Abstract
Several species of alphaviruses have been previously described in the Americas, some of which are associated with encephalitis and others are associated with arthralgia. Venezuelan equine encephalitis virus (VEEV) and eastern equine encephalitis virus (EEEV) are endemic to Venezuela, with the former being responsible for major outbreaks of severe and often fatal disease in animals and humans. The aim of this study was to analyze the genetic diversity of Venezuelan alphaviruses isolated during two decades (1973–1999) of surveillance in northern Venezuela. Phylogenetic analysis indicated the circulation of a VEEV subtype IAB strain 8 years after the last reported outbreak. Thirteen strains within two subclades of South American lineage III of EEEV were also found in Venezuela. Considerable genetic variability was observed among Venezuelan Una virus strains, which were widely distributed among the clades. The first Venezuelan Mayaro sequence was also characterized.
Introduction
The genus Alphavirus includes approximately 30 species of positive-strand RNA viruses, displays a nearly worldwide distribution, and belongs to the Family Togaviridae. Alphavirus infection can cause a variety of diseases in humans and domesticated animals. The genus includes eight genetic/antigenic complexes, of which five have been described in the New World.1 Venezuelan equine encephalitis virus (VEEV) and eastern equine encephalitis virus (EEEV; the species Madariaga virus in South America2) are enzootic in Venezuela and other parts of South America, with the former responsible for major equine-amplified outbreaks of severe, often fatal disease in animals and humans (mainly subtypes IAB and IC) as well as endemic disease resulting from enzootic spillover (subtype ID).1 Among the arthralgic alphaviruses, only Mayaro virus (MAYV) occurs in the New World.
Several field studies have been performed in Venezuela for alphavirus surveillance.3,4 VEEV has previously been detected during epidemics occurring in 1938, 1969, and 1973.5 However, little information is available on the molecular characterization of VEEV and other alphaviruses during interepidemic periods in the country. To further characterize South American alphaviruses and their genetic diversity, we studied strains isolated during two decades of surveillance in Venezuela.
Methods
The virus strains used in this study are listed in Table 1. The viruses were isolated primarily by the use of sentinel hamsters at seven locations in five states from northern Venezuela. Alphavirus isolates were propagated in Vero cells as previously described.6 Viral RNA was extracted with Trizol LS (Invitrogen, Carlsbad, CA), and reverse transcription polymerase chain reaction (RT-PCR) amplification of the 3′–non-coding region (3′-NCR) was performed with primers α10247A and T25-MLu6 to generate amplicons of approximately 1,200 nucleotides (nt). Purified amplicons were sequenced by dye-terminator sequencing using a 23 ABI 3730XLs (Macrogen, Seoul, Korea) using both amplification primers to allow sequencing of both complementary DNA (cDNA) strands and correct base calling. The complete genome sequence of VEEV strain Pan66640 was determined using subtype IAB-specific primers (sequences available on request). Each region was sequenced using four primers covering both cDNA strands.
Table 1.
Venezuelan alphavirus isolates obtained in this study
| Strain name (virus) | Passage history before sequencing | Source of isolate | State, region, and year of isolation |
|---|---|---|---|
| SAPan27014 (EEEV) | VERO2, BHK1 | Hamster | Zulia, Catatumbo, 1975 |
| SAPan35022 (EEEV) | VERO2, BHK1 | Hamster | Zulia, Catatumbo, 1976 |
| SAPan35093 (EEEV) | VERO1, BHK1 | Hamster | Zulia, Catatumbo, 1976 |
| SAPan66643 (EEEV) | VERO2, BHK1 | Hamster | Zulia, Goajira, 1981 |
| SAPan 66649 (EEEV) | VERO1, BHK1 | Hamster | Zulia, Goajira, 1981 |
| SAPan 66652 (EEEV) | VERO1, BHK1 | Hamster | Zulia, Goajira, 1981 |
| SA251349 (EEEV) | SMB5, VERO2, BHK1 | Equine | Guarico, Las Mercedes, 1996 |
| SAPan27029 (EEEV) | VERO1, BHK1 | Hamster | Zulia, Catatumbo, 1975 |
| SAPan66061 (EEEV) | VERO1, BHK1 | Hamster | Zulia, Goajira, 1981 |
| SAPan66367 (EEEV) | VERO2, BHK1 | Hamster | Zulia, Goajira, 1981 |
| 207546 (EEEV) | SMB4, BHK1 | Equine | San Felipe, Yaracuy, 1978 |
| 254762 (EEEV) | SMB6, BHK1 | Equine | Falcón, Tucacas, 1984 |
| SADelirio (EEEV) | SMB5, VERO2, BHK1 | Equine | Zulia, Sur del Lago, 1976 |
| Pan36080 (VEEV) | VERO2, BHK1 | Hamster | Zulia, Catatumbo, 1976 |
| 204381(VEEV) | SMB4, VERO1, BHK1 | Mosquito | Delta Amacuro, 1973 |
| AB66640 (VEEV) | VERO2, BHK1 | Hamster | Zulia, Goajira, 1981 |
| MAYPAR (MAYV) | VERO2, BHK1 | Hamster | Zulia, Catatumbo, 1999 |
| ZPC526 (UNAV) | VERO2, BHK1 | Hamster | Zulia, 1997 |
| PAR18983 (UNAV) | VERO2, BHK1 | Mosquito | Zulia, Catatumbo, 1976 |
BHK = baby hamster kidney; SMB = suckling mouse brain; VERO = African green monkey kidney.
