Summary
Protein conformational change is analyzed by finding the minimalist backbone torsion angle rotations that superpose crystal structures within experimental error. Of several approaches to enforcing parsimony during flexible least-squares superposition, an -norm restraint provided greatest consistency with independent indications of flexibility from NMR relaxation dispersion and chemical shift perturbation in arginine kinase, and four previously studied systems. Crystallographic cross-validation shows that the dihedral parameterization describes conformational change more accurately than rigid-group approaches. The rotations that superpose the principal elements of structure constitute a small fraction of the raw (φ, ψ)-differences that also reflect local conformation and experimental error. Substantial long-range displacements can be mediated by modest dihedral rotations, accommodated even within α-helices and β-sheets without disruption of hydrogen bonding at the hinges. Consistency between ligand-associated and intrinsic motions (in the unliganded state) implies that induced changes tend to follow low-barrier paths between conformational sub-states that are in intrinsic dynamic equilibrium.
Introduction
It has long been recognized that proteins are not static. Under physiological conditions, conformational transitions are common and are critical, for example, to enzyme specificity, allosteric regulation and molecular motors. A variety of subunit, domain, sub-domain and loop transitions have been characterized by comparing structures stabilized in different states (Echols et al., 2003; Rashin et al., 2010). However, our understanding of the mechanics and evolution of motion has been limited by how well the hinges of rotations can be defined. Early analyses simply examined the differences in backbone dihedrals or pseudo-dihedral angles (Levitt, 1976), but limitations soon became apparent. In triose phosphate isomerase (TIM), for example, the “lid” hinge was clear in one subunit, but not the other, due to mostly “background” differences of 20° RMS pervading the protein (Joseph et al., 1990). Even at higher resolution, “background” differences, often averaging 10° or more (Table S1), obscure the transitions to be analyzed. More subtle changes can be analyzed by identifying groups of atoms that move near-rigidly between two crystal structures (eg. Abyzov et al., 2010; Huang et al., 1993; Schneider, 2002). Rotation/translation (RT) transformations can be calculated between corresponding groups (Kabsch, 1976), reduced to a screw axis, or approximated as a pure rotation (Wriggers and Schulten, 1997). Hinge residues can be proposed by proximity to such axes if the rigid-group assumption holds well and translational components are small (Gerstein et al., 1994; Wriggers and Schulten, 1997).
Hinge identification is a mathematically challenging “inverse” problem, where causes (dihedral rotations) are sought for observations (atomic displacements). It is ill-posed: many sets of dihedral rotations yield similar conformational transitions. The current work examines how direct optimization of torsion angle rotations, without prior rigid-group analysis, might improve the conditioning, in contrast to other methods which are sensitive to user-defined settings in the clustering of atoms into rigid groups. Conditioning is often improved by elimination of inconsequential parameters. Others have shown that polypeptides are often adequately described with pseudo-dihedral angles. The Cα—C and N—Cα of consecutive amino acids, (i-1, i) are within 10° of parallel, leading Diamond (Diamond, 1965) to refactor backbone torsion angles (ψi–1, φi) as pseudo-dihedrals (θi = ψi–1, + φi, τi = ψi–1 – φi). The set of θi approximates the protein fold (of interest here), but τi mostly affects peptide carbonyl orientations which will be ignored here. Continuing with the same strategy, we will explore methods used for other ill-posed problems to select and optimize only the subset of model variables that are most critical: parameter filtering (or subset selection), and regularization with - or -Norm restraints. In linear optimization, use of an -Norm is termed ridge regression, while the -Norm constitutes the widely-used LASSO technique (Tibshirani, 1996, 2011). The acronym stands for “Least Absolute Selection and Shrinkage Operator”, with “selection” referring to finding the model parameters of most consequence, and shrinkage referring to decrease in the total magnitude of parameter change. Approaches like LASSO limit the degrees of freedom and impose parsimony upon the solution. The LASSO method has recently been extended to non-linear optimization, as required for flexible superposition of molecules, with both ad hoc and theoretically justified rationalizations, and has been applied in varied fields outside structural biology (Rasouli, 2014; Tateishi et al., 2010; Yamada et al., 2013).
Judging which computational approach best models real conformational change is not trivial. Residual coordinate differences (RMSD) are not an appropriate measure, because RMSD is expected to increase with additional constraint or restraint. Manually annotated conformational changes are useful for comparison, but subjective. NMR data provide independent yardsticks, but not necessarily a one-to-one correspondence to molecular flexing. Chemical shift (CS) perturbations reflect changes in the local chemical environment, including, but not limited to conformation change or direct influence of ligand-binding (Osborne et al., 2003; Zuiderweg, 2002). NMR relaxation exchange contributions (Rex), quantified from relaxation dispersion experiments, reflects dynamic changes in CS occurring on μs-to-ms time scales (Boehr et al., 2006). Thus, while the NMR measurements are likely to reflect different types of conformational changes, there is no guarantee that every conformational change will be reflected in the NMR data, and the NMR data might also reflect the propagated effects of remote changes. Finally, crystallographically-inferred changes are usually ligand-induced, whereas NMR Rex reflects intrinsic equilibrium motional fluctuations. The induced and intrinsic motions may, but need not be related (Niu et al., 2011). Thus, one expects imperfect correlation between measurable Rex or CS perturbation and hinges identified from crystallographic atomic coordinates. In this work, we choose from the handful of model systems where the relevant NMR data are available in the literature or through our own efforts, and seek computer algorithms that identify direct links between the crystallographic and NMR data that are intuitive.
Using the LASSO method, we find that, with parsimonious rotations of backbone dihedrals, the conformational changes between different states of the same structure can often be more accurately described than with rotations / translations of rigid group atom clusters. Torsion angles that are rotated during the flexible superposition are, on average, 2.3 residues from sites exhibiting NMR relaxation exchange in arginine kinase, suggesting that the superposition is indicating loci of real flexibility. Substantial domain reorientations are possible with very modest hinge rotations that need not be constrained to flexible linkers or loops. Thus, the new computational approach provides fresh insights into the ways that proteins undergo functionally important conformational changes.
Results
The section opens with an in-depth analysis of arginine kinase on forming a transition state analog complex (AK; Niu et al., 2011), the system used to optimize the computational algorithms. It concludes with applications to ligand-induced transitions in four other enzymes.
The sum of pseudo-torsion angle differences between the substrate-bound and -free AK structures, Σi Δ|θi | = 3456°, is much larger than the rotations needed to superimpose subdomain-sized fragments. A preliminary estimate of the latter, 247°, comes from the rotations needed to superimpose consecutive fragments of DynDom (Hayward and Lee, 2002) quasi-rigid groups. Indeed, comparisons between structures expected to be very similar suggest that much of the dihedral variance between paired coordinate sets results from experimental error in the structures or specifics of a crystallization environment. For example, the mean pseudo-torsion angle difference is 8 ± 16° between triose phosphat e isomerase and a single site mutant (Rozovsky et al., 2001); 8 ± 19° between RNase crys tal forms (PDBids 3MZQ, 2G8Q; Berger et al., 2010; Leonidas et al., 2006); and 9 ± 11° betw een independently-determined methotrexate complexes of dihydrofolate reductase (DHFR; PDBids 1RG7, 1DDS; Dunbar et al., 1997; Sawaya and Kraut, 1997). Such differences, whether from experimental error or local conformational variation, will obscure key hinge rotations that are less than 20 or 30° (Figure S1). Methods are needed to flexibly align structures using only the minimal fraction of dihedral rotations required to superpose the principal elements of structure.
