Abstract
Systemic lupus erythematosus is a multi-system disease characterized by wide-spread DNA methylation changes. To identify epigenetic susceptibility loci for lupus nephritis, genome-wide DNA methylation changes in naïve CD4+ T cells were compared between two sets of lupus patients with and without a history of renal involvement. A total of 56 lupus patients (28 with renal involvement and 28 without renal involvement), and 56 age-, sex-, and ethnicity-matched healthy controls were included in our study. We identified 191 CG sites and 121 genes that were only differentially methylated in lupus patients with but not without a history of renal involvement. The tyrosine kinase gene TNK2 involved in cell trafficking and tissue invasion, and the phosphatase gene DUSP5 which dephosphorylates and inhibits the ERK signaling pathway, were among the most hypomethylated. Independent of disease activity, renal involvement is characterized by more robust demethylation in interferon regulated genes differentially methylated in both sets of lupus patients with and without renal involvement (fold change 1.4, P = 0.0014). The type-I interferon master regulator gene IRF7 is only hypomethylated in lupus patients with renal involvement. IRF-7 is an upstream transcription factor that regulates several loci demethylated only with renal involvement such as CD80, HERC5, IFI44, IRF7, ISG15, ISG20, ITGAX, and PARP12 (P = 1.78 × 10−6). Among the CG sites only hypomethylated with renal involvement, CG10152449 in CHST12 has a sensitivity of 85.7% and a specificity of 64.3% for stratifying lupus patients for a history of renal involvement (P = 0.0029). Our data identified novel epigenetic susceptibility loci that are differentially methylated with renal involvement in lupus. These loci will help better understand lupus nephritis, and provide a proof of principle for the potential applicability of specific methylation changes as predictors for specific organ involvement in lupus.
Keywords: Lupus, Nephritis, Epigenetics, Methylation, Biomarker, EWAS
1. Introduction
Systemic lupus erythematosus is a chronic relapsing autoimmune disease characterized by the production of autoantibodies to nuclear antigens. The disease course is influenced by ~55 confirmed genetic predisposing loci and a number of environmental triggers [1,2]. DNA methylation changes, particularly in T cells, also play an important role in the pathogenesis of the disease [3]. Indeed, recent genome-wide DNA methylation studies have identified a number of differentially methylated loci in lupus, and characterized several differentially methylated genes and pathways that provided novel insight into disease pathogenesis [4–6]. DNA methylation studies in lupus T cells also suggest that dynamic DNA methylation changes can provide potential novel biomarkers to monitor disease activity [4]. We have previously provided evidence that interferon-regulated genes are hypomethylated in naïve CD4+ T cells in lupus patients, and this hypomethylation precedes active transcription of interferon-regulated genes [5]. These data suggested that interferon-regulated genes are epigenetically poised for transcription, therefore providing a mechanism to explain type-1 interferon hyper-responsiveness in lupus [5].
Renal involvement is estimated to occur in up to 60% of lupus patients, and its incidence and severity varies between patients. Lupus nephritis also tends to be more common and more severe in African-American compared to European-American lupus patients, and patients of African-American, Asian, or Hispanic ethnicity are more likely to progress to end-stage kidney disease from lupus nephritis [7]. Lupus nephritis is a major cause of morbidity and mortality in lupus patients, but diagnosis and monitoring remains challenging. Further, despite aggressive immunosuppressive therapy that is often complicated by serious adverse events, the incidence of end-stage kidney disease from lupus nephritis has not declined [7,8]. This emphasizes the need for a better understanding of the pathogenesis of lupus nephritis and the development of novel biomarkers to predict and monitor renal involvement in lupus patients.
In this study we performed a genome-wide DNA methylation study in naïve CD4+ T cells from lupus patients with and without renal involvement. We compared the methylation patterns in each disease subset with normal healthy age, sex, and ethnicity matched controls. We focused on naïve CD4+ T cells to be able to capture epigenetic susceptibility loci for renal involvement in lupus that pre-date T cell activation and differentiation. This approach avoids identifying epigenetic changes that might result from T cell activation and differentiation in lupus patients, and identifies epigenetic changes that more likely predispose to rather than result from the inflammatory environment in lupus. Our data identify a unique DNA methylation pattern that distinguishes lupus patients with a history of renal involvement from patients with no renal involvement. We also suggest an interferon methylation index (IMI) and report that lupus patients with renal involvement can be characterized by significantly lower DNA methylation levels across interferon-regulated genes. We then identified a specific CG site with reasonable sensitivity and specificity for renal involvement in lupus patients.
2. Materials and methods
2.1. Lupus patients and controls
We studied 56 female lupus patients and 56 normal healthy controls matched for age, sex, and ethnicity. All lupus patients included in this study fulfill the American College of Rheumatology (ACR) classification criteria for SLE, and were recruited from the University of Michigan rheumatology clinics, Oklahoma Medical Research Foundation, and Henry Ford Health System rheumatology clinics. All study participants signed an informed consent approved by the institutional review boards of our institutions. Our lupus cohort included 28 female lupus patients who met the ACR criterion for renal involvement as defined by the ACR criteria for classification of systemic lupus erythematous (persistent proteinuria > 0.5 g per day or > than 3 + if quantitation not performed, or the presence of cellular casts including red cell, hemoglobin, granular, tubular, or mixed casts) [9,10], and 28 female lupus patients who had no evidence for renal involvement. Both renal and non-renal lupus patients groups included 17 European-American and 11 African-American lupus patients. Healthy female controls were matched for age (+/− 5 years) and ethnicity for each patient (Table 1). Basic demographic information, SLEDAI scores, and SLEDAI criteria present at the time of blood draw and enrollment in our study for each patient are presented in Supplementary Table 1.