Nucleotide sequences were deposited into the GenBank database under accession numbers KF770732–KF770750 and KJ410017. Sequences were aligned with representative homologous alphavirus sequences available in GenBank, and Bayesian phylogenies were inferred using the generalized time reversible + I + Γ4 nucleotide substitution model.
Results and Discussion
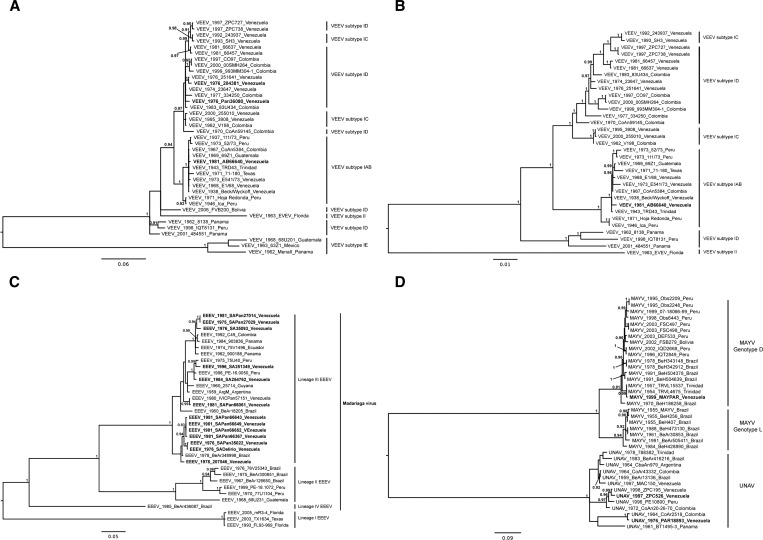
In total, 19 Venezuelan alphavirus sequences were studied (Figure 1 and Table 1): 13 EEEV, 3 VEEV, 1 MAYV, and 2 UNA virus (UNAV) strains were compared with sequences obtained previously from Venezuela and other countries. The E1/3′-NCR genome region was selected for amplification and sequencing to simultaneously take advantage of the conserved primer binding sequences within the alphavirus genus and produce a fragment that is phylogenetically informative given the sequence length.6 Indeed, phylogenetic analysis of this region discriminated the VEEV complex viruses with high bootstrap support (Figure 2A ). Strains VEEVIDPAn36080 and VEEVID 204381 were closely related to isolates from Venezuela and Colombia collected during the same time period. Strain VEEV PAn66640, isolated from a sentinel hamster in the Guajira region, Zulia State during a non-epidemic/epizootic period, displayed 0.1–0.9% nucleotide divergence from all other IAB isolates analyzed.
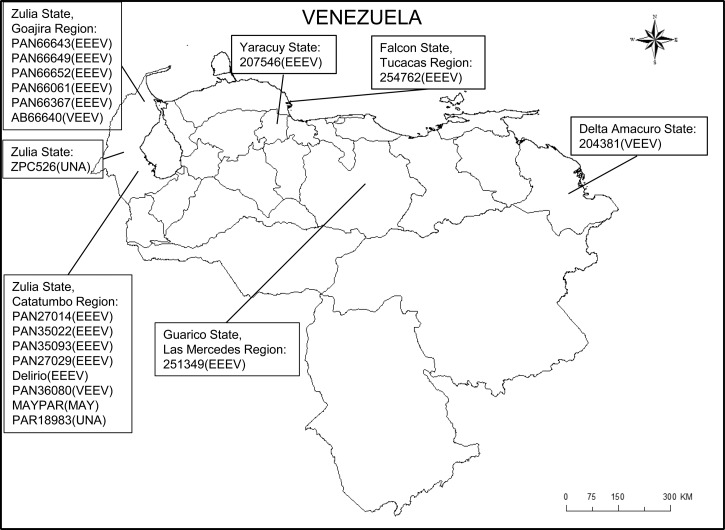
Figure 1.
