNG-3-C9Cy) maintain complete protein solubilization at 55 °C. No protein was detectable after incubation at 65°C for any of the test conditions. Of the new agents, MNG-3-C9Cy gave the most comparable results with the original MNG-3-C10. Note that these two MNG agents (i.e., MNG-3-C10 and MNG-3-C9Cy) are the most hydrophobic test detergents. Detergent efficacy order for the MelBSt was similar to that obtained for the LeuT; MNG-3-C10 had the best properties of the straight alkyl chain-containing MNGs (the first set), followed by MNG-3-C9 and MNG-3-C8, and MNG-3-C9Cy was more effective than MNG-3-C8Cy of the cyclohexane-containing MNGs (the second set). Also note that, in contrast with the results for LeuT, there is little difference in detergent stabilization efficacy between these two sets of test MNG agents for this protein. The large and small difference in the stabilization efficacy between the two sets observed for LeuT and MelBSt, respectively, may be due in part to variations in assay temperature (25 °C vs. 55 °C).
Figure 3.
SDS-PAGE and Western blotting analysis of MelBSt. The same amount of the membrane containing MelBSt was treated with each detergent for the designated time period (10 min or 90 min). After detergent exposure, samples were analyzed by SDS-16%PAGE and the amount of solubilized protein was estimated using Western blotting with anti-Histidine tag antibody. To differentiate detergent efficacy, incubation temperature varied from 45 °C to 55 °C to 65 °C. Protein aggregation was estimated by analyzing the samples before (−) and after (+) ultracentrifugation. An untreated membrane sample (“Memb”) was included as a control.
Since it is known that detergent stabilization efficacy is MP-specific, a detergent displaying the best behavior is generally different for each MP.8c,d A similar variation was found in this study. The original compound (MNG-3-C10) was superior to the other MNG agents in the stabilization of LeuT and MelBSt proteins whereas MNG-3-C8 and MNG-3-C8Cy were more promising than the other test MNGs for the LHI-RC complex. This protein specificity is likely to mainly result from variation in MP properties. Some proteins with large hydrophobic and small hydrophilic domains are typically much more prone to aggregation. In contrast, MP complexes with multiple quaternary structures are much more likely to suffer from subunit dissociation, leading to denaturation. Thus, detergent stabilization efficacy would be dependent on protein sensitivity to aggregation or loss of tertiary/quaternary structure (i.e., denaturation), the two main causes of membrane protein degradation in aqueous solutions.
It would be important to discuss in detail the specific characteristics of the MNGs with respect to the individual protein preference. To achieve the best stabilization efficacy, detergent characteristics should be in harmony with the tendency of MPs to degrade. Of most importance is the detergent hydrophobicity since this property dictates the binding strength of a detergent for the hydrophobic segment of MPs. Such association strength between detergent molecules and MPs will impact protein stability either favorably or unfavorably, depending on protein propensity for aggregation and denaturation. Specifically, a strongly binding detergent could promote protein denaturation because it energetically favors protein-detergent interactions over interactions amongst protein subunits or protein secondary structures. However, the same phenomenon could also reduce protein aggregation, particularly for more hydrophobic proteins. Conversely, a weakly binding detergent could limit loss of subunits from a complex but could result in higher levels of aggregation via preferential nonspecific association of proteins. Thus, detergent hydrophobicity has different effects on protein stability depending on the sensitivity of the target protein to aggregation and denaturation. Based on this correlation between detergent hydrophobicity and MP characteristics, we could select a set of detergent candidates of the many available; weakly-binding detergents would be the first choice for denaturation-prone MPs while strongly-binding detergents would be more appropriate for aggregation-sensitive MPs. Thus, depending on the propensity of the membrane protein to aggregate and/or denature, a different type of detergent could be used to facilitate MP study; trial and error approaches involving a large number of detergents could be significantly avoided by this approach.
