Abstract
Enteropathogenic Escherichia coli (EPEC) remain one the most important pathogens infecting children and they are one of the main causes of persistent diarrhea worldwide. Historically, typical EPEC (tEPEC), defined as those isolates with the attaching and effacement (A/E) genotype (eae+), which possess bfpA+ and lack the stx- genes are found strongly associated with diarrheal cases. However, occurrence of atypical EPEC (aEPEC; eae+ bfpA- stx-) in diarrheal and asymptomatic hosts has made investigators question the role of these pathogens in human disease. Current epidemiological data is helping answering the question whether EPEC is mainly a foe or an innocent bystander during infection.
Keywords: enteropathogenic E. coli; EPEC; epidemiology; diarrhea; asymptomatic, pathogenic E. coli
Introduction
Diarrhea is one of the global leading causes of death in children less than 5 years of age, especially in low-income countries (1, 2), accounting for 800,000 fatalities per year worldwide. EPEC is a major cause of infantile diarrhea in developing countries (Figure 1). It was first described in 1955 when a number of E. coli strains, epidemiologically associated with outbreaks in 1940s and 1950s, was described (3). A hallmark phenotype of EPEC is the induction of a distinctive histopathology known as the attaching and effacing (A/E) lesion, which is characterized by the effacement of the intestine microvilli and the intimate attachment of the bacteria to the host epithelial surface (4). After entering the gastrointestinal tract, EPEC adhere to the mucosa of the small and large intestines and at least three steps for pathogenesis have been described (5). The initial step includes adherence to the host cell. After attachment, a type III secretion system would be used to inject virulence factors in the host cell. Finally, an intimate bacterial attachment and pedestal formation is observed. The initial definition of EPEC indicated that this pathotype is part of the diarrheagenic E. coli strains that have the ability to produce the A/E lesion without producing Shiga toxin (stx-) (6, 7).
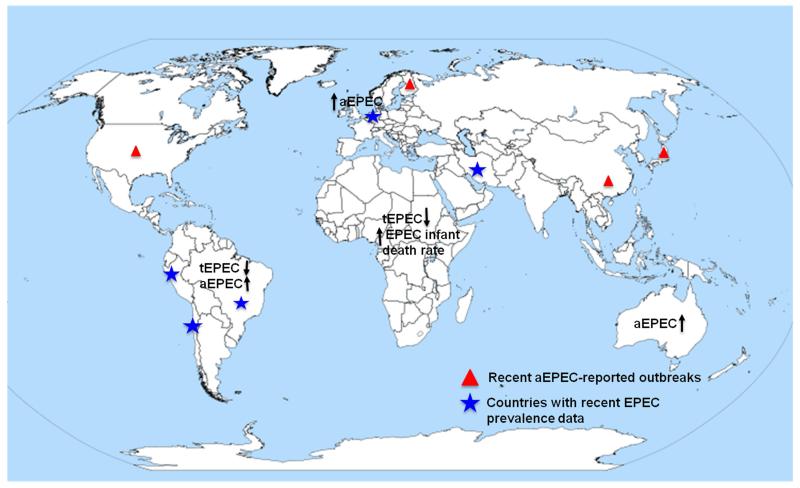
Figure 1.
Distribution of recent worldwide epidemiological studies of enteropathogenic Escherichia coli (EPEC). The arrows represent the increase or decrease of tEPEC and aEPEC cases per geographical region. The blue stars depicted those countries with increase EPEC prevalence reported in recent years. The red triangles represent countries with recent reported aEPEC outbreaks. aEPEC, atypical EPEC; tEPEC, typical EPEC.
