Abstract
Polyethylene glycol (PEG)-based hydrogels, with variable stiffness, are widely used in tissue engineering to investigate substrate stiffness effects on cell properties. Transcriptome analysis is a critical method for understanding cell physiology. However, significant RNA degradation was observed in the process of isolating and purifying RNA from cells encapsulated in the PEG hydrogel, thus precluding purification of high quality RNA. Here, we describe a simple protocol that prevents RNA degradation and improves the quality and yield of RNA isolated from cells cultured in PEG hydrogels. This modification produces high quality total RNA suitable for RNA sequencing and microarray analysis.
3D cultures are well established tools to mimic cell behavior in living organisms more precisely than standard 2D tissue culture plates. They are widely used to model the response of normal and tumor cells to tissue microenvironments [1, 2]. One of the most important factors affecting cells behavior, including proliferation and regulation of signaling pathways, is the mechanical properties of the tissue. PEG hydrogels are used extensively as a matrix for cell encapsulation because they provide enormous flexibility in designing matrices with tunable mechanical properties for the analysis of matrix-dependent cellular behavior [3]. RNA expression profiling by microarray hybridization or RNA sequencing (RNA-seq) are the most powerful and widely used approaches for global analysis of cellular responses. Both approaches require purification of high quality RNA from cells or tissues. However, the extraction of RNA from cells encapsulated in PEG gels results in mostly degraded RNA (low RNA integrity number (RIN)) [4, 5].
Cells were encapsulated in the hydrogel as in [2]. Briefly, the hydroxy-terminated PEG was functionalized with acrylate groups by the reaction of acryloyl chloride with PEG hydroxy end-groups as previously described [2]. 30 mg of the functionalized PEG macromer was dissolved in 270 μL of the initiator solution (0.5% initiator in PBS) by heating to 50°C and vortexing for 5 min. Next, a MDA-MB-231 cell suspension in 100 μL PBS was added to the hydrogel precursor solution and mixed gently with a glass rod. The suspension of cells in the precursor solution were degassed and UV irradiated with a mercury long wavelength (365 nm) UV lamp (UVP, Upland, CA) for 10 min as described [2]. Initially, the cellular RNA was isolated with a combination of TRIzol reagent (Life Technologies) extraction and column purification as described previously [6] (see Figure 1 for details). However, this standard procedure failed to provide high quality RNA with RIN > 7 suitable for microarray analysis or preparation of RNA-seq libraries [5] (Figure 1). Ribonuclease activity is a common cause of degradation of cellular RNAs. To prevent RNase-driven degradation, the gels were treated with RNAlater solution (Ambion) which is 70% ammonium sulfate and prevents RNA degradation by in-cell precipitation of riboprotein complexes. However, the RNase treatment provided only a minimal effect on RNA integrity (data not shown) suggesting that the gel components, rather than cellular RNases, caused RNA degradation. Therefore as a control, cell-free hydrogel in an amount equivalent to our gel-embedded cells was added to the TRIzol reagent and this TRIzol solution was mixed with previously purified high-quality total RNA (RIN > 7). The total RNA purified from the gel-containing TRIzol was significantly degraded while no RNA degradation was observed in the control TRIzol reagent (without gel) (Supplemental Fig S1, A,B ). Since guanidine thiocyanate in concentrations used in TRIzol solution effectively inhibits any RNase activity, the results provide further support for the effect of gel components on RNA degradation. Moreover, incubation of purified RNA with hydrogel components (PEGDA macromer and photoinitiator) caused significant RNA degradation in a concentration-dependent manner (Supplemental Fig S1, C-G). Therefore, the results suggest that components of the PEG gel affected RNA stability in TRIzol. The mechanism of RNA degradation by the gel components in TRIzol reagent is unclear; however, recent studies suggest that the acidic conditions in TRIzol (pH ~ 4.5 ) [7] can accelerate degradation of acrylate-functionalized PEG gels [8].