It would be valuable to know whether a target MP is aggregation-prone or denaturation-prone. This information could be obtained by experimental estimation of the relative amount of aggregated and denatured MPs. However, this approach may not be fully relevant because protein denaturation and aggregation are likely to be closely associated. For instance, protein aggregation could be accelerated upon protein denaturation because a large hydrophobic surface buried inside the interior of the native conformation becomes exposed to the external environment. In this circumstance, a large amount of aggregated proteins will be apparent although protein denaturation is the initial event for degradation. Conversely, protein aggregation can induce protein denaturation as well. The set of MNG-3 agents presented here could play a role in this regard. The straight alkyl chain MNGs, MNG-3-C10, MNG-3-C9 and MNG-3-C8, exhibit increased hydrophobicity with increasing alkyl chain length. As discussed above, such MNG hydrophobicity would determine detergent efficacy order for protein stabilization depending on MP propensity for aggregation and denaturation. Thus, the following conclusion from our current results can be reached. The stability of LeuT and MelBSt increased with detergent hydrophobicity (MNG-3-C10>MNG-3-C9>MNG-3-C8), suggesting that these MPs undergo structural degradation mainly via protein aggregation rather than protein denaturation. In contrast, the other MP, LHI-RC complex, was most stable in the least hydrophobic agent (MNG-3-C8), followed by more hydrophobic MNGs (MNG-3-C9 and MNG-3-C10), and can therefore be regarded as denaturation-sensitive. This MP classification is further supported by the data obtained for the cyclohexane-bearing MNGs, MNG-3-C8Cy and MNG-3-C9Cy; LeuT and MelB were favorably stabilized by the more hydrophobic MNG-3-C9Cy while LHI-RC complex was more stable when solubilized in the less hydrophobic MNG-3-C8Cy. This preliminary assessment for the relative sensitivity of target MPs to aggregation or denaturation could be particularly valuable because currently there is no simple way to access such information as described above. Thus, these MNGs may be useful for MP characterization. Note that it is likely there are a number of MPs having comparable sensitivity to both protein denaturation and aggregation. In this case, it would not be possible to determine the most suitable detergent for MP stabilization based on detergent hydrophobicity.
It is very difficult to obtain clear evidences on whether the target MPs investigated here are denaturation- or aggregation-sensitive but the 3D structural information and/or oligomeric states of those proteins could provide some clues on this topic. For example, the crystal structures of LeuT and MelB indicate that major parts of these proteins are embedded inside the lipid bilayer with the large hydrophobic surface area relative to the hydrophilic parts exposed to an aqueous medium.21b,22 This information strongly suggests that both LeuT and MelB could be aggregation-prone, which may be further supported by the fact that these MPs crystallize as individual dimeric forms. On the other hand, LHI-RC complex from R. capsulatus comprises 30–40 subunits having multiple tertiary and quaternary structures along with substantially large hydrophilic surface area. This protein was isolated as a monomeric complex23 and use of strong detergents such as LDAO, Triton X-100, and Fos-Choline-12 led to structural degradation during extraction from the membranes,11b,24 which could be a symptom of denaturation-sensitivity. Although these pieces of information are not conclusive, they include an implication on protein propensity to denature and aggregate for each membrane protein. The results inferred from this information are consistent with our current estimation on membrane protein propensity by the use of MNGs.
Conclusions
The synthetic variants of MNG-3 showed superior behaviors toward the stabilization of selected MPs as compared to DDM, the most widely used detergent for membrane protein research.9c Despite a large number of amphiphile studies, the origin for the variation in detergent stabilization efficacy for a specific target MP has rarely been discussed in detail. Here we have taken the first step to providing a clue for this fundamental question based on the correlation between detergent hydrophobicity and protein propensity for aggregation and denaturation observed in the present study. Furthermore, we have proposed a convenient method to qualitatively characterize MP propensity for aggregation and denaturation by utilizing the current MNG set. Therefore, the MNG agents are not only more favorable than conventional detergents in terms of membrane protein stabilization, but could also be useful as a resource for protein characterization in terms of their tendency for aggregation and denaturation. This mutual benefit in both detergent efficacy evaluation and target MP characterization is conceptually new and will undoubtedly help advance MP research.
Supplementary Material
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) funded by the Korean government (MSIP) (grant number 2008-0061891 and 2013R1A2A2A03067623 to P.S.C., K.H.J, H.J.L and J.Y.G), Biotechnology and Biological Sciences Research Council grant BB/K017292/1 to B.B., the National Science Foundation (grant MCB-1158085), the Norman Hackerman Advanced Research Program (grant 010674-0034-2009), and the National Institutes of Health (grant R01 GM095538) to L.G., the Danish Council for Independent Research – Sapere Aude, The Lundbeck Foundation and the UNIK Center for Synthetic Biology to C.J.L.s.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
Contributor Information
Jin Woong Kim, Email: KJWoong@hanyang.ac.kr.