Table 1.
Basic demographic information for the patients and healthy controls included in this study.
| Renal | Non-renal | P Value | ||
|---|---|---|---|---|
| Patients | Number | 28 | 28 | 1 |
| Age (mean ± SD) | 39.82 ± 10.71 | 41.07 ± 14.70 | 0.72 | |
| Sex (% female) | 100% | 100% | 1 | |
| Ethnicity (EA, AA) | 17,11 | 17,11 | 1 | |
| SLEDAI (mean ± SD) | 3.36 ± 3.80 | 3.21 ± 3.18 | 0.88 | |
| Controls | Number | 28 | 28 | 1 |
| Age (mean ± SD) | 40.07 ± 10.92 | 41.07 ± 13.73 | 0.76 | |
| Sex (% female) | 100% | 100% | 1 | |
| Ethnicity (EA, AA) | 17,11 | 17,11 | 1 |
EA, European-American; AA, African-American, SLEDAI, Systemic Lupus Erythematosus Disease Activity Index.
2.2. Naïve CD4+ T cell isolation and DNA extraction
Fresh peripheral blood samples (80 ml) were collected from each participant. Density gradient centrifugation (Ficoll) was used to isolate peripheral blood mononuclear cells (PBMCs). Naïve CD4+ T cells were isolated using indirect labeling and magnetic bead separation with the Naïve CD4+ T Cell Isolation kit II (Miltenyi Biotec, Cambridge, MA) as previously described [5]. Naïve CD4+ T cell purity was confirmed by flow cytometry using fluorochrome-conjugated antibodies against CD3, CD4, CD45RA, and CD28 and was over 95%. DNA was extracted from each sample using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA), followed by sodium bisulfite treatment using the EZ DNA Methylation kit (Zymo Research, Irvine, CA) for DNA methylation studies.
2.3. DNA methylation profiling
Analysis of genome-wide DNA methylation in naïve CD4+ T cells isolated from lupus patients with and without renal involvement, and healthy matched controls was performed using the Infinium HumanMethylation450 BeadChip Kit (Illumina). This platform allowed for simultaneous assessment of DNA methylation levels in over 485,000 DNA methylation sites in over 99% of RefSeq genes and 96% of CpG islands. An average of 17 CG sites per gene is included on the array to cover the promoter, 5′UTR, first exon, 3′UTR, and CG sites within the gene body. Other regions covered include miRNA promoter regions and ~3000 non-CG methylation sites.
2.4. Statistical and bioinformatics analysis
Data processing and data analysis were performed as previously described in multiple studies by our group. Briefly, following quality control assessment measures, probe intensity values were normalized and used to determine the average methylation level on each DNA methylation site in each sample as represented by beta values (β). We excluded from the analysis any probe with a known genetic variant within 10bp of its 3′ end. Probes that had a detection P value (detection above array background) of ≥0.05 were also excluded. Case-control analysis was performed to determine differentially methylated sites between lupus patients with and without renal involvement and their matched healthy controls. Differentially methylated sites in this study were defined as methylation sites with an absolute difference in beta value (delta beta) of at least 0.1 and a P value of <0.01 after correction for multiple testing using a Benjamini and Hochberg false discovery rate of 5%.
The variation in the estimate of β is a function of β, and was estimated by Illumina for all values of β by repeatedly measuring loci with known methylation fractions ranging from 0 to 1, and then fitting a parabola to the standard deviation as a function of β (GenomeStudio Methylation Module User Guide, Illumina, USA). The standard deviation estimate is then calculated by
P values for differential DNA methylation were calculated using the formula:
where z is the two-sided tail probability of the standard normal distribution.
Differentially methylated sites identified were then mapped within the genome to determine differentially methylated genes in both lupus subsets, and further investigated by bioinformatic analysis using Ingenuity Pathway Analysis (IPA) and the Database for Annotation, Visualization, and Integrated Discovery (DAVID) [11].
3. Results
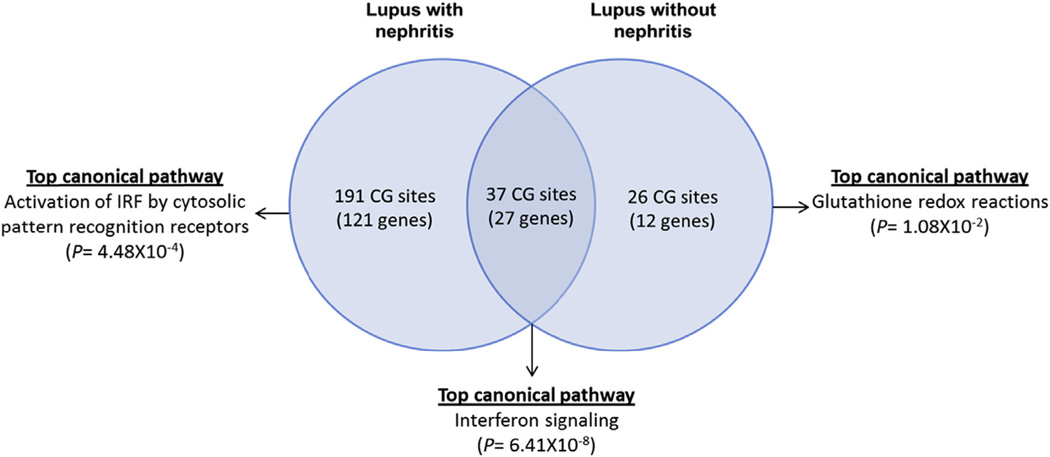
Genome-wide DNA methylation changes were compared between two sets of lupus patients with and without renal involvement and healthy age, sex, and ethnicity matched controls. We identified 228 differentially methylated CG sites in patients with renal involvement compared to only 63 differentially methylated CG sites in lupus patients with no renal involvement. There were 37 shared differentially methylated CG sites in patients with and without renal involvement. Lupus patients with renal involvement showed differential DNA methylation levels in 191 unique CG sites that are not differentially methylated in the same number of lupus patients of similar ethnicity but without renal involvement. These sites represent 121 differentially methylated genes in lupus patients with but not without renal involvement (Fig. 1), (Supplementary Tables 2 and 3, and 4).
