Abstract
RpBphP2 and RpBphP3, two tandem bacteriophytochromes from the photosynthetic bacterium R. palustris, share high sequence identity but exhibit distinct photoconversion behavior. Unlike the canonical RpBphP2, RpBphP3 photoconverts to an unusual nearred-absorbing (Pnr) state; both are required for synthesis of light harvesting complexes under low-light conditions. Here we report the crystal structures of the photosensory core modules of RpBphP2 and RpBphP3. Despite different quaternary structures, RpBphP2 and RpBphP3 adopt nearly identical tertiary structures. The RpBphP3 structure reveals “tongue-and-groove” interactions at the interface between the GAF and PHY domains. A single mutation in the PRxSF motif at the GAF-PHY interface abolishes light-induced formation of the Pnr state in RpBphP3, possibly due to altered structural rigidity of the chromophore-binding pocket. Structural comparisons suggest that long-range signaling involves structural rearrangement of the helical spine at the dimer interface. These structures together with mutational studies provide insights into photoconversion and long-range signaling mechanism in phytochromes.
Introduction
Phytochromes are a superfamily of photoreceptors that perceive ambient light signals and mediate a wide range of important physiological processes in plants, fungi and bacteria (Rockwell et al., 2006). With the recent discovery of cyanobacteriochromes (CBCRs), this family of bilin-based photoreceptors confers light sensitivity over the entire UV/visible range of the solar spectrum (Rockwell et al., 2012). Plant phytochromes and bacteriophytochromes (BphPs) are highly homologous in their photosensory core modules (PCM), in which three modular domains - PAS (Per-ARNT-Sim), GAF (cGMP phosphodiesterase/adenylate cyclase/FhlA transcriptional activator) and PHY (phytochrome-specific)(Rockwell et al., 2006) - share a common PAS fold. Typically, a histidine kinase (HK) domain is located at the C-terminus of BphPs that undergoes light-dependent auto-phosphorylation. BphP often partners with a downstream response regulator to regulate gene expression in response to light via the two-component signaling pathway. In plants, the C-terminal region of phytochromes consists of two additional PAS domains followed by a histidine-kinase related domain (HKRD), and light responses are mediated via light-dependent interactions with phytochrome-interacting factors (PIFs) and COP1 (constitutively photomorphogenesis 1), a central regulator of light signaling pathways in plants (Li et al., 2011).
Phytochromes utilize linear tetrapyrroles as chromophores to perceive light signals via reversible photoconversion between red-absorbing (Pr) and far-red-absorbing (Pfr) states. The crystal structures of the photosensory core modules (PCMs) determined for several BphPs reveal extensive structural similarities despite different origins and distinct signaling states (Essen et al., 2008; Yang et al., 2008, 2009; Bellini and Papiz, 2012a; Mailliet et al., 2011; Takala et al., 2014; Burgie et al., 2014). The modular photosensory domains are linearly arranged in both sequence and space, but they are integrated via a “figure-of-eight” knot between the PAS and GAF domains and a long, extended arm of the PHY domain. These two unusual structural features bring together functional regions of the PAS and PHY domains close to the chromophore embedded in the GAF domain (Essen et al., 2008; Yang et al., 2008). In particular, the arm of the PHY domain shields the opening of the chromophore-binding site.
BphPs exhibit diverse photoconversion behaviors. The canonical BphPs adopt the Pr state (λmax~700nm) in the dark and photo-convert to the Pfr (λmax~750nm) state. However, bathy-BphPs such as PaBphP from P. aeruginosa, RpBphP1 and RpBphP5 from R. palustris adopt the Pfr state in the dark and photo-convert to the Pr state (Tasler et al., 2005; Njimona and Lamparter, 2011; Bellini and Papiz, 2012a). While the dark-adapted Pr or Pfr structures have been readily crystallized, the crystal structure of a photoactive BphP in a homogeneous, light state remains to be a rarity. One exception is DrBphP, for which both dark Pr and light Pfr structures of different diffraction quality have been reported (Takala et al., 2014). Extensive spectroscopic data and structural studies establish that Pr/Pfr photoconversion in both the forward and reverse directions involves isomerization around the C15=C16 double bond in the bilin chromophore (Rockwell et al., 2006, 2012). Other major differences between the Pr and Pfr structures reside in the arm region of the PHY domain, where a structural segment containing the conserved PRxSF sequence motif appears to undergo transition from a β-strand to an α-helix during Pr/Pfr photoconversion (Takala et al., 2014; Yang et al., 2009). Two key questions remain. What structural features within the PCM affect the photoconversion behavior of BphPs? How do structural signals propagate from the PCM to the HK domain, where they ultimately generate a biological signal?
Here we present the crystal structures of the PCMs in two highly homologous BphPs from the photosynthetic bacterium R. palustris - RpBphP2 and RpBphP3. RpBphP2 and RpBphP3 are encoded by two tandem genes (rpa3015 and rpa3016) in the same operon and interact with the same downstream response regulator RPA3017 (Giraud et al., 2005). Although both adopt the Pr state (λmax ~700 nm) in the dark, upon illumination at 700 nm RpBphP2 exhibits canonical photochemistry and photo-converts to the Pfr state (λmax ~750 nm) while RpBphP3 converts to an unusual Pnr state (λmax ~650 nm). Their contrasting photochemical properties despite high sequence homology allow us to identify key interactions that govern photoconversion via comparative structural and mutagenesis studies.
We also aim to identify structural elements and/or motifs that are important for transmitting structural signals over long distances. The long linker helices between modular domains have been denoted signaling helices and implicated in signal propagation from sensory to effector domains (Anantharaman et al., 2006; Möglich et al., 2009). Different BphPs seem to share general signaling principles – irrespective of their dark-adapted state, the Pr state always displays higher HK activity than the Pfr state (Giraud et al., 2005; Psakis et al., 2011; Yang et al., 2011). In this work, we compare BphPs in different photochemical states and/or with different dimeric scaffolds, to explore possible conformational rearrangements associated with long-range signaling that enables light-dependent regulation of the spatially distant HK activity.
Results
Crystal structure of RpBphP2-PCM
Two crystal structures of RpBphP2 were independently determined in the dark-adapted Pr state using different constructs. Both contain the identical protein sequence spanning residues 1-505 of RPA3015 from R. palustris CGA009 but carry the 6xHis affinity tag at different termini, hereafter denoted RpBphP2-Ntag and RpBphP2-Ctag. The crystal structure of RpBphP2-Ctag was determined in space group P61 by molecular replacement (PHASER in CCP4) using the chromophore-binding domain of RpBphP3 (PDB ID 2OOL) as a search model, and refined at 3.4 Å resolution to the final R-factor and free R-factor of 0.236 and 0.292, respectively (PHENIX; Table 1). RpBphP2-Ntag contains 20 additional residues at the N-terminus including the tag sequence. The initial model of RpBphP2-Ntag was built based on electron densities via phase combination of single-wavelength anomalous dispersion (SAD) from selenomethionine-containing crystals and molecular replacement (Fig. S1a). The final model was refined at 3.25 Å resolution to the final R-factor and free R-factor of 0.25 and 0.31, respectively (Table 1).
Table 1.