Currently, the EPEC pathotype is subdivided into typical EPEC (tEPEC) and atypical EPEC (aEPEC) strains. This classification is initially based on the presence of EAF (EPEC adherence factor) plasmid (pEAF) (8). The bpf and per are two important loci encoded on the plasmid, with bfp encoding the type IV bundle forming pilus (BFP), which promotes bacterial microcolony formation (9). The per operon encodes a transcriptional activator called the plasmid encoded regulator of the Locus for Enterocyte Effacement (LEE) pathogenicity island (10, 11). tEPEC strains are more homogeneous in their virulence traits than aEPEC. Most of the typical strains produce the virulence factors encoded by the LEE region and EAF plasmid (8). aEPEC might possess enteroaggregative heat stable toxin (EAST1) and other potential virulence factors not encoded in the LEE, such as a hemolysin (8). EPEC belongs to specific O:H serotypes and at least 13 O groups are representative of these strains: O26, O39, O55, O86, O88, O103, O111, O114, O119, O125ac, O126, O127, O128ab, O142, O145, O157, and O158 (12). Some aEPEC strains (e.g. O55:H7) are more closely related to LEE-positive Shiga toxin-producing E. coli (e.g. STEC O157:H7) in their genetic characteristics and virulence properties. The tEPEC and aEPEC strains also have different adherence patterns. While tEPEC strains display the localized adherence pattern, the atypical strains can produce a localized-like adherence, a diffuse adherence, or an aggregative adherence pattern (8). tEPEC are rarely found in animals and humans are the major reservoir (8). aEPEC are present in both healthy and disease animals and humans (13).
Diarrheal cases caused by EPEC varied from subclinical to fatal infections (14). The tEPEC strains can cause abundant secretory diarrhea with mucus and significant losses of water and electrolyte in the feces (15). In addition, EPEC may lead to severe malabsorption of nutrients, which would progress to nutritional aggravation and persistence of diarrhea (16). Studies in volunteers demonstrated a large bacterial inoculum (109 to 1010) during short incubation periods (12 to 24 h) is able to induce diarrhea in adults (17). For aEPEC, the role as a diarrheagenic pathogen in disease is controversial. The pathogenesis of aEPEC seems to be related to the serotypes of aEPEC (18). For example, in the case of aEPEC O128:H2, after administering to 15 adult volunteers, none of them became ill (17). It has also been shown that a tEPEC O127:H6 strain without EAF plasmid was less virulent for adult volunteers than the wild type strain (19). However, there are also plenty of reports showing that aEPEC causes outbreaks linked to diarrhea (Figure 1). In a Japanese daycare center, the only diarrheagenic pathogen isolated from patients was aEPEC O55:HNM and these clones showed indistinguishable pulsed field gel electrophoresis patterns (20). E. coli O111:B4 was responsible for a diarrheal outbreak including 611 pupils and 39 adults in Finland (21). E. coli O39:NM was also associated with an outbreak involving more than 100 adults in the United States (22). An aEPEC EC3605 caused an outbreak in 75 students (ages 12 to 15) in Japan (23) and another aEPEC strain O127a:K63 was isolated from a 2010 food poisoning outbreak involving 112 adults in China. This strain displayed multidrug resistance to quinolones and extended spectrum cephalosporins (24).
Some aEPEC are strongly associated with acute disease, but some strains are also associated with persistent diarrhea (25-28). Santona et al identified 28 aEPEC strains among 402 E. coli strains isolated from feces of children with acute diarrhea in Italy (29). A study carried out in Australia compared aEPEC infected patients with other diarrheal agents (28). They found that patients infected with aEPEC experienced mild, non-dehydrating, non-inflammatory diarrhea, that was not associated with fever, vomiting or abdominal pain; however, the duration of diarrhea was longer. Strains of aEPEC were also found to be the most common pathogens among children with persistent diarrhea (diarrhea lasting for more than 14 days) in Australia (43%) and Norway (22%) (5). The exact mechanism of how EPEC causes diarrhea is not well established. However, it is postulated that the extensive disruption of the intestinal microvilli may lead to decrease in absorptive surfaces, affecting absorptive channels, thereby contributing to diarrhea (30). Other mechanisms that may participate in the diarrheal process include the effect of type III secretion system (T3SS) effectors on the intestinal cell. The T3SS Tir, Map, EspF, and EspG play a role in water and ion channel transport activity of intestinal epithelial. Also, EspF, EspG, and Map disrupt the tight junctions and enhance intestinal permeability, which may lead to diarrhea (31).