Subset selection is a conceptually simple approach to fixing the less consequential parameters. With repeated batches of least-squares coordinate alignment, the model parameters were ranked, then either a growing number of the most consequential parameters were added, or the least consequential were fixed at their starting values. Several criteria for ranking were tested (see Supplementary Information) including partial derivatives of the alignment residual, summed shift vectors and, finally, parameter changes during a prior LASSO-restrained refinement (see below). Subset selection could be used to eliminate the large number of dihedral parameters that had very little impact, but, when iterated to narrow down to small numbers of key dihedrals, selection bias was evident (Figure S2 & Table S2). Selection bias is the ill-conditioned dependence of the final answer on selections made in prior iterations and the exact criteria used to make them. It is a well-known issue with subset selection methods, and occurred with all of the metrics that we tested for parameter ranking. The most stable ranking was by magnitude of LASSO-restrained parameter change, and this was well-conditioned when used to fix the large number (75%) of dihedrals of negligible consequence. Subset selection was relegated to the role of eliminating inconsequential parameters for efficiency, but different approaches were needed to distinguish high and moderate impact parameters.
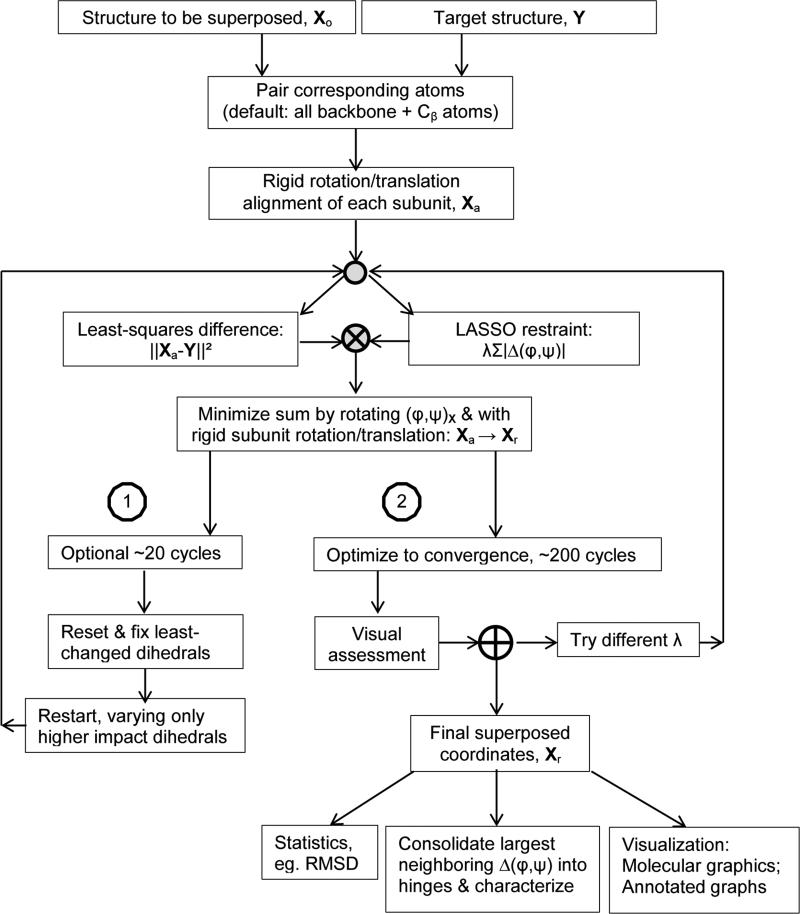
More successful was coordinate refinement with an -Norm (LASSO) restraint on the sum of dihedral rotations. By comparison to the more familiar least-squares -Norm, the -Norm gave greater contrast between large and small rotations, and lower coordinate RMSD for a given total dihedral change. This is consistent with extensive experience in linear least squares, comparing ridge regression (-Norm) with LASSO (-Norm), and recent findings that the same holds true in non-linear optimization (Rasouli, 2014; Tateishi et al., 2010; Yamada et al., 2013). It is noted that the theoretical justifications of least-squares regularization in terms of solutions of minimal error rest on an assumption, that often does not hold, of normally distributed errors. In fact, there is an extensive history of optimization with other types of objective function (Branham Jr., 1982) with -Norm minimization going by many names such as least absolute deviation (LAD) or least absolute error (LAE). In the LASSO technique, the objective function is mixed, combining least squares () fitting of the observed data with a weighted () restraint on absolute changes in the model parameters. With a weight, λ = 2 (Eqn. 1), the -penalty accounts for only 4% of the objective function change in AK, but reduces the total pseudo-dihedral change from 3456° to 287°, close to the 247° expected from rigid-group a nalysis. The RMSD decreases from 3.1 to 0.70 Å, an improvement on the 0.92 Å rigid-group RMSD. The overall approach is summarized in Figure 1.
Figure 1.
Overview of superposition through optimization of parsimonious dihedral angle changes.
The choice of λ is subjective. With greater latitude (lower λ), RMSD decreases, but with diminishing returns (Table S3). Subdomains are approximately overlaid even at high λ (Figure S4). As λ is lowered, domain overlay improves slightly, but interactive graphics show mostly local loop changes. There is not a statistic that clearly indicates the best λ. Rather, structures superposed with different λ are compared visually, choosing the lowest λ where domain-scale changes still predominate.
In test cases, the flexible superposition can be judged in two ways. Later, we will ask whether the sites of greatest dihedral change are consistent with NMR data. First, we ask whether the dihedral parameterization provides a more accurate representation of conformational change than rigid-group in terms of accuracy of the resulting atomic coordinates. This can be done through cross-validation to the crystallographic test set data measured for the target structure, but not used in its determination. Thus, Table 1 compares Rfree for various AK atomic models that have been derived from the transition state structure, modeling the change towards the substrate-free state with various parameterizations. In all cases, the model is compared to test set diffraction data of the substrate-free form (Niu et al., 2011) that has not been used in either the starting (TSA) or target (apo) structure determinations. All of the values are high, corresponding to Rfree for unrefined models, because none of the parameterizations are refining side chain configuration, B-factors or including explicit solvent. Relative to completely rigid subunit superimposition, DynDom's dynamic domains (Hayward and Lee, 2002) achieve modest improvement (Rfree decreasing from 0.54 to 0.51). ESCET's (Schneider, 2002) definition of conformationally invariant regions is substantially better (Rfree = 0.46). Our parsimonious optimization of φ, ψ angles does even better with Rfree between 0.43 and 0.45, depending on λ (5 and 2 respectively). Thus, with approximately the same degrees of freedom, parameterizing conformational change through parsimonious dihedral rotations has the potential to be more accurate than available methods for rotation-translation of rigid clusters of atoms.