Data collection and structure refinement statistics of RpBphP2-PCM crystals.
| Crystal | RpBphP2-Ctag | RpBphP2-Ntag (native) | RpBphP2-Ntag (SeMet) |
|---|---|---|---|
| Resolution (Å) | 50–3.4 (3.49–3.40)/50–4.4* | 50–3.25 (3.3–3.25) | 50–3.5 (3.56–3.5) |
| Completeness (%) | 66 (11.5)/97.7* | 99.5 (99.8) | 94 (60) |
| Rmerge | 0.041(0.21)/0.035* | 0.030 (0.39) | 0.07 (0.34) |
| Redundancy | 4.5 (1.2)/5.5* | 10.0 (9.3) | 9.5 (5.5) |
| I/σI | 14.8 (1.9)/21.9* | 25.5(1.6) | 25.7 (2.75) |
| Space group | P61 | P61 | P61 |
| Cell parameters (Å) | a=b=176.79; c=95.91 | a=b=172.15 c=95.22 |
a=b=174.62 c=95.72 |
| beamline | APS 21-IDG | APS 14-BMC | APS 19-ID |
| Refinement | |||
| R-factor | 0.236 (0.32) | 0.25 (0.34) | |
| Free R-factor | 0.292 (0.37) | 0.31 (0.38) | |
| Resolution (Å) | 20–3.4 (3.66–3.40) | 20–3.25 (3.37–3.25) | |
| Geometry | |||
| r.m.s.d bond length(Å) | 0.012 | 0.017 | |
| r.m.s.d. bond angle(°) | 2.36 | 2.32 | |
| Structure | |||
| Protein | Chain A, B | Chain A, B | |
| Ligand/water | 2 BV/18 waters | 2 BV/0 waters | |
| Ramachandran stats | |||
| Favored | 93% | 90.7% | |
| Allowed/Disallowed | 6.6%/0.4% | 8.5%/0.8% | |
| PDB ID | 4R6L | 4S21 |
Data collection statistics are reported in two different resolution ranges to reflect the anisotropic nature of the dataset.
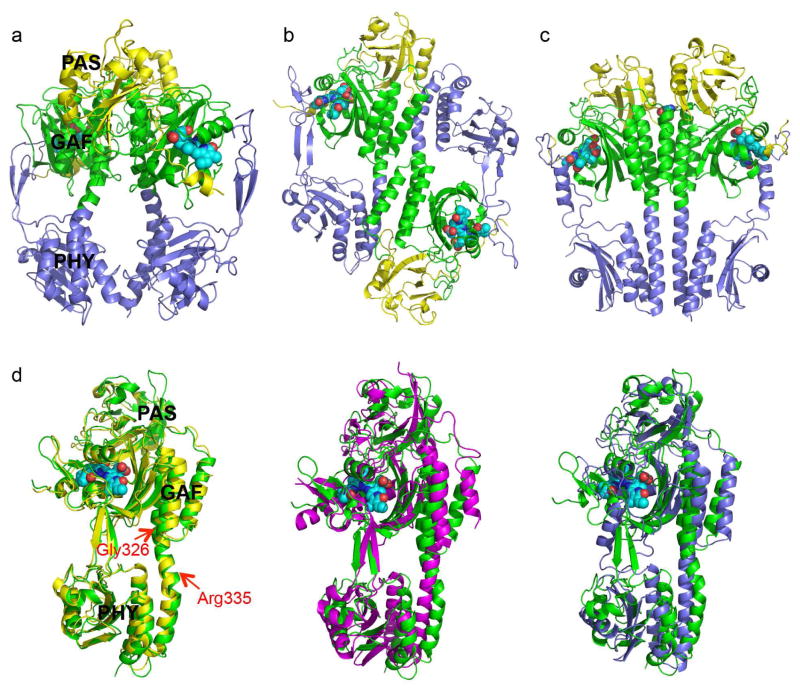
Despite differences in crystallization conditions and constructs, both the RpBphP2-Ntag and RpBphP2-Ctag structures adopt the same space group P61 with nearly identical cell parameters and molecular packing, in which two monomers form a parallel dimer in the asymmetric unit. This suggests that the affinity tags do not influence either the mode of dimerization or the inter-molecular packing. Structural alignment based on one monomer reveals subtle displacements in the other (Fig. S2a). Both crystals exhibit anisotropic diffraction with high R-merge factors and/or incompleteness in high-resolution shells (Table 1). The overall quality of electron densities is better in one monomer (chain A) than the other (chain B), probably due to subtle differences in molecular packing. We hereafter use RpBphP2-Ctag to describe and discuss the RpBphP2 structure unless mentioned otherwise.
RpBphP2 forms a head-to-head, tail-to-tail parallel dimer and buries a surface area of 1323Å2 at the dimer interface (PISA (Krissinel and Henrick, 2007)) (Fig. 1a). The PAS, GAF and PHY domains from each RpBphP2 monomer are juxtaposed via a helical spine along the dimer interface. An unusually large gap is present at the dimer interface between the GAF and PHY domains (Fig. 1a). This sharply contrasts with the crystal structure of PaBphP in the Pfr state, in which the parallel GAF-PHY linker helices are nearly straight (Fig. 1c) and form a more extended, tight dimer interface that buries a significantly larger surface area of 1893Å2 (PISA (Krissinel and Henrick, 2007)).
Fig. 1. Crystal structures of RpBphP2 and RpBphP3.
a) Ribbon diagram of the parallel dimer of RpBphP2-Ctag in the Pr state. The PAS (yellow), GAF (green) and PHY (blue) domains are juxtaposed along the dimer interface, where a large void is found. The BV chromophores are shown in cyan spheres. b) Ribbon diagram of the anti-parallel dimer of RpBphP3 in the Pr state. c) Parallel dimer structure of PaBphP in the Pfr state exhibits a tighter dimer interface (PDB ID: 3NHQ). d) Pairwise comparison of the tertiary structures of BphP monomers: RpBphP3 (green; as reference); RpBphP2 (yellow), Cph1 (magenta) and PaBphP (blue). The red arrows mark the locations of kink at the GAF-PHY linker helix (RpBphP3 numbering). Related to Figure S1 and Figure S2a.
Crystal structure of RpBphP3-PCM
The RpBphP3-PCM construct includes residues 1-521 of RPA3016 from R. palustris CGA009 and a 6xHis affinity tag at the C-terminus. RpBphP3-PCM was crystallized in the dark at 20°C with a final protein concentration of 10 mg/ml and 0.1M magnesium formate. The crystal structure was determined in space group P3121 by molecular replacement (PHASER(McCoy et al., 2007)) using the crystal structure of a shorter RpBphP3 construct lacking the PHY domain as a search model (PDB ID: 2OOL(Yang et al., 2007)). The initial model for the PHY domain was manually built based on the density-modified electron densities derived from a partial MR solution. The N-terminal 24 residues of the PAS domain and seven residues from the C-terminal His-tag are not visible in the electron density map. The arm region spanning residues 454-494 shows well-resolved electron density (Fig. S1b). The final model was refined at 2.85Å resolution to the R-factor and free R-factor of 0.185 and 0.26, respectively (Table 2).
Table 2.
Data collection and refinement statistics of the crystal structures of RpBphP3-PCM and RpBphP3-CBD.
| Crystal | RpBphP3-PCM | RpBphP3-CBD |
|---|---|---|
| Resolution (Å) | 50–2.85 (2.95–2.85) | 50–2.1 (2.2–2.1) |
| Completeness (%) | 98.2 (84.4) | 97.0(80.2) |
| Rmerge | 0.053 (0.63) | 0.045 (0.49) |
| Redundancy | 19.1 (6.4) | 5.8 (2.5) |
| I/σI | 52.7(1.05) | 25.7(1.0) |
| Space group | P3121 | P321 |
| Cell parameters (Å) | a=b=143.31 c=120.51 |
a=b=151.18 c=75.75 |
| Beamline | APS 21-IDG | APS 21-IDG |
| Refinement | ||
| R-factor | 0.185 (0.361) | 0.174 (0.281) |
| Free R-factor | 0.259 (0.436) | 0.217 (0.354) |
| Resolution(Å) | 20–2.85 (2.93–2.85) | 20–2.1 (2.2–2.1) |
| Geometry | ||
| r.m.s.d bond length(Å) | 0.024 | 0.012 |
| r.m.s.d bond angle(°) | 2.468 | 1.588 |
| Structure | ||
| Protein | 2 molecules | 2 molecules |
| Ligand | 2 biliverdin (BV) | 2 biliverdin (BV) |
| Water | 23 | 252 |
| Ramachandran plot | ||
| Favored | 94.2% | 97.5% |
| Allowed | 5.1% | 2.5% |
| Disallowed | 0.7% | 0% |
| PDB ID | 4R70 | 2OOL (updated)* |
In contrast to the parallel dimer of RpBphP2, two RpBphP3 monomers form a head-to-tail, anti-parallel dimer with a buried surface area of 1993Å2 at the dimer interface (Fig. 1b). The GAF domain of one monomer is packed against the PHY domain of the other via a helical bundle and the two PAS domains are located at opposite ends of the dimer (Fig. 1b). The tertiary structures of the monomers of RpBphP2 and RpBphP3 are nearly identical with an overall r.m.s.d. over the main chain atoms of 461 aligned residues of 1.56Å (Fig. 1d). The arm of the PHY domain resembles the equivalent “tongue” of Cph1 (Essen et al., 2008; Mailliet et al., 2011), in which two anti-parallel β-strands in an extended conformation adopt an overall right-handed twist. The biliverdin chromophore adopts the ZZZssa configuration as expected for the Pr state (Fig. S1c). As in the RpBphP2 structure, the linker helix spanning the GAF and PHY domains contains two significant kinks at similar locations, one at Gly326 near the junction between the GAF and PHY domains and the other near Arg335 where the helix is partially unwound (RpBphP3 numbering) (Fig. 1).
