Abstract
The ability to escape apoptosis is a hallmark of cancer-initiating cells and a key factor of resistance to oncolytic therapy. Here, we identify FAM96A as a ubiquitous, evolutionarily conserved apoptosome-activating protein and investigate its potential pro-apoptotic tumor suppressor function in gastrointestinal stromal tumors (GISTs). Interaction between FAM96A and apoptotic peptidase activating factor 1 (APAF1) was identified in yeast two-hybrid screen and further studied by deletion mutants, glutathione-S-transferase pull-down, co-immunoprecipitation and immunofluorescence. Effects of FAM96A overexpression and knock-down on apoptosis sensitivity were examined in cancer cells and zebrafish embryos. Expression of FAM96A in GIST and histogenetically related cells including interstitial cells of Cajal (ICCs), ‘fibroblast-like cells’ (FLCs) and ICC stem cells (ICC-SCs) was investigated by Northern blotting, reverse transcription—polymerase chain reaction, immunohistochemistry and Western immunoblotting. Tumorigenicity of GIST cells and transformed murine ICC-SC stably transduced to re-express FAM96A was studied by xeno- and allografting into immunocompromised mice. FAM96A was found to bind APAF1 and to enhance the induction of mitochondrial apoptosis. FAM96A protein or mRNA was dramatically reduced or lost in 106 of 108 GIST samples representing three independent patient cohorts. Whereas ICCs, ICC-SCs and FLCs, the presumed normal counterparts of GIST, were found to robustly express FAM96A protein and mRNA, FAM96A expression was much reduced in tumorigenic ICC-SCs. Re-expression of FAM96A in GIST cells and transformed ICC-SCs increased apoptosis sensitivity and diminished tumorigenicity. Our data suggest FAM96A is a novel pro-apoptotic tumor suppressor that is lost during GIST tumorigenesis.
Keywords: GIST, ICC, FAM96A, apoptosis, tumor suppressor
Introduction
Tumor persistence due to incomplete apoptotic response to oncolytic treatment and the consequent need for life-long medical therapy represent a significant clinical problem in gastrointestinal stromal tumors (GISTs), the most common mesenchymal tumors of the gastrointestinal tract.1–5 70–80% of GISTs harbor oncogenic activating mutations in the receptor tyrosine kinase (RTK) KIT.6 These tumors are histogenetically related to interstitial cells of Cajal (ICCs),7 KIT+, KIT-dependent regulators of gastrointestinal motility, and their KITlowCD44+CD34+ stem cells (ICC-SCs).1 5–8% of GISTs exhibit oncogenic mutations in the platelet-derived growth factor receptor α (PDGFRA) gene, a close homolog of KIT expressed by ICC-SC1 and the so-called ‘fibroblast-like cells’ (FLCs), a functionally distinct class of interstitial cells.8, 9 Targeting mutant KIT or PDGFRA proteins with RTK inhibitors, such as imatinib mesylate, is effective in GIST patients with advanced disease. However, this treatment is almost never curative due to the survival of cells for which KIT/PDGFRA blockade is not cytotoxic and the emergence of secondary drug resistance in many patients.1, 2, 4
Reduced apoptotic response likely underlies disease progression and therapy resistance that develops in most GIST patients treated with RTK inhibitors.4 The mitochondrial apoptosis pathway involves cytochrome c-induced formation of the apoptosome, a heptamer of apoptotic peptidase activating factor 1 (APAF1; the mammalian ortholog of the Caenorhabditis elegans protein CED-4), and subsequent recruitment and activation of caspase-9.10 The intrinsic apoptosis pathway can be triggered by a plethora of stimuli including radiation and drugs employed in cancer chemotherapy.11 However, ineffective activation of caspase-9 has been observed in a variety of tumors.12 Here, we show that FAM96A (family with sequence similarity 96, member A), a recently identified member of the cytosolic iron-sulfur (Fe/S) protein assembly machinery and regulator of cellular iron homeostasis,13 is a novel pro-apoptotic APAF1-binding protein facilitating the effective induction of cell death via the mitochondrial apoptosis pathway. While FAM96A is expressed in ICCs, ICC-SCs and FLCs, its expression is downregulated in GISTs and tumorigenic ICC-SCs. Re-establishment of FAM96A expression enhanced apoptosis sensitivity and inhibited tumor growth in vitro and in vivo, indicating a key role for FAM96A loss in GIST oncogenesis and the limited apoptosis sensitivity characteristic of this cancer.
Materials and Methods
Patient Samples
All studies were approved by the Institutional Review Boards of the Mayo Clinic, Rochester, MN, USA and other collaborating institutions. Northern blot membranes loaded with total RNA from adult human tissues were purchased from RNWAY Laboratories, Seoul, Republic of Korea (Human Tissue Combination B). The comparative genomic hybridization (CGH) database Progenetix (http://www.progenetix.org/cgi-bin/pgHome.cgi) containing data from 31,915 anonymized human malignancies (June 2014)14 was analyzed at the German Cancer Research Center in Heidelberg, Germany, and at the Institute of Molecular Life Sciences, University of Zurich, Switzerland. mRNA expression studies were performed in anonymized GIST samples and corresponding normal tissues obtained from the archives of the Institutes of Pathology of the Medical University of Graz, Austria, and the University of Heidelberg, Germany. Tissue microarrays containing anonymized GIST tumor samples were from the Institute for Pathology, University Hospital Erlangen, Germany. De-identified GIST histological sections were obtained from the Mayo Clinic biorepositories through the Clinical Core of the Center for Cell Signaling in Gastroenterology. De-identified human gastric tissues were obtained as surgical excess tissue from patients undergoing bariatric surgery at the Mayo Clinic. The Mayo Clinic Institutional Review Board waived the need for written informed consent from the patients in both protocols.
Cell Preparations, Cultures and Transfections
ICCs, ICC-SCs and FLCs were identified by flow cytometry (FCM) in the hematopoietic marker-negative fraction of dissociated gastric corpus+antrum tunica muscularis as KIT+CD44+CD34−, KITlowCD44+CD34+ and KIT−CD44−CD34−PDGFRA+ cells, respectively, and isolated by fluorescence-activated cell sorting (FACS) using previously published protocols1 with modifications (see Supplementary Material section, Supplementary Table 1 and Supplementary Figure 1). HEK293T human embryonic kidney cells (ACC-635, German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany), RKO human colorectal carcinoma cells (ATCC CRL-2577), and GIST48 and GIST882 human GIST cells (kindly provided by J. A. Fletcher, Boston, MA, USA) were cultured as described in the Supplementary Material section. D2211B, 2xSCS70 and 2xSCS2F10 murine ICC-SCs lines were maintained as described previously.1 Cell transfections were performed using either Lipofectamine™ 2000 (Life Technologies GmbH, Darmstadt, Germany) or polyethylenimine (Polysciences, Inc., Warrington, PA, USA) according to the manufacturer’s instructions.
Animals and Tumor Xenograft and Allograft Experiments
Experiments were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals. All protocols were IACUC-approved. Subcutaneous xenograft and allograft experiments were performed by injecting the lentivirally transduced tumor cells into the left flank of 8- to 12-week-old nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (host strain for human RKO and GIST882 cells; Charles River, Sulzfeld, Germany) or athymic NCr-nu/nu mice (host strain for mouse 2xSCS70 cells; Harlan Laboratories Indianapolis, IN, USA). RKO cells (4×106) and 2xSCS70 cells (5×106) were injected in 100 μL growth factor-reduced Matrigel (Becton Dickinson, Franklin Lakes, NJ, USA) diluted 1:1 with phosphate-buffered saline (PBS); GIST882 cells (7.7×106) were injected in 100 μLPBS. Tumor volumes were monitored 2–3 times a week for 5–8 weeks by measuring length and width with calipers and calculated as follows: tumor volume [mm3] = length × (width)2/2.