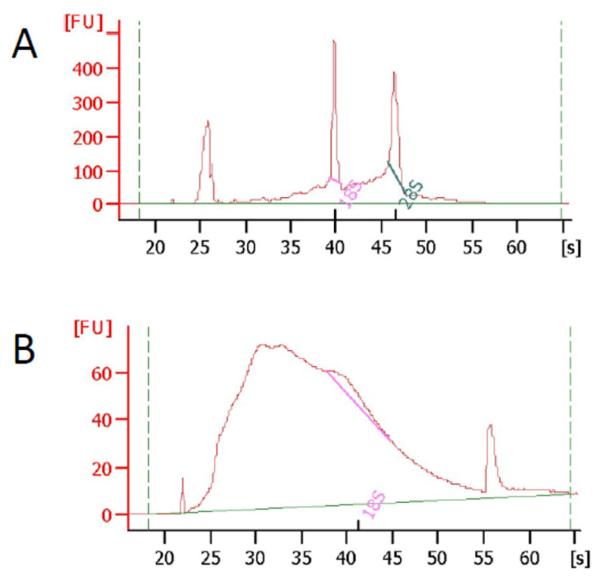
Figure 1. RNA degradation during purification from PEG hydrogels.
Total RNA was purified using a combined TRIzol® /column purification protocol. MDA-MB-231 cells were embedded in PEG hydrogels (1 million cells/mL) (B) control MDA-MB-231 cells plated on 60 mm tissue culture plates were lysed with 1 mL TRIzol (Life Technologies) and homogenized using pellet pestle (Kimble-Kontes, Vineland, NJ) followed by centrifugation. The aqueous phase was collected and total RNA was purified with RNeasy plus mini kit (Qiagen) according to the manufacturer’s protocol. RNA integrity was analyzed using RNA 6000 Pico Kit on Agilent Bioanalyzer.
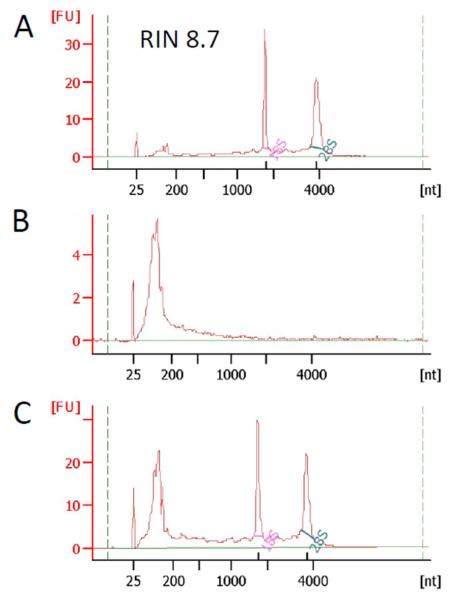
To overcome the effect of gel components on RNA stability, RNA exposure to the gel-containing TRIzol was limited by grinding the gel in liquid nitrogen followed by immediate lysis in TRIzol and column purification. As a result, RNA quality significantly improved. While total RNA purified from the gels with high concentration of encapsulated cells (4 million/mL) was of good quality (RIN > 7), RNAs purified from gels with low concentration cells (<0.5 million/m/L) showed significant degradation (Figure 2A,B). These results suggest that the large amounts of r- and tRNAs in the gel samples with a high concentration of encapsulated cells acted as competitive inhibitors for the gel’s RNA degradation activity. Therefore, the addition of heterologous tRNAs may protect the cellular RNA extracted from the cells encapsulated in the gel. As an external RNA, commercially available S. cerevisiae transfer RNA (Sigma-Aldrich, # R8508) was added. Short (70-90 nucleotide) tRNAs are mainly washed out during column purification steps (RNAs <200 nucleotides are excluded with Qiagen RNeasy kit [9] ) and the remaining amount can be eliminated during steps in the RNA-seq library constructions. Indeed, addition of S. cerevisiae tRNA drastically improved the quality of RNAs purified from cells encapsulated in PEG hydrogels (Fig 2C).
Figure 2. Addition of heterologous tRNA protects cellular RNA from degradation.
0.25 mL of PEG hydrogels with encapsulated cells (A – 2 millions/mL, B,C- 0.5 millions /mL), homogenized in liquid nitrogen and lyzed in TRIzol (A,B) or TRIzol with 5 mg/mL of E.coli tRNA ((Sigma, cat # R-8508). RNA integrity was analyzed using RNA 6000 Pico Kit on Agilent Bioanalyzer.