Notably, the crystal structures of RpBphP3-PCM and the shorter RpBphP3-CBD construct (residues 1-337) (Yang et al., 2007) can be well aligned in their common PAS and GAF domains, despite quite different modes of dimerization. RpBphP3-CBD forms parallel dimers in the crystal lattice whereas RpBphP3-PCM forms anti-parallel dimers (Fig. 3a). Not surprisingly, subtle structural differences are present in regions of the GAF domain (Tyr272, Asp216, Leu207 and Tyr185) that make direct contact with the PHY domain. Specifically, Tyr272 moves closer to the chromophore in the PCM structure by about 1.3Å and forms a hydrogen bond (2.7 Å) with the side chain of Asp216. This interaction is absent in the CBD structure. Rings A and D are concomitantly displaced in the same direction by ~0.8Å (Fig. 3b).
Figure 3. Structural comparisons between the CBD and PCM constructs of RpBphP3.
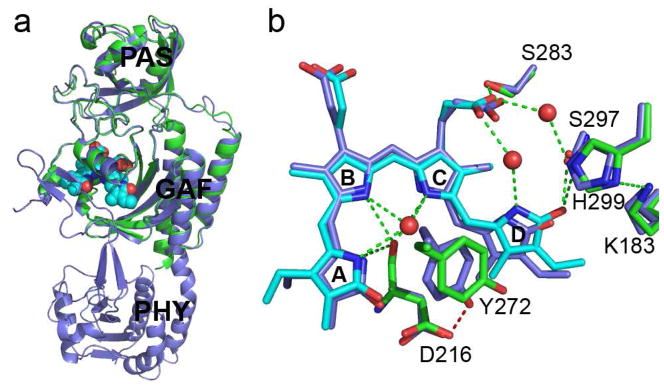
a) Superposition of the RpBphP3-PCM (blue) and RpBphP3-CBD (green/cyan) monomers shows nearly identical tertiary structures in the PAS and GAF domains; b) Structural differences in the chromophore-binding pocket between PCM (cyan and green) and CBD (blue) of RpBphP3. Dashed green lines represent hydrogen bonds. Water molecules are marked in red spheres. Related to Figure S1c.
The tertiary structures of various BphPs are quite similar. The overall r.m.s. deviation between the main chain atoms of the aligned residues is 3.2 Å between RpBphP3 and Cph1, and ~4.7 Å between RpBphP3 and PaBphP, respectively. The differences among the structures of RpBphP2, RpBphP3, Cph1 and PaBphP (Fig. 1d) lie mainly in the arm and the long helix linking the GAF and PHY domains. As a result, the relative orientation of the GAF and PHY domains differs in these structures.
The dimer scaffolds
RpBphP2 and RpBphP3 adopt completely different quaternary structures despite their nearly identical tertiary structures (Fig. 1). Such parallel and antiparallel arrangements have been observed in various crystal structures of BphPs that lack the C-terminal HK domains. Specifically, Cph1, Cph1-T263F, Cph2 and RpBphP1 (Essen et al., 2008; Mailliet et al., 2011; Anders et al., 2013; Bellini and Papiz, 2012a) adopt anti-parallel scaffolds, while DrBphP, RpBphP3-CBD, PaBphP (Wagner et al., 2005, 2007; Yang et al., 2007, 2008, 2009) and the recently reported plant AtPhyB (Burgie et al., 2014) form parallel dimers.
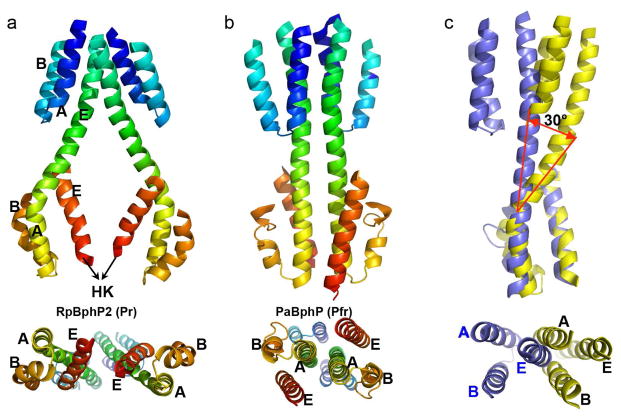
We denote the three helices in the GAF domain that are bundled at the dimer interface as GAF-hA/hB/hE; and the three equivalent helices in the PHY domain as PHY-hA/hB/hE (Fig. 2a). Each 3-helix bundle is stabilized largely via hydrophobic interactions. The GAF-hE and PHY-hA helices fuse to form a long, continuous but curved helix which we denote the GAF-PHY linker helix. This nomenclature emphasizes the topological relationship and equivalency among interfacial helices from different modular domains. In the parallel framework of RpBphP2, two 3-helix bundles including two GAF-PHY linker helices further aggregate to form a 6-helix bundle at the dimer interface in the GAF domain (Fig. 2). The GAF-PHY linker helices are well separated as they enter the PHY domain, in which direct contacts between two PHY-hE helices bring together two rather twisted 3-helix bundles (Fig. 2a). Although equivalent helices at the dimer interface of RpBphP2 and PaBphP structures are comparable in length, structural alignment according to the 3-helix bundle of the PHY domain reveals a large rotation of about 30° of the GAF domain (Fig. 2c), which apparently arises from the striking curve of the GAF-PHY linker helix in RpBphP2.
Fig. 2. Helices at the dimer interface.
Interfacial helices in the parallel dimer structures of RpBphP2 (a) and PaBphP (b) are shown in rainbow colors from blue to red corresponding to the residue order from the N- to C- termini. Equivalent helices A, B and E in the GAF and PHY domains are labeled. Lower panels represent the bottom views of the PCM dimer, as seen from the HK domain. c) Structural alignment of interfacial helices in RpBphP2 (yellow) and PaBphP (blue) according to the 3-helix bundle of the PHY domain. The lower panel represents the top view of the superposition.
Our RpBphP2 parallel dimer structure bears a significant resemblance to the cryo-EM structure of the full-length DrBphP in the Pr state, which also displays a striking hole at the dimer interface (Li et al., 2010). Li and colleagues reported that the PHY domain must be rotated by about 30° from its position in the PaBphP structure to account for the cryoEM densities associated with the PHY domain of DrBphP. This rotation is consistent with structural differences between RpBphP2 and PaBphP (Fig. 2c). In other words, DrBphP and RpBphP2 adopt very similar dimeric scaffolds in the Pr state despite differences in sequence, biological source and the methods by which the scaffolds were determined.