Fig. 1.
A Venn diagram depicting shared and unique differentially methylated CG sites and genetic loci in lupus patients with and without renal involvement compared to matched healthy controls, as well as the top canonical pathways representing these groups of genes.
Of the 37 CG sites (27 genes) that are differentially methylated in both groups of lupus patients with and without renal involvement, 20 CG sites are located in 15 interferon-regulated gene loci. The majority of these CG sites (19 out of 20) are significantly hypomethylated in both patients groups with and without renal involvement compared to matched healthy controls. Bioinformatic analysis of the 27 shared differentially methylated genes highlights interferon signaling as the most significant unifying canonical pathway (P = 6.41 × 10−8) and interferon alpha 2 as the most significant upstream regulator in these genes (P = 6.77 × 10−23) (Table 2). Shared hypermethylated genes between lupus patients with and without renal involvement include the SMAD family member signaling transducer gene SMAD3, and BAG3 which plays a distinct role in non-canonical autophagy.
Table 2.
Upstream regulator analysis in differentially methylated genes shared in lupus patients with and without renal involvement, differentially methylated genes only in patients with renal involvement, and differentially methylated genes only in patients without renal involvement.
| Upstream regulator | P Value | Target differentially methylated genes | |
|---|---|---|---|
| Shared | IFNA2 | 6.77E-23 | BAG3,CMPK2,DDX58,EIF2AK2,HERC6,IFI44L,IFIT1,IFIT3,LY6E,MX1,PARP9,PLSCR1,STAT1,USP18 |
| IFNL1 | 1.31E-19 | DDX58,EIF2AK2,HERC6,IFI44L,IFIT1,IFIT3,MX1,PLSCR1,STAT1,USP18 | |
| STAT1 | 7.22E-16 | CMPK2,EIF2AK2,HERC6,HLA-DRB5,IFIT1,IFIT3,LY6E,MX1,SMAD3,STAT1,USP18 | |
| IFNG | 2.54E-15 | CMPK2,DDX58,DTX3L,EIF2AK2,HERC6,HLA-DRB1,HLA-DRB5,IFI44L,IFIT1,IFIT3,LY6E,MX1,PARP9,SMAD3,STAT1,USP18 | |
| IRF7 | 4.67E-14 | CMPK2,DDX58,IFI44L,IFIT1,IFIT3,MX1,PLSCR1,STAT1,USP18 | |
| Renal involvement only | MAPK1 | 9.58E-08 | ARSB,BST2,CCDC82,EHD1,HERC5,IFI44,IRF7,ISG15,ISG20,PARP12,PPT2,RCAN3 |
| IRF7 | 1.78E-06 | CD80,HERC5,IFI44,IRF7,ISG15,ISG20,ITGAX,PARP12 | |
| IFNL1 | 2.03E-06 | BST2,CD80,HERC5,IFI44,ISG15,ISG20 | |
| TLR3 | 3.53E-06 | CD80,HERC5,IFI44,IRF7,ISG15,ISG20,NFE2L3,NFKB1,ZNF644 | |
| PAF1 | 3.88E-06 | HERC5,IFI44,ISG15,ISG20,LRRN3 | |
| No renal involvement only | KLF8 | 1.58E-03 | USP44 |
| EIF3A | 3.68E-03 | RPA2 | |
| ANGPT2 | 4.27E-03 | GSTT1,RPA2 | |
| miR-218 | 5.78E-03 | MDGA1 | |
| TSPYL5 | 1.20E-02 | GABRP |
Bolded and unbolded genes are hypermethylated and hypomethylated, respectively, in lupus patients compared to healthy controls.
Next, we asked if the degree of DNA demethylation in interferon-regulated genes is different in lupus patients with and without renal involvement. We measured demethylation in interferon-regulated genes by calculating the average level of DNA methylation (beta) across all 19 demethylated CG sites located in interferon-regulated gene loci shared between patients with and without renal involvement. This interferon methylation index (IMI) represents a global assessment for the degree of DNA demethylation in interferon-regulated genes in lupus patients, and was significantly lower in lupus patients with renal disease compared to lupus patients without renal involvement (IMI renal = −0.22, IMI non-renal = −0.16, P = 0.0014). To determine if specific CG sites among these 19 sites used to calculate IMI are more predictive for renal involvement in lupus patients, we calculated the difference in Δβ between lupus patients with and without renal involvement at each of these CG sites individually (Table 3). The methylation site CG21549285 in MX1 demonstrates the largest divergence between patients with and without renal involvement among all 19 demethylated CG sites located in interferon-regulated genes, with a Δ(Δβ) = −0.13, representing a 1.6 fold difference (Δβ renal = −0.36, Δβ non-renal = −0.23, P = 0.011). The methylation fraction (β) in this CG site in lupus patients with renal involvement was 0.29 ± 0.15 and in patients without renal involvement 0.42 ± 0.19 (P = 0.0061).