Statistical Analyses
Data are expressed as mean ± SEM or median [interquartile range] of data from independent biological replicates and analyzed by Student’s t test, one-way ANOVA (with post-hoc multiple comparisons) or nonparametric alternatives. P<0.05 was considered significant.
Additional experimental procedures are described in the Supplementary Material section.
Results
FAM96A Interacts with the Pro-Apoptotic Adaptor Protein APAF1
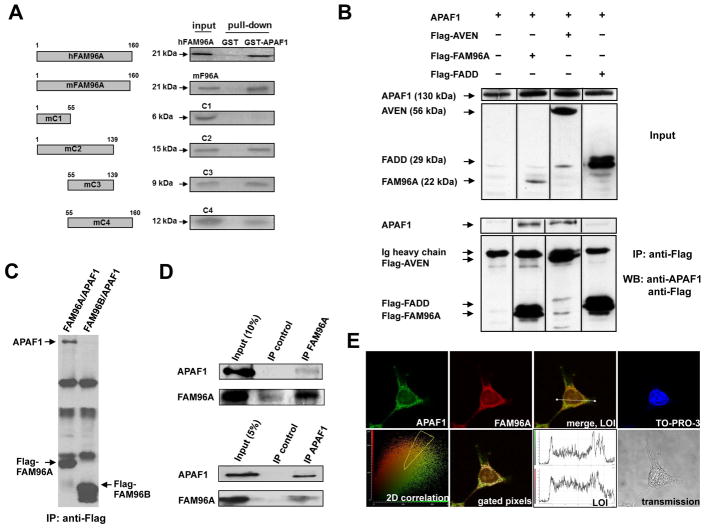
In a yeast two-hybrid screen utilizing a mouse thymoma cDNA library as prey and the pro-apoptotic Caenorhabditis elegans CED-4 protein, an ortholog of APAF1, as bait we identified FAM96A as a potential APAF1-binding factor (for additonal proteins identified by yeast-two-hybrid screening with APAF1 as the bait see Supplementary Table 2). We confirmed the interaction of FAM96A with APAF1 by repeating the interaction analysis with a human APAF1 bait fusion protein instead of CED-4. Human FAM96A, a 160-aa protein containing a “domain of unknown function” (DUF59; aa 33–121; refs.15–18 and our unpublished sequence and structure alignments) has recently been identified as a member of the cytosolic Fe/S protein assembly machinery and regulator of cellular iron homeostasis.13 FAM96A is detected as a ~21-kDa band in protein gels (Figure 1A–D). The ability of in-vitro translated, full-length human (h) and mouse (m) FAM96A to bind GST-hAPAF1SΔWD40 (hAPAF1S isoform lacking the C-terminal WD40 repeats fused with glutathione-S-transferase [GST]) was confirmed by GST pull-down (Figure 1A). The aa 55–139 region containing a large portion of the DUF59 domain was sufficient and necessary for in-vitro co-precipitation with GST-hAPAF1SΔWD40 (Figure 1A). To demonstrate the interaction between FAM96A and APAF1 in the cellular context, HEK293T cells were transiently co-transfected with Flag-hFAM96A and hAPAF1S. Immunoprecipitation with an anti-Flag antibody revealed specific binding of overexpressed FAM96A to APAF1S (Figure 1B). In contrast, APAF1 did not co-immunoprecipitate with Flag-tagged FAM96B, a homolog of FAM96A, expressed in HEK293T cells (Figure 1C). Moreover, we detected co-immunoprecipitation of endogenous hFAM96A and hAPAF1 proteins in HEK293T cell lysates using either a self-raised anti-FAM96A antiserum (Supplementary Figure 2) or an anti-APAF1 antibody (Figure 1D). Finally, subcellular co-localization of hFAM96A and endogenous full-length hAPAF1 expressed in HEK293T cells was suggested by confocal immunofluorescent microscopy (Figure 1E). Most APAF1 and FAM96A expression and co-localization was observed in the cytoplasm. Collectively, our results indicate a specific, physiological interaction between FAM96A and APAF1.
Figure 1.
FAM96A interacts with APAF1. (A) Human and mouse full-length FAM96A and mouse FAM96A deletion mutants (C1–C4) were in-vitro translated and labeled with 35[S]-methionine. After incubation with bacterially expressed GST-APAF1SΔWD40 or GST alone, glutathione-sepharose beads were used to precipitate GST fusion proteins and their complexing partners. The C3 mutant containing mFAM96A aa 55–139 was sufficient and necessary for in-vitro APAF1 binding. (B) Flag-tagged hFAM96A was co-transfected into 2×106 HEK293T cells with hAPAF1S. Immunoprecipitation (IP) with anti-Flag-coupled agarose beads and subsequent Western blot analysis showed co-immunoprecipitation of APAF1S with Flag-FAM96A and the APAF1-binding protein Flag-AVEN (caspase activation inhibitor; positive control) but not with the cytosolic adaptor protein Flag-FADD (negative control). (C) HEK293T cells were co-transfected with Flag-FAM96A or Flag-FAM96B together with APAF1. Following protein lysate preparation, immunoprecipitation with anti- Flag-coupled agarose beads and subsequent Western blot analysis showed co-immunoprecipitation of APAF1 with Flag-FAM96A but not with FAM96B. (D) Upper panel: IP of endogenous FAM96A in HEK293T cells using a self-raised rabbit anti-mFAM96A antiserum that cross-reacts with hFAM96A (see Supplementary Material section and Supplementary Figure 2) and rabbit IgG as negative control. Co-immunoprecipitated endogenous APAF1 protein was detected by Western blotting. Lower panel: Endogenous FAM96A was co-immunoprecipitated with endogenous APAF1 in HEK293T cell lysates. An anti-FLIP antibody served as a negative isotype IP control. (E) Detection of endogenous APAF1 (using rat anti-APAF1 and Alexa Fluor [AF] 488-anti-rat IgG; green) and FAM96A (using rabbit anti-mFAM96A and AF546-anti-rabbit IgG antibodies; red) in a HEK293T cell. Co-localization is indicated by the yellow color in the merged image. The lower left panel shows a 2D correlation diagram in which the pixels positive for both FAM96A and APAF1 were gated (yellow polygon) for representation in the panel labeled “gated pixels”, where white color indicates co-localization in the cytoplasm. The distributions of green (APAF1) and red (FAM96A) fluorescence intensities along the line in the merged image (LOI; line-of-interest) are shown in the upper and lower panels, respectively, of the panel labeled LOI. Lower right panel: transmission microscopic image of the analyzed cell.
FAM96A Is a Ubiquitously Expressed Gene
The FAM96A gene has been identified in at least 50 eukaryotic species ranging from slime mold (Dictyostelium discoideum) to humans (http://www.ncbi.nlm.nih.gov/gene). We examined the distribution of FAM96A expression by hybridizing Human Tissue Combination B Northern blot membranes loaded with total RNA from various adult human and mouse tissues (RNWAY Laboratories) with a 32[P]-labeled mFAM96A cDNA probe. Ubiquitous expression of FAM96A mRNA was detected in both human and mouse tissues (Supplementary Figure 3). Particularly high expression of the single 1.2 kb FAM96A transcript was observed in the kidney.
