Abstract
Odorant Receptor (OR) genes and proteins represent more than 2% of our genome and 4% of our proteome 25 and constitute the largest sub-group of G Protein-Coupled Receptors (GPCRs). The mechanism underlying OR activation remains poorly understood, as they do not share some of the highly conserved motifs critical for activation of non-olfactory GPCRs. By combining site-directed mutagenesis, heterologous expression, and molecular dynamics simulations that capture the conformational change of constitutively active mutants, we tentatively identified crucial residues for the function of these receptors using the mouse MOR256-3 (Olfr124) as a model. The toggle-switch for sensing agonists in-volves a highly conserved tyrosine residue in helix VI. The ionic-lock is located between the `DRY' motif in helix III and a positively charged `R/K' residue in helix VI. This study provides an unprecedented model that captures the main mechanisms of odorant receptor activation.
Keywords: Olfactory receptor, GPCR, heterologous expression, site-directed mutagenesis, molecular modeling
INTRODUCTION
The strategy used by mammals to sense odorant molecules is a combinatorial code based on the differential activation of a large family of Odorant Receptors (ORs).1 One of the major functions of these receptors is to transmit external signals from the environment (odorant molecules) to the nervous system. Furthermore, these proteins are also expressed in non-olfactory tissues, high-lighting their role beyond odor detection and potential as drug targets.2 The OR genes and proteins represent more than 2% of our genome and 4% of our proteome.3 ORs are seven trans-membrane (TM) helix proteins constituting the largest sub-group of the class-A G protein-coupled receptor (GPCR) family.
GPCRs play critical roles in cellular signal transduction. The initial activation relies on conformational switches between inactive and active states, which depend on both the nature of the receptor and the eventually bound ligand.4 Upon agonist binding, the receptor switches from an inactive to an active form that couples with the intra-cellular G protein to trigger signal transduction. Inspired from experimental structural data of some GPCRs in active and inactive states, Molecular Dynamics (MD) simulations have been adopted to reveal the atomic-level steps involved in GPCR activation. These models have successfully recapitulated activation of the β2-adrenergic, rhodopsin, muscarinic, and A2A receptors,4c,5 suggesting that this tool is well suited to decipher OR activation. From a mechanistic perspective, GPCR activation is notably associated with the opening of a cleft between the intracellular Parts of transmembrane domains 3 and 6 (TM3 and TM6)6. Several motifs are shown to be important for their activation, e.g., the conserved DRY motif in TM3, the CWxP motif in TM6, and the NPxxY motif in TM7.4d
ORs have a low sequence identity with other class-A GPCRs. They nonetheless show the same highly conserved motifs within most TM domains, suggesting a conserved general mechanism for their function.7 ORs also contain some specific motifs, considered as hallmarks for their identification, such as MAYDRYVAICxPLxY in TM3 or KAFSTCxSH in TM68. However, the CWxP motif that plays the role of toggle-switch for GPCR activation is lacking in ORs. Also, although the DRY motif remains highly conserved, the negatively charged residue (E/D in non-olfactory GPCRs) of TM6 facing the DRY motif and involved in the ionic lock between TM3 and TM6 is to date unidentified in ORs. No crystallographic structure of an OR is available and most mechanistic studies rely on the use of molecular modeling.7a
When combined with site-directed mutagenesis data, molecular modeling has led to identification of some specific residues for ligand binding. To date, most studies have focused on the binding cavity, which is consistently made up of residues within TM3, TM5, TM6 and TM7.9 In some ORs, the location of a copper ion as co-factor for detection of sulfur compounds involves residues belonging to the canonical binding site. This supports a conserved activation mechanism, while the metal would only play a role in ligand affinity,9f,10 although it does not completely rule out that some ORs function as metalloproteins.11 The ion-odorant complex might be detected as a single ligand, whose presence may be sensed through a similar mechanism as for all ORs.