Geographic location of the Venezuelan alphavirus strains analyzed in this study.
Figure 2.
Phylogenetic analyses of alphavirus isolates. A midpoint rooted Bayesian Markov chain Monte Carlo phylogeny was constructed based on (A) 810 nt of the E1/3′-NCR genome region for VEEV, (B) a structural polyprotein ORF (26S) for VEEV, (C) 810 nt of the E1/3′-NCR for EEEV, and (D) 993 nt of the E1/3′-NCR for MAYV/UNAV. Venezuelan sequences generated in this study are highlighted in bold. Tip labels include the year of isolation, strain name, and country of isolation. Clade credibility values over 90% are shown adjacent to their corresponding nodes. The scale bar represents the percentage of nucleotide sequence divergence.
The complete coding sequence and structural polyprotein open reading frame (ORF) sequence analysis (approximately 4,000 nt) for PAn66640 confirmed its relationship to IAB strains, displaying ≥ 99% identity with others in this VEEV subtype (Figure 2B). As in previous analyses with subgenomic ORF sequences,7 the relationships among most IAB strains were not robust, which was indicated by the inconsistent groupings obtained with the E1/3′-NCR versus complete ORF sequences. The nucleotide differences for PAn66640 compared with other IAB strains were all unique, ruling out laboratory contamination. Thus, strain PAn66640 represents a subtype IAB-like VEEV strain isolated during an interepizootic period; it may represent cryptic circulation up to 8 years after the last reported IAB Venezuelan outbreak in 1973. Another possibility is that this strain represents an enzootic progenitor lineage that gave rise to the IAB strains in the early 20th century. However, a basal position within the IAB clade (at least the main portion, excluding the 1942 and 1946 Peruvian strains) would be expected for such a progenitor, and none of our analyses showed this relationship.
Several hypotheses have been postulated to explain the interepizootic maintenance of IAB and IC VEEV strains: (1) periodic generation from enzootic progenitors, (2) maintenance of epizootic strains within enzootic viral subpopulations, (3) presence of cryptic IAB and IC transmission cycles that emerge periodically, (4) reactivation of latent infections, and (5) administration of improperly inactivated vaccines. Epizootic strains characterized using serologic methods have been collected during interepizootic periods.8 An epizootic strain of VEEV was collected in Guatemala in an enzootic habitat in 1969, but this occurred during the 1969–1971 Central/North American outbreak.9 Navarro and others10 also detected post-epizootic persistence of a VEEV subtype IC strain in Venezuela on cattle ranches 5 years after the apparent end of the 1995 outbreak. Additional surveillance in western Venezuela to determine if IAB strains continue to circulate there is warranted.
Sequence analysis of our EEEV strains allowed their classification into the four previously described subtypes8,10 and showed the presence of two subclades inside South American lineage III, which includes all Venezuelan isolates (Figure 2C). Lineages II–IV are now reclassified as M. virus.6 Newly delineated subclade A comprised isolates from Panama, Venezuela, and Peru, and subclade B comprised the remaining Venezuelan isolates. EEESAPAn35022, EEESADELIRIO, and EEESAPAn66061 were isolated in 1976, 1976, and 1981, respectively, and displayed 100% sequence identity in the region analyzed. The same was true for isolates EEEPan27011, EEEPan27014, and EEEPan27029, which were collected from sentinel hamsters exposed only a few meters apart. The South American EEEV group displayed 0–5.3% nucleotide sequence diversity. The Venezuelan strains were closely related within each subclade and were not grouped geographically or temporally (Figure 2C). The presence of two subclades of Venezuelan EEEV strains may reflect independent evolution for long time periods.
The MAYV sequences studied were grouped within two previously described genotypes.11 Genotype L comprises isolates exclusively from Brazil, whereas genotype D comprises strains from several parts of South America, including Brazil (Figure 2D). Our Venezuelan isolate grouped within genotype D and was most closely related to isolates from Trinidad. Little information is available in Venezuela about human infections by MAYV. Serological studies in rural parts of northern South America and the Amazon Basin indicate that human infections are relatively common.12,13 Mayaro fever typically occurs in individuals with a history of recent activity in forest habitats.14,15 Torres and others16 reported the only known human cases of Mayaro in Venezuela. The disease was found in a household consisting of four adults who spent the night in a rural area of Miranda State.