If the parallel dimer structures of RpBphP2 and PaBphP indeed represent the Pr and Pfr states respectively, the helical bundles at the dimer interface must undergo significant structural rearrangements during Pr/Pfr photoconversion. Specifically, the three-helix bundles of the PHY domains in the Pr state would untwist while the GAF-PHY linker helices straighten, as in the Pfr structure of PaBphP (Fig. 2b). In the full-length context, the C-terminal helix (PHY-hE) as well as the DHp segment of the HK domain would move concomitantly. It is noteworthy that the helical hairpin structure of the DHp domain is expected to promote parallel dimerization (Casino et al., 2009).
We further propose a model for the full-length RpBphP2 based on the parallel scaffold of the RpBphP2-PCM structure and the crystal structure of a homologous HK domain (PDBID: 3DGE; Casino et al., 2009) (Fig. S2b). In this model, we extend the C-terminal PHY-hE helix from RpBphP2 and fuse it to the N-terminal helix of the DHp segment of the HK domain. The relative spatial and angular positioning of the PCM relative to the HK domain is determined by the length of the PHY-HK linker helix, discussed further below. Surface representation of this full-length model reveals three main structural features (Fig. S2b). First, a large gap exists at the dimer interface between the GAF and PHY domains. Second, the RpBphP2 monomers wrap around each other in a left-handed manner. Third, the catalytic domain of HK is distant from the core of the PHY domain. These features agree well with the parallel model for full length DrBphP derived from cryo-EM images (Li et al., 2010) with one important discrepancy. Although the 3D reconstructed maps suggest an overall twist in a left-handed manner, Li et al proposed a right-handed model for DrBphP, perhaps due to difficulties in resolving topological crossovers with a low-resolution cryoEM map (Fig. 2 and Fig. 4 of Li et al., 2010). Nevertheless, the models of full-length RpBphP2 and DrBphP derived from quite different experimental approaches share significant similarities, which suggests that the parallel dimeric scaffold with a gap between the PCMs represents the biological assembly for BphPs in the Pr state.
The GAF-PHY interface
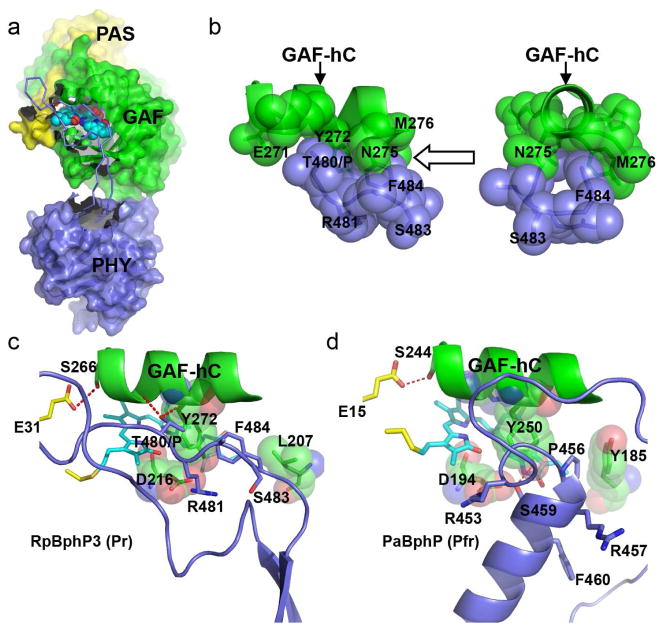
The presence of the PHY domain is important for Pr/Pfr photoconversion and for formation of the Pnr state in RpBphP3 (Yang et al., 2007). The crystal structure of RpBphP3 reveals a well-defined arm of the PHY domain that directly interacts with the GAF domain and shields the opening of the chromophore-binding pocket (Fig. 4a). The GAF-PHY interface is extensive and buries a surface area of ~833Å2, in which 13 inter-domain distances are less than 3.2Å (Contact in CCP4 (Winn et al., 2011)) (Table S1). We also note that Thr480, which in RpBphP3 replaces Pro in the PRxSF motif, forms hydrogen bonds with both the main chain carbonyl oxygen of Thr268 and the main chain nitrogen of Tyr272. A space-filling representation shows a “tongue-and-groove” interaction, where the side chains of Thr480 and Phe484 in the PRxSF motif snugly fit into grooves between the large side chains of Glu271/Tyr272 and Asn275/Met276 in the GAF-hC helix (Fig. 4b). In comparison, only five GAF-PHY inter-domain distances are less than 3.2Å in the WT Cph1 structure (2VEA), none of which is near the PRxSF motif (Table S1). The Cph1 and RpBphP3 structures are highly comparable in the PRxSF region, except that Pro471 of Cph1 is unable to form any hydrogen bonds. Furthermore, in the high-resolution crystal structure of Cph1-Y263F(Mailliet et al., 2011), Phe475 in the PRxSF motif does not interact with the GAF-hC helix perhaps due to repulsive clashes arising from the Y263F mutation. It is plausible that intimate interactions between the GAF-hC and the PRxSF region of the PHY domain contribute to formation of the unusual Pnr state of RpBphP3 in contrast to formation of the Pfr state of canonical phytochromes such as Cph1 and RpBphP2.
Figure 4. The GAF-PHY interface.
a) In the Pr structure of RpBphP3-PCM, a long extended arm spanning residues 454-494 (in ribbon representation) protruding from the core of the PHY domain (blue) shields a large surface area of the GAF domain (green) near the chromophore (in cyan). b) “Tongue-and-groove” contacts between the side chains of the GAF-hC helix (green) and the PRxSF motif (blue) in two orthogonal views as indicated by the arrow. c) In RpBphP3-PCM in the Pr state, the PRxSF motif adopts an extended conformation, in which Thr480 in place of Pro makes direct interactions with the GAF-hC helix (hydrogen bonds are marked in red dashed lines). Residues Tyr272, Asp216 and Leu207 in the GAF domain are shown in spheres. d) In PaBphP in the Pfr state, the PRxSF motif is part of a 3-turn helix, in which side chains adopt very different orientations relative to the GAF-hC helix than in RpBphP3-PCM. Related to Figure S3 and Figure S5.
The GAF-hC helix is a highly conserved structural element among BphPs. This helix, positioned roughly parallel to rings A/D, contains two highly conserved residues (His269 and Tyr272 of RpBphP3) whose side chains approach the α-face of the chromophore. His269 interacts with the chromophore via a pyrrole water that forms hydrogen bonds with nitrogens in rings A/B/C(Anders et al., 2013; Essen et al., 2008; Wagner et al., 2005; Yang et al., 2008). Two conserved residues at its N-terminus, Ser266 of the GAF domain and Glu31 of the PAS domain, form a hydrogen bond that stabilizes the figure-of-eight knot. At its C-terminus is the ring D cavity, surrounded by residues from both the GAF and PHY (Phe484) domains (Fig. 4c). Interestingly, F484W in RpBphP3 seems to affect formation of the dark-adapted Pr state while equivalent mutations do not affect the photoconversion in RpBphP2 and PaBphP (Fig. S3).
We propose that replacement of Pro by Thr480 in RpBphP3 at the “tongue-and-groove” interface contributes to formation of the unusual Pnr state. To test this hypothesis, we made single mutants at corresponding sites in RpBphP3 (Thr480), RpBphP2 (Pro465) and PaBphP (Pro456) (Table S2; Fig. S3). T480P in RpBphP3 abolishes formation of the Pnr state and enables limited formation of the normal Pfr state. P456T in PaBphP destabilizes the dark-adapted Pfr state. However, P465T in RpBphP2 does not affect Pr/Pfr photoconversion; and in particular does not favor formation of a Pnr state. This suggests that Thr480 in the PRxSF motif of RpBphP3 indeed contributes to the Pnr formation. However, the single substitution Pro->Thr is not sufficient to significantly alter Pr/Pfr photoconversion.