Table 1.
Comparison of dihedral and rigid-group parameterizations of conformational change through crystallographic cross-validation.
| Model | R | Rfree |
|---|---|---|
| Arginine kinase, substrate-free (apo-AK), full refined, 1.7 Å resolution (Niu et al., 2011) | 0.189 | 0.241 |
| apo-AK, without residues disordered in transition state analog structure (TSA), waters & TLS-based anisotropic B-factors | 0.262 | 0.287 |
| TSA structure (Yousef et al., 2000) superposed as a rigid subunit | 0.534 | 0.538 |
| TSA structure (Yousef et al., 2000) superposed with DynDom (Hayward and Lee, 2002), default parameters yielding 2 quasi-rigid domains | 0.505 | 0.510 |
| TSA-superposed, manually varying DynDom parameters to yield 3 or 4 dynamic domains | 0.510±0.001 | 0.520±0.003 |
| TSA-superposed by ESCET distance difference matrix analysis (Schneider, 2002), yielding 5 rigid groups | 0.457 | 0.458 |
| TSA-superposed, rigid clusters combined from DynDom and ESCET and manually consolidated into consensus 5 groups | 0.466 | 0.458 |
| TSA-superposed by parsimonius dihedral rotation with tight restraint, λ = 5.0 | 0.453 | 0.450 |
| TSA-superposed by parsimonius dihedral rotation, looser restraint, λ = 2.0 | 0.431 | 0.435 |
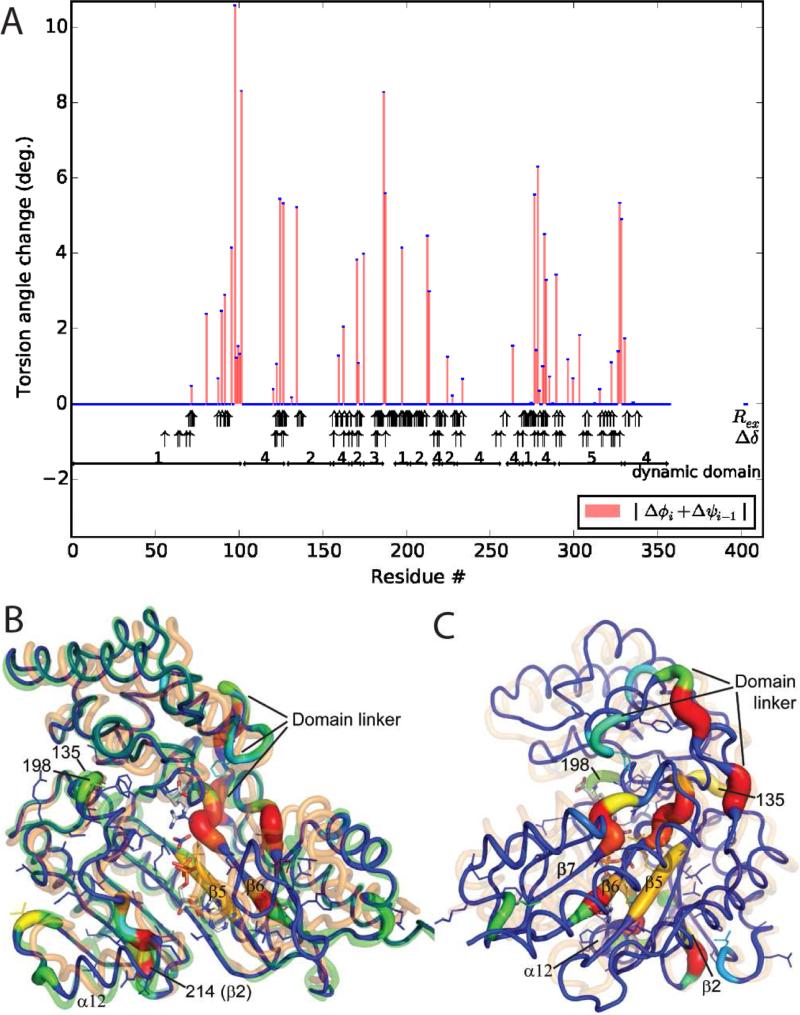
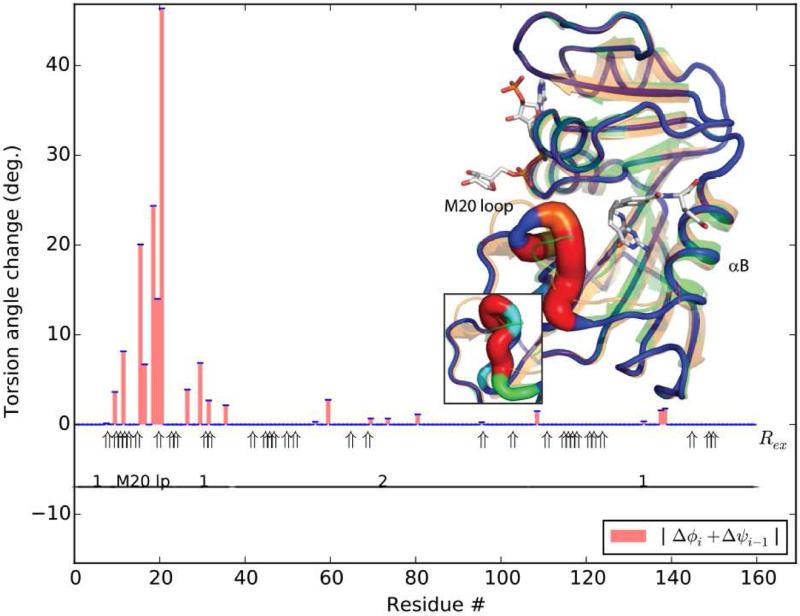
Consistency is high between torsion angles rotated most during superposition and NMR measured of change (Figure 2). Perfect consistency cannot be expected. Like structure superposition, NMR chemical shift perturbations (Δδ) reflect substrate-associated changes, but are available only for 70% of AK's backbone, because assignable, non-degenerate resonances are needed for both structures (Davulcu et al., 2005; Davulcu et al., 2013). Additionally, residues close to the substrates are excluded from analysis to avoid confusion between direct interactions and effects mediated through conformational change. Furthermore, Δδ reflects diverse changes in structure and environment, not just the (φ, ψ) rotations being analyzed. Thus, it is not surprising that the largest dihedral rotations are, on average 7 residues from significant Δδ (≥ 0.5 ppm). By contrast, Rex, a probe of dynamics, is measurable for > 90% of backbone nitrogens either in substrate-free or TSA form (Davulcu et al., 2009 and this work). Not all motions are observable; those outside the μs-ms timescale or with excited-state populations below 0.5% are difficult to detect (Boehr et al., 2006). Between the 50 non-zero pseudo-dihedral rotations and the 61 experimentally significant Rex, there are 17 exact matches, and an average separation of 2.3 residues between closest pairs. This level of agreement is striking, given that Rex samples intrinsic dynamics that may not include all of the crystallographically-observed substrate-induced changes. Furthermore, although Lipari-Szabo analysis indicates ns timescale motions (Davulcu et al., 2009) and a hinge was implicated near 102/103 by dynamic domain analysis (Niu et al., 2011), neither Rex nor Δδ is seen near residues 96-102, perhaps because the backbone nitrogens of prolines 100 and 101 are not NMR-observable. If this region is excluded, the average separation between changing (φ, ψ) and the nearest Rex or Δδ is 1.5 residues.