In contrast to the close genetic relatedness of the other Venezuelan alphaviruses within each species, a high level of genetic diversity was found among our UNAV isolates (Figure 2D). No genotypic grouping has been proposed for UNAV diversity,11 but at least three clades can be distinguished based on our analyses. Our Venezuelan isolates fell within two of these clades, with UNAPAr18983 closely related to a strain circulating in Colombia, UNAZPC195HAM and UNAZPC526 closely related to each other but quite distinct from all strains previously sequenced, and UNAMAC150HAM closely related to isolates from Brazil and Colombia. UNAV infection has been detected in sentinel hamsters, and antibodies were found in birds, horses, and humans during epidemiological surveys in Zulia State, Venezuela.17 The exact geographical distribution and potential disease association of UNAV remain unknown. Previous studies of UNAV genetics suggested that its diversity reflects its ability to colonize new ecological niches.11 After discrete enzootic foci are established, the virus is maintained in independent evolutionary lineages. These patterns of genetic diversity may be influenced by both vector and enzootic vertebrate hosts.
In conclusion, phylogenetic analysis of Venezuelan alphaviruses suggested cryptic, post-epizootic circulation of subtype IAB VEEV 8 years after the last reported outbreak caused by the strain. Two distinct clades within lineage III of closely related EEEV strains within the newly designated species M. virus were also delineated. EEEV and UNAV genetic diversity was also described, with the former showing geographic clustering suggestive of regionally independent evolution and the latter showing widely distributed clades, indicating greater virus mobility.
ACKNOWLEDGMENTS
The American Society of Tropical Medicine and Hygiene (ASTMH) assisted with publication expenses.
Footnotes
Financial support: This work was supported by Grant S1-2002000338 from Fondo Nacional de Ciencia y Ttecnologìa (FONACIT), Organización Panamericana de la Salud (OPS) Grant 2003, and the Defense Threat Reduction Agency under an Interagency Agreement with Lawrence Livermore National Laboratory through Contract DTRA10027IA-2359–BASIC. A.J.A. is supported by the James W. McLaughlin Endowment Fund.
Authors' addresses: Gladys Medina, INIA, CENIAP, Maracay, Aragua, Venezuela, E-mail: gladicitamedina1@hotmail.com. Domingo J. Garzaro, Miguel Barrios, and Flor H. Pujol, IVIC, CMBC, Caracas, Miranda, Venezuela, E-mails: dgarzaro@gmail.com, mbarrios_3709@hotmail.com, and fhpujol@gmail.com. Albert J. Auguste, Institute for Human Infections and Immunity and Department of Pathology, University of Texas Medical Branch, Galveston, TX, E-mail: aj1augus@utmb.edu. Scott C. Weaver, Department of Pathology, University of Texas Medical Branch, Galveston, TX, E-mail: sweaver@utmb.edu.
References
- 1.Weaver SC, Winegar R, Manger ID, Forrester NL. Alphaviruses: population genetics and determinants of emergence. Antiviral Res. 2012;94:242–257. doi: 10.1016/j.antiviral.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrigo NC, Adams AP, Weaver SC. Evolutionary patterns of eastern equine encephalitis virus in North versus South America suggest ecological differences and taxonomic revision. J Virol. 2010;84:1014–1025. doi: 10.1128/JVI.01586-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salas RA, Garcia CZ, Liria J, Barrera R, Navarro JC, Medina G, Vasquez C, Fernandez Z, Weaver SC. Ecological studies of enzootic Venezuelan equine encephalitis in northcentral Venezuela. Am J Trop Med Hyg. 2001;64:84–92. doi: 10.4269/ajtmh.2001.64.84. [DOI] [PubMed] [Google Scholar]
- 4.Walder R, Suarez OM, Calisher CH. Arbovirus studies in the Guajira region of Venezuela: activities of eastern equine encephalitis and Venezuelan equine encephalitis viruses during an interepizootic period. Am J Trop Med Hyg. 1984;33:699–707. doi: 10.4269/ajtmh.1984.33.699. [DOI] [PubMed] [Google Scholar]