Based on our results, the following procedure was established for high quality RNA purification from cells encapsulated in PEG hydrogels: (i) Flash-freeze the crosslinked gels with encapsulated cells (prepared from 0.5 mL of gel solution) using liquid nitrogen; (ii) Grind the gel (using mortar and pestle ) in liquid nitrogen; (iii) Quickly transfer the ground gel to a Dounce Homogenizer by liquid nitrogen chilled spatula with 1 mL of TRIzol containing S. cerevisiae tRNA (5 μg/ml, Sigma, # R8508) and mix for 5 – 6 cycles; (iv) Incubate for 5 min at ambient conditions followed by centrifugation (10,000 g, 10 min at 2-4 oC); (v) and continue purification using the TRIzol / RNeasy hybrid protocol [6].
The procedure was utilized for purification of total RNAs from gels encapsulating MDA-MB-232, PC3 and DU145 cell lines. The purified RNAs were successfully used for the preparation of RNA-seq libraries. S. cerevisiae tRNAs are removed on multiple steps of library construction due to short size (less than 100 bp) and lack of polyadenylation. Additionally traces of yeast tRNA sequences are not aligned to the genome of mammalian cells, therefore they will not affect RNA-seq results. Our modification of the total RNA purification procedure from cells encapsulated in PEG hydrogels significantly improved gene expression profiling of tissue microenvironments modeled with PEG hydrogels.
Supplementary Material
ACNOWLEDGMENTS
This study was supported by NIGMS grant 1P20GM109091-01 and NIH grant P30 GM103336 (MS), NSF grant CBET1403545 and NIH grant AR063745 (EJ) and SC ASPIRE pilot award (HJ, EJ, MS)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Yang X, Sarvestani SK, Moeinzadeh S, He X, Jabbari E. Effect of CD44 binding peptide conjugated to an engineered inert matrix on maintenance of breast cancer stem cells and tumorsphere formation. PloS one. 2013;8:e59147. doi: 10.1371/journal.pone.0059147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yang X, Sarvestani SK, Moeinzadeh S, He X, Jabbari E. Three-dimensional-engineered matrix to study cancer stem cells and tumorsphere formation: effect of matrix modulus. Tissue engineering. Part A. 2013;19:669–684. doi: 10.1089/ten.tea.2012.0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mabilleau G, Stancu IC, Honore T, Legeay G, Cincu C, Basle MF, Chappard D. Effects of the length of crosslink chain on poly(2-hydroxyethyl methacrylate) (pHEMA) swelling and biomechanical properties. Journal of biomedical materials research. Part A. 2006;77:35–42. doi: 10.1002/jbm.a.30618. [DOI] [PubMed] [Google Scholar]
- [4].Kirchner B, Paul V, Riedmaier I, Pfaffl MW. mRNA and microRNA purity and integrity: the key to success in expression profiling. Methods in molecular biology (Clifton, N.J.) 2014;1160:43–53. doi: 10.1007/978-1-4939-0733-5_5. [DOI] [PubMed] [Google Scholar]
- [5].Chen EA, Souaiaia T, Herstein JS, Evgrafov OV, Spitsyna VN, Rebolini DF, Knowles JA. Effect of RNA integrity on uniquely mapped reads in RNA-Seq. BMC research notes. 2014;7:753. doi: 10.1186/1756-0500-7-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Garagorri N, Fermanian S, Thibault R, Ambrose WM, Schein OD, Chakravarti S, Elisseeff J. Keratocyte behavior in three-dimensional photopolymerizable poly(ethylene glycol) hydrogels. Acta biomaterialia. 2008;4:1139–1147. doi: 10.1016/j.actbio.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nature protocols. 2006;1:581–585. doi: 10.1038/nprot.2006.83. [DOI] [PubMed] [Google Scholar]
- [8].Browning MB, Cereceres SN, Luong PT, Cosgriff-Hernandez EM. Determination of the in vivo degradation mechanism of PEGDA hydrogels. Journal of biomedical materials research. Part A. 2014;102:4244–4251. doi: 10.1002/jbm.a.35096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].RNeasy® Plus Mini Handbook Qiagen. 2010.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.