Figure 2.
Flexible superposition of AK TSA on the substrate-free form. Two rounds of selection eliminated the 75% least consequential dihedrals. Refinement continued for 200 cycles with an -norm λ = 5. (A) Changes in pseudo-torsion angles. Annotation includes: dynamic domain designation (Hayward and Lee, 2002; Niu et al., 2011); observation of NMR relaxation exchange (Rex, wide arrows) in the substrate-free (Davulcu et al., 2009) or –bound (this work) states; and NMR chemical shift perturbation (Δδ, thin arrows) between substrate-free and -bound forms, excluding residues close to substrates. (B) Starting from the AK TSA structure (orange), the parsimoniously superposed model (color- and thickness-coded by magnitude of pseudo-dihedral rotation, blue to red) is now well fit to the substrate-free form (green). Transition state analogs are shown as a stick model. Side chains are shown for residues with backbone nitrogen Rex to highlight proximity to changing dihedrals. (See also Figure S5) (C) The TSA and superposed structures are rotated 90° to highlight flexing of the β-sheet.
Of existing hinge-determination methods, DynDom (Hayward and Lee, 2002) and MolMovDB (Echols et al., 2003) are perhaps the most prevalent. Hinges predicted using these methods with default parameters are also close to sites of Rex, differing by 2.0 and 3.0 residues respectively. The main difference is seen when the comparison is inverted, asking how close sites of Rex are to the nearest hinge: the respective averages of 21 and 11 residues are much greater than the 2.2 residues coming from our new approach. Thus, while all three approaches identify hinges from the crystallographic coordinates that are approximately consistent with the NMR data, the prior rigid-group approaches are identifying only a subset of the sites that are implicated in the NMR data (and recognized in the new approach). For DynDom, non-default rigid-group clustering parameters yield better statistics: hinges within an average 1.2 residues of Rex sites, and Rex within 7 residues of hinges, but these parameters were chosen to mimic the clusters of ESCET (Schneider, 2002), then manually adjusted for consistency with the NMR Rex. This shows that the rigid clustering is the main limitation of prior hinge definition methods, but, even with the manually curated parameters, some of the hinges remain uncharacterized.
Consider the anatomy of the substrate-induced changes revealed by our new approach to superposition. Total rotations are calculated for putative hinges by (1) clustering neighboring pseudo-dihedrals and (2) concatenating their rotation matrices (Table S4). Residues 90 to 102, the flexible linker between N- and C-domains (Figure 2 B/C), have a combined 20.2° rotation, consistent with a 21.2° rotation between dynamic domains 1 and 4 (Niu et al., 2011). Residues 277-9 (12.0°) and 283-4 (7.0°), before and in β-strand 6, have nearly co-linear rotations totaling 18.6°. In adjacent β-strand 5, residues 125-7 rotate 10.7°, while on the other side of β-strand 6, residues 328 to 331, following β-strand 7, undergo a 9.9° rotation. Thus, three regions separated in linear sequence, but adjacent in 3D space, exhibit a torsional flexing parallel to the strands of the β-sheet. The next largest rotations are in 171-6 (7.4°) and 187-8 (7.5°) in the loops flanking α-helix 12. Rotations at 135 (5.2°) and 198 (4.2°) a re in loops adjacent to each other. The last large rotation is in residues 213-4 (6.1°) at the C -terminal end of β-strand 2, on the opposite edge of the β-sheet. It is remarkable that enzyme closure comes through individually modest rotations within a β-sheet, but the conclusion is supported by co-localization of intrinsic motions implied by Rex.
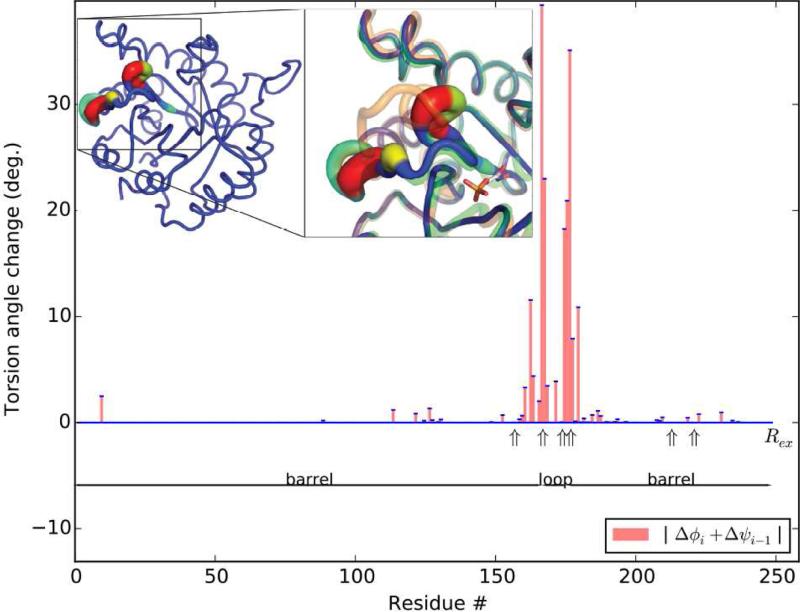
Parsimonious superposition was applied to other systems where NMR data allowed validation. First was triose phosphate isomerase, on binding an intermediate analog (TIM, PDBids 1YPI & 7TIM; Davenport et al., 1991; Lolis et al., 1990). The largest rotation (34°) and 91% of the total change are in a loop (161-180) that folds over the active site (Figure 3). No other change exceeds 2.5°. Raw pseudo-dihedral differences are widespread and much larger (averaging 9° vs. 0.8°), obscuring the loop with 3-fold greater changes elsewhere. The parsimony restraint does not limit convergence, the RMSD of 0.48Å approaching the experimental accuracy of 0.44Å (backbone/Cβ) judged by comparing mutant and wild-type substrate-free structures. Hinge analysis shows composite 40° and 41° rotations in the loop at 167-9 and 175-7 (Figure 3B). Five of the top eight NMR Rex (Massi et al., 2006) are within the loop, and two more (residues 213 and 221) are in the helix against which the loop packs. Consistency between the hinge analysis and NMR affirms the computational analysis and implies use of the protein's intrinsic flexibility in substrate-induced changes.
Figure 3.
Superposition of the substrate free triose phosphate isomerase (inset orange) on the complex (green) with an intermediate analog (stick model), using λ=0.3, reduces the RMSD from 1.1 to 0.48 Å. The parsimoniously superposed model is color- and thickness-coded as in Figure 2. NMR Rex observations are concentrated in the loop or at proximal sites (213, 221) directly affected by the loop's conformational change.
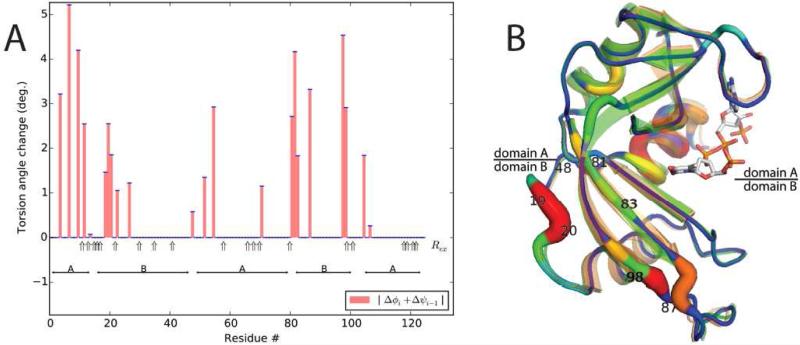
Changes in RNase A on binding a dinucleotide substrate analog are more subtle (RMSD = 0.75Å; PDBids 7RSA & 1U1B; Beach et al., 2005; Wlodawer et al., 1988), especially with substrate-free crystal structures differing by 0.77Å (Berger et al., 2010; Leonidas et al., 2006). Raw pseudo-dihedral differences reveal the hinge near residue 20, but otherwise defy rationalization, averaging 5.4°. Parsimonious super position (λ=1.0) yields an RMSD of 0.44 Å with average rotations of 0.4°, and hinges revealed that are consistent with rigid-group analysis (Figure 4A). NMR Rex measurements of intrinsic motion (Beach et al., 2005) agree about equally well with both analyses, although there are some differences (e.g. residue 70) with more dispersed flexing in the new analysis. Even though the rotations are small, they are physically plausible. Thus, hinges near residues 87 and 98 that are close in 3D space (Figure 4B) have compatible 3.3° and 4.3° rotations consistent with hinged rotation of the intervening loop.
Figure 4.
Superposition of substrate-free bovine RNaseA on a dinucleotide complex. (A) Consistency of dihedral changes with NMR Rex, after optimization with λ=1; (B) The superposed model (color-coded by dihedral change) fits the complex (orange) with an RMSD of 0.44 Å.
Dihydrofolate reductase (DHFR) is challenging with an RMSD of only 0.67 Å between open and closed states (PDBids 5DFR & 1RX2; Bystroff and Kraut, 1991; Sawaya and Kraut, 1997), although only part of the M20 loop is resolved in the open form. Parsimonious superimposition yields RMSD = 0.53 Å, with a combined rotation of 7° at residues 36, 39 and 41 (following helix B) as helix C slides over the β sheet. The M20 loop is fully resolved in the occluded state (PDBid 1RX6; Sawaya and Kraut, 1997), increasing the RMSD vs. the closed state to 1.25 Å. Our algorithm is poorly suited to describing the reconfiguration of the M20 loop (Figure 5) that includes a 180° rotation in ψ14 (Sawaya and Kraut, 1997). A more faithful representation is achieved with a weak parsimony restraint (λ = 0.3), but the more stringent restraint (λ = 1.0) shows hinges in other parts more clearly. There is a 10° bending within helix B that moves the loop sub-domain; a 2° rotation at the start of the next loop (at the site of the 7° change between open and closed forms); and 2° rotations in two β-strands, where, retrospectively, kinking can be observed in the structures.
Figure 5.
Starting from the closed E. coli DHFR structure (orange), parsimonious superposition (color-coded by dihedral change) is fit to the occluded form (green). Optimized using λ=1.0 (graph and full molecule), the RMSD is 0.85 Å. With λ=0.3 (inset), RMSD drops to 0.74 Å through increased latitude to fit the local configuration of loop M20. Rex is shown for 3 complexes, both occluded and closed (Osborne et al., 2001). Rex in the FG and GH loops, near residues 120 and 148 respectively are thought to reflect changing interactions with the M20 loop (Osborne et al., 2001) due to motions of the loop and rotation of dihedrals near helix B (i.e. long-range effects), while those near residue 45 might reflect transient cofactor dissociation (Osborne et al., 2001), so the absence of large dihedral changes in these regions is not unexpected.
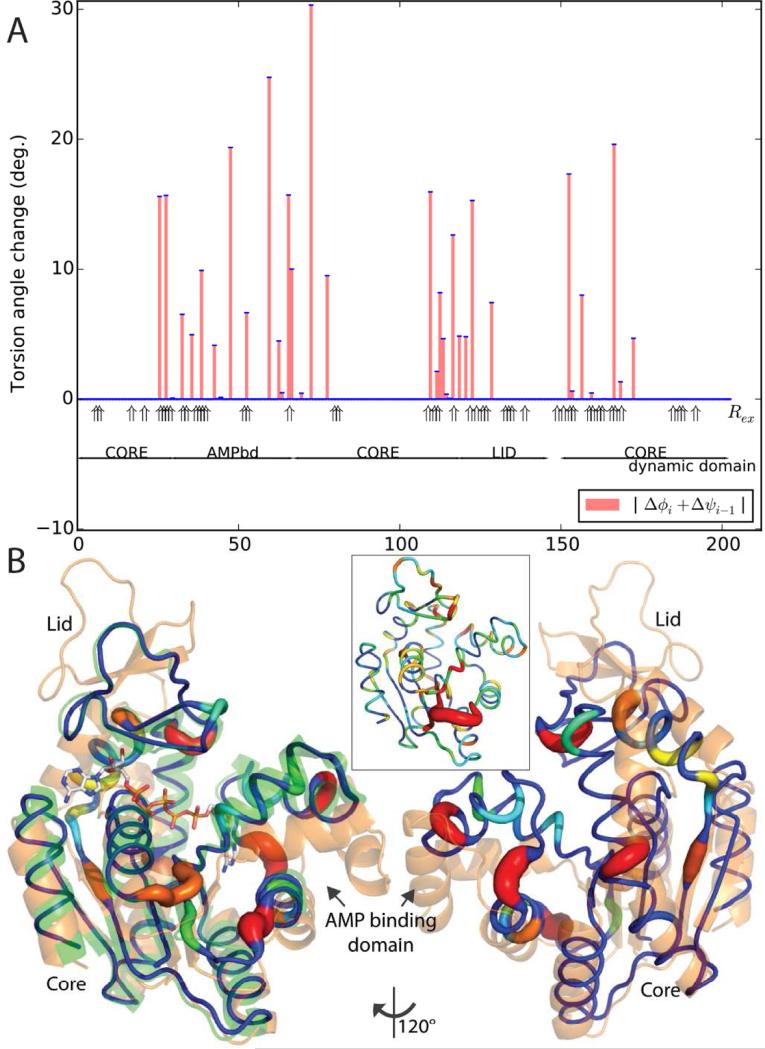
Adenylate kinase (AdK) exhibits substantial differences between the apo form and a bisubstrate analog complex with a backbone/Cβ RMSD of 5.9 Å (Henzler-Wildman et al., 2007). The “lid” and AMP-binding domains (Matsunaga et al., 2012) fold in over the substrates with rotations of 40° and 41°, but classical rigid-group modeling yields only approximate superposition (RMSD = 2.7 Å). Our flexible superposition yields an RMSD of 0.79 Å. Hinges are apparent that were obscured in the raw dihedral differences which were dispersed (Figure 6) and, on average, 7-fold greater (10.4°/residue). Th e hinges are within 6 residues of those identified (Henzler-Wildman et al., 2007) by large dihedral differences close to rotation axes between DynDom dynamic domains (Hayward and Lee, 2002). The new analysis reveals the importance of articulated hinges. The outer hinge for the AMP binding domain pairs a 27° rotation at residues 26-28 with a 31° rotation at pseudo-dihedral 73 which is neighboring in 3D space. There are several rotations within the intervening domain, most prominently, the paired 19° and 25° rotations in neighboring 48 and 60 as the distal loop folds over the substrates. The lid domain also moves through articulated hinges, a 48° rotation in residues 110-123 paired with a 38° rotation in 153-167, as these neighboring seg ments run antiparallel behind the active site cleft. The residues of greatest change are in register, 110 with 167 and 123 with 154. Quasi-rigid group analysis had placed the hinges at the ends of helix 8, but actually, residue 167 is in the middle of the helix at a kink, that, in retrospect, clearly becomes larger in the substrate-free structure (Figure 6B).
Figure 6.
Superimposition of the substrate-free A. aeolicus adenylate kinase on a bisubstrate analog complex. (A) The parsimony-restrained dihedral changes (λ=1.0) are compared to NMR Rex and domain designations; (B) The superposed structure (colored by dihedral change) has converged upon the substrate complex (green) from the substrate-free starting structure (orange). The inset is colored according to the raw pseudo-dihedral differences between the substrate-free and -bound structures (not parsimoniously refined), from which it is unclear where the hinges are.
Correspondence between AdK pseudo-dihedrals changing on substrate-binding and NMR Rex, implying intrinsic substrate-free dynamics, is good, but imperfect (Figure 6A) (Wolf-Watz et al., 2004). For the AMP-binding domain, the N-terminal hinge shows Rex. The C-terminal hinge does not, but an adjacent strand does (residues 80/81), perhaps due to changes in the environment. Not all, but most of the domain's other large torsion rotations are at sites of Rex. For the lid domain, both articulated hinges show multiple Rex, and the Rex at residues 166-7 maps exactly to the kink in helix 8. The number of residues with Rex is noticeably elevated in the four regions identified as articulated hinges.
Discussion
Torsion angles are understood to be the most variable parameters of protein structure, but their use in describing conformational change has fallen out of favor. The challenges of reliably divining the most critical changes have led to the preeminence of analyses that cluster atoms into quasi-rigid groups (Hayward and Lee, 2002; Rashin et al., 2009; Schneider, 2002) or that model flexibility as elastic bulk deformation (Tama et al., 2002). Stereochemical distortions in both approaches undermine detailed interpretation of large conformational changes. Advantages of the new approach include maintenance of ideal stereochemistry with the constraint that rotations of atoms are about covalent bonds.
We find that the sum of rotations needed to superimpose structures within experimental accuracy is often 10-fold less than the observed ϕ, ψ differences. The excess may reflect real local differences in conformation, but wide distribution throughout the structures of single site mutants suggests that dihedral differences are heavily influenced by experimental errors in crystal structures, including both the dependence of torsion angles on experimental errors in the underlying atomic coordinates, and the subtle influences of extrinsic factors, such as different crystal environments, upon conformation. Thus, we now understand why simple analyses of torsion angle differences are generally informative only for the largest of hinged rotations. The new approach allows analysis down to rotations of ~1°, because, with the parsimony restraint, conformational transitions are modeled with the minimal dihedral rotations responsible for gross superposition.
It is clear that large multi-Ångström conformational changes are possible with modest torsion angle rotations that do not require the presence of glycines and are possible within secondary structures that, intuitively, might be excluded from consideration. Indeed, our analysis, supported by experimentally observed Rex, shows hinged rotations in adjacent strands of the β-sheet in arginine kinase, and in helices of DHFR and adenylate kinase, all of which can occur without serious disruption of the hydrogen bonding structure.
It is surprising that rotations of fewer than 3% of torsion angles usually suffice to superpose structures within the experimental errors. Subset selection illustrated the potential ill-posedness, rotations about different (nearly parallel) bonds yielding similar conformations. Conditioning depends on how ill-posed problems are formulated, so it is not surprising that hinges determined from rigid group analysis are sensitive to the parameters of atom clustering. Better conditioning is expected from the direct optimization of torsion angles, and from an -norm parsimony restraint that both selects the most consequential parameters and shrinks their total change (Tibshirani, 2011).
Beyond theoretical arguments, are we closer to reality? Although the NMR paints an incomplete picture, it is valuable as an independent cross-validating reference. It is striking how the new analysis improves the consistency between the crystallography and NMR by detecting hinges that are passed over in rigid-group analysis. Even though incompatibilities are expected between the crystallographic and NMR approaches, hinges and sites of relaxation exchange agree in AK remarkably well with an average discrepancy < 2.5 residues.
In contrast to prior approaches, the new method embodies an a priori constraint that conformational change be described as rotations about bonds, thereby maintaining ideal stereochemistry. A posteriori, we see that dihedral rotations separated in primary sequence often come together in 3D space with quantitatively matching composite rotations. This is consistent with hinged rotations of loops and domains, which is highly plausible, but not constrained. As seen in AdK, domains are often moving with articulated hinges, a rotation within a rotating domain, which is difficult to resolve by quasi-rigid analysis. Some transformations, previously regarded as complex (non-rigid), can be broken down into component rotations with the articulated hinge points pairing up in 3D space.
The new approach reveals greater consistency between substrate-induced changes measured crystallographically and intrinsic flexibility reflected in NMR Rex measurements. In a set of representative enzymes, the hinge points of induced changes are mostly within a couple of residues of sites implicated in intrinsic dynamics, a small difference considering the complementary origins of the crystallographic and NMR measurements. Such consistency supports the postulate that substrate-associated motions take advantage of flexibility that has been selected at specific hinge locations through the protein (Ma and Nussinov, 2010). The Rex data do not inform us whether the intrinsic changes are as large in magnitude as those ligand-induced, suggesting only that modes of flexibility are qualitatively similar. Thus, the substrate-associated conformational changes appear to proceed in low-dimensional dihedral space, passing along low barrier paths about which intrinsic dynamics is naturally occurring under physiological conditions, as observed through NMR Rex.
Experimental Procedures
Torsion angle structure refinements are usually implemented for best local improvement by optimizing a moving window of residues, with deformable bonds at the window limits subject to stereochemical restraints to the rest of the chain which is fixed. By contrast, we needed a global optimization that best reflected the long-range effects of small torsional rotations. To determine which bonds can be rotated and the atomic positions that depend on each, graph theory is used to find connected components (chains of bonded atoms) and articulation edges (rotatable bonds) in undirected graphs. To improve the efficiency of backbone refinement, side-chain atoms are connected topologically to the Cα atoms.
On each iteration of refinement, to avoid error accumulation, the current set of torsion angle rotations is applied to the matrix of original coordinates Xo, yielding rotated coordinates, Xr. Aligned coordinates, Xa, come from rigid alignment of Xr to the target coordinates Y, using the Kabsch algorithm (Kabsch, 1976). The unrestrained least-squares objective function is then Ou = ∥Xa - Y∥2. For gradient descent optimization, analytic approximations are calculated for the partial derivatives of Ou with respect to each of the torsion angle parameters. Partial derivatives are also calculated for the Kabsch alignment operator with respect to the rotated coordinates, Xr. Partial derivatives of Xr with respect to the torsion angles, and those of Xa with respect to the alignment operator, can be computed easily. The chain rule then allows us to forecast, and incorporate into the partial derivatives of the objective function, the effect of the rigid alignment as it changes with torsion angle rotations. With internal coordinates, there is an ambiguity, usually solved heuristically, of how much the coordinates should be rotated each side of a torsion angle. Our approach breaks the ambiguity by constraining dihedral operators to maintain least-squares alignment, and calculates partials consistent with this constraint to improve convergence.
Consider regularization of the objective function such that it is parsimonious in terms of both the number (P) of parameters, βi, that are variable, and the aggregate magnitude of their changes. These characteristics, technically termed variable selection and shrinkage, are often achieved in linear regression via the LASSO approach, minimizing the -penalized objective function (Tibshirani, 1996, 2011):
| Equation 1 |
Tibishirani and others have noted that the LASSO approach is superior to -Norms in driving β to zero for inconsequential parameters. The approach is analogous to compressed sensing in signal reconstruction, where the –Norm yields the sparsest solution with fewest non-zero terms (Donoho, 2006a; Donoho, 2006b). Here, it is extended to the non-linear dependence of atoms on torsion angle changes when superposing on a fixed structure :
| Equation 2 |
where the -Norm restrains the pseudo-torsion angle, ϕ. In our implementation, equation 2 is minimized using the L-BFGS quasi-Newton optimizer (Byrd et al., 1995).
For interpretation, rotations of individual dihedrals are collected into hinges. Hinges span residues whose pseudo-dihedral rotations exceed a user-specified threshold, that are separated by gaps less than a user-defined limit. The combined hinge rotation is calculated by concatenating the rotation matrices for the successive dihedrals followed by Eigen analysis.
Measurement of the NMR relaxation dispersion for the AK TSA complex follows that of apo AK (Davulcu et al., 2009) and is described in the Supplementary Methods.
Supplementary Material
Highlights.
Protein conformational change is characterized as minimal torsion angle rotations.
Imposition of parsimony improves consistency with NMR measures of dynamics.
Large conformational changes are assembled from small dihedral rotations.
Changes on ligand-binding follow modes of motion seen in the intrinsic dynamics.
Acknowledgements
This research was supported by NIH R01 GM77643. For software licensing see http://xtal.ohsu.edu/.
Abbreviations and symbols
- AK
arginine kinase
- AdK
adenylate kinase
- CS
chemical shift
- DHFR
dihydrofolate reductase
- NMR
nuclear magnetic resonance
- Rex
relaxation exchange
- RMSD
root mean square deviation
- TIM
triose phosphate isomerase
- TSA
transition state analog
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abyzov A, Bjornson R, Felipe M, Gerstein M. RigidFinder: a fast and sensitive method to detect rigid blocks in large macromolecular complexes. Proteins. 2010;78:309–324. doi: 10.1002/prot.22544. [DOI] [PubMed] [Google Scholar]
- Beach H, Cole R, Gill ML, Loria JP. Conservation of mus-ms enzyme motions in the apo- and substrate-mimicked state. J Am Chem Soc. 2005;127:9167–9176. doi: 10.1021/ja0514949. [DOI] [PubMed] [Google Scholar]
- Berger MA, Decker JH, Mathews II. Diffraction study of protein crystals grown in cryoloops and micromounts. J Appl Crystallogr. 2010;43:1513–1518. doi: 10.1107/S0021889810040409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chem Rev. 2006;106:3055–3079. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- Branham RL., Jr. Alternatives to least squares. Astronomical Journal. 1982;87:928–937. [Google Scholar]
- Byrd RH, Lu P, Nocedal J. A Limited Memory Algorithm for Bound Constrained Optimization. SIAM Journal of Scientific and Statistical Computing. 1995;16:1190–1208. [Google Scholar]
- Bystroff C, Kraut J. Crystal structure of unliganded Escherichia coli dihydrofolate reductase. Ligand-induced conformational changes and cooperativity in binding. Biochemistry. 1991;30:2227. doi: 10.1021/bi00222a028. [DOI] [PubMed] [Google Scholar]
- Davenport RC, Bash PA, Seaton BA, Karplus M, Petsko GA, Ringe D. Structure of the triosephosphate isomerase-phosphoglycolohydroxamate complex: an analogue of the intermediate on the reaction pathway. Biochemistry. 1991;30:5821–5826. doi: 10.1021/bi00238a002. [DOI] [PubMed] [Google Scholar]
- Davulcu O, Clark SA, Chapman MS, Skalicky JJ. Main chain (1)H, (13)C, and (15) N resonance assignments of the 42-kDa enzyme arginine kinase. J Biomol NMR. 2005;32:178. doi: 10.1007/s10858-005-6731-8. [DOI] [PubMed] [Google Scholar]
- Davulcu O, Flynn PF, Chapman MS, Skalicky JJ. Intrinsic domain and loop dynamics commensurate with catalytic turnover in an induced-fit enzyme. Structure. 2009;17:1356–1367. doi: 10.1016/j.str.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davulcu O, Niu X, Brüschweiler-Li L, Brüschweiler R, Skalicky JJ, Chapman MS. Backbone resonance assignments of the 42 kDa enzyme arginine kinase in the transition state analogue form. Biomolecular NMR Assignments. 2013:1–4. doi: 10.1007/s12104-013-9512-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond R. A Theoretical Study of the Least-Squares Refinement of Flexible Long-Chain Molecules, with Special Reference to α-Helical Structures. Acta Cryst. 1965;19:774–789. [Google Scholar]
- Donoho DL. Compressed sensing. Information Theory, IEEE Transactions on. 2006a;52:1289–1306. [Google Scholar]
- Donoho DL. For most large underdetermined systems of linear equations the minimal l1-norm solution is also the sparsest solution. Communications on Pure and Applied Mathematics. 2006b;59:797–829. [Google Scholar]
- Dunbar J, Yennawar HP, Banerjee S, Luo J, Farber GK. The effect of denaturants on protein structure. Protein Science. 1997;6:1727–1733. doi: 10.1002/pro.5560060813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols N, Milburn D, Gerstein M. MolMovDB: analysis and visualization of conformational change and structural flexibility. Nucleic Acids Res. 2003;31:478–482. doi: 10.1093/nar/gkg104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M, Lesk AM, Chothia C. Structural mechanisms for domain movements in proteins. Biochemistry. 1994;33:6739–6749. doi: 10.1021/bi00188a001. [DOI] [PubMed] [Google Scholar]
- Hayward S, Lee RA. Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J Mol Graph Model. 2002;21:181–183. doi: 10.1016/s1093-3263(02)00140-7. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, et al. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- Huang ES, Rock EP, Subbiah S. Automatic and accurate method for analysis of proteins that undergo hinge-mediated domain and loop movements. Curr Biol. 1993;3:740–748. doi: 10.1016/0960-9822(93)90021-f. [DOI] [PubMed] [Google Scholar]
- Joseph D, Petsko GA, Karplus M. Anatomy of a conformational change: hinged “lid” motion of the triosephosphate isomerase loop. Science. 1990;249:1425–1428. doi: 10.1126/science.2402636. [DOI] [PubMed] [Google Scholar]
- Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallographica. 1976;A32:922–923. [Google Scholar]
- Leonidas DD, Maiti TK, Samanta A, Dasgupta S, Pathak T, Zographos SE, Oikonomakos NG. The binding of 3′-N-piperidine-4-carboxyl-3′-deoxy-ara-uridine to ribonuclease A in the crystal. Bioorg Med Chem. 2006;14:6055–6064. doi: 10.1016/j.bmc.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Levitt M. A simplified representation of protein conformations for rapid simulation of protein folding. J Mol Biol. 1976;104:59–107. doi: 10.1016/0022-2836(76)90004-8. [DOI] [PubMed] [Google Scholar]
- Lolis E, Alber T, Davenport RC, Rose D, Hartman FC, Petsko GA. Structure of yeast triosephosphate isomerase at 1.9-A resolution. Biochemistry. 1990;29:6609–6618. doi: 10.1021/bi00480a009. [DOI] [PubMed] [Google Scholar]
- Ma B, Nussinov R. Enzyme dynamics point to stepwise conformational selection in catalysis. Curr Opin Chem Biol. 2010;14:652–659. doi: 10.1016/j.cbpa.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massi F, Wang C, Palmer AG., 3rd Solution NMR and computer simulation studies of active site loop motion in triosephosphate isomerase. Biochemistry. 2006;45:10787–10794. doi: 10.1021/bi060764c. [DOI] [PubMed] [Google Scholar]
- Matsunaga Y, Fujisaki H, Terada T, Furuta T, Moritsugu K, Kidera A. Minimum Free Energy Path of Ligand-Induced Transition in Adenylate Kinase. PLoS Comput Biol. 2012;8:e1002555. doi: 10.1371/journal.pcbi.1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X, Bruschweiler-Li L, Davulcu O, Skalicky JJ, Bruschweiler R, Chapman MS. Arginine Kinase: Joint Crystallographic and NMR RDC Analyses Link Substrate-Associated Motions to Intrinsic Flexibility. J Mol Biol. 2011;405:479–496. doi: 10.1016/j.jmb.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne MJ, Schnell J, Benkovic SJ, Dyson HJ, Wright PE. Backbone dynamics in dihydrofolate reductase complexes: role of loop flexibility in the catalytic mechanism. Biochemistry. 2001;40:9846–9859. doi: 10.1021/bi010621k. [DOI] [PubMed] [Google Scholar]
- Osborne MJ, Venkitakrishnan RP, Dyson HJ, Wright PE. Diagnostic chemical shift markers for loop conformation and substrate and cofactor binding in dihydrofolate reductase complexes. Protein Sci. 2003;12:2230–2238. doi: 10.1110/ps.03219603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashin AA, Rashin AH, Jernigan RL. Protein flexibility: coordinate uncertainties and interpretation of structural differences. Acta Crystallogr D Biol Crystallogr. 2009;65:1140–1161. doi: 10.1107/S090744490903145X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashin AA, Rashin AHL, Jernigan RL. Diversity of Function-Related Conformational Changes in Proteins: Coordinate Uncertainty, Fragment Rigidity, and Stability. Biochemistry. 2010;49:5683–5704. doi: 10.1021/bi100110x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasouli M. Term selection for an induction motor via nonlinear Lasso. 2014:90740I–90740 I-90715. [Google Scholar]
- Rozovsky S, Jogl G, Tong L, McDermott AE. Solution-state nmr investigations of triosephosphate isomerase active site loop motion: ligand release in relation to active site loop dynamics. J Mol Biol. 2001;310:271–280. doi: 10.1006/jmbi.2001.4673. [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Kraut J. Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- Schneider TR. A genetic algorithm for the identification of conformationally invariant regions in protein molecules. Acta Crystallogr D Biol Crystallogr. 2002;58:195–208. doi: 10.1107/s0907444901019291. [DOI] [PubMed] [Google Scholar]
- Tama F, Wriggers W, Brooks CL., 3rd Exploring global distortions of biological macromolecules and assemblies from low-resolution structural information and elastic network theory. J Mol Biol. 2002;321:297–305. doi: 10.1016/s0022-2836(02)00627-7. [DOI] [PubMed] [Google Scholar]
- Tateishi S, Matsui H, Konishi S. Nonlinear regression modeling via the lasso-type regularization. Journal of Statistical Planning and Inference. 2010;140:1125–1134. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society B. 1996;58:267–288. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso: a retrospective. Journal of the Royal Statistical Society B. 2011;73:273–283. [Google Scholar]
- Wlodawer A, Svensson LA, Sjolin L, Gilliland GL. Structure of phosphate-free ribonuclease A refined at 1.26 A. Biochemistry. 1988;27:2705–2717. doi: 10.1021/bi00408a010. [DOI] [PubMed] [Google Scholar]
- Wolf-Watz M, Thai V, Henzler-Wildman K, Hadjipavlou G, Eisenmesser EZ, Kern D. Linkage between dynamics and catalysis in a thermophilic-mesophilic enzyme pair. Nat Struct Mol Biol. 2004;11:945–949. doi: 10.1038/nsmb821. [DOI] [PubMed] [Google Scholar]
- Wriggers W, Schulten K. Protein domain movements: detection of rigid domains and visualization of hinges in comparisons of atomic coordinates. Proteins. 1997;29:1–14. [PubMed] [Google Scholar]
- Yamada M, Jitkrittum W, Sigal L, Xing EP, Sugiyama M. High-Dimensional Feature Selection by Feature-Wise Kernelized Lasso. Neural Computation. 2013;26:185–207. doi: 10.1162/NECO_a_00537. [DOI] [PubMed] [Google Scholar]
- Yousef M, Somasundaram T, Gattis J, Pruett P, Gabriel FF, Chapman M. Protein Society Annual Meeting. Protein Society; San Diego: 2000. Refinement of Arginine Kinase Transition State Analogue Complex (AK-TSAC) to 1.2A : a deeper insight of a mechanistic question. [Google Scholar]
- Zuiderweg ER. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.