Claus J. Loland, Email: cllo@sund.ku.dk.
Lan Guan, Email: lan.guan@ttuhsc.edu.
Bernadette Byrne, Email: b.byrne@imperial.ac.uk.
Pil Seok Chae, Email: pchae@hanyang.ac.kr.
Notes and references
- 1.Fagerberg L, Jonasson K, Von Heijne G, Uhlen M, Berglund L. Proteomics. 2010;10:1141–1149. doi: 10.1002/pmic.200900258. [DOI] [PubMed] [Google Scholar]
- 2.Overington JP, Al-Lazikani B, Hopkins AL. Nat. Rev. Drug Discovery. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 3.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. Nature. 1985;318:618–624. doi: 10.1038/318618a0. [DOI] [PubMed] [Google Scholar]
- 4. http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html. [Google Scholar]
- 5.a) White SH, Wimley WC. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]; b) Bowie JU. Curr. Opin. Struct. Biol. 2001;11:397–402. doi: 10.1016/s0959-440x(00)00223-2. [DOI] [PubMed] [Google Scholar]; c) Lacapere JJ, Pebay-Peyroula E, Neumann JM, Etchebest C. Trends Biochem. Sci. 2007;32:259–270. doi: 10.1016/j.tibs.2007.04.001. [DOI] [PubMed] [Google Scholar]; d) Loll PJ. Acta Cryst. 2014;F70:1576–1583. doi: 10.1107/S2053230X14025035. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Landreh M, Robinson CV. J. Physiol. 2015;593:355–362. doi: 10.1113/jphysiol.2014.283150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marsh D. Biochem. Biophys. Acta. 2008;1778:1545–1575. doi: 10.1016/j.bbamem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Anglin TC, Conboy JC. Biophys. J. 2008;95:186–193. doi: 10.1529/biophysj.107.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Møller JV, le Maire M. J. Biol. Chem. 1993;268:18659–18672. [PubMed] [Google Scholar]; b) Privé GG. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]; c) Caffrey M, Li D, Dukkipati A. Biochemistry. 2012;51:6266–6288. doi: 10.1021/bi300010w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Proc. Natl. Acad. Sci. U. S. A. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Newstead S, Ferrandon S, Iwata S. Protein Sci. 2008;17:466–472. doi: 10.1110/ps.073263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Q, Tao H, Hong W-X. Methods. 2011;55:318–323. doi: 10.1016/j.ymeth.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) McQuade DT, Quinn MA, Yu SM, Polans AS, Krebs MP, Gellman SH. Angew. Chem. Int. Ed. 2000;39:758–761. [PubMed] [Google Scholar]; b) Chae PS, Wander MJ, Bowling AP, Laible PD, Gellman SH. ChemBioChem. 2008;9:1706–1709. doi: 10.1002/cbic.200800169. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chae PS, Kruse AC, Gotfryd K, Rana RR, Cho KH, Rasmussen SGF, Bae HE, Chandra R, Gether U, Guan L, Kobilka BK, Loland CJ, Byrne B, Gellman SH. Chem. Eur. J. 2013;19:15645–15651. doi: 10.1002/chem.201301423. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Chae PS, Cho KH, Wander MJ, Bae HE, Gellman SH, Labile PD. Biochim. Biophys. Acta. 2014;1838:278–286. doi: 10.1016/j.bbamem.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Breyton C, Chabaud E, Chaudier Y, Pucci B, Popot J-L. FEBS Lett. 2004;564:312–318. doi: 10.1016/S0014-5793(04)00227-3. [DOI] [PubMed] [Google Scholar]; f) Chae PS, Gotfryd K, Pacyna J, Miercke LJW, Rasmussen SGF, Robbins RA, Rana RR, Loland CJ, Kobilka B, Stroud R, Byrne B, Gether U, Gellman SH. J. Am. Chem. Soc. 2010;132:16750–16752. doi: 10.1021/ja1072959. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Lee SC, Bennett BC, Hong W-X, Fu Y, Baker KA, Marcoux J, Robinson CV, Ward AB, Halpert JR, Stevens RC, Stout CD, Yeager MJ, Zhang Q. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E1203–E1211. doi: 10.1073/pnas.1221442110. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Howell SC, Mittal R, Huang L, Travis B, Breyer RM, Sanders CR. Biochemistry. 2010;49:9572–9583. doi: 10.1021/bi101334j. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Chae PS, Rasmussen SGF, Rana RR, Gotfryd K, Kruse AC, Nurva S, Gether U, Guan L, Loland CJ, Byrne B, Kobilka BK, Gellman SH. Chem. Eur. J. 2012;18:9485–9490. doi: 10.1002/chem.201200069. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Chae PS, Rasmussen SGF, Rana RR, Gotfryd K, Chandra R, Goren MA, Kruse AC, Nurva S, Loland CJ, Pierre Y, Drew D, Popot J-L, Picot D, Fox BG, Guan L, Gether U, Byrne B, Kobilka B, Gellman SH. Nat. Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]; k) Cho KH, Byrne B, Chae PS. ChemBioChem. 2013;14:452–455. doi: 10.1002/cbic.201200759. [DOI] [PubMed] [Google Scholar]; l) Chae PS, Rana RR, Gotfryd K, Rasmussen SGF, Kruse AC, Cho KH, Capaldi S, Carlsson E, Kobilka BK, Loland CJ, Gether U, Banerjee S, Byrne B, Lee JK, Gellman SH. Chem. Commun. 2013;49:2287–2289. doi: 10.1039/c2cc36844g. [DOI] [PMC free article] [PubMed] [Google Scholar]; m) Cho KH, Bae HE, Das M, Gellman SH, Chae PS. Chem. Asian J. 2014;9:632–638. doi: 10.1002/asia.201301303. [DOI] [PubMed] [Google Scholar]; n) Matar-Merheb R, Rhimi M, Leydier A, Huche F, Galian C, Desuzinges-Mandon E, Ficheux D, Flot D, Aghajari N, Kahn R, Di Pietro A, Jault JM, Coleman AW, Falson P. PLoS One. 2011;6:e18036–e18036. doi: 10.1371/journal.pone.0018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Schafmeister CE, Meircke LJW, Stroud RM. Science. 1993;262:734–738. doi: 10.1126/science.8235592. [DOI] [PubMed] [Google Scholar]; b) McGregor C-L, Chen L, Pomroy NC, Hwang P, Go S, Chakrabartty A, Privé GG. Nat. Biotechnol. 2003;21:171–176. doi: 10.1038/nbt776. [DOI] [PubMed] [Google Scholar]; c) Zhao X, Nagai Y, Reeves PJ, Kiley P, Khorana HG, Zhang S. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17707–17712. doi: 10.1073/pnas.0607167103. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Tao H, Lee SC, Moeller A, Roy RS, Siu FY, Zimmermann J, Stevens RC, Potter CS, Carragher B, Zhang Q. Nat. Methods. 2013;10:759–761. doi: 10.1038/nmeth.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Tribet C, Audebert R, Popot J-L. Proc. Natl. Acad. Sci. U. S. A. 1996;93:15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Popot J-L, et al. Annu. Rev. Biophys. 2011;40:379–408. doi: 10.1146/annurev-biophys-042910-155219. [DOI] [PubMed] [Google Scholar]; c) Nath A, Atkins WM, Sligar SG. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]; d) Orwick-Rydmark M, Lovett JE, Graziadei A, Lindholm L, Hicks MR, Watts A. Nano Lett. 2012;12:4687–4692. doi: 10.1021/nl3020395. [DOI] [PubMed] [Google Scholar]
- 14.a) Kellosalo J, Kajander T, Kogan K, Pokharel K, Goldman A. Science. 2012;337:473–476. doi: 10.1126/science.1222505. [DOI] [PubMed] [Google Scholar]; b) Frick A, Eriksson UK, de Mattia F, Oberg F, Hedfalk K, Neutze R, Grip WJ, Deen PMT, Tornroth-Horsefield S. Proc. Natl. Acad. Sci. U. S. A. 2014;111:6305–6310. doi: 10.1073/pnas.1321406111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Rasmussen SGF, Choi H-J, Fung JJ, Pardon E, Casarosa P, Chae PS, DeVree BT, Rosenbaum DM, Thian FS, kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rosenbaum DM, Zhang C, Lyons J, Holl R, Aragao D, Arlow DH, Rasmussen SGF, Choi H-J, DeVree BT, Sunahara RK, Chae PS, Gellman SH, Dror RO, Shaw DE, Weis WI, Caffrey M, Gmeiner P, Kobilka BK. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, Rosemond E, Green HF, Liu T, Chae PS, Dror RO, Shaw DE, Weis WI, Wess J, Kobilka BK. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Haga K, Kruse AC, Asada H, Kobayashi TY, Shiroishi M, Zhang C, Weis WI, Okada T, Kobilka BK, Haga T, Kobayashi T. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Granier S, Manglik A, Kruse AC, Kobilka TS, Thian FS, Weis WI, Kobilka BK. Nature. 2012;485:400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) White JF, Noinaj N, Shibata Y, Love J, Kloss B, Xu F, Gvozdenovic-Jeremic J, Shah P, Shiloach J, Tate CG, Grisshammer R. Nature. 2012;490:508–513. doi: 10.1038/nature11558. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Kruse AC, Ring AM, Manglik A, Hu J, Hu K, Eitel K, Hubner H, Pardon E, Valant C, Sexton PM, Christopoulos A, Felder CC, Gmeiner P, Steyaert J, Weis WI, Garcia KC, Wess J, Kobilka BK. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Ring AM, Manglik A, Kruse AC, Enos MD, Weis WI, Garcia KC, Kobilka BK. Nature. 2013;502:575–579. doi: 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Miller PS, Aricescu AR. Nature. 2014;512:270–275. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]; k) Rollauer SE, Tarry MJ, Graham JE, Jaaskelainen M, Jager F, Johnson S, Krehenbrink M, Liu SM, Lukey MJ, Marcoux J, McDowell MA, Rodriguez F, Roversi P, Stansfeld PJ, Robinson CV, Sansom MS, Palmer T, Hcgbom M, Berks BC, Lea SM. Nature. 2012;492:210–214. doi: 10.1038/nature11683. [DOI] [PMC free article] [PubMed] [Google Scholar]; l) Karakas E, Furukawa H. Science. 2014;344:992–997. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]; m) Suzuki H, Nishizawa T, Tani K, Yamazaki Y, Tamura A, Ishitani R, Dohmae N, Tsukita S, Nureki O, Fujiyoshi Y. Science. 2014;344:304–307. doi: 10.1126/science.1248571. [DOI] [PubMed] [Google Scholar]; n) Rasmussen SGF, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah STA, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]; o) Shukla AK, et al. Nature. 2014;512:218–222. doi: 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chattopadhyay A, London E. Anal. Biochem. 1984;139:408–412. doi: 10.1016/0003-2697(84)90026-5. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Reeve J, Wang Y, Thomas RK, Wang J, Yan H, Phys J. Chem. B. 2005;109:16070–16074. doi: 10.1021/jp0523874. [DOI] [PubMed] [Google Scholar]
- 18.Deckert G, et al. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 19.a) Hart HE, Greenwald EB. Mol. Immunol. 1979;16:265–267. doi: 10.1016/0161-5890(79)90065-8. [DOI] [PubMed] [Google Scholar]; b) Quick M, Javitch JA. Proc. Natl. Acad. Sci., USA. 2007;104:3603–3608. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laible PD, Kirmaier C, Udawatte CS, Hofman SJ, Holten D, Hanson DK. Biochemistry. 2003;42:1718–1730. doi: 10.1021/bi026959b. [DOI] [PubMed] [Google Scholar]
- 21.a) Guan L, Nurva S, Ankeshwarapu SP. J. Biol. Chem. 2011;286:6367–6374. doi: 10.1074/jbc.M110.206227. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ethayathulla AS, Yousef MS, Amin A, Leblanc G, Kaback HR, Guan L. Nat. Commun. 2014;5:3009. doi: 10.1038/ncomms4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 23.Crouch LI, Jones MR. Biochem. Biophys. Acta. 2012;1817:336–352. doi: 10.1016/j.bbabio.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Chae PS, Wander MJ, Cho KH, Laible PD, Gellman SH. Mol. BioSyst. 2013;9:626–629. doi: 10.1039/c3mb25584k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.