Clues about residues potentially involved in the OR activation mechanism were tentatively proposed but they still remain to be assessed by means of in vitro experiments and long scale MD simulations.12 Residues involved in the dynamic process that converts an inactive OR structure into an active one are still elusive. This article is a step forward in their identification. The mouse receptor MOR256-3 (also named 0lfr124 or SR1), a broadly-tuned receptor,13 is the focus of the current study by a joint approach that combines molecular modeling, site-directed mutagenesis and heterologous expression. The MOR256-3 sequence contains the hallmarks of mammalian ORs, with typical highly conserved motifs in all TMs (Figure S1 and Table S1). Their conservations were assessed by a thorough sequence analysis on 396 human and 1111 mouse ORs. We provide a body of evidence for the functional role of several of these motifs within OR sequence. Based on an experimental observation of mutant ORs with either increased basal activity (ligand-independent receptor activation) or locked into a constitutively active state, we have built a structural model that captures this active state while the wild-type (wt) OR stays in an inactive form. This offers the opportunity to decipher the strategy used by ORs to detect agonists without being subjected to the difficult task of finding the accurate position of the ligand within the binding site. We show that in ORs, the highly conserved Y residue of the FYG motif in TM6 acts as the toggle-switch. Also, the ionic-lock involves the D and R residues of the DRY motif in TM3 and a conserved positive residue in TM6. This is the first report of a homology OR model that evolves between active and inactive states.
RESULTS
Residues within the binding cavity control ligand specificity and basal activity
To gain insights into the OR activation mechanism, a 3D atomic computational model of MOR256-3 was built by homology modeling using an alignment and a multi-template approach shown to be consistent with experimental affinity data on OR-ligand pairs.7a, 14 Most amino acid residues we identified as belonging to the binding site (made up of residues from the upper parts towards the extracellular side) of the helices of TM3, TM5, TM6 and TM7) are consistent with previous studies (ref. 15 and references therein). Residues 1043.32, 1083.36, 2035.43, and 2526.48 are notably pointing into the binding site, as shown in Figure 1, where the superscript numbers are the Ballesteros-Weinstein notation in the alignment (Figures S1 and S2). These residues have been identified in ligand recognition in many studied ORs so far, including copper-mediated ORs.9C–f
Figure 1.
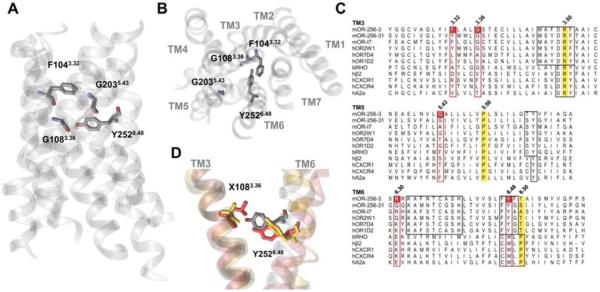
Binding cavity residues that interact with agonists according to the model. (A) & (B) Front view and top view, respectively, of MOR256-3 highlighting selected residues belonging to the binding cavity. (C) Alignments highlighting equivalent roles of certain residues within TM3, TM5 and TM6 in olfactory and non-olfactory GPCRs. The Ballesteros-Weinstein notation is shown for each TM and residues corresponding to the reference position (x.50, with x the TM number) are highlighted in yellow in the alignment. Conserved motifs in ORs and GPCRs are boxed in black. Boxed in red are the residues corresponding to those mutated in this study. (D) The position of Y2526.48 shifts as a function of residue X1083.36; wt (wild type, G108) is shown in gray, X108= A (G108A) in yellow and X108=L (G108L) in red.
Notice that residues 1043.32, 1083.36, and 2526.48 make consensus contacts with ligands across class-A GPCRs.7b To assess the functional role of residues of interest in MOR256-3 and its mutants, both basal activity and responses elicited by a set of chemically diverse odorants were measured (Figure 2; for dose-response curves, see Figure S3).
Figure 2.
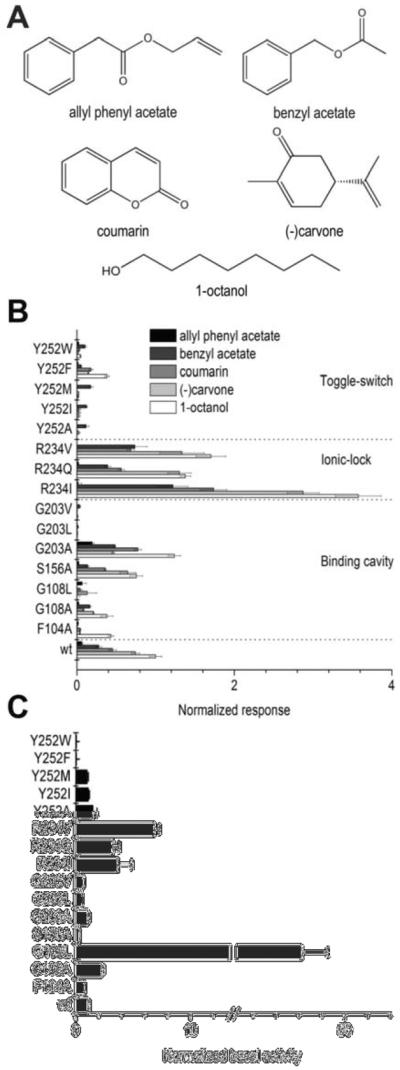
Odorant-evoked responses and basal activities of MOR256-3 receptors. (A) Structures of odorants tested in this study. (B) MOR256-3 wild-type (wt) and mutant responses to odorants. All data are normalized to the wt response to 1-octanol. (C) Basal activities of the same receptors, normalized to that of the wt OR. The five odorants are ranked based on the responses of the wt receptor from largest to smallest. Mutations include residues presumably involved in the binding cavity, in the ionic-lock, and in the toggle-switch. Data for each OR are averaged from three repeats on the same 96-well plate except for WT, averaged from 12 plates with three repeats on each (mean ± s.e.m.).
Five odorants ranging from strong to weak agonists were selected to cover agonists with a range of potency (Figure 2A). As predicted by the model, the F104A3.32 mutant indeed displays altered agonist recognition by modifying the selectivity of the receptor in vitro. In this mutant OR, the response to all agonists tested is generally decreased and the receptor only responds moderately to octanol, which is the only odorant that lacks a π-cloud.
Response of this mutant to odorants suggests that F1043.32 contributes to stabilizing bound ligands through an interaction between its aromatic cycle and double bonds present in odorants. The G203A5.43 mutant presents a short hydrophobic side-chain that does not dramatically modify accessibility to the cavity but is likely to contribute to van der Waals contacts that slightly increase the response to odorants. Upon increase of the side-chain size (G203V or G203L), the mutants do not respond to odorants anymore highlighting that this position is also within the binding cavity. In accordance with the model, the S156A4.57 mutation has no influence on odorant recognition, as its side-chain is located outside the binding cavity. This model also identifies other residues contributing to receptor selectivity, as reported by Yu et al.16 Remarkably, without ligand stimulation, G108A and G108L display unique behaviors. G108A shows a basal activity that is twice higher as the wt, while G108L has a basal activity ~45 times higher (Figure 2C). Interestingly, similar modulation upon mutating G108 has been observed in MOR256-31 (Figure S4) confirming that this effect is not specific to our model MOR256-3.
When stimulated with each odorant, G108A is still able to discriminate between weak and strong agonists but with much weaker responses than the wt. G108L, however, is virtually unresponsive to agonists (Figure 2B). These data suggest that the MOR256-3 G108A mutant favors the active state while the G108L mutant is locked into a constitutively active state.
The FYG motif in TM6 is associated with the toggle-switch for sensing agonists
Position 1083.36 is not highly conserved among ORs but it is represented in more than 85% ORs by a small residue (see Table S1), suggesting that this part of the binding cavity must be accessible to odorants. In the G108A or G108L mutants, the side chain is pointing towards Y2526.48 where a Y/F residue is conserved in more than 92% in human and mouse ORs. These two residues are reported to form a ligand-binding cradle across class-A GPCRs.7b Interestingly, Y/F6.48 is aligned with the tryptophan residue W6.48 of the highly conserved CWxP motif in non-olfactory class-A GPCRs (Figure 1C), reported as a toggle-switch for receptor activation.17 Here, the side chain of residue 108 in the mutants is likely to play the role of an artificial agonist which interacts with the side-chain of Y/F2526.48 (Figure 1D). Accordingly, we tested substitutions at position 252 with several different amino-acids. MOR256-3 Y252A, Y252I and Y252M mutants, although expressed at the cell surface (Figure S5), do not exhibit any statistically significant in vitro response upon odorant stimulation (Figure 2B and Figure S3; note that all odorant responses are corrected for the expression efficiency). These data are consistent with the role of Y/F2526.48 as a toggle-switch that triggers activation of the receptor upon agonist binding.9d, 18 The difference between tyrosine and phenylalanine was also investigated. Consistent with an F conserved in ~25% mammalian ORs at position 252, the Y252F mutant shows responses to some odorants. Its responsiveness is however decreased by 70% compared with the wt. Contrasting with non-olfactory GPCRs, when position 6.48 is a tryptophan residue (not found in native ORs), the OR becomes almost non-responsive (~5% of the wt response) (Figure 2A).
Molecular dynamics simulations model active and inactive states and identify the ionic-lock residues
GPCR activation is associated with a conformational change involving the ionic-lock between TM3 and TM6.6b MD simulations performed on models of the wt, G108A and G108L free of agonists reveal interesting structural features. In the wt, one systematically observes a structure showing the hallmarks of an inactive state, with the bottom (intracellular side) of the helix of TM6 close to that of TM3.
In the G108A and G108L models, the simulations converge towards a structure where TM6 has shifted from its initial position and moved ~8Å outward (Figure 3), which is the signature of GPCR activation.6b For these two mutants, all four independent simulations converge to an alternative model of the MOR256–3 receptor that closely resembles crystallographic structures of GPCRs in an active state (Figure 3A, B).
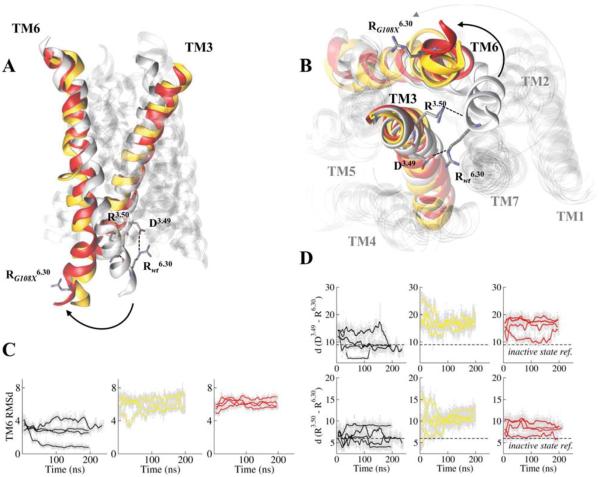
Figure 3.
MOR256-3 wt systematically reports an inactive state, while mutations at position 108 evolve toward active states. (A) Comparison between typical structures of MOR256-3 wt (white), G108A (yellow) and G108L (red) mutants. (B) Structures of the mutants have the intracellular part of TM6 shifting outward while that of the wt is close to other TMs. (C) The Root Mean Square deviation (RMSd) of TM6 (residues 234 to 253) with respect to its reference position in the wt structure reveals a systematic shift in the mutants (RMSd ≈ 6Å) but not in the wt (RMSd ≈ 3Å). (D) The ionic-lock between R6.30 and D3.49 as well as between R6.30 and R3.50 is closed in the wt (d(O_D3.49…N_R6.30) ~ 9 Å and d(N_R3.50…O_R6.30) ~ 6 Å) while it is open in the mutants (the distance becomes ~17 Å and 9 Å, for d(O_D3.49…N_R6.30) and d(N_R3.50…O_R6.30), respectively).
The wt model systematically presents a double interaction between the D3.49 and R3.50 (conserved at 98% and 88%, respectively) of the DRY motif in TM3 on one part, and the R6.30 side-chain and backbone in TM6 on the other part. A positive residue (R/K) at this position in TM6 is highly conserved in ORs (more than 75%, see Table S1) and aligned with the residue involved in the ionic-lock in non-olfactory GPCRs (Figure 1C). The interaction between TM6 and TM3 at the ionic-lock involves the side-chain and the backbone of R6.30 and the side chains of D3.49 and R3.50, respectively. These interactions are observed during three out of the four simulations of the wt system, as shown in Figure 3C, D. The four independent simulations performed for each G108X mutant systematically report a typical structure where the interactions between TM3 and TM6 are broken. Very early in the equilibration phases of the mutant receptors, TM6 shifts outward relative to TM3 (see arrows in Figure 2A, B) while it stays in its initial position in the wt. An analysis of the root mean square deviation of TM6 heavy atoms with respect to their average position in the wt reveals a large structural drift of TM6 while the rest of the edifice remains similar to the starting structure (Figure S6).
This conformational switch is in all cases associated with a break of the hydrogen bond between D3.49 and R6.30. The distance between the closest H-bond donor and acceptor atoms within these residues is ~17Å in the mutants while it is ~9 Å in the wt. Such a distance evolution (an increase of 8 Å) is consistent with those measured in experimental structures.4d
The second interaction between the R3.50 side-chain and the R6.30 backbone oxygen atom is also largely weakened, with a distance 3 Å larger in the mutants compared with the wt, in line with experimental data on rhodopsin.6b In the G108X mutants, the R6.30 side-chain has shifted towards the intracellular part of the receptor and is solvated by bulk water. D3.49 forms a hydrogen bond with either Y132IL3 or/and R3.50 which has also broken its interaction with TM6, consistent with MD simulations performed on X-ray structures of GPCRs.5a, 19 Experimentally, when position 6.40 is modified to a non-polar residue that prevents any interaction with TM3 (R234I or R234V mutants), one observes an increase of the response to odorants together with high basal activity, consistent with a shift of the conformational equilibrium of the receptor towards the active state. In the R234Q mutant, the charged DTM3 – RTM6 ionic-lock is altered from an ionic interaction into a hydrogen bond and the receptor now exhibits similar, although weaker, increase of basal activity and responsiveness (Figure 2B, C). In all cases, the ranking of the odorants remains mostly unchanged, confirming that this modification is taking place at residues involved in OR activation rather than recognition of odorants.
DISCUSSION
A model for OR activation
The mammalian olfactory system uses a combinatorial strategy based on a large family of ORs to sense odorant compounds. As for all GPCRs, when a receptor is activated by an agonist, the coupling to the G protein occurs, while in its inactive state, a receptor does not trigger the biochemical cascade leading to neuron membrane depolarization. At the atomic-level, although the mechanism is becoming more and more precisely understood for nonolfactory GPCRs, that of ORs remains elusive. In this study, using in vitro observations of mouse OR mutants in a constitutively active state, we provide insights into the way specific amino acid residues lead to the activation of an OR.
Several motifs are highly conserved in class-A GPCRs. The three-residue E/DRY motif in TM3 is involved in a so-called ionic-lock with a residue in TM6. In ORs, this DRY motif is also highly conserved although the D residue is predominant with respect to the R (Table S1), contrary to what is seen in other non-olfactory GPCRs. Indeed, in non-olfactory GPCRs, TM3 interacts with TM6 through the positively charged R3.50 of the DRY motif and a negatively charged E6.30 (as observed in rhodopsin,20 β121 and β2-adrenergic,22 human D3-dopamine,23 human H1-histamine,24 human M2-muscarinic,25 and A2A adenosine receptors).26 In the inactive state structure, this ionic-lock is closed while in the active state it is open. Chemokine receptors CXCR4 and CXCR1 stand as an exception since they exhibit a positively charged residue at position 6.30.27 In the crystal structure of these molecules, no ionic lock is observed but instead a charge dipole interaction exists involving the R3.50 side chain and the backbone of TM6 at residue in position 6.30. The case of ORs appears related to those of CXCRs. Our model is indeed in line with this structural feature with a strong hydrogen bond between R3.50 and the backbone of R6.30. In addition, we show that the D3.49 residue is engaged in a hydrogen bond with an arginine (R) residue in TM6 to stabilize the TM3-TM6 interaction, justifying its high degree of conservation in ORs. The R residue of the DRY motif is also engaged in a strong interaction with the backbone of residue 6.30. Considering MOR256-3 as a prototype of mammalian ORs (because of its conserved residues with all other mammalian ORs in this motif), a double ionic-lock seems prevalent between TM3 and TM6 in this family.
It is very likely that the active state of ORs is highly similar to that of class-A GPCRs as revealed by X-Ray crystallography. Our molecular models are in full accordance with a conserved mechanism of activation. In the multiple MD simulations we recover—without any constraint—a cleft between TM3 and TM6 in OR mutants with a higher basal activity or in a constitutively active state. Although MD simulations have already reported active and inactive GPCR structures, they were all based on experimental data.4a, 4c, 5
The model of an active OR was made possible by the unique behavior associated to mutations at position 1083.36. Notice that a small perturbation at position 1073.35 in MOR256-8 strongly increases the basal activity and affect responsiveness of the mutant OR, confirming that this part of the receptor is crucial for activation.16 Position 1083.36 in TM3 is associated with a small residue that may allow space for agonist binding deep into the pocket. The nature of this residue affects responsiveness of the receptor, as previously reported for a hOR1A2 A108G mutant.9b Position 108 faces Y2526.48, which is highly conserved in ORs and interestingly aligned with the toggle switch residue (W6.48 of the `CWxP' motif) found in non-olfactory GPCRs. Consequently, Y2526.48 can be considered to be the toggle-switch in ORs. This residue has been speculated to be involved in OR activation by agonists but has not been clearly assessed.9c, 9d Our current data confirm this hypothesis, similar to what has been shown in the A3 adenosine receptor.18 Site-directed mutations suggest that the toggle-switch should share physico-chemical properties with the associated OR ligands. Airborne odorants are more hydrophobic than non-olfactory GPCR ligands. Accordingly, based on sequence analysis, the transmission switch for agonists within the cavity of an OR is an aromatic residue (Y/F) at position 6.48. The tryptophan cycle cannot play the role of toggle-switch in ORs, because of either too-large hydrophilicity or a too-bulky character. In the first case, the interaction with agonists wouldn't be favored. Note that the contribution of residue 6.48 to the free energy of binding is computed to be important when agonists are bound to hOR1G1.14,28
Once the agonist is bound within the OR cavity, the activation process propagates by creating a drift of TM6 which will, as for other GPCRs, open a cleft at the intracellular part of the bundle to favor G protein coupling. Interestingly, the part of TM6 that moves outward involves the highly conserved KAFSTCxSH motif, consistent with both the role of the serine residue (S) in the change from the active to the inactive conformation and more generally the contribution of this motif to the receptor conformation.29
The multidisciplinary approach used here is promising in elucidating the activation process of receptors with unknown experimental structures. In this article, we focused on residues belonging to TM3, TM5 and TM6. These residues studied in MOR256-3 are highly conserved in human and mouse ORs. Since MOR256-3 is broadly tuned, there is a possibility that the proposed mechanism only applies to broadly-tuned receptors. However, the identified residues are conserved in both broadly-tuned and narrowly-tuned ORs suggesting that the mechanism may apply to all ORs independent of their tuning properties. Further investigations with narrowly-tuned receptors are required to distinguish these two possibilities
Other parts of the OR are also surely important for OR activation by fulfilling the network of amino acids involved in the process from ligand binding to G protein coupling. This is for example the case of the conserved NPxxY motif within TM7.7b, 30 This approach nevertheless provides a fruitful working model for OR activation based on site-directed mutagenesis and molecular dynamics simulations, which is of high importance for predicting olfactory sensory neuron responses upon ligand stimulation.
Supplementary Material
ACKNOWLEDGMENT
We thank Elise Bruguera for comments and edits. This work was supported by grants from the National Institute on Deafness and Other Communication Disorders, National Institute of Health (DC011554 and DC006213 to M.M., and DC005782 and DC012095 to H.M.), APEX Region PACA to J. G. (OLFACTOME) and from the Fondation Roudnitska under the aegis of Fondation de France to C.A.D.M. C.A.D.M. also thanks GIRACT for a PhD bursary.
Footnotes
Supporting Information Alignment of GPCR sequences, full dose-response curves, supplementary figures and structures of the OR models. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- (1).Buck L, Axel R. Cell. 1991;65:175. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- (2).Flegel C, Manteniotis S, Osthold S, Hatt H, Gissel-mann G. PLoS ONE. 2013;8:655368. doi: 10.1371/journal.pone.0055368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Niimura Y. Curr Genomics. 2012;13:103. doi: 10.2174/138920212799860706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).(a) Dror RO, Arlow DH, Maragakis P, Mildorf TJ, Pan AC, Xu H, Borhani DW, Shaw DE. Proc Natl Acad Sci U S A. 2011;108:18684. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kruse AC, Ring AM, Manglik A, Hu J, Hu K, Eitel K, Hubner H, Pardon E, Valant C, Sexton PM, Christopoulos A, Felder CC, Gmeiner P, Steyaert J, Weis WI, Garcia KC, Wess J, Kobilka BK. Nature. 2013;504:101. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Nygaard R, Zou Y, Dror Ron O., Mildorf Thomas J., Arlow Daniel H., Manglik A, Pan Albert C., Liu Corey W., Fung Juan J., Bokoch Michael P., Thian Foon S., Kobilka Tong S., Shaw David E., Mueller L, Prosser RS, Kobilka Brian K. Cell. 2013;152:532. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Trzaskowski B, Latek D, Yuan S, Ghoshdastider U, Debinski A, Filipek S. Curr Med Chem. 2012;19:1090. doi: 10.2174/092986712799320556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).(a) Laricheva EN, Arora K, Knight JL, Brooks CL. J Am Chem Soc. 2013;135:10906. doi: 10.1021/ja4042687. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Miao Y, Nichols SE, Gasper PM, Metzger VT, McCammon JA. Proc Natl Acad Sci U S A. 2013;110:10982. doi: 10.1073/pnas.1309755110. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Li J, Jonsson AL, Beuming T, Shelley JC, Voth GA. Am Chem Soc. 2013;135:8749. doi: 10.1021/ja404391q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).(a) Gether U, Kobilka BK. J Biol Chem. 1998;273:17979. doi: 10.1074/jbc.273.29.17979. [DOI] [PubMed] [Google Scholar]; (b) Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. Proc Natl Acad Sci U S A. 2008;105:7439. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).(a) Charlier L, Topin J, de March CA, Lai PC, Crasto CJ, Golebiowski J. In: In Olfactory Receptors. Crasto CJ, editor. Humana Press; New York: 2013. [Google Scholar]; (b) Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Nature. 2013;494:185. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- (8).(a) Zozulya S, Echeverri F, Nguyen T. Genome Biol. 2001;2:1. doi: 10.1186/gb-2001-2-6-research0018. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Man O, Gilad Y, Lancet D. Protein Sci. 2004;13:240. doi: 10.1110/ps.03296404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).(a) Abaffy T, Malhotra A, Luetje CW. J Biol Chem. 2007;282:1219. doi: 10.1074/jbc.M609355200. [DOI] [PubMed] [Google Scholar]; (b) Schmiedeberg K, Shirokova E, Weber H-P, Schilling B, Meyerhof W, Krautwurst D. J Struct Biol. 2007;159:400. doi: 10.1016/j.jsb.2007.04.013. [DOI] [PubMed] [Google Scholar]; (c) Baud O, Etter S, Spreafico M, Bordoli L, Schwede T, Vogel H, Pick H. Biochemistry. 2011;50:843. doi: 10.1021/bi1017396. [DOI] [PubMed] [Google Scholar]; (d) Katada S, Hirokawa T, Oka Y, Suwa M, Touhara K. J Neurosci. 2005;25:1806. doi: 10.1523/JNEUROSCI.4723-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Gelis L, Wolf S, Hatt H, Neuhaus E, Gerwert K. Angew. Chem. Int. Ed. Engl. 2012;51:1274. doi: 10.1002/anie.201103980. [DOI] [PubMed] [Google Scholar]; (f) Sekharan S, Ertem Mehmed Z., Zhuang H, Block E, Matsunami H, Zhang R, Wei Jennifer N., Pan Y, Batista Victor S. Biophys J. 2014;107:L5. doi: 10.1016/j.bpj.2014.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) de March CA, Kim S-K, Antonczak S, Goddard WA, Golebiowski J. Protein Sci. 2015 doi: 10.1002/pro.2717. 10.1002/pr0.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Duan X, Block E, Li Z, Connelly T, Zhang J, Huang Z, Su X, Pan Y, Wu L, Chi Q. Proc Natl Acad Sci U S A. 2012;109:3492. doi: 10.1073/pnas.1111297109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wang J, Luthey-Schulten ZA, Suslick KS. Proc Natl Acad Sci U S A. 2003;100:3035. doi: 10.1073/pnas.262792899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).(a) Kim S-K, Goddard W., III J Comput Aided Mol Des. 2014;28:1. doi: 10.1007/s10822-014-9793-4. [DOI] [PubMed] [Google Scholar]; (b) Lai PC, inger MS, Crasto CJ. Chem Senses. 2005;30:781. doi: 10.1093/chemse/bji070. [DOI] [PubMed] [Google Scholar]; (c) Lai PC, Guida B, Shi J, Crasto CJ. hem Senses. 2014;39:107. doi: 10.1093/chemse/bjt060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Grosmaitre X, Fuss SH, Lee AC, Adipietro KA, Matsunami H, Mombaerts P, Ma M. J Neurosci. 2009;29:14545. doi: 10.1523/JNEUROSCI.2752-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Topin J, de March CA, Charlier L, Ronin C, Antonczak S, Golebiowski J. Chem Eur J. 2014;20:10227. doi: 10.1002/chem.201402486. [DOI] [PubMed] [Google Scholar]
- (15).de March CA, Golebiowski J. Biochimie. 2014;107:3. doi: 10.1016/j.biochi.2014.06.006. [DOI] [PubMed] [Google Scholar]
- (16).Yu Y, de March CA, Ni MJ, Adipietro KA, Golebiowski J, Matsunami H, Ma M. submitted.
- (17).Deupi X, Standfuss J. Curr Opin Chem Biol. 2011;21:541. doi: 10.1016/j.sbi.2011.06.002. [DOI] [PubMed] [Google Scholar]
- (18).Gao Z-G, Chen A, Barak D, Kim S-K, Müller CE, Jacobson KA. J Biol Chem. 2002;277:19056. doi: 10.1074/jbc.M110960200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Dror RO, Arlow DH, Borhani DW, Jensen MØ, Piana S, Shaw DE. Proc Natl Acad Sci U S A. 2009;106:4689. doi: 10.1073/pnas.0811065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. J Mol Biol. 2004;342:571. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- (21).Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GF. Nature. 2008;454:486. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. Science. 2007;318:1258. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Chien EY, Liu W, Zhao Q, Katritch V, Han GW, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, Stevens RC. Science. 2010;330:1091. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Shimamura T, Shiroishi M, Weyand S, Tsujimoto H, Winter G, Katritch V, Abagyan R, Cherezov V, Liu W, Han GW, Kobayashi T, Stevens RC, Iwata S. Nature. 2011;475:65. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Haga K, Kruse AC, Asada H, Yurugi-Kobayashi T, Shiroishi M, Zhang C, Weis WI, Okada T, Kobilka BK, Haga T, Kobayashi T. Nature. 2012;482:547. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Lebon G, Warne T, Edwards PC, Bennett K, Langmead CJ, Leslie AG, Tate CG. Nature. 2011;474:521. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).(a) Park SH, Das BB, Casagrande F, Tian Y, Nothnagel HJ, Chu M, Kiefer H, Maier K, De Angelis AA, Marassi FM, Opella SJ. Nature. 2012;491:779. doi: 10.1038/nature11580. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wu B, Chien EYT, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Science. 2010;330:1066. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Charlier L, Topin J, Ronin C, Kim S-K, Goddard W, Efremov R, Golebiowski J. Cell Mol Life Sci. 2012;4205 doi: 10.1007/s00018-012-1116-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Kato A, Katada S, Touhara K. J Neurochem. 2008;107:1261. doi: 10.1111/j.1471-4159.2008.05693.x. [DOI] [PubMed] [Google Scholar]
- (30).(a) Kohlhoff KJ, Shukla D, Lawrenz M, Bowman GR, Konerding DE, Belov D, Altman RB, Pande VS. Nat Chem. 2014;6:15. doi: 10.1038/nchem.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Audet M, Bouvier M. Cell. 2012;151:14. doi: 10.1016/j.cell.2012.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.