FAM96A Facililtates Efficient Induction of Mitochondrial Apoptosis
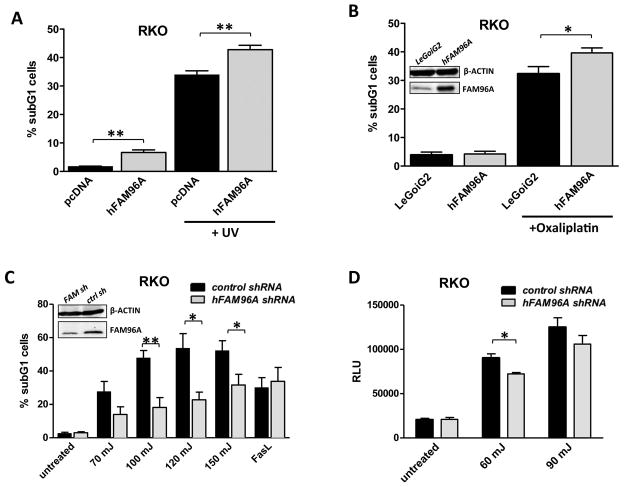
We initially analyzed the role of FAM96A in apoptosis by transfecting hFAM96A into RKO human colon carcinoma cells and quantifying cell death induced by irradiation with ultraviolet (UV) light (254 nm) or exposure to anti-cancer chemotherapeutic agents, which activate the mitochondrial apoptosis pathway.19, 20 FAM96A overexpression accentuated cell death upon UV irradiation (Figure 2A) and significantly increased apoptosis induced by oxaliplatin (Figure 2B). Conversely, lentiviral short hairpin RNA (shRNA)-mediated knock-down of FAM96A diminished UV-induced apoptosis in human RKO cells without affecting Fas ligand (FasL)-induced extrinsic apoptosis (Figure 2C). Caspase-9 activation was reduced in FAM96A knock-down RKO cells 6 hours after UV irradiation (Figure 2D), further supporting a pro-apoptotic function of FAM96A in the mitochondrial apoptosis pathway.
Figure 2.
FAM96A facilitates mitochondrial apoptosis in human cancer cells. (A) RKO human colon carcinoma cells were transiently transfected either with hFAM96A or the empty vector, pcDNA3.1, prior to UV irradiation (120 mJ/cm2). Six hours later, the cells were fixed with ethanol and incubated with propidium iodide (PI). Apoptosis was quantified by analyzing the sub-G1 cell fraction by FCM. **, P≤0.005; n=4. (B) RKO cells (2×105) stably transduced with either hFAM96A or the empty vector, LeGoiG2, were treated with 100μM oxaliplatin for 20 hours. Apoptosis was quantified by FCM. FAM96A expression was determined by Western blotting (inset). *, P<0.05; n=6. (C) RKO cells were stably transduced either with a lentiviral hFAM96A shRNA construct (FAM sh) or a non-targeting control shRNA (ctrl sh). The cells were exposed to various levels of UV irradiation or FasL (40 ng/μl (Alexis Corporation, Lausen, Switzerland) plus 1 μg/μl anti-Flag antibody (M2, Sigma-Aldrich) plus 0.1 μg/μl Cycloheximide (Sigma-Aldrich)), and apoptosis was quantified by FCM. Knock-down efficiency was determined by Western blotting (inset). *, P<0.05; **, P≤0.005; n=5. (D) Knock-down of FAM96A in RKO cells led to decreased Caspase-9 activation detected by Promega Caspase-Glo® 9 Assay 6 hours after irradiation with 60 mJ and 90 mJ UV light (control: scrambled shRNA). RLU, relative luminescence unit. Knock-down efficiency is shown in C. *, P<0.05; n=3. The results indicate facilitation of mitochondrial but not extrinsic apoptosis by FAM96A.
To generalize and further validate our findings, we injected 1–2-cell zebrafish embryos with antisense morpholino oligonucleotides specifically blocking translation of zebrafish FAM96A (zFAM96A) before inducing apoptosis by UV irradiation (Supplementary Figure 4). Knockdown of zFAM96A expression profoundly reduced the number of apoptotic cells detected by terminal deoxynucleotidyl transferase-mediated biotin-dUTP nick-end labeling (TUNEL) relative to both uninjected embryos and embryos injected with a morpholino bearing 5 mismatch mutations. This effect was indistinguishable from the anti-apoptotic effect of morpholino-induced knock-down of tp53, an established mediator of DNA damage-induced apoptosis, used as a positive control. Thus, FAM96A supports efficient induction of DNA damage-triggered apoptosis in both mammals and zebrafish.
FAM96A Expression Is Diminished in GISTs
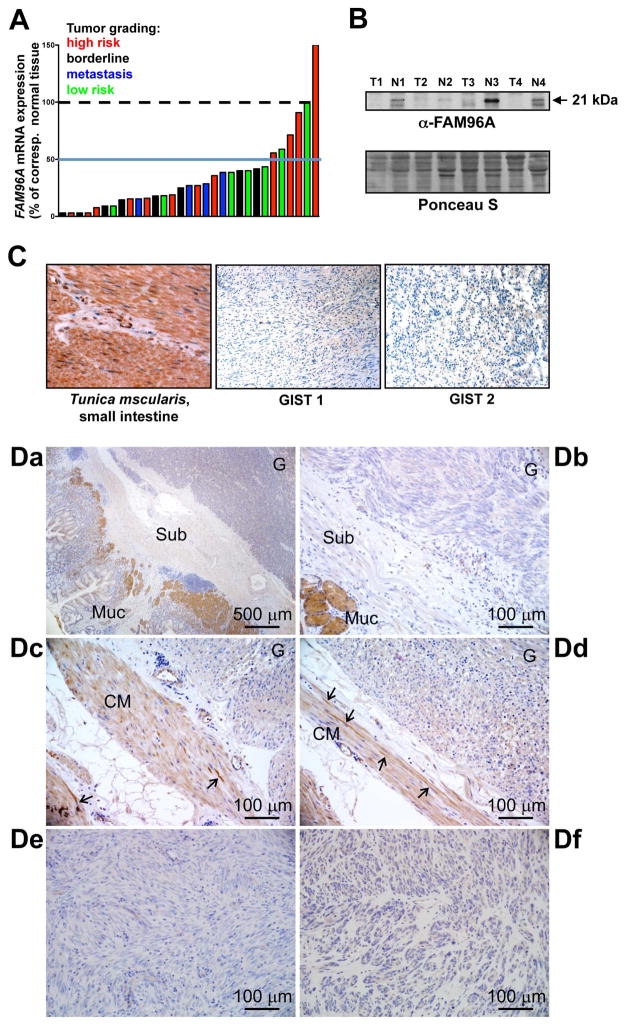
Tumor cells often escape apoptotic stimuli by acquiring mutations that lead to the inactivation or epigenetic silencing of pro-apoptotic tumor suppressors such as TP53.21 We investigated whether FAM96A could also be subject to silencing or genomic deletion in tumors. First, we identified cancers with genomic deletions affecting the FAM96A locus by screening the public CGH database Progenetix (http://www.progenetix.org/) which at the time of last access (June 2014) included 31,915 cases of most human malignancies. 22 This database permits the genomic localization of cytogenetic changes with a minimal resolution of approximately 10 Mb. We detected a decrease in copy number of the genomic sub-region corresponding to the FAM96A locus in several different cancers (Supplementary Table 3). The highest percentage of tumors with a deleted FAM96A locus (15q22; hg19: chr15:62151818-62173260) was observed in GISTs with 165 of the 334 samples represented in the database (49.4%) showing loss, suggesting that reduced FAM96A may play a particularly important role in GIST oncogenesis.