The arm structures of RpBphP2, RpBphP3, Cph1 and AtPhyB in the Pr state are very similar: all form two anti-parallel β-strands in contrast to the 3-turn helix in the Pfr structures of PaBphP and RpBphP1. In the PaBphP structure, this helix is juxtaposed to the chromophore in a nearly perpendicular direction (Bellini and Papiz, 2012a; Yang et al., 2008) (Fig. 4d). The PRxSF motif, which is distant from the GAF-hC helix in the Pfr state, is part of the 3-turn helix, in which the side chain of Ser459 interacts with that of the highly conserved Asp194 at the β-face of the chromophore (Yang et al., 2008) (Fig. 4c, d). It is noteworthy that the side chain dispositions of the PRxSF motif are markedly different in the RpBphP3/RpBphP2 (Pr) and PaBphP (Pfr) structures (Fig. 4c, d). Although these differences are found in BphP structures of different biological origin and sequence, their relevance to light-induced structural changes during Pr/Pfr photoconversion (Anders et al., 2013) has been corroborated by recent direct comparison of two DrBphP crystal structures determined in the dark and illuminated states (Takala et al., 2014).
The PHY-HK linker helix
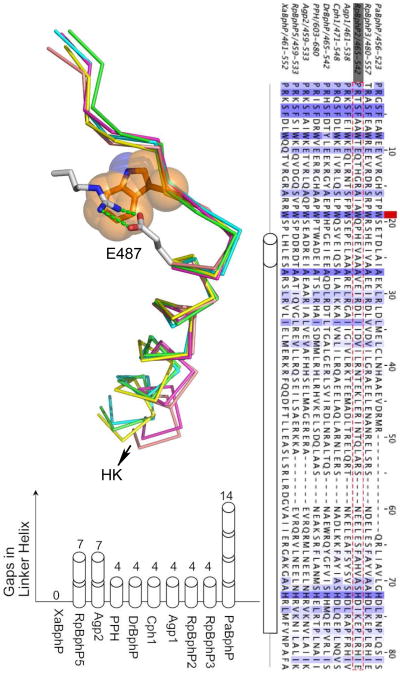
Secondary structure predictions suggest that the PHY domain is covalently connected to the HK domain via a long, continuous helix which we denote the PHY-HK helix (Fig. 5b). Interestingly, the predicted PHY-HK linker helix varies among BphPs in both sequence and length, where “length” is defined as the number of residues between the highly conserved Trp483 (numbering in RpBphP2) and the phospho-acceptor histidine (His532) near the C-terminus of the linker helix (Fig. 5b, Fig. S4). Remarkably, we note that the length differences are always multiples of one α-helical turn (Fig. 5c). If we use XaBphP from Xanthomonas axonopodis as the length reference with a total of 54 amino acids, the PHY-HK linkers in classical BphPs such as RpBphP2, DrBphP and Cph1 are shorter by four residues, roughly equivalent to one helical turn. In bathy BphPs such as PaBphP, Agp2 and RpBphP5, these linkers are shorter by multiples of seven residues, roughly equivalent to two α-helical turns. That is, the angular orientations of phospho-accepting histidines relative to the sensor domains are conserved, not the spatial distance between the sensor and effector domains (Fig. S4). The length variations suggest that direct contacts between the PHY domain and the catalytic domain of HK are not required for effective signal transmission. In other words, auto-phosphorylation of HK is directly and structurally coupled to the photosensory domains via the long PHY-HK linker helix, in which the dimer interface acts as an angular reference frame to assist precise positioning of the histidine side chain relative to the active site of ATPase. This closely resembles the situation of blue-light sensor HKs in which LOV domains are covalently coupled to the C-terminal HK domains (Möglich et al., 2009). We propose that such conservation in relative angular orientation between the sensor and effector domains is a general feature among sensor histidine kinases no matter whether the sensor domain is LOV (YF1) or PAS-GAF-PHY (BphPs) or heme-binding PAS (FixL)(Ayers and Moffat, 2008; Diensthuber et al., 2013; Möglich et al., 2009; Yang et al., 2008).
Fig. 5. The PHY-HK linker helix.
a) Superposition of five crystal structures in the junction region between the arm of the PHY domain and the HK domain. RpBphP2 (PDB 4R6L; green), RpBphP3 (PDB 4R70; yellow), Cph1 (2VEA; magenta), PaBphP (3C2W; blue) and RpBphP1 (4EH0; orange). The side chain of the conserved Trp residue (Trp483 of RpBphP2) is shown in orange, and the side chains of Arg439 and Glu487 are in grey with stick. Green dashed lines mark the salt bridges. b) Sequence alignment between the PRxSF motif and the phospho-accepting histidine in 10 representative BphPs. The red square marks the position of the conserved Trp483 in RpBphP2 (in gray box and sequence is highlighted red dashed). Gaps are filled by short dashes. A long tube represents the continuous PHY-HK linker helix according to the secondary structure prediction. The length of the short tube measures one α-helical turn. c) Length differences in the PHY-HK linker helices are represented by gaps in the sequence alignment as measured by the number of helical turns. Related to Figure S4.
Trp483 is both strictly conserved among all BphPs and strategically located at the junction between the arm of the PHY domain and the PHY-HK linker helix. Other structural features in the vicinity of Trp483 are also conserved among the Pr and Pfr structures. In particular, a salt bridge is formed between two conserved residues: Arg439 from the central β–strand of the PHY domain preceding the arm and Glu487 located in the first, N-terminal turn of the PHY-HK linker helix (Fig. 5a; Table S2). In RpBphP2, both Arg439 and Glu487 adopt extended side chain conformations such that electron densities of their side chains lie parallel to the structural segment in which Trp483 is located. This salt bridge in effect shortcuts the entire arm structure (Fig. 6). It is noteworthy that Asp replaces Glu in bathy BphPs such as PaBphP, RpBph1, RpBphP5 and Agp2; and hence yields a shorter Arg-Asp salt bridge in the PaBphP and RpBphP1 structures (Table S2). This conserved salt bridge and Trp483 are located at the junction between two important “action centers” of the BphP structure, namely the chromophore binding pocket and the active site of HK. At one end, the arm of the PHY domain directly interacts with the chromophore embedded in the GAF domain via the PRxSF motif (Yang 2008, Essen, 2008, Yang 2012). At the other end, the PHY-HK linker helix dictates the angular orientation of the phospho-accepting histidine. We denote the feature consisting of the conserved Trp483 and the Arg439-Glu487 salt bridge as the R-W-E/D motif. We speculate that the local structure of this R-W-E/D motif does not change significantly upon photoconversion; rather, it acts as a critical pivot that transmits light-induced structural signals from the chromophore to the active site of the spatially remote HK domain (Fig. 6).
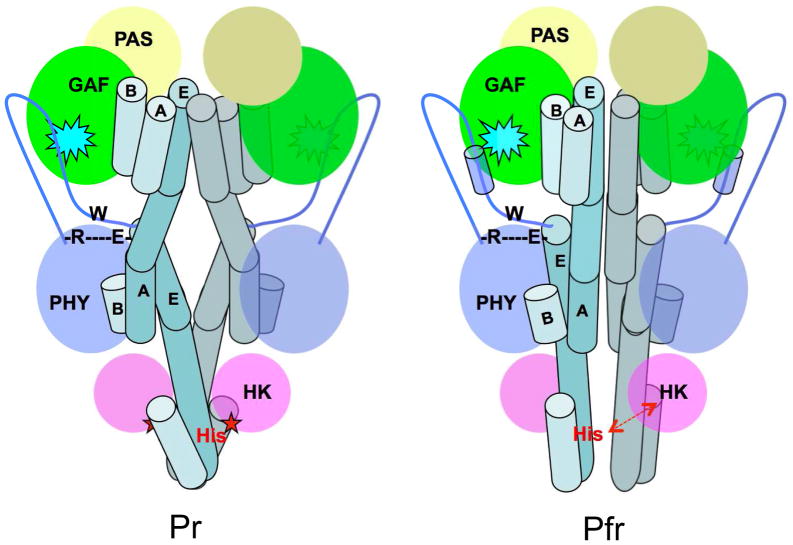
Fig. 6. Long-range signaling mechanism in BphPs.