- 5.Moncayo AC, Medina G, Kalvatchev Z, Brault A, Barrera R, Boshell J, Ferro C, Freier J, Navarro JC, Salas R, Siger J, Vasquez C, Walder R, Weaver S. Genetic diversity and relationships among Venezuelan equine encephalitis virus field isolates from Colombia and Venezuela. Am J Trop Med Hyg. 2001;65:738–746. doi: 10.4269/ajtmh.2001.65.738. [DOI] [PubMed] [Google Scholar]
- 6.Powers A, Brault A, Shirako Y, Strauss E, Kang W, Strauss J, Weaver S. Evolutionary relationships and systematic of the alphaviruses. J Virol. 2001;75:10118–10131. doi: 10.1128/JVI.75.21.10118-10131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weaver SC, Pfeffer M, Marriott K, Kang W, Kinney RM. Genetic evidence for the origins of Venezuelan equine encephalitis virus subtype IAB outbreaks. Am J Trop Med Hyg. 1999;60:441–448. doi: 10.4269/ajtmh.1999.60.441. [DOI] [PubMed] [Google Scholar]
- 8.Kinney RM, Trent DW, France JK. Comparative immunological and biochemical analyses of viruses in the Venezuelan equine encephalitis complex. J Gen Virol. 1983;64:135–147. doi: 10.1099/0022-1317-64-1-135. [DOI] [PubMed] [Google Scholar]
- 9.Sherer WF, Anderson K, Pancake BA, Dickerman RW, Ordoñez JV. Search for epizootic-like Venezuelan encephalitis virus enzootic habitats in Guatemala during 1969–1971. Am J Epidemiol. 1976;103:576–588. doi: 10.1093/oxfordjournals.aje.a112262. [DOI] [PubMed] [Google Scholar]
- 10.Navarro JC, Medina G, Vasquez C, Coffey L, Wang E, Suárez A, Biord H, Salas M, Weaver S. Postepizootic persistence of Venezuelan equine encephalitis virus, Venezuela. Emerg Infect Dis. 2005;11:1907–1915. doi: 10.3201/eid1112.050533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Powers A, Aguilar P, Chandler L, Brault A, Meakins T, Wattss D, Wattss D, Russel K, Olson J, Vasconcelos P, Travassos A, Weaver S, Tesh R. Genetic relationships among Mayaro and UNA virus suggest distinct patterns of transmission. Am J Trop Med Hyg. 2006;75:461–469. [PubMed] [Google Scholar]
- 12.Forshey BM, Guevara C, Laguna-Torres VA, Cespedes M, Vargas J, Gianella A, Vallejo E, Madrid C, Aguayo N, Gotuzzo E, Suarez V, Morales AM, Beingolea L, Reyes N, Perez J, Negrete M, Rocha C, Morrison AC, Russell KL, Blair PJ, Olson JG, Kochel TJ. NMRCD Febrile Surveillance Working Group Arboviral etiologies of acute febrile illnesses in western South America, 2000–2007. PLoS Negl Trop Dis. 2010;4:e787. doi: 10.1371/journal.pntd.0000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinheiro F, LeDuch J. Mayaro virus disease. In: Monath TP, editor. The Arbovirus. Epidemiology and Ecology. Boca Raton, FL: CRC Press; 1988. pp. 137–150. [Google Scholar]
- 14.Tesh RB, Watts DM, Russell KL, Damodaran C, Calampa C, Cabezas C, Ramirez G, Vasquez B, Hayes C, Rosi C, Powers A, Hice C, Chandler L, Cropp B, Karabatsos N, Roehring J, Gubler D. Mayaro virus disease: an emerging mosquito-borne zoonosis in tropical South America. Clin Infect Dis. 1999;28:67–73. doi: 10.1086/515070. [DOI] [PubMed] [Google Scholar]
- 15.Pinheiro F, Freitas R, Travassos J, Travassos A, Gabbay Y, Mello W, LeDuc J. An outbreak of MAYARO virus disease in Belterra, Brazil. Am J Trop Med Hyg. 2004;30:674–681. doi: 10.4269/ajtmh.1981.30.674. [DOI] [PubMed] [Google Scholar]
- 16.Torres J, Russell K, Vasquez C, Barrera R, Tesh R, Salas R, Watts D. Family cluster of Mayaro fever, Venezuela. Emerg Infect Dis. 2004;10:1304–1306. doi: 10.3201/eid1007.030860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walder R, Suarez OM, Calisher CH. Arbovirus studies in southwestern Venezuela during 1973–1981. II. Isolations and further studies of Venezuelan and eastern equine encephalitis, Una, Itaqui, and Moju viruses. Am J Trop Med Hyg. 1984;33:483–491. [PubMed] [Google Scholar]