Table 3.
CG sites hypomethylated in patients with and without renal involvement and that are located in interferon-regulated genes.
| Gene | CG site | Renal involvement | No renal involvement | P Value | ||||
|---|---|---|---|---|---|---|---|---|
| Methylation fraction (beta) | Methylation fraction (beta) | |||||||
| Patients | Controls | Delta beta | Patients | Controls | Delta beta | |||
| CMPK2 | cg01028142 | 0.59 | 0.83 | −0.25 | 0.71 | 0.86 | −0.15 | 0.03 |
| DDX58 | cg14286514 | 0.44 | 0.56 | −0.12 | 0.44 | 0.56 | −0.12 | 0.97 |
| EIF2AK2 | cg14126601 | 0.41 | 0.54 | −0.13 | 0.48 | 0.60 | −0.12 | 0.79 |
| HERC6 | cg14212360 | 0.57 | 0.76 | −0.19 | 0.63 | 0.75 | −0.12 | 0.04 |
| IFI44L | cg00855901 | 0.24 | 0.41 | −0.17 | 0.30 | 0.42 | −0.13 | 0.13 |
| IFI44L | cg03607951 | 0.42 | 0.67 | −0.26 | 0.53 | 0.70 | −0.17 | 0.04 |
| IFI44L | cg05696877 | 0.28 | 0.50 | −0.22 | 0.33 | 0.53 | −0.20 | 0.58 |
| IFI44L | cg06872964 | 0.30 | 0.50 | −0.20 | 0.38 | 0.52 | −0.14 | 0.10 |
| IFI44L | cg17980508 | 0.41 | 0.60 | −0.19 | 0.49 | 0.61 | −0.12 | 0.04 |
| IFIT1 | cg05552874 | 0.51 | 0.76 | −0.25 | 0.62 | 0.78 | −0.16 | 0.05 |
| IFIT3 | cg06188083 | 0.21 | 0.37 | −0.16 | 0.26 | 0.39 | −0.13 | 0.23 |
| LY6E | cg14392283 | 0.54 | 0.88 | −0.33 | 0.69 | 0.91 | −0.22 | 0.10 |
| MX1 | cg21549285 | 0.29 | 0.65 | −0.36 | 0.42 | 0.64 | −0.23 | 0.01 |
| MX1 | cg22862003 | 0.47 | 0.69 | −0.22 | 0.56 | 0.70 | −0.14 | 0.02 |
| PARP9;DTX3L | cg08122652 | 0.49 | 0.73 | −0.25 | 0.57 | 0.75 | −0.18 | 0.99 |
| PARP9;DTX3L | cg22930808 | 0.50 | 0.77 | −0.27 | 0.61 | 0.80 | −0.19 | 0.10 |
| PLSCR1 | cg06981309 | 0.37 | 0.61 | −0.24 | 0.48 | 0.64 | −0.16 | 0.05 |
| STAT1 | cg14951497 | 0.29 | 0.40 | −0.11 | 0.29 | 0.42 | −0.13 | 0.57 |
| USP18 | cg14293575 | 0.26 | 0.44 | −0.18 | 0.29 | 0.47 | −0.19 | 0.91 |
P values represent the difference between delta beta in lupus patients with renal involvement compared to delta beta in patients without renal involvement.
Out of the 191 differentially methylated CG sites unique to lupus patients with renal involvement, 64 were hypomethylated and 127 were hypermethylated compared to healthy matched controls (Supplementary Table 2). There were 121 genes differentially methylated only in lupus patients with renal involvement. The two most hypomethylated genes are TNK2 and DUSP5, and the two most hypermethylated genes are MAN1C1 and PLEKHA1. Other genes hypermethylated only in lupus patients with renal involvement include CD47 and CD247.
Lupus patients with renal involvement demonstrate robust DNA demethylation in other interferon-regulated genes not demethylated in patients without renal disease. These include ISG15, ISG20, IFI44, PARP12, and BST2 among others (Table 4). In addition, the interferon regulatory factor family member gene IRF7 was demethylated only in lupus patients with renal involvement. Regulation by IRF-7 is shared between genes differentially methylated only in lupus patients with renal involvement (P = 1.78 × 10−6). Therefore, these findings emphasize a more robust evidence for DNA demethylation in interferon-regulated genes in lupus patients with renal disease and suggest an important role for type-I interferon signature in lupus nephritis.
Table 4.