Schematic illustration of the BphP parallel dimer structures in the Pr (left) and Pfr (right) states. Important structural elements involved in long-range signaling are highlighted: the BV chromophore embedded in the GAF domain (cyan spike), the arm (blue line with the tube representing a helix) of the PHY domain, the R-W-E/D motif, and the phospho-accepting histidine residue (red), and the long linker helices (in teal tubes). Interfacial helices in one monomer are labeled according to the crystal structures of RpBphP2-PCM (Pr) and PaBphP-PCM (Pfr). The core of each modular domain is colored as the following: PAS (yellow), GAF (green), PHY (blue) and HK (magenta). Straightening of the linker helices in the helical spine affects relative positioning of the catalytic ATPase (magenta oval) and His. Specifically, activation of HK auto-phosphorylation is marked by a red star when ATPase is sufficiently close to His. Deactivation of HK is indicated with a red double-arrow as His separates from the active site of ATPase.
Discussion
Parallel and anti-parallel dimers of BphPs
Both parallel and anti-parallel dimers of BphPs are formed under a wide range of crystallization conditions. Examples of parallel dimers are RpBphP2, AtPhyB and DrBphP in the Pr state, wild type PaBphP in the Pfr state and the PaBphP-Q188L mutant in a mixed Pr/Pfr state (Bellini and Papiz, 2012b; Burgie et al., 2014; Takala et al., 2014; Yang et al., 2008, 2009). All CBD constructs with wild type sequences forms parallel dimers (Wagner et al., 2005; Yang et al., 2007). Examples of anti-parallel arrangements are RpBphP3 and Cph1 in the Pr state, and RpBphP1 in the Pfr state (Bellini and Papiz, 2012a; Essen et al., 2008; Mailliet et al., 2011). No obvious correlation exists between crystallization conditions and quaternary structures in the crystal lattices. These results suggest a few general conclusions. First, parallel and anti-parallel dimerization are both mediated by helices at the dimer interface. Second, aggregation of helical bundles at the dimer interface is not sequence-specific, nor does it discriminate between GAF and PHY domains. Such pairing can occur between the same domains (either GAF or PHY) in parallel dimers, or between the GAF and PHY domains in anti-parallel dimers. In the RpBphP1 structure, only the C-terminal PAS domain is involved in dimerization (Bellini and Papiz, 2012a). Since the GAF and PHY domains adopt similar tertiary fold with 3-helix bundles, the parallel dimers of BphP-PCMs evidently do not have any significant thermodynamic advantages over the anti-parallel dimers in the absence of the HK domain. Monomers of isolated PAS domains have been shown to form quite different dimer interfaces (Ayers and Moffat, 2008).
It is plausible that the parallel dimer represents the biological assembly in a full-length BphP. First, similar parallel scaffolds have been captured by different experimental approaches, as evident in the RpBphP2, AtPhyB, DrBphP and PaBphP structures by crystallography (Bellini and Papiz, 2012b; Burgie et al., 2014; Takala et al., 2014; Yang et al., 2008, 2009) and the full-length DrBphP model by cryo-EM(Li et al., 2010). Second, most crystal structures containing the HK domain adopt parallel scaffold via coiled-coil helical bundles in the DHp domain (Casino et al., 2009; Diensthuber et al., 2013). Third, the parallel dimer of full length BphP is expected to bury significantly larger surface areas than its anti-parallel counterparts, in which only the helical bundles in the GAF and PHY domains could contribute to dimerization.
Given their high sequence homology and nearly identical monomeric structures, the parallel or anti-parallel dimer scaffolds are also compatible with heterodimers of RpBphP2 and RpBphP3, if indeed such heterodimers exist. Giraud et al showed that RpBphP2 and RpBphP3 are products of two tandem genes in the same operon and both interact with the same response regulator RPA3017 (Giraud et al., 2005). They further demonstrated that both genes are required for regulating the LH4 synthesis under various light conditions in R. palustris. Although many signaling proteins are chemically homodimers, they display conformational heterodimers in various states (Diensthuber et al., 2013; Neiditch et al., 2006; Yang et al., 2008). Naturally occurring chemical heterodimers are also widespread among regulatory proteins such as the DNA-binding protein Fos/Jun (Rauscher et al., 1988) and the mammalian circadian clock protein CLOCK:BMAL1 (Huang et al., 2012). Although the structures of RpBphP2 and RpBphP3 are largely similar in the dark Pr state, the putative chemical heterodimer of RpBphP2 and RpBphP3 would offer the unique, light state of Pfr|Pnr, differing from the Pfr|Pfr and Pnr|Pnr illuminated states in the RpBphP2 and RpBphP3 homodimers. Experimental data are needed to demonstrate the existence of such heterodimers and if found, to characterize their roles in in vivo functions.
Structural basis for the Pnr state in RpBphP3
What is the structural basis of the unusual Pnr state of RpBphP3? RpBphP3 and RpBphP2 show very similar features in their dark-minus-light difference FTIR spectra (Giraud et al., 2005). Mutations in the 15Za pocket suggest that the chromophore undergoes a similar overall “flip-and-rotate” motion in forming the Pnr state in RpBphP3 and the Pfr state in RpBphP2 (Yang et al., 2009). This suggests that the Pnr and Pfr states have similar chromophore-protein interactions around rings A/B/C, and differ only in ring D disposition (Fig. S5).
The crystal structure of RpBphP3 reveals an unusual “tongue-and-groove” interface between the GAF-hC helix and the PRxSF motif in the PHY domain. Two single mutations in the vicinity of this interface, L207Y and T480P, separately abolish the Pnr state and enable formation of the Pfr state (Yang et al., 2007) (Fig. 4c, Fig. S3), again indicating that this “tongue-and-groove” interface is associated with the unusual Pr/Pnr photoconversion.
We therefore advance a hypothesis to explain how the unusual Pnr state is formed. Upon 15Za/15Ea isomerization, the chromophore undergoes “flip-and-rotate” motions (Yang et al., 2009). The ring C propionate group acquires new protein anchors in the 15Za pocket. As the initially strained chromophore relaxes further, residues surrounding ring D must rearrange to accommodate its 15Ea configuration. These rearrangements involve motion in the arm of the PHY domain and/or in nearby bulky residues such as Phe212 and Tyr185. When Pr/Pfr photoconversion occurs, the fully relaxed chromophore adopts the ZZEssa configuration and protein interactions around ring D similar to those observed in PaBphP(Yang et al., 2008) (Fig. S5). However in RpBphP3, the PHY arm interacts more strongly with the GAF domain and hinders rearrangement of the ring D pocket. Further chromophore relaxation is hindered by a cavity that is still optimized for the Pr state. As a result, ring D disengages at least partly from the extended conjugation system of rings A/B/C. The shorter conjugated system leads to blue-shifted absorption in the Pnr state (Fig. S5). If the “tongue-and-groove” interface is disrupted in the single mutants such as T480P and L207Y, the arm is dislodged from the GAF-hC helix, allowing formation of the Pfr state to occur. Clearly, further investigation is needed to confirm many details of this hypothetical model.
Long-range signaling mechanism
Signaling proteins often consist of multiple modular domains linearly connected via long, signaling helices in a “mix-and-match” manner (Anantharaman et al., 2006). In BphPs, the PAS, GAF and PHY domains and the C-terminal HK domain are attached to a central helical bundle like “beans on a stalk”, where direct interactions between the core domains are limited. The helical stalks evidently mediate such long-range signaling. But how do light-induced molecular events at the chromophore in the core of the GAF domain affect phosphorylation of a histidine residue located in the spatially remote HK domain, approximately ~90Å distant along the same “stalk”?