Some aEPEC isolates have been linked to bloody diarrhea. A study in Germany from January 1995 till June 2007 found that aEPEC strains were isolated from 18 (15.3%) of 118 patients with bloody diarrhea and from 141 (1.3%) of 10,550 patients with non-bloody diarrhea. Bielaszewska et al found that these strains were originally STEC O103 isolates that lost the Shiga toxin phage (32). Finally, aEPEC may become the precursor for stx-positive isolates. Sekse et al showed that aEPEC O103:H25 can be converted to STEC by a stx bacteriophage infection and become more virulent (33).
Recent epidemiological reports of EPEC
Currently, EPEC is estimated to be responsible for 5-10% of pediatric diarrhea in developing countries such as Brazil, Chile, Peru, and Iran (5). For many decades, tEPEC have been considered to be strongly associated with infantile diarrhea in developing countries. In several studies conducted in Latin America, tEPEC was found to be the main cause of endemic diarrhea in children less than one year of age. The frequency of tEPEC infection drops with increase in the age group and adults rarely experience tEPEC episodes (5). This may be due to development of immunity or the loss of receptors interacting with some specific adhesins. Although tEPEC were major agents of acute diarrhea in infants until the 1990s, there is a clear decline in many of these countries (8). The Global Enteric Multicenter Study (GEMS) was a population-based case control study including seven countries in Africa and Asia with the goal to identify the etiology, burden, and mortality to acute, moderate to severe diarrhea in children less than five years of age (34). At most GEMS study sites, tEPEC strains were not among the leading pathogens that cause acute moderate and severe diarrhea. The reasons for the decline in cases are not known, but it may due to improvements in public health measures such as: active interventions, therapy, sanitary conditions and control of hospital infections (8, 12). However, tEPEC infection seems to be associated with a 2.8-fold increased risk of death among infants ages 0-11 months (34).
Atypical EPEC continue to be frequently detected in both developing countries and industrialized countries nowadays (35). They are often associated with diarrhea and in some countries; they have outnumbered tEPEC infections. Studies from 13 developing countries showed that aEPEC isolates were responsible for 78% (131/169) of all EPEC cases in children less than five years old (5). Wheeler et al identified 142 aEPEC strains and only one tEPEC among 2,774 samples isolated from symptomatic children in the UK (36, 37). In another study, 61 EPEC strains were isolated from stool samples of symptomatic persons from 2008 to 2011 in Australia (38), where 95.1% (58/61) were aEPEC. In 2009, aEPEC strain O76 was associated with a nursery outbreak in Finland (39). Further, Sakkejha et al studied 109 EPEC isolates detected in England from 2010 to 2012, with 93% of the patients reporting diarrheal episodes and 32% bloody diarrhea. The study found that aEPEC were more common and were associated with a wider variety of serogroups than tEPEC (40).
Overall, according to 266 studies published between 1990 and 2002, EPEC are still among the most important pathogens causing diarrhea (35). As such, in 2014 an European, multi-center, prospective quarterly point prevalence study of community acquired diarrhea (EUCODI) showed that EPEC is highly prevalent during both the first (Jan. 2014) and the second (April 2014) rounds of the survey (Spina A, Kerr K, Cormican M, et al. Acute gastroenteritis and the spectrum of pathogens: results of the EUCODI-study 2014. Clin Microb Dis 2015; this issue). However, there are important regional and temporal variations. In Asia, a separate study found that EPEC (no information about whether the isolates were tEPEC or aEPEC) was responsible for 3.2% of 648 diarrhea samples in children were less than 5 years old in an Indian hospital (41).
Asymptomatic hosts carrying EPEC
Although there is a significant strong association between EPEC and infant diarrhea, many studies have found EPEC, especially aEPEC, in asymptomatic controls (5). The recent EUCODI study shows that EPEC was the most frequent pathogen detected in mixed infections (Spina A, Kerr K, Cormican M, et al. Acute gastroenteritis and the spectrum of pathogens: results of the EUCODI-study 2014. Clin Microb Dis 2015; this issue). A study in the Netherlands collected 5,197 samples from 29 child-care centers, with 95.4% of samples from children who had no gastroenteritis symptoms at time of sampling and EPEC isolates were most prevalent in asymptomatic samples (19.9%) (42). Another survey in Peruvian children isolated EPEC with a similar frequency from children with diarrhea (7.6%) and those from asymptomatic controls (9.9%) (43). A study in Mexico found that although aEPEC is most prevalent among diarrheagenic E. coli, most isolates are from asymptomatic carriers (25). A recent survey in Germany demonstrated that EPEC has a similar high prevalence (17.4%) in both control and diarrheal patients (Spina A, Kerr K, Cormican M, et al. Acute gastroenteritis and the spectrum of pathogens: results of the EUCODI-study 2014. Clin Microb Dis 2015; this issue).
It has been proposed that at least three reasons exist explaining the frequency of EPEC in symptomatic and control patients. The first one is host susceptibility. The precise mechanism leading to the diarrhea is not fully understood (44). Enteropathogens may initiate pathogenesis by binding to the host surface specific receptors, including sugar moieties as well as proteins. The susceptibility to infection may be associated with presence or lack of receptors (45). Non-specific host barriers may also prevent bacterial pathogenesis. The intact barriers, such as the intestinal microbiota, mucus layer, and epithelial cell layer may prevent diarrheal episodes (45). The immune status of the host prevents clinical illness but does not prevent intestinal colonization. Experiments carried out with ETEC and Shigella have shown that in endemic areas, where individuals are repetitively exposed to enteropathogens, individuals might carry pathogens without suffering from diarrheal episodes (46).
It is well known that secretory immunoglobulin A (sIgA) antibodies from intestine and breast milk as well as human breast milk oligosaccharides can prevent enterocyte colonization or mucosal invasion by enteropathogens without killing the bacteria (47). Breastfeeding can prevent diarrhea in infants and toddlers and protection is due to the presence of sIgA or non-specific factors, such as lactoferrin and enterotoxin-binding oligosaccharides. In endemic areas, the colostrum of puerperal women is rich in sIgA against EPEC (48-50). A study in Peru showed that EPEC prevalence increased with age within the first two years of age (51). EPEC was found in 3% of diarrhea samples in children < 6 months old, in 11% of children 6-12 months old, and in 16% of children 13-24 months old. Small infants may be protected from symptomatic EPEC infection due to breastfeeding. In addition, children may acquire natural immunity in some developing countries where EPEC is highly endemic (5). Host age also plays a role for carrying bacteria asymptomatically. Neter et al showed that almost 100% of children developed LPS specific antibodies against three of the most common O serogroups by the age of twelve (3). Opintan et al found that although EPEC is one of the most common pathogens recovered from healthy individuals 3 years and older, and it has not been detected in healthy infants less than two years old (52).
The second reason is linked to bacterial factors. EPEC strains are heterogeneous serotypes that include different clones or genetic lineages (8). Some strains cause diarrhea more frequently than others at the same challenge inoculum. EPEC strain E2348/69 causes more severe diarrhea than strain E74/68 (45). Several studies have identified certain virulence genes significantly associated with diarrhea. Afset et al used DNA microarray to analyze aEPEC strains isolated from children with and without diarrhea. Genomic DNA was hybridized against 242 different oligonucleotide probes. They found O-island 122 (OI-122), carrying efa1/lifA and several other genes, significantly associated with diarrhea. In contrast, the phylogenic marker gene yhaA was negatively associated with diarrhea. Children with diarrhea were infected with OI-122 efa1/lifA positive, yhaA negative strains, while children without diarrhea had aEPEC strains which were OI-122 efa1/lifA negative, yhaA positive (53). Afset et al further compared the phylogenetic ancestry and diarrhea association of aEPEC strains (54). Fifty six aEPEC strains were divided into 4 phylogenetic groups (B1, A, D, B2) and they found a borderline significant association with diarrhea for the phylogenetic groups B1 and D. Wang et al also tried to distinguish aEPEC from diarrheic patients and healthy controls. Multiplex real-time PCR was used to examine the intimin gene typing, phylogenetic grouping and the virulence profile of isolated aEPEC strains. After examining 159 strains from 679 samples, their results indicated that aEPEC, particularly those from phylogenetic groups B1 or D, virulence group Ia, or intimin typing β1 and γ1, induce diarrhea in humans (55). Contreras et al also characterized a collection of EPEC strains obtained from a study in Peru using PCR-RFLP analysis. