Dear Editor,
microRNAs (miRNAs) have been frequently reported to play critical roles in tumorigenesis and to have great potential for the development of molecular cancer therapeutics. miRNAs are transcribed as long primary transcripts whose maturation occurs through sequential endonucleolytic steps that yield precursor miRNA (pre-miRNA) intermediates and then the mature miRNAs. Each pre-miRNA has two arms, 5′ arm and 3′ arm, and are denoted with -5p and -3p suffixes in its name 1. Depending on the tissue or cell type, both arms can be processed to become functional mature miRNAs 2-4. The two arms have different sequences; therefore, they either target different mRNAs or the same mRNA but at different sites in the mRNA's 3′ untranslated region. Until recently, in most miRNA studies, researchers usually reported the functions of only one arm (i.e., either -5p or -3p), rather than both. However, recent in vitro and in vivo studies 2-4 showed that miR-5p and miR-3p (hereafter referred to miR-5p/-3p) mediated synergistic regulations could enhance a more effective suppression of tumor-suppressor pathways or activation of oncogenic pathways in a specific cancer. For example, the synergistic effect of miR-17-5p/-3p pair was investigated in hepatocellular carcinoma (HCC) 3 and prostate tumor 4. In HCC, miR-17-5p repressed the expression of tumor suppressor gene PTEN, and miR-17-3p repressed the expression of genes Vimentin and GalNT7; whereas in prostate tumor, miR-17-5p and miR-17-3p co-regulated the tumor suppressor gene TIMP3 and repressed its expression. In both cancers, the abundant expression of miR-17-5p/-3p pair enhanced cell proliferation, migration, and invasion by silencing the above mentioned direct target genes. The synergistic effect of several other miR-5p/-3p pairs has been examined in specific cancer type (Supporting Information Table S1). However, there is no systematic examination about how frequently miR-5p/-3p pairs are dysregulated across multiple cancer types and involved in synergistic regulation.
To explore this important issue, we used the publicly available, large-scale miRNA-Seq expression data for multiple cancer types that were generated by The Cancer Genome Atlas (TCGA) project (https://tcga-data.nci.nih.gov/tcga/). We extracted miRNA-Seq expression profiles from 3778 clinically-documented high-quality tumor and normal samples in 10 cancer types (Supporting Information Table S2) and identified differentially expressed miR-5p/-3p pairs in each cancer (Supporting Information Data S1 and Table S3). This massive amount of data made it possible for us not only to pinpoint abnormally expressed miR-5p/-3p pairs in a specific cancer type, but also to assess whether these pairs follow any dysregulation pattern in multiple cancers (i.e., pan-cancer).
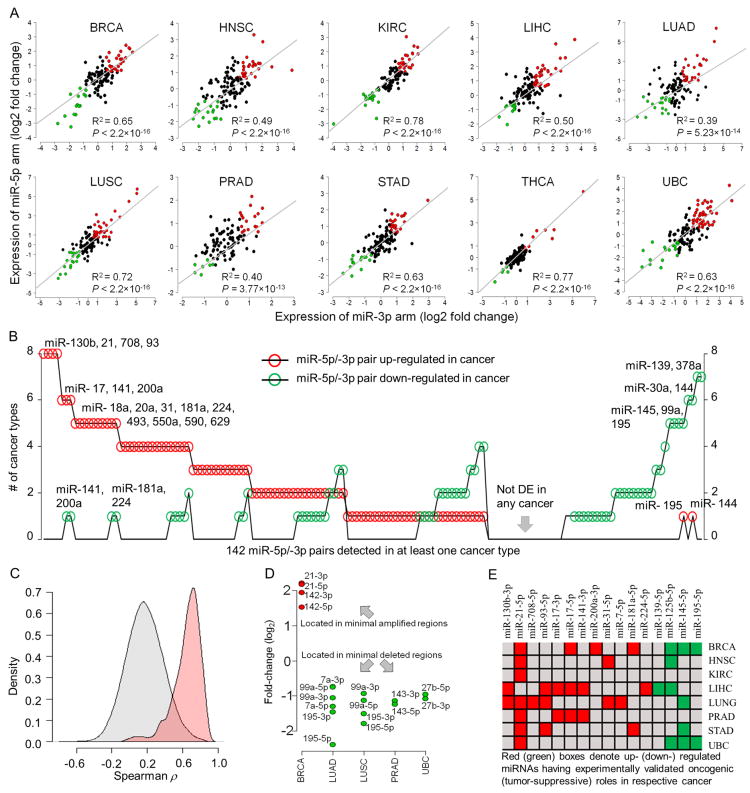
Interestingly, we found a strong trend that miR-5p/-3p pairs follow a concordant dysregulation pattern, i.e., both arms were either up- or down-regulated in cancer compared to normal tissue samples (Fig. 1A). We identified 23 miR-5p/-3p pairs that have consistent up- or down-regulation in five or more cancer types, suggesting their potentiality in pan-cancer regulation (labeled miR-5p/-3p pairs in Fig. 1B). For example, miR-130b-5p/-3p, miR-21-5p/-3p, miR-708-5p/-3p, and miR-93-5p/3p were up-regulated in eight cancer types. We used >1.5 fold-change and adjusted P-value < 0.05 (adjusted by Benjamini-Hochberg method 5) to denote differentially expressed miRNAs; this is because a 1.5-fold change of miRNA expression may have significant impact on cellular process 6. Notably, miR-17-5p/-3p and miR-31-5p/-3p pairs were up-regulated in six and five cancer types, respectively. The up-regulation of miR-17-5p/-3p pair in HCC 3 and prostate cancer 4, and the up-regulation of miR-31-5p/-3p pair in head and neck cancer 7 were confirmed by qRT-PCR in previous studies (Supporting Information Table S1). We also found that miR-5p/-3p pairs had stronger expression correlation (P-value < 2.2×10-16, one-sided Kolmogorov-Smirnov test) than that of the mature miRNA pairs that were randomly selected from different pre-miRNAs with the requirement of concordant dysregulation of the two miRNAs (Fig 1C). The observed concordant dysregulation might in many cases be a direct consequence of the deregulating mechanisms, for example, the two arms of the same precursor have very close proximity (< 30 nucleotides) in the human genome so that both arms are always located in minimally deleted/amplified regions 8 (Fig. 1D) or regulated by common upstream regulators (e.g. transcription factors). For instance, transcription factor SOX9 regulates both miR-202-5p and miR-202-3p expression during mouse testis differentiation 9. Previous reports, obtained from the OncomiRDB database 10 and Supporting Information Table S1, suggest that the well-studied arm of several pre-miRNAs (labeled in Fig. 1B) has oncogenic or tumor suppressive roles in one or more cancer types (Fig. 1E). Hence, future experimental works are warranted to uncover the oncogenic/tumor suppressive potential of the less studied arms.
Figure 1. Concordant dysregulation of miR-5p/-3p pairs in cancer: potential causes and consequences.