The helical “stalks” display distinct arrangements at the dimer interface in the Pr and Pfr states, exemplified by the RpBphP2 and PaBphP structures (Fig. 2). First, the GAF-PHY linker helix is rather curved in the Pr state of RpBphP2 but is nearly straight in the Pfr state of PaBphP. Concomitantly, the PHY-HK helices are positioned differently at the dimer interface (Fig. 2). The phospho-accepting histidine is located near the C-terminal end of the same PHY-HK linker helix. It is plausible that changes in the relative positions of these helices at the dimer interface would affect the HK activity, in which the level of auto-phosphorylation is usually higher in the Pr state than in the Pfr state (Giraud et al., 2005; Psakis et al., 2011; Yang et al., 2011). Another major difference between the Pr and Pfr states lies in the arm structure, where the PRxSF motif adopts different conformations (Fig. 4c, d). The R-W-E/D structure motif that connects the PRxSF motif and the PHY-HK helix may serve as a pivot or signaling route that channels light-induced structural changes from the chromophore to the HK domain (Fig. 6).
Long-range signaling mechanism in BphPs is likely to involve tertiary structural changes following photoconversion in the chromophore region, such as the “strand-to-helix” transformation in the arm structure and rearrangements in the helical spine. The rigid R-W-E/D structure may serve as a pivot to transduce structural changes from the GAF domain to the PHY domain, which affects the PHY-HK helix. Our preliminary data showed that the RpBphP3-PCM crystals did not exhibit any photo-activity, that is, no formation of the Pnr state was detected judged by micro-spectroscopy on crystals. This suggests that formation of the Pnr state is coupled with structural change(s) in the protein region that are prohibited by the crystal lattice. It is yet to be determined whether these structural changes are associated with the strand-to-helix conversion in the arm.
Signal transduction via long linker helices seems to be a general strategy for long-range signaling. The GAF-PHY and PHY-HK linker helices in BphPs are topologically equivalent to the Jα helix of the LOV domains originally found in phototropin (Harper et al., 2003) and correspond to the putative signaling helix widely found in the sequences of modular signaling proteins (Anantharaman et al., 2006). For YF1, a chimeric photoreceptor, torque motions in linker helices in the dimer interface have been suggested to be responsible for light-dependence of the effector HK activity (Möglich et al., 2009). We also note that Asp or Glu in the DIT motif identified at the N-terminus of the Jα helix in LOV domains is also found to form salt bridge(s) with Arg or Lys near a conserved Trp in LOV and other PAS domains such as EcDos (Tuckerman et al., 2009). Given that similar modular architectures are widespread among signaling proteins in all kingdoms of life, our findings in BphPs may shed light on general principles for long-range signal transduction.
Experimental Procedures
Cloning, mutagenesis and purification
Both RpBphP2-Ctag (residues 1-506) and RpBphP3 (residues 1-521) were PCR-amplified from R. palustris strain CGA009 genomic DNA (ATCC) and then cloned into pET24 vectors (Novagen) between restriction sites NdeI/HindIII and NdeI/XhoI, respectively. Both constructs carry a 6xhistine affinity tag at their C-termini. The RpBphP2-Ntag construct was cloned into pET28c between restriction sites NdeI and HindIII, and includes 20 additional residues at its N-terminus (Yang et al., 2007). The Seleno-Met RpBphP2-Ntag was produced according to the published protocol (Doublié, 1997). Site-directed mutagenesis was carried out using the QuikChange Site-directed Lightening mutagenesis kit (Strategene). All BphP proteins including wild types and mutants were co-expressed with heme oxygenase and purified as described (Yang et al., 2007).
Crystallization and data collection
RpBphP2-Ctag was crystallized in the dark at 20°C in hanging drops containing 10mg/ml protein, 12% polyacrylic acid, 0.1M MgCl2 and 0.1M Hepes (pH7.0). The crystals were cryo-protected under double-filtered blue/green light in mother liquor containing 20–30% glycerol. Diffraction data were collected at LS-CAT 21-IDG beamline station of the Advanced Photon Source (APS). The RpBphP2-Ntag crystals grew in the dark at 16°C under a crystallization condition containing 10mg/ml protein, 1.4 M sodium acetate, 0.1M sodium cacodylate, pH 6.5. Diffraction data at 100 K were collected at SBC 19-ID and BioCARS 14-IDB beamlines of APS. The SeMet diffraction data at the wavelength of 0.978Å was collected at the cryogenic temperature of 100 K.
RpBphP3-PCM crystals were obtained at 20°C in the dark using the hanging drop vapor diffusion method under the condition of 10mg/ml protein and 0.1M magnesium formate. The crystals were cryo-protected under blue/green safe light in 20–30% glycerol mixed with mother liquor. Diffraction data at 100 K were collected at the LS-CAT 21-IDG. All images were indexed, integrated, and scaled using HKL2000 (Otwinowski and Minor, 1997).
Structure determination and refinement
The crystal structure of RpBphP3-PCM was determined in space group P3121 by molecular replacement (PHASER in CCP4 (Winn et al., 2011)) using the RpBphP3-CBD structure (PDB ID 2OOL) as the search model. The initial model for the PHY domain was built based on a non-crystallographic symmetry (NCS) averaged electron density map following density modification (Resolve in PHENIX(Adams et al., 2010)) and guided by the structure of the PHY domain of Cph1 (PDB ID 2VEA). The final model at 2.85Å resolution was refined to a R-factor of 0.185 and a free R-factor of 0.259 (PHENIX(Adams et al., 2010)).
The crystal structure of RpBphP2-Ctag was determined by molecular replacement (PHASER in CCP4 (McCoy et al., 2007)) using the chromophore-binding domain of RpBphP3 (PDB ID 2OOL) as the search model. The initial model for the PHY domain was built (Coot (Emsley and Cowtan, 2004)) based on electron density derived from several rounds of density modification including NCS averaging (DM in CCP4 and Resolve in Phenix) guided by both Cph1 and PaBphP structures (PDB ID: 2VEA and 3C2W). The final model of RpBphP2-Ctag was refined (PHENIX) to 3.4Å resolution, which includes two molecules and two BV chromophores in the asymmetric unit. The RpBphP3-PCM structure was also used to validate the details of the RpBphP2-Ctag structure given its limited resolution.
Initial phases for the RpBphP2-Ntag structures were obtained using molecular replacement (PHASER in CCP4) using RpBphP3-CBD as a search model. This partial MR solution was used to identify the SeMet sites in the single-wavelength anomalous dispersion (SAD) data (PHENIX). The initial model was built based on the figure-of-merit-weighted Fo map derived from phase combination of MR and SAD solutions (CCP4). Initial structure refinement was carried out using Refmac5 (CCP4). The final model of the RpBphP2-Ntag structure was rebuilt (Coot) using the refined RpBphP2-Ctag structure as a reference and refined to 3.25Å resolution. All structures are illustrated using PyMol (http://pymol.org).
The final coordinates of the crystal structures of RpBphP2-Ctag, RpBphP3-PCM and RpBphP2-Ntag have been deposited to Protein Data Bank under accession numbers 4R6L, 4R70 and 4S21, respectively.
UV-visible absorption spectroscopy
UV-visible spectra from 900 to 230nm of the wild type and mutant proteins were recorded in solution at room temperature with a Shimazu UV-1650 PC spectrophotometer. Illumination for measuring photoactivities of these samples was provided by filtered fiber optic light.
Supplementary Material
Highlights.
RpBphP2 and RpBphP3 adopt similar tertiary structures but different dimer scaffolds.
Residues at the GAF-PHY interface modulate photo-conversion properties.
A single mutation (T480P) in RpBphP3 enables canonical photoconversion.
Angular orientation between PHY and HK domains is crucial for long-range signaling.
Acknowledgments
We thank Ying Pigli for help in cloning. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). Use of BioCARS was supported by NIH under grant number RR007707. Structural Biology Center is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. This work was supported by National Institutes of Health grants GM036452 (to KM) and R01EY024363 (to KM and XY).