Hypomethylated CG sites in genes differentially methylated in lupus patients with renal involvement but not lupus patients without renal involvement.
| CG site | Methylation fraction (beta) | Delta beta |
P Values | Gene | |
|---|---|---|---|---|---|
| Patients | Controls | ||||
| cg15065340 | 0.43 | 0.67 | −0.24 | 8.23E-35 | TNK2 |
| cg03290131 | 0.39 | 0.57 | −0.18 | 8.23E-35 | DUSP5 |
| cg01079652 | 0.67 | 0.82 | −0.15 | 8.23E-35 | IFI44 |
| cg17052170 | 0.60 | 0.74 | −0.15 | 6.41E-32 | LOC100133669 |
| cg05994974 | 0.46 | 0.61 | −0.15 | 1.43E-25 | PARP12 |
| cg12110437 | 0.20 | 0.34 | −0.14 | 8.09E-29 | LOC100133669 |
| cg23264429 | 0.49 | 0.63 | −0.14 | 4.44E-22 | STAMBPL1 |
| cg10819238 | 0.74 | 0.88 | −0.13 | 8.23E-35 | SBNO2 |
| cg16526047 | 0.47 | 0.60 | −0.13 | 3.27E-20 | ISG15 |
| cg13909895 | 0.32 | 0.45 | −0.13 | 1.12E-20 | ARSA |
| cg20062691 | 0.56 | 0.69 | −0.12 | 1.16E-19 | ISG15 |
| cg04788999 | 0.56 | 0.68 | −0.12 | 3.12E-19 | ISG15 |
| cg08926253 | 0.61 | 0.74 | −0.12 | 1.96E-21 | IRF7 |
| cg13458803 | 0.45 | 0.57 | −0.12 | 4.75E-16 | CD80 |
| cg21878650 | 0.31 | 0.43 | −0.12 | 6.62E-17 | ADAMTS6 |
| cg23084016 | 0.37 | 0.48 | −0.12 | 1.05E-15 | ALOX5 |
| cg04742550 | 0.46 | 0.58 | −0.12 | 2.25E-15 | ITGAX |
| cg20167074 | 0.59 | 0.71 | −0.12 | 6.45E-18 | S100A10 |
| cg24506221 | 0.15 | 0.27 | −0.11 | 5.62E-24 | GSTM1 |
| cg13801402 | 0.54 | 0.65 | −0.11 | 8.05E-16 | BCL2L15 |
| cg14181576 | 0.75 | 0.86 | −0.11 | 7.01E-31 | FGR |
| cg13092901 | 0.29 | 0.41 | −0.11 | 1.07E-15 | TYMP;SCO2 |
| cg22544881 | 0.59 | 0.70 | −0.11 | 2.56E-16 | FLJ43663 |
| cg03763873 | 0.10 | 0.21 | −0.11 | 4.09E-28 | EPSTI1 |
| cg13130398 | 0.69 | 0.80 | −0.11 | 2.53E-21 | RABGAP1L |
| cg01329005 | 0.17 | 0.28 | −0.11 | 1.83E-20 | BST2 |
| cg18020065 | 0.41 | 0.52 | −0.11 | 3.70E-13 | RASA3 |
| cg19649900 | 0.82 | 0.92 | −0.11 | 8.23E-35 | SBNO2 |
| cg12510708 | 0.66 | 0.77 | −0.11 | 8.24E-19 | NFE2L3 |
| cg15085899 | 0.74 | 0.85 | −0.11 | 6.69E-27 | NCOR2 |
| cg03906115 | 0.49 | 0.60 | −0.11 | 3.58E-13 | LTBP1 |
| cg25969878 | 0.40 | 0.51 | −0.11 | 1.47E-12 | STK32C |
| cg02334775 | 0.50 | 0.61 | −0.11 | 7.26E-13 | ISG20 |
| cg19754622 | 0.75 | 0.86 | −0.11 | 1.71E-27 | STK32C |
| cg01613294 | 0.55 | 0.65 | −0.11 | 1.48E-13 | APOL3 |
| cg06679494 | 0.27 | 0.38 | −0.11 | 3.84E-14 | MIR497;MIR195 |
| cg04936619 | 0.24 | 0.34 | −0.10 | 3.14E-15 | C17orf75 |
| cg18049167 | 0.61 | 0.71 | −0.10 | 1.61E-14 | PPT2 |
| cg19955928 | 0.61 | 0.71 | −0.10 | 2.28E-14 | HDAC4 |
| cg21188533 | 0.61 | 0.72 | −0.10 | 3.17E-14 | CACNA1D |
| cg02160608 | 0.68 | 0.79 | −0.10 | 1.06E-17 | PSD4;LOC440839 |
| cg02215171 | 0.46 | 0.56 | −0.10 | 3.40E-11 | HERC5 |
| cg11103390 | 0.47 | 0.57 | −0.10 | 3.19E-11 | ECHDC3 |
| cg26802063 | 0.46 | 0.56 | −0.10 | 4.61E-11 | ARSB |
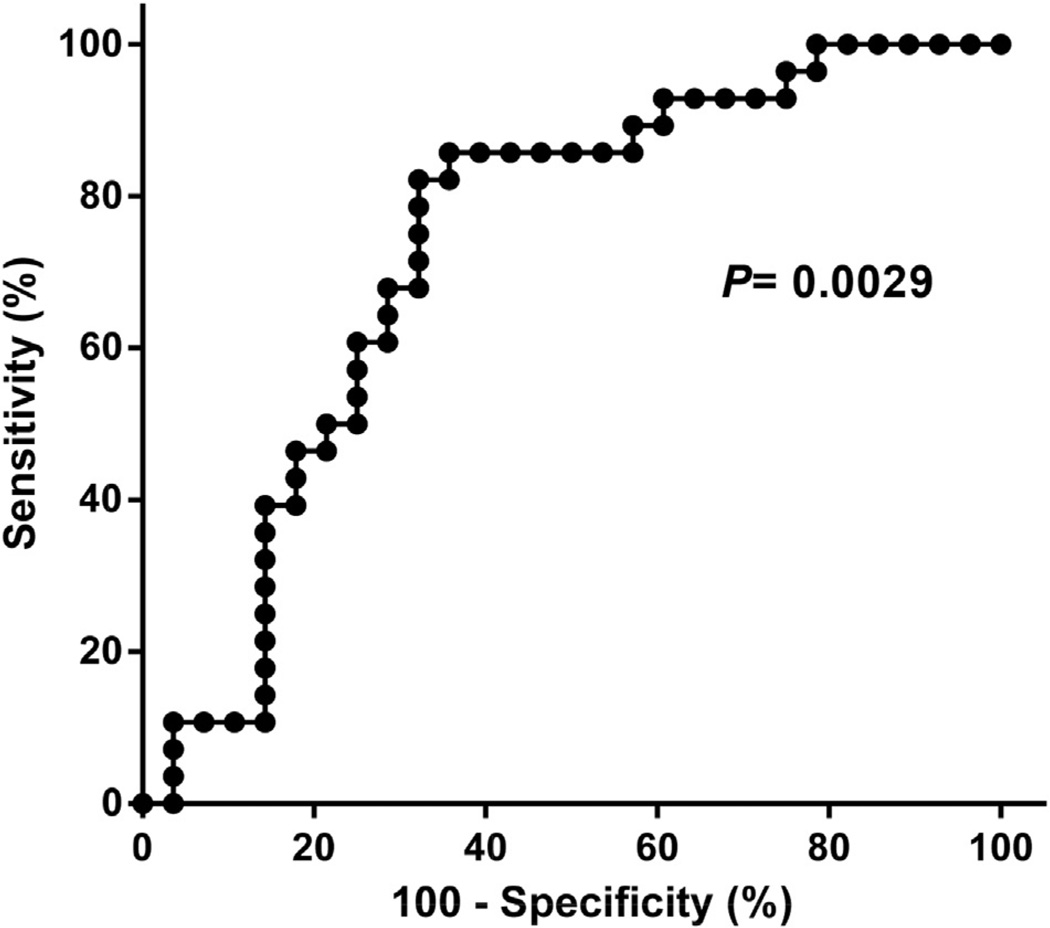
using receiver operator characteristic (ROC) curve analysis (P for area under the ROC curve = 0.0029) (Fig. 2).
Next, we determined if any of the CG sites that are differentially methylated in lupus patients with renal involvement compared to controls (but not differentially methylated in patients without renal involvement) can accurately discriminate lupus patients with renal involvement from patients with no evidence of renal involvement. We divided our cohort of 56 lupus patients into “training” and “testing” sets. Each set included 28 lupus patients, half with lupus renal involvement. We determined the sensitivity and specificity for detecting renal involvement using a test cutoff value equal to the mean + SD in 56 healthy controls. Among CG sites differentially methylated only in lupus patients with renal involvement, the training set identified 4 CG sites with a sensitivity of >85% and a specificity of >55% for renal involvement in lupus patients. We then tested the sensitivity and specificity of these 4 CG sites in our “testing” set of patients with and without renal involvement. The CG site CG10152449 in the gene CHST12 had the best sensitivity and specificity in our testing set with a sensitivity of 85.7% and a specificity of 71.4%. Combining both training and testing sets we obtain a sensitivity of 85.7% and a specificity of 64.3% for this CG site to detect renal involvement in lupus patients. This was confirmed using receiver operator characteristic (ROC) curve analysis (P for area under the ROC curve = 0.0029) (Fig. 2).
Fig. 2.
Receiver operator characteristic (ROC) curve showing the sensitivity and specificity of the methylation level (beta) on CG10152449 for lupus renal involvement.
4. Discussion
DNA methylation is generally a repressive epigenetic mark that alters chromatin accessibility and silences gene expression. Mapping DNA methylation changes in health and disease can inform about disease-associated chromatin structure, and identify epigenetically modified genetic targets that can help to better understand disease pathogenesis and potentially identify novel targets for therapy [5,12–15]. Further, the dynamic nature of DNA methylation changes makes them appealing targets to explore as disease biomarkers [13,16].
In this study we focused on exploring genome-wide DNA methylation changes in a group of lupus patients with and without a history of renal involvement as defined by the ACR criteria for classification of systemic lupus erythematous [9,10]. The goal of the study was to identify and characterize unique and shared DNA methylation changes in naïve CD4+ T cells in the two groups of lupus patients compared to matched healthy controls. We studied naïve CD4+ T cells to identify epigenetic changes in T cells that are more likely inherent to the pathogenesis of lupus, and minimize the effect of the disease environment and chronic T cell stimulation and anergy, which could alter the epigenome in antigen-experienced T cell pools.
We revealed a number of epigenetic changes that are unique to lupus patients with renal involvement. In total, 191 CG sites and 121 genes were differentially methylated in lupus patients with but not without renal involvement. Consistent with our previous data, lupus patients with or without renal involvement demonstrate a robust demethylation in interferon-regulated genes [5]. Importantly, however, we demonstrate that this demethylation is significantly more robust in lupus patients with renal involvement (fold change 1.4, P = 0.0014). There was no difference in disease activity as measured by SLEDAI scores between patients with and without renal involvement at the time of enrolment in our study (P = 0.88), suggesting that increased demethylation of interferon-regulated genes in lupus patients with renal involvement is not due to differences in disease activity. Further, we have previously demonstrated that demethylation of interferon-regulated genes in naïve CD4+ T cells in lupus patients does not correlate with disease activity [5].
Among the most hypomethylated genes in lupus patients with but not without renal involvement are TNK2 (tyrosine kinase, non-receptor, 2) and DUSP5 (dual specificity phosphatase 5). TNK2 encodes a non-receptor tyrosine kinase that modulates a number of downstream effector molecules, and is involved in cell trafficking, endocytosis, cell migration, and tissue invasion [17]. Whether demethylation of TNK2 plays a role in T cell migration and invasion of kidney tissues in lupus nephritis remains to be determined. DUSP5 encodes a phosphatase that is capable of dephosphorylating phosphoserine/threonine and phosphotyrosine residues, with the highest relative activity reported for dephosphorylating mitogen-activated protein kinase/extracellular signal-regulated kinase molecules (MAPK/ERK) [17]. DUSP5 dephosphorylates and inhibits the ERK signaling pathway in T cells [18]. Indeed, ERK signaling is defective in T cells from lupus patients, and inducing an ERK signaling defect in T cells using a transgenic murine approach resulted in reduced DNA methyltransferase 1 (DNMT1) expression, T cell DNA demethylation, and autoimmunity in vivo [19,20]. Taken together, DUSP5 demethylation might contribute to a defective ERK signaling pathway in lupus T cells, thereby inducing T cell autoreactivity.
A CG site located in the loci encoding microRNAs miR-497 and miR-195 was hypomethylated in lupus patients with renal involvement. Both microRNAs target the MAPK/ERK signaling gene MAPK3 (also known as ERK1). Several other MAPK signaling-regulated genes were also differentially methylated only in lupus patients with renal involvement but not lupus patients with no evidence of renal disease. Indeed, MAPK1 is the most significantly identified upstream regulator in unique genes with altered DNA methylation in lupus patients with renal involvement (P = 9.58 × 10−8) (Table 2). Genes hypermethylated only in lupus patients with renal involvement include CD47 which has been recently shown to regulate the immunosuppressive function of VEGF in T cells [21], and CD247 which encodes the T cell receptor zeta chain and is known to be down-regulated in lupus T cells [22].
A number of interferon-regulated genes are demethylated only in lupus patients with renal involvement. The gene encoding interferon-regulatory factor 7 (IRF7), which has been previously identified as a lupus susceptibility gene [23], is demethylated only with renal involvement in lupus patients in naïve CD4+ T cells. At the transcriptional level and in total CD4+ T cells, IRF-7 expression levels are increased in lupus patients with or without renal involvement [6]. The lupus risk allele in IRF7 is associated with increased activity of IRF-7 as reflected by a 2-fold increase in interferon-regulated gene expression [23]. The demethylation we identified in this locus is also consistent with increased transcriptional accessibility. Subsequent studies have suggested the association between IRF7 lupus risk allele and renal disease in lupus patients [24]. IRF-7 is a critical regulator that orchestrates the entire type-I interferon response [25], and our data indicate that IRF-7 is an upstream regulator of multiple genes differentially methylated only in lupus patients with renal involvement (P = 1.78 × 10−6). Taken together, these data suggest a potential for genetic and epigenetic alterations in the same genetic locus to predispose to a specific disease manifestation, in this case lupus nephritis. Indeed, these data support a role for IRF-7 in lupus nephritis, and pose the hypothesis that inhibiting IRF-7 might provide a potential therapeutic benefit for renal involvement in lupus.
We provided evidence to suggest that specific DNA methylation changes might have the potential to be developed into disease biomarkers. In a heterogeneous disease like lupus, there is a dire need for the identification of novel biomarkers that can aid in predicting disease manifestations, disease activity, and response to therapy. We demonstrated that a CG site located in CHST12 was hypomethylated in lupus patients with renal involvement and showed a relatively high sensitivity and a reasonable specificity for identifying lupus patients with a history of renal disease. These data provide a proof of principle only, and certainly require independent validation in a large cohort. CHST12 encodes carbohydrate (chondroitin 4) sulfotransferase 12, and is localized to the golgi membrane. It belongs to the sulfotransferase 2 family and modifies N-acetylgalactosamine residues that are constituents of chondroitin. Chondroitin sulfate is a proteoglycan present in cartilage, extracellular matrices, and on the cell surface. Expression profiles in normal human tissues suggest that the highest levels of CHST12 mRNA expression occur in immune cells, including T cells [17], though the exact function of this gene in immune responses remains unknown.
5. Conclusion
We performed the most extensive DNA methylation study in naïve CD4+ T cells in lupus and characterized DNA methylation changes in lupus patients with renal involvement. Our data identified unique loci that are epigenetically altered in the presence of renal involvement in lupus; some can be potentially useful as novel therapeutic targets. Importantly, our study provides a proof of principle for exploring specific DNA methylation changes as biomarkers for specific disease manifestations in lupus. Follow up studies in larger cohorts, other cell types, and using a longitudinal approach are warranted.
Supplementary Material
Acknowledgment
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01AI097134.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jaut.2015.05.003.
Footnotes
Conflict of interest
None of the authors has any financial conflict of interest to report.
References
- 1.Deng Y, Tsao BP. Advances in lupus genetics and epigenetics. Curr. Opin. Rheumatol. 2014;26:482–492. doi: 10.1097/BOR.0000000000000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mak A, Tay SH. Environmental factors, toxicants and systemic lupus erythematosus. Int. J. Mol. Sci. 2014;15:16043–16056. doi: 10.3390/ijms150916043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altorok N, Sawalha AH. Epigenetics in the pathogenesis of systemic lupus erythematosus. Curr. Opin. Rheumatol. 2013;25:569–576. doi: 10.1097/BOR.0b013e328364206f. [DOI] [PubMed] [Google Scholar]
- 4.Jeffries MA, Dozmorov M, Tang Y, Merrill JT, Wren JD, Sawalha AH. Genome-wide DNA methylation patterns in CD4+ T cells from patients with systemic lupus erythematosus. Epigenetics Off. J. DNA Methylation Soc. 2011;6:593–601. doi: 10.4161/epi.6.5.15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coit P, Jeffries M, Altorok N, Dozmorov MG, Koelsch KA, Wren JD, et al. Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naive CD4+ T cells from lupus patients. J. Autoimmun. 2013;43:78–84. doi: 10.1016/j.jaut.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao M, Liu S, Luo S, Wu H, Tang M, Cheng W, et al. DNA methylation and mRNA and microRNA expression of SLE CD4+ T cells correlate with disease phenotype. J. Autoimmun. 2014;54:127–136. doi: 10.1016/j.jaut.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Maroz N, Segal MS. Lupus nephritis and end-stage kidney disease. Am. J. Med. Sci. 2013;346:319–323. doi: 10.1097/MAJ.0b013e31827f4ee3. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz N, Goilav B, Putterman C. The pathogenesis, diagnosis and treatment of lupus nephritis. Curr. Opin. Rheumatol. 2014;26:502–509. doi: 10.1097/BOR.0000000000000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheumatism. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 10.Hochberg MC. Updating the American college of rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheumatism. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 11.Da Wei Huang BTS, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 12.Altorok N, Coit P, Hughes T, Koelsch KA, Stone DU, Rasmussen A, et al. Genome-wide DNA methylation patterns in naive CD4+ T cells from patients with primary Sjogren's syndrome. Arthritis & Rheumatol. 2014;66:731–739. doi: 10.1002/art.38264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes T, Ture-Ozdemir F, Alibaz-Oner F, Coit P, Direskeneli H, Sawalha AH. Epigenome-wide scan identifies a treatment-responsive pattern of altered DNA methylation among cytoskeletal remodeling genes in monocytes and CD4+ T cells from patients with Behcet's disease. Arthritis & Rheumatol. 2014;66:1648–1658. doi: 10.1002/art.38409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altorok N, Tsou PS, Coit P, Khanna D, Sawalha AH. Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann. Rheumatic Dis. 2014 May 8; doi: 10.1136/annrheumdis-2014-205303. http://dx.doi.org/10.1136/annrheumdis-2014-205303. pii: annrheumdis-2014-205303 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeffries MA, Donica M, Baker LW, Stevenson ME, Annan AC, Humphrey MB, et al. Genome-wide DNA methylation study identifies significant epigenomic changes in osteoarthritic cartilage. Arthritis & Rheumatol. 2014;66:2804–2815. doi: 10.1002/art.38762. [DOI] [PubMed] [Google Scholar]
- 16.Guo Y, Sawalha AH, Lu Q. Epigenetics in the treatment of systemic lupus erythematosus: potential clinical application. Clin. Immunol. 2014;155:79–90. doi: 10.1016/j.clim.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Stelzer G, Dalah I, Stein TI, Satanower Y, Rosen N, Nativ N, et al. In-silico human genomics with genecards. Hum. Genomics. 2011;5:709–717. doi: 10.1186/1479-7364-5-6-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovanen PE, Rosenwald A, Fu J, Hurt EM, Lam LT, Giltnane JM, et al. Analysis of gamma c-family cytokine target genes. Identification of dualspecificity phosphatase 5 (DUSP5) as a regulator of mitogen-activated protein kinase activity in interleukin-2 signaling. J. Biological Chem. 2003;278:5205–5213. doi: 10.1074/jbc.M209015200. [DOI] [PubMed] [Google Scholar]
- 19.Sawalha AH, Jeffries M, Webb R, Lu Q, Gorelik G, Ray D, et al. Defective T-cell ERK signaling induces interferon-regulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes Immun. 2008;9:368–378. doi: 10.1038/gene.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strickland FM, Hewagama A, Lu Q, Wu A, Hinderer R, Webb R, et al. Environmental exposure, estrogen and two X chromosomes are required for disease development in an epigenetic model of lupus. J. Autoimmun. 2012;38:J135–J143. doi: 10.1016/j.jaut.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaur S, Chang T, Singh SP, Lim L, Mannan P, Garfield SH, et al. CD47 signaling regulates the immunosuppressive activity of VEGF in T Cells. J. Immunol. 2014;193:3914–3924. doi: 10.4049/jimmunol.1303116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nambiar MP, Enyedy EJ, Fisher CU, Krishnan S, Warke VG, Gilliland WR, et al. Abnormal expression of various molecular forms and distribution of T cell receptor zeta chain in patients with systemic lupus erythematosus. Arthritis Rheumatism. 2002;46:163–174. doi: 10.1002/1529-0131(200201)46:1<163::AID-ART10065>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 23.Fu Q, Zhao J, Qian X, Wong JL, Kaufman KM, Yu CY, et al. Association of a functional IRF7 variant with systemic lupus erythematosus. Arthritis Rheumatism. 2011;63:749–754. doi: 10.1002/art.30193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawasaki A, Furukawa H, Kondo Y, Ito S, Hayashi T, Kusaoi M, et al. Association of PHRF1-IRF7 region polymorphism with clinical manifestations of systemic lupus erythematosus in a Japanese population. Lupus. 2012;21:890–895. doi: 10.1177/0961203312439333. [DOI] [PubMed] [Google Scholar]
- 25.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.