BRCA: breast invasive carcinoma; HNSC: head and neck squamous cell carcinoma; KIRC: kidney renal clear cell carcinoma; LIHC: liver hepatocellular carcinoma; LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma; PRAD: prostate adenocarcinoma; STAD: stomach adenocarcinoma; THCA: thyroid carcinoma; and UBC: urothelial bladder carcinoma. A) miR-5p/-3p pairs display a general trend of concordant dysregulation in cancer. The red, green, and black dots represent significantly up-regulated, significantly down-regulated, and not significantly dysregulated miR-5p/-3p pairs in cancer, respectively. Regression lines and corresponding R2 values indicate that differential expression levels of miR-5p arms have strong concordance (all P-values <10-12) with miR-3p arms in 10 types of cancer. B) Several miR-5p/-3p pairs had consistent up- (16 labeled miRNAs with red circles) or down- (7 labeled miRNAs with green circles) regulations in five or more cancers. X-axis shows the 142 miR-5p/-3p pairs detected in at least one cancer type. Y-axis shows the number of cancer types in which miR-5p/-3p pairs were abnormally expressed. C) Density plots revealed that miR-5p/-3p pairs (pink shaded area) had stronger expression correlations than any two concordantly dysregulated miRNAs that were randomly selected from different pre-miRNAs (grey shaded area). D) Several concordantly up- or down-regulated miR-5p/-3p pairs are located in minimally amplified or deleted chromosomal regions. E) Sixteen up- or down-regulated miRNAs (shown in Fig. 1B) have oncogenic or tumor-suppressive roles in respective cancer type, as reported in in-vitro and in-vivo studies.
While we first time reported that the concordant dysregulation of miR-5p/-3p pair is a common feature in cancer, several issues remain for future investigation. First, whether and how miR-5p/-3p pairs are under selective pressure to be concordantly dysregulated in cancer? Second, what are the specific subcellular systems or signaling pathways synergistically controlled by miR-5p/-3p pairs? Third, how can concordant dysregulation of miR-5p/3p pairs help us develop more accurate miRNA-based molecular therapeutic strategies? Fourth, whether this concordant dysregulation is universal in other phenotype or in other organisms remains for further investigation.
Yours sincerely,
Ramkrishna Mitra
Jingchun Sun
Zhongming Zhao
Supplementary Material
Acknowledgments
This work was partially supported by National Institutes of Health (NIH) grants (R01LM011177, R03CA167695, and P30CA68485), and Ingram Professorship Funds (to ZZ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supporting Information: Additional Supporting Information may be found in the online version of this article.
References
- 1.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uchino K, Takeshita F, Takahashi RU, Kosaka N, Fujiwara K, Naruoka H, Sonoke S, Yano J, Sasaki H, Nozawa S, Yoshiike M, Kitajima K, et al. Therapeutic effects of microRNA-582-5p and -3p on the inhibition of bladder cancer progression. Mol Ther. 2013;21:610–9. doi: 10.1038/mt.2012.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shan SW, Fang L, Shatseva T, Rutnam ZJ, Yang X, Du W, Lu WY, Xuan JW, Deng Z, Yang BB. Mature miR-17-5p and passenger miR-17-3p induce hepatocellular carcinoma by targeting PTEN, GalNT7 and vimentin in different signal pathways. J Cell Sci. 2013;126:1517–30. doi: 10.1242/jcs.122895. [DOI] [PubMed] [Google Scholar]
- 4.Yang X, Du WW, Li H, Liu F, Khorshidi A, Rutnam ZJ, Yang BB. Both mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasion. Nucleic Acids Res. 2013;41:9688–704. doi: 10.1093/nar/gkt680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J R Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 6.Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang KW, Kao SY, Wu YH, Tsai MM, Tu HF, Liu CJ, Lui MT, Lin SC. Passenger strand miRNA miR-31* regulates the phenotypes of oral cancer cells by targeting RhoA. Oral Oncol. 2013;49:27–33. doi: 10.1016/j.oraloncology.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wainwright EN, Jorgensen JS, Kim Y, Truong V, Bagheri-Fam S, Davidson T, Svingen T, Fernandez-Valverde SL, McClelland KS, Taft RJ, Harley VR, Koopman P, et al. SOX9 regulates microRNA miR-202-5p/3p expression during mouse testis differentiation. Biol Reprod. 2013;89:34. doi: 10.1095/biolreprod.113.110155. [DOI] [PubMed] [Google Scholar]
- 10.Wang D, Gu J, Wang T, Ding Z. OncomiRDB: a database for the experimentally verified oncogenic and tumor-suppressive microRNAs. Bioinformatics. 2014;30:2237–8. doi: 10.1093/bioinformatics/btu155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.