Footnotes
Author contributions
X.Y. initiated and designed research, solved crystal structures and analyzed data; E.A.S., W.O. and E.D. contributed to the RpBphP2-Ntag structure; J.K. grew crystals and made mutants; X.Y. and K.M wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Balaji S, Aravind L. The signaling helix: a common functional theme in diverse signaling proteins. Biol Direct. 2006;1:25. doi: 10.1186/1745-6150-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders K, Daminelli-Widany G, Mroginski MA, von Stetten D, Essen LO. Structure of the Cyanobacterial Phytochrome 2 Photosensor Implies a Tryptophan Switch for Phytochrome Signaling. J Biol Chem. 2013;288:35714–35725. doi: 10.1074/jbc.M113.510461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers RA, Moffat K. Changes in Quaternary Structure in the Signaling Mechanisms of PAS Domains. Biochemistry (Mosc) 2008;47:12078–12086. doi: 10.1021/bi801254c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini D, Papiz MZ. Structure of a Bacteriophytochrome and Light-Stimulated Protomer Swapping with a Gene Repressor. Structure. 2012a;20:1436–1446. doi: 10.1016/j.str.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Bellini D, Papiz MZ. Dimerization properties of the RpBphP2 chromophore-binding domain crystallized by homologue-directed mutagenesis. Acta Crystallogr D Biol Crystallogr. 2012b;68:1058–1066. doi: 10.1107/S0907444912020537. [DOI] [PubMed] [Google Scholar]
- Burgie ES, Bussell AN, Walker JM, Dubiel K, Vierstra RD. Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc Natl Acad Sci. 2014;111:10179–10184. doi: 10.1073/pnas.1403096111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casino P, Rubio V, Marina A. Structural Insight into Partner Specificity and Phosphoryl Transfer in Two-Component Signal Transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- Diensthuber RP, Bommer M, Gleichmann T, Möglich A. Full-Length Structure of a Sensor Histidine Kinase Pinpoints Coaxial Coiled Coils as Signal Transducers and Modulators. Structure. 2013;21:1127–1136. doi: 10.1016/j.str.2013.04.024. [DOI] [PubMed] [Google Scholar]
- Doublié S. [29] Preparation of selenomethionyl proteins for phase determination. In: Charles J, Carter W, editors. Methods in Enzymology. Academic Press; 1997. pp. 523–530. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Essen LO, Mailliet J, Hughes J. The structure of a complete phytochrome sensory module in the Pr ground state. Proc Natl Acad Sci. 2008;105:14709–14714. doi: 10.1073/pnas.0806477105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Zappa S, Vuillet L, Adriano JM, Hannibal L, Fardoux J, Berthomieu C, Bouyer P, Pignol D, Verméglio A. A New Type of Bacteriophytochrome Acts in Tandem with a Classical Bacteriophytochrome to Control the Antennae Synthesis in Rhodopseudomonas palustris. J Biol Chem. 2005;280:32389–32397. doi: 10.1074/jbc.M506890200. [DOI] [PubMed] [Google Scholar]
- Harper SM, Neil LC, Gardner KH. Structural Basis of a Phototropin Light Switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- Huang N, Chelliah Y, Shan Y, Taylor CA, Yoo SH, Partch C, Green CB, Zhang H, Takahashi JS. Crystal Structure of the Heterodimeric CLOCK:BMAL1 Transcriptional Activator Complex. Science. 2012;337:189–194. doi: 10.1126/science.1222804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of Macromolecular Assemblies from Crystalline State. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Li H, Zhang J, Vierstra RD, Li H. Quaternary organization of a phytochrome dimer as revealed by cryoelectron microscopy. Proc Natl Acad Sci. 2010;107:10872–10877. doi: 10.1073/pnas.1001908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Wang H, Deng X-W. Phytochrome Signaling Mechanisms. Arab Book Am Soc Plant Biol. 2011;9 doi: 10.1199/tab.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailliet J, Psakis G, Feilke K, Sineshchekov V, Essen LO, Hughes J. Spectroscopy and a High-Resolution Crystal Structure of Tyr263 Mutants of Cyanobacterial Phytochrome Cph1. J Mol Biol. 2011;413:115–127. doi: 10.1016/j.jmb.2011.08.023. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A, Ayers RA, Moffat K. Design and Signaling Mechanism of Light-Regulated Histidine Kinases. J Mol Biol. 2009;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM. Ligand-Induced Asymmetry in Histidine Sensor Kinase Complex Regulates Quorum Sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njimona I, Lamparter T. Temperature Effects on Agrobacterium Phytochrome Agp1. PLoS ONE. 2011;6:e25977. doi: 10.1371/journal.pone.0025977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. [20] Processing of X-ray diffraction data collected in oscillation mode. In: Charles J, Carter W, editors. Methods in Enzymology. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Psakis G, Mailliet J, Lang C, Teufel L, Essen LO, Hughes J. Signaling Kinetics of Cyanobacterial Phytochrome Cph1, a Light Regulated Histidine Kinase. Biochemistry (Mosc) 2011;50:6178–6188. doi: 10.1021/bi200612d. [DOI] [PubMed] [Google Scholar]
- Rauscher FJ, Voulalas PJ, Franza BR, Curran T. Fos and Jun bind cooperatively to the AP-1 site: reconstitution in vitro. Genes Dev. 1988;2:1687–1699. doi: 10.1101/gad.2.12b.1687. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC. Phytochrome Structure and Signaling Mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Lagarias JC. Red/Green Cyanobacteriochromes: Sensors of Color and Power. Biochemistry (Mosc) 2012;51:9667–9677. doi: 10.1021/bi3013565. [DOI] [PubMed] [Google Scholar]
- Takala H, Björling A, Berntsson O, Lehtivuori H, Niebling S, Hoernke M, Kosheleva I, Henning R, Menzel A, Ihalainen JA, et al. Signal amplification and transduction in phytochrome photosensors. Nature. 2014;509:245–248. doi: 10.1038/nature13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasler R, Moises T, Frankenberg-Dinkel N. Biochemical and spectroscopic characterization of the bacterial phytochrome of Pseudomonas aeruginosa. FEBS J. 2005;272:1927–1936. doi: 10.1111/j.1742-4658.2005.04623.x. [DOI] [PubMed] [Google Scholar]
- Tuckerman JR, Gonzalez G, Sousa EHS, Wan X, Saito JA, Alam M, Gilles-Gonzalez MA. An Oxygen-Sensing Diguanylate Cyclase and Phosphodiesterase Couple for c-di-GMP Control. Biochemistry (Mosc) 2009;48:9764–9774. doi: 10.1021/bi901409g. [DOI] [PubMed] [Google Scholar]
- Wagner JR, Brunzelle JS, Forest KT, Vierstra RD. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature. 2005;438:325–331. doi: 10.1038/nature04118. [DOI] [PubMed] [Google Scholar]
- Wagner JR, Zhang J, Brunzelle JS, Vierstra RD, Forest KT. High Resolution Structure of Deinococcus Bacteriophytochrome Yields New Insights into Phytochrome Architecture and Evolution. J Biol Chem. 2007;282:12298–12309. doi: 10.1074/jbc.M611824200. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Stojković EA, Kuk J, Moffat K. Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion. Proc Natl Acad Sci. 2007;104:12571–12576. doi: 10.1073/pnas.0701737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kuk J, Moffat K. Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: Photoconversion and signal transduction. Proc Natl Acad Sci. 2008;105:14715–14720. doi: 10.1073/pnas.0806718105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kuk J, Moffat K. Conformational differences between the Pfr and Pr states in Pseudomonas aeruginosa bacteriophytochrome. Proc Natl Acad Sci. 2009;106:15639–15644. doi: 10.1073/pnas.0902178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Ren Z, Kuk J, Moffat K. Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome. Nature. 2011;479:428–432. doi: 10.1038/nature10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.