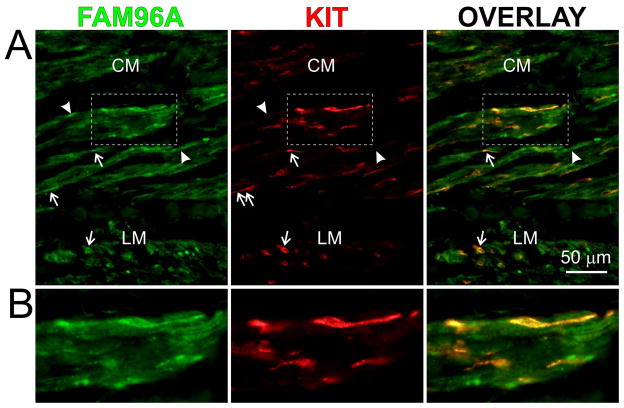
Therefore, we next quantified FAM96A mRNA in 27 primary gastric GISTs, 4 metastases from primary gastric GISTs and 14 corresponding normal stomach tissues by real-time reverse transcription—polymerase chain reaction (RT-PCR; Figure 3A). 25 of the 31 GIST samples (81%) showed a >2-fold reduction in FAM96A mRNA (median fold downregulation: 3.7; range: 33 – −1.5). The decrease in FAM96A mRNA did not correlate with tumor risk for progression (low risk, intermediate risk, high risk or metastatic23). In four additional pairs of gastric GISTs and corresponding normal stomach tissues from the same patients, we also detected reduced expression of FAM96A protein by Western immunoblotting (Figure 3B). To analyze the expression of FAM96A protein in greater detail, we performed immunohistochemistry in two sets of GIST samples. The first set, archived at the University of Erlangen, Germany, included 53 GIST samples represented in a tissue microarray (source: gastric: 36, small bowel: 13, colon: 1, mesentery/peritoneum: 3; histotype: spindle-cell: 27, epithelioid/mixed: 26; risk for progression: very low: 6, low: 15; intermediate: 9, high: 13, no data: 10). No FAM96A protein was detected using the self-raised anti-FAM96A antiserum in any of the GIST samples irrespective of their anatomic site, risk group or morphology, whereas diffuse immunostaining was observed in normal small intestinal tunica musclaris (Figure 3C). A second set of GISTs obtained from the Mayo Clinic biorepositories included samples from 10 patients with 2–24-month neoadjuvant imatinib therapy24 and 10 imatinib-naïve GISTs (see demographic and clinicopathological information in Supplementary Table 4). Immunostaining with a commercial antibody (HPA040459, Sigma-Aldrich, St. Louis, MO, USA) also indicated no or very low FAM96A immunoreactivity in the GIST tissues by blind scoring (median: 0; range: 0–1 on a scale of 0–3) regardless of anatomic site, histotype, mutation or neoadjuvant treatment (Figure 3D), whereas mucosal glands present in the sections displayed strong immunostaining as expected (Figure 3Da, b). Weaker, diffuse cytoplasmic immunolabeling was also observed in the adjacent smooth muscles, which also contained some strongly positive, bipolar, elongated cells morphologically resembling ICCs (Figure 3Dc, d). To verify these cells as ICCs, we performed double immunofluorescent labeling for FAM96A and KIT in 5 normal human gastric tissues. These studies verified the diffuse staining in the smooth muscle cells and revealed that a subset of the bipolar, spindle-shaped cells were KIT+ ICCs (Figure 4). Unlike ICC, KIT+ mast cells of the gastric tunica mucosa did not stain for FAM96A (Supplementary Figure 5).
Figure 3.
FAM96A expression is decreased in GIST. (A) FAM96A mRNA in 31 gastric GISTs (21 frozen and 10 paraffin-embedded samples) was compared to 14 normal stomach mucosal samples obtained from the same patients by real-time RT-PCR. Expression data were normalized to the average expression value of the normal tissues. 29 of the 31 tumors showed reduced FAM96A mRNA irrespective of tumor grading (green bars: low risk, blue: borderline, red: high risk, black: metastasis). (B) Western blot analysis of FAM96A protein expression in four pairs of GISTs (T) and corresponding normal samples (N). The tumor samples showed reduced FAM96A expression. (C) FAM96A immunohistochemistry performed in a tissue microarray containing 53 individual GIST samples using a self-raised antibody. Brown color indicates FAM96A immunoreactivity in the tunica muscularis of the small intestine. The complete loss of FAM96A immunoreactivity in a representative spindle-cell (GIST 1) and an epithelioid GIST (GIST 2) is shown at high magnification (200x). (D) FAM96A immunohistochemistry performed using the commercial antibody HPA040459 (see Supplementary Table 2 for patient and sample information). Da, GIST-2; Db, GIST-16; Dc-d, GIST-6; De, GIST-3; Df, GIST-18. Note no or very low FAM96A immunoreactivity in the tumors (labeled G in Da-d); strong staining in mucosal glands (Muc in Da-b), weaker staining in circular smooth muscle cells (CM in Dc-d) and strong staining in interstitial cells (arrows in Dc-d). Sub, submucosa.
Figure 4.
FAM96A is expressed in KIT+ and KIT− interstitial cells. (A) FAM96A and KIT immunofluorescence in normal human gastric tunica muscularis. (B) Enlarged view of the tissue area identified by the boxes in A. A subset of spindle-shaped, KIT+ ICC (arrows) displayed stronger FAM96A immunoreactivity than smooth muscle cells in the circular and longitudinal layers (CM and LM, respectively). Some KIT− cells with interstitial cell-like morphology (likely FLC) also expressed FAM96A (arrowheads).
FAM96A Is Expressed in Mouse Interstitial Cell Lineages and Is Downregulated in Tumorigenic KitlowCd44+Cd34+ ICC-SCs
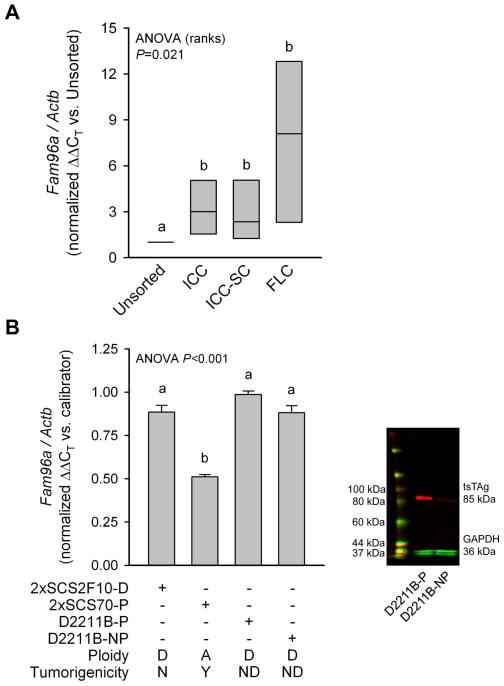
To obtain independent verification of FAM96A expression in interstitial cell lineages, we measured FAM96A mRNA in ICCs, ICC-SCs and FLCs purified from the gastric tunica muscularis of BALB/c mice by FACS1 (Supplementary Figure 1 and Supplementary Table 1). Median FAM96A expression was ~2.4–3 times higher in ICCs and ICC-SCs and 8 times higher in FLCs than in unfractionated cells (Figure 5A). Next, we correlated FAM96A expression in ICC-SC lines with their in-vivo tumorigenicity in NCr-nu/nu mice (Figure 5B). One of the lines examined was an Immortomouse-derived cell line we previously found to give rise to large malignant tumors expressing GIST markers in NCr-nu/nu mice 43–57 days after the grafting of 5×106 cells/mouse (2xSCS70-P, where P indicates that the cells were maintained in the presence of the temperature-sensitive SV40 large T antigen [tsTAg]).1 Expression of FAM96A in these cells was significantly lower than in 2xSCS2F10-D cells, the diploid parent line of the C57BL/6J-derived 2xSCS2F10 cells described in ref.1 Diploid 2xSCS2F10-D cells were not oncogenic as inoculations of NCr-nu/nu mice with up to 9.4×106 cells/mouse failed to cause detectable tumors for up to 116 days (n=2 mice/group). The reduced FAM96A expression in the 2xSCS70-P line was not due to the tsTAg as FAM96A mRNA remained significantly higher in diploid, Immortomouse-derived D2211B cells (the parent line of 2xSCS70 cells) irrespective of the presence or absence of tsTAg (right panel in Figure 5B). Thus, loss of FAM96A expression is detectable in transformed, tumor-initiating ICC-SCs and may occur early in GIST oncogenesis.
Figure 5.
FAM96A is expressed in murine interstitial cells and precursors and is downregulated in tumorigenic ICC-SC. (A) FAM96A mRNA in FACS-purified ICCs, ICC-SCs and FLCs. Gastric muscles were obtained from 31 BALB/c mice (age: 10–20 days) and pooled into four cohorts for FACS. Groups not sharing the same superscript are significantly different by post-hoc multiple comparisons (Student-Newman-Keuls method). FAM96A expression was significantly higher in ICC, ICC-SC and FLC than in unfractionated cells. (B) FAM96A mRNA in tumorigenic and non-tumorigenic ICC-SC lines. 2xSCS2F10-D, 2xSCS70-P, D2211B-P and D2211B-NP cells are precursors or derivatives of the spontaneously transformed, aneuploid ICC-SC lines previously found to cause malignant, GIST marker-positive tumors in NCr-nu/nu mice.1 2xSCS2F10-D cells are diploid parent cells of the C57BL/6J-derived 2xSCS2F10 cells reported in ref.1 2xSCS2F10-D cells did not form tumors in NCr-nu/nu mice. The aneuploid 2xSCS70-P cells are direct descendants of the tumorigenic, Immortomouse-derived cells reported in ref.1 These cells were maintained under conditions permissive (-P) for the SV40 tsTAg. D2211B-P and D2211B-NP cells are diploid parent cells of the 2xSCS70 lines, and they were maintained under conditions permissive (-P) or nonpermissive (-NP) for the tsTAg. Right panel: verification of the loss of tsTAg expression in D2211B-NP cells after 5 days of culturing at 39.5°C in the absence of interferon-γ by Western blotting. A, aneuploid; D, diploid; N, no; Y, yes; ND, not determined. Groups not sharing the same superscript are significantly different by post-hoc multiple comparisons (Holm-Sidak method). Note reduced FAM96A expression in the tumorigenic 2xSCS70-P cell line. The presence or absence of tsTAg did not significantly affect FAM96A expression.
FAM96A Is a Pro-Apoptotic Tumor Suppressor
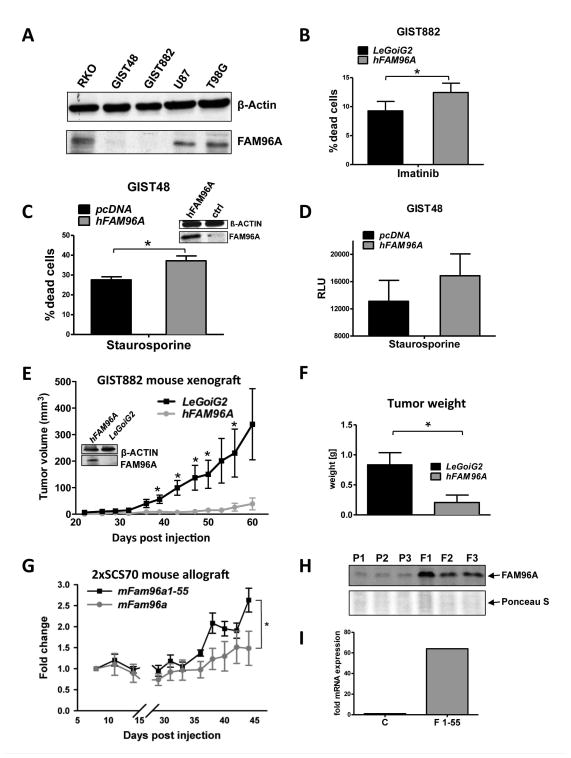
To obtain mechanistic evidence of a role for FAM96A as a tumor suppressor, we investigated the effects of hFAM96A re-expression in GIST and related cell lines in vitro and in tumor models in vivo. Similarly to GIST patient samples, FAM96A protein was barely detectable in the KIT-dependent GIST cell lines GIST882 (imatinib-sensitive)25 and GIST48 (imatinib-resistant) by Western immunoblotting (Figure 6A). Re-expression of hFAM96A by lentiviral transduction significantly enhanced cell death in GIST882 cells upon treatment with the KIT/PDGFRA kinase inhibitor imatinib (Figure 6B). The amount of dead cells (Fig. 6C) and caspase-9 activity (Fig. 6D) also increased in FAM96A-transfected GIST48 cells in response to the nonselective protein kinase inhibitor staurosporine in comparison to empty vector-transfected control cells, supporting a pro-apoptotic function of FAM96A in the mitochondrial apoptosis pathway in GISTs.
Figure 6.
Re-expression of FAM96A in GIST and related cell lines increases apoptosis sensitivity and inhibits tumorigenicity. (A) FAM96A protein expression in GIST and other cancer cell lines detected by Western blotting. GIST48, imatinib-resistant GIST cells; GIST882, imatinib-sensitive GIST cells; U87 and T98G, glioblastoma cells; β-actin, loading control. Both GIST lines displayed severely reduced levels of endogenous FAM96A expression. (B) GIST882 cells lentivirally transduced either with hFAM96A or the empty vector, LeGoiG2, were incubated with 10μM imatinib mesylate for 24 hours. Re-expression of FAM96A was confirmed by immunoblotting (see Figure 6E, inset); cell death was assessed by FCM using 7-AAD. *, P≤0.05; n=4. (C) GIST48 cells were transiently transfected with either hFAM96A or the empty vector, pcDNA3.1. FAM96A re-expression was confirmed by immunoblotting (see Fig. 6D, inset). Apoptosis was induced by incubating the cells with 1μM staurosporine for 16 hours. The proportion of dead cells was determined by FCM using PI exclusion. *, P≤0.05; n=3. (D) Caspase-9 activity was measured in hFAM96A- and empty vector-transfected GIST48 cells 16 hours after treatment with 1μM staurosporine using the Promega Caspase-Glo® 9 Assay. n=3. (E) 7.7×106 GIST882 cells lentivirally transduced either with hFAM96A or the LeGoiG2 empty vector were subcutaneously xenografted into NOD/SCID mice. Overexpression of hFAM96A reduced the frequency of successful engraftment from 8/9 to 5/9 and significantly reduced tumor volumes at 39, 43, 47, 50 and 56 days post-injection (P<0.05). Western blot analysis confirmed stable FAM96A protein overexpression in the FAM96A-transduced GIST882 cells before injection into the mice (inset). (F) The weights of hFAM96A-expressing tumors were significantly reduced upon excision at 60 days post-injection (P<0.05). (G) Tumorigenic 2xSCS70 murine ICC-SC cells were transduced with full-length mFAM96A or the deletion mutant mFAM96A aa1-55, which does not bind APAF, and allografted subcutaneously into NCr-nu/nu mice (5×106 cells/mouse, n=6 mice/group). Overexpression of full-length mFAM96A significantly reduced tumor growth (P<0.05). (H) Verification of stable FAM96A protein overexpression by Western blotting in three tumors each derived from either non-transduced parental (P1–P3) or FAM96A-transduced (F1–F3) 2xSCS70 cells. Ponceau S staining was used as loading control. (I) Because FAM96A aa1-55 is not detected by the self-raised anti-FAM96A antiserum used in this study, we demonstrated overexpression of FAM96A aa1-55 mRNA using quantitative real time RT-PCR. C represents a control pool of RNA isolated from 5 non-transduced parental tumors; F1-55 is a pool of RNA isolated from 5 FAM96A aa1-55-transduced tumors.
To test the effects of hFAM96A on GIST tumorigenicity, we injected 7.7×106 GIST882 cells stably transduced either with hFAM96A-containing or empty lentiviral vector subcutaneously into the flanks of NOD/SCID mice. Whereas 8 of the 9 xenografts of empty vector-transduced GIST882 cells formed a growing tumor, only 5 of the 9 inoculations with the hFAM96A-expressing cells took hold. Furthermore, the tumors formed by the hFAM96A-expressing GIST882 cells only displayed minimal growth (Figure 6E), resulting in significantly lower tumor weights at 60 days after inoculation (Figure 6F). Similarly, tumorigenic 2xSCS70 cells (5×106) transduced to express full-length mFAM96A displayed significantly reduced tumor growth in NCr-nu/nu mice relative to cells transduced with the deletion mutant mFam96a aa1-55 (Figure 6G–I), which does not bind APAF1 (mC1 in Figure 1A). Of note, the FAM96A-transduced RKO cells that showed increased apoptosis in response to oxaliplatin in vitro (Figure 2B) also failed to grow following xenografting into NOD/SCID mice (Supplementary Figure 6).
Discussion
Here, we report that FAM96A, a recently identified member of the cytosolic Fe/S protein assembly machinery,13 is also an evolutionarily conserved pro-apoptotic protein and demonstrate that its genomic deletion or epigenetic silencing in GIST cells and their precursors plays a key role in GIST oncogenesis. Evidence supporting a role for FAM96A as a tumor suppressor for GISTs was generated by several complementary approaches in both human and mouse cells and tissues. Importantly, we found that while normal human and mouse ICCs and murine KitlowCd44+Cd34+ ICC-SCs, the presumed cellular sources of GISTs, express FAM96A mRNA or protein, FAM96A protein expression detected by immunohistochemistry was severely reduced or missing in every tumor in two independent cohorts of human GIST samples containing 53 and 20 tumors each. Furthermore, FAM96A mRNA was reduced at least twofold in 25 of 31 additional GIST samples tested. Similarly, a significant reduction of FAM96A was also observed in transformed, tumorigenic ICC-SCs indicating that loss of FAM96A may occur early in GIST oncogenesis. Our tumor xenograft experiments with the FAM96A-negative GIST882 cell line and allografts of transformed murine ICC-SCs demonstrated that the re-introduction of FAM96A expression inhibited both the engraftment of tumor cells and tumor growth. Together with our finding that GIST cell lines re-expressing FAM96A display increased apoptosis sensitivity, these data strongly support a tumor suppressor function of FAM96A in GISTs. The decrease in FAM96A expression in GISTs was near-universal and, consequently, did not correlate with tumor prognosis suggesting an early loss of apoptosis sensitivity in GIST tumorigenesis. Loss of FAM96A expression in GISTs appears to be due to more than one factor. CGH data listed in the Progenetix database indicate that ~50% of GISTs may have a deletion of the FAM96A locus. In the remaining cases, FAM96A may be epigenetically silenced. Therefore, elucidating the mechanisms of FAM96A repression may lead to new ways to fully restore apoptosis sensitivity in a substantial subset of GIST patients.
While the proposed pro-apoptotic function of FAM96A was supported by both knock-down and overexpression experiments, it is of note that the former indicated more robust effects (see Figure 2C vs. Figures 2A–B). This discrepancy was likely due the lack of a linear relationship between expression levels and apoptosis – i.e., whereas FAM96A knock-down can effectively reduce apoptosis sensitivity, increasing FAM96A expression beyond a certain level may not be able to increase the efficiency of apoptosome assembly and caspase-9 activation any further. Still, in contrast to the relatively modest increase in apoptosis following FAM96A overexpression in vitro, we detected a robust negative effect of FAM96A overexpression on tumor growth in the GIST882 mouse xenograft experiment (Figure 6E). This difference likely reflects the much longer duration of the in-vivo studies, which allows the translation of the smaller changes in apoptosis rates into larger differences in the weights and volumes of tumors derived from the exponentially growing cells.
Our data suggest that FAM96A may exert its pro-apoptotic function by interacting with APAF1, which establishes a critical scaffold for caspase-9 activation.10 The oligomerization of the APAF1 protein at the onset of mitochondrial apoptosis is triggered by the binding of cytosolic cytochrome c, which relieves the auto-inhibition of the APAF1 molecule imposed by its C-terminus. FAM96A is an evolutionarily highly conserved protein, suggesting that it performs important ancient functions. Indeed, FAM96A has recently been identified to be responsible for the stability and maturation of iron regulatory proteins as cytosolic Fe/S protein assembly machinery member 2A (CIA2A).13 FAM96A and its close homolog FAM96B (CIA2B) interact independently with the WD40-repeat protein CIAO1/CIA1, an iron sulfur cluster assembly factor important for the synthesis of vital iron-sulfur proteins.26, 27 Whether this role is related to FAM96A’s pro-apoptotic function remains to be investigated. However, our preliminary expression analysis of key proteins involved in the regulation of iron homeostasis revealed no significant differences upon overexpression of FAM96A in GIST882 cells (Supplementary Figure 7).
KIT/PDGFRA RTK inhibitors such as imatinib mesylate have been the mainstay of treatment of nonresectable primary or recurrent and metastatic GISTs, which are known to be universally resistant to conventional chemo- or radiation therapy.3 Front-line imatinib treatment has extended median survival of patients with advanced GISTs to ~5 years, and adjuvant imatinib has been shown to reduce the risk of recurrence after curative-intent surgery.24 However, the treatment must be maintained indefinitely due to the persistence of GIST cells that cannot be eradicated by KIT/PDGFRA RTK inhibitors alone, which facilitates the emergence of secondary drug resistance that eventually afflicts most patients with advanced GISTs.3 The “inherent resistance” that underlies persistent disease may reflect the pre-existence of stem cell-like clones relying on RTKs different from KIT and PDGFRA.1 Evasion of oncolytic therapy may also occur by the activation of “escape” mechanisms such as autophagy,4 ATP-dependent multidrug resistance channels,28 ineffective immune cell-mediated tumor clearance29 and by reversible exit from the cell cycle due to the activation of the APCCDH1–SKP2–p27Kip1 signaling axis30 and the DREAM complex.2 These mechanisms may further limit cytotoxic responses in cells with stem cell-like characteristics without influencing initial tumor responses31 and thereby contribute to the continuing presence of viable cells in imatinib-treated GISTs.3
A central cell survival mechanism in GISTs is oncogenic KIT/PDGFRA signaling via the phosphatidylinositol-4,5-bisphosphate 3-kinase pathway.3 Whereas cell cycle regulators such as the cyclin-dependent kinase inhibitor genes CDKN2A and CDKN1B are often inactivated in malignant GISTs,3 little is known about the mutations in tumor suppressors that directly control cell survival. For example, inactivating mutations in TP53 have only been detected in 3% of GISTs.5 Thus, identifying novel apoptosome-activating molecules like FAM96A that could facilitate apoptosis in response to cancer treatment is of critical importance and may aid the development of therapies targeting the restoration of mitochondrial apoptosis in GISTs. Intriguingly, FAM96A protein expression may also be reduced in tumors other than GISTs. By immunohistochemical analysis of multi-tumor arrays we found that FAM96A was downregulated in bladder carcinoma compared to the corresponding normal tissues, suggesting that FAM96A acts as a rather general tumor suppressor (unpublished). This conclusion is further supported by our experiments demonstrating diminished tumorigenicity of RKO colon carcinoma cells upon FAM96A overexpression.
Supplementary Material
Brief description of novelty and impact of the work.
Acquired resistance to apoptosis is a hallmark of tumorigenesis and therapy resistance. Here, the authors show that FAM96A is an APAF1-associated pro-apoptotic protein which is profoundly reduced in most gastrointestinal stromal tumors (GISTs). Re-establishment of FAM96A expression increased apoptosis sensitivity and inhibited tumor growth in vivo. These results indicate an important role for FAM96A as a molecular GIST marker and tumor suppressor and suggest that identification of novel apoptosome-activating proteins may aid the development of therapies targeting the restoration of mitochondrial apoptosis in GISTs.
Acknowledgments
Funding:
Supported by grants from the German Research Foundation (DFG ZO 110/1-2 and ZO 110/5-1), the Georg-Speyer-Haus (funded by the German Federal Ministry of Health [BMG] and the Ministry of Higher Education, Research and the Arts of the state of Hessen [HMWK]), the LOEWE Center for Cell and Gene Therapy Frankfurt, the LOEWE Schwerpunkt Oncogenic Signaling Frankfurt, the Austria Genome-Programme GEN-AU and the Lore Saldow Fund, The Life Raft Group (Wayne, NJ, USA) and the National Institute of Diabetes, Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA (R01 DK058185 and P30 DK084567).
We are thankful to Dr. Michael Baudis (Institute of Molecular Life Sciences, University of Zurich, Switzerland) for help with the Progenetix database. We also thank Dr. Jason T. Lewis (Department of Pathology and Laboratory Medicine, Mayo Clinic, Rochester, Minnesota, USA) for providing blind scores of FAM96A expression in GIST samples. FAM96A staining and genotyping of GIST samples were performed at the Mayo Clinic Pathology Research Core (Director: Dr. Thomas J. Flotte) and in the Cytopathology and Molecular Anatomic Pathology Laboratory (Director: Dr. Andre M. Oliveira), respectively. GIST48 and GIST882 cells were kindly provided by Jonathan A. Fletcher (Department of Pathology, Brigham and Women’s Hospital, Boston, MA, USA) and Sebastian Bauer (Sarcoma Center, West German Cancer Center, University of Essen, Medical School, Essen, Germany). The excellent technical assistance of Bettina Strohmeier (Institute of Pathology, Medical University of Graz, Austria), Rudolf Jung (Institute for Pathology, University Hospital Erlangen, Germany) and Laurie A. Popp (Mayo Clinic) is gratefully acknowledged. We thank Florian Greten (Georg-Speyer-Haus, Frankfurt, Germany) for critical evaluation of the manuscript.
Abbreviations
- aa
amino acid(s)
- AF
Alexa Fluor
- APAF1
apoptotic peptidase activating factor 1
- ATCC
American Type Culture Collection
- AVEN
Aventin, caspase activation inhibitor
- b
base
- bp
base pair
- CDKN2A
cyclin-dependent kinase inhibitor 2A
- CDKN1B
cyclin-dependent kinase inhibitor 1B
- CGH
comparative genomic hybridization
- DUF
domain of unknown function
- FACS
fluorescence-activated cell sorting
- FasL
Fas Ligand
- FCM
flow cytometry
- Fe/S
iron-sulfur
- FLC
fibroblast-like cell
- GIST
gastrointestinal stromal tumor
- GST
glutathione-S-transferase
- h
human
- ICC
interstitial cell of Cajal
- ICC-SC
ICC stem cell
- KIT
KIT receptor tyrosine kinase
- m
mouse/murine
- PBS
phosphate-buffered saline
- PDGFRA
plateled-derived growth factor receptor α
- PI
propidium iodide
- RTK
receptor tyrosine kinase
- RT-PCR
reverse transcription—polymerase chain reaction
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- shRNA
short hairpin RNA
- tsTAg
temperature-sensitive SV40 large T antigen
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling
- UV
ultraviolet
- z
zebrafish
Footnotes
Disclosures:
No conflicts.
Author Contributions:
B.S., R.Pi., S.B.M.F., K.V., S.B., J.J., R.B.-K., J.W., I.Z., J.H., V.D., M.H., M.N., S.J., R.J.R., A.A., S.S., M.R.B., Y.H., D.T.A., S.A.S.: acquisition, analysis and interpretation of data; K.Z. and R.Pe.: material support; acquisition, analysis and interpretation of data; K.M.R.L. and M.L.K.: provision of critical research material; T.O.: experimental design, analysis and interpretation of data, drafting and critical revision of the manuscript for important intellectual content; M.Z.: experimental design, analysis and interpretation of data, drafting of the manuscript.
References
- 1.Bardsley MR, Horvath VJ, Asuzu DT, Lorincz A, Redelman D, Hayashi Y, Popko LN, Young DL, Lomberk GA, Urrutia RA, Farrugia G, Rubin BP, et al. Kitlow stem cells cause resistance to Kit/platelet-derived growth factor alpha inhibitors in murine gastrointestinal stromal tumors. Gastroenterology. 2010;139:942–52. doi: 10.1053/j.gastro.2010.05.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boichuk S, Parry JA, Makielski KR, Litovchick L, Baron JL, Zewe JP, Wozniak A, Mehalek KR, Korzeniewski N, Seneviratne DS, Schoffski P, Debiec-Rychter M, et al. The DREAM complex mediates GIST cell quiescence and is a novel therapeutic target to enhance imatinib-induced apoptosis. Cancer Res. 2013;73:5120–9. doi: 10.1158/0008-5472.CAN-13-0579. [DOI] [PubMed] [Google Scholar]
- 3.Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer. 2011;11:865–78. doi: 10.1038/nrc3143. [DOI] [PubMed] [Google Scholar]
- 4.Gupta A, Roy S, Lazar AJ, Wang WL, McAuliffe JC, Reynoso D, McMahon J, Taguchi T, Floris G, Debiec-Rychter M, Schoffski P, Trent JA, et al. Autophagy inhibition and antimalarials promote cell death in gastrointestinal stromal tumor (GIST) Proc Natl Acad Sci U S A. 2010;107:14333–8. doi: 10.1073/pnas.1000248107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henze J, Muhlenberg T, Simon S, Grabellus F, Rubin B, Taeger G, Schuler M, Treckmann J, Debiec-Rychter M, Taguchi T, Fletcher JA, Bauer S. p53 modulation as a therapeutic strategy in gastrointestinal stromal tumors. PLoS One. 2012;7:e37776. doi: 10.1371/journal.pone.0037776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubin BP, Heinrich MC, Corless CL. Gastrointestinal stromal tumour. Lancet. 2007;369:1731–41. doi: 10.1016/S0140-6736(07)60780-6. [DOI] [PubMed] [Google Scholar]
- 7.Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, Muhammad Tunio G, Matsuzawa Y, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–80. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 8.Grover M, Bernard CE, Pasricha PJ, Parkman HP, Abell TL, Nguyen LA, Snape W, Shen KR, Sarr M, Swain J, Kendrick M, Gibbons S, et al. Platelet-derived growth factor receptor alpha (PDGFRalpha)-expressing “fibroblast-like cells” in diabetic and idiopathic gastroparesis of humans. Neurogastroenterol Motil. 2012;24:844–52. doi: 10.1111/j.1365-2982.2012.01944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurahashi M, Zheng H, Dwyer L, Ward SM, Koh SD, Sanders KM. A functional role for the ‘fibroblast-like cells’ in gastrointestinal smooth muscles. J Physiol. 2011;589:697–710. doi: 10.1113/jphysiol.2010.201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenner D, Mak TW. Mitochondrial cell death effectors. Curr Opin Cell Biol. 2009;21:871–7. doi: 10.1016/j.ceb.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Fadeel B, Ottosson A, Pervaiz S. Big wheel keeps on turning: apoptosome regulation and its role in chemoresistance. Cell Death Differ. 2008;15:443–52. doi: 10.1038/sj.cdd.4402265. [DOI] [PubMed] [Google Scholar]
- 12.Ledgerwood EC, Morison IM. Targeting the apoptosome for cancer therapy. Clin Cancer Res. 2009;15:420–4. doi: 10.1158/1078-0432.CCR-08-1172. [DOI] [PubMed] [Google Scholar]
- 13.Stehling O, Mascarenhas J, Vashisht AA, Sheftel AD, Niggemeyer B, Rosser R, Pierik AJ, Wohlschlegel JA, Lill R. Human CIA2A-FAM96A and CIA2B-FAM96B integrate iron homeostasis and maturation of different subsets of cytosolic-nuclear iron-sulfur proteins. Cell Metab. 2013;18:187–98. doi: 10.1016/j.cmet.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berrar D, Dubitzky W, Solinas-Toldo S, Bulashevska S, Granzow M, Conrad C, Kalla J, Lichter P, Eils R. A database system for comparative genomic hybridization analysis. IEEE Eng Med Biol Mag. 2001;20:75–83. doi: 10.1109/51.940052. [DOI] [PubMed] [Google Scholar]
- 15.Almeida MS, Herrmann T, Peti W, Wilson IA, Wuthrich K. NMR structure of the conserved hypothetical protein TM0487 from Thermotoga maritima: implications for 216 homologous DUF59 proteins. Protein Sci. 2005;14:2880–6. doi: 10.1110/ps.051755805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen KE, Richards AA, Ariffin JK, Ross IL, Sweet MJ, Kellie S, Kobe B, Martin JL. The mammalian DUF59 protein Fam96a forms two distinct types of domain-swapped dimer. Acta Crystallogr D Biol Crystallogr. 2012;68:637–48. doi: 10.1107/S0907444912006592. [DOI] [PubMed] [Google Scholar]
- 17.Mas C, Chen KE, Brereton IM, Martin JL, Hill JM. Backbone resonance assignments of the monomeric DUF59 domain of human Fam96a. Biomol NMR Assign. 2013;7:117–20. doi: 10.1007/s12104-012-9390-1. [DOI] [PubMed] [Google Scholar]
- 18.Ouyang B, Wang L, Wan S, Luo Y, Lin J, Xia B. Solution structure of monomeric human FAM96A. J Biomol NMR. 2013;56:387–92. doi: 10.1007/s10858-013-9746-6. [DOI] [PubMed] [Google Scholar]
- 19.Kulms D, Schwarz T. Molecular mechanisms of UV-induced apoptosis. Photodermatol Photoimmunol Photomed. 2000;16:195–201. doi: 10.1034/j.1600-0781.2000.160501.x. [DOI] [PubMed] [Google Scholar]
- 20.Li K, Li Y, Shelton JM, Richardson JA, Spencer E, Chen ZJ, Wang X, Williams RS. Cytochrome c deficiency causes embryonic lethality and attenuates stress-induced apoptosis. Cell. 2000;101:389–99. doi: 10.1016/s0092-8674(00)80849-1. [DOI] [PubMed] [Google Scholar]
- 21.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Baudis M, Cleary ML. Progenetix.net: an online repository for molecular cytogenetic aberration data. Bioinformatics. 2001;17:1228–9. doi: 10.1093/bioinformatics/17.12.1228. [DOI] [PubMed] [Google Scholar]
- 23.Miettinen M, Lasota J. Gastrointestinal stromal tumors: pathology and prognosis at different sites. Semin Diagn Pathol. 2006;23:70–83. doi: 10.1053/j.semdp.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Dematteo RP, Ballman KV, Antonescu CR, Maki RG, Pisters PW, Demetri GD, Blackstein ME, Blanke CD, von Mehren M, Brennan MF, Patel S, McCarter MD, et al. Adjuvant imatinib mesylate after resection of localised, primary gastrointestinal stromal tumour: a randomised, double-blind, placebo-controlled trial. Lancet. 2009;373:1097–104. doi: 10.1016/S0140-6736(09)60500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bauer S, Yu LK, Demetri GD, Fletcher JA. Heat shock protein 90 inhibition in imatinib-resistant gastrointestinal stromal tumor. Cancer Res. 2006;66:9153–61. doi: 10.1158/0008-5472.CAN-06-0165. [DOI] [PubMed] [Google Scholar]
- 26.Gari K, Leon Ortiz AM, Borel V, Flynn H, Skehel JM, Boulton SJ. MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science. 2012;337:243–5. doi: 10.1126/science.1219664. [DOI] [PubMed] [Google Scholar]
- 27.Stehling O, Vashisht AA, Mascarenhas J, Jonsson ZO, Sharma T, Netz DJ, Pierik AJ, Wohlschlegel JA, Lill R. MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science. 2012;337:195–9. doi: 10.1126/science.1219723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Theou N, Gil S, Devocelle A, Julie C, Lavergne-Slove A, Beauchet A, Callard P, Farinotti R, Le Cesne A, Lemoine A, Faivre-Bonhomme L, Emile JF. Multidrug resistance proteins in gastrointestinal stromal tumors: site-dependent expression and initial response to imatinib. Clin Cancer Res. 2005;11:7593–8. doi: 10.1158/1078-0432.CCR-05-0710. [DOI] [PubMed] [Google Scholar]
- 29.Edris B, Willingham S, Weiskopf K, Volkmer AK, Volkmer JP, Muhlenberg T, Weissman IL, van de Rijn M. Use of a KIT-specific monoclonal antibody to bypass imatinib resistance in gastrointestinal stromal tumors. Oncoimmunology. 2013;2:e24452. doi: 10.4161/onci.24452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Perdreau SA, Chatterjee P, Wang L, Kuan SF, Duensing A. Imatinib mesylate induces quiescence in gastrointestinal stromal tumor cells through the CDH1-SKP2-p27Kip1 signaling axis. Cancer Res. 2008;68:9015–23. doi: 10.1158/0008-5472.CAN-08-1935. [DOI] [PubMed] [Google Scholar]
- 31.Chao MP, Majeti R, Weissman IL. Programmed cell removal: a new obstacle in the road to developing cancer. Nat Rev Cancer. 2012;12:58–67. doi: 10.1038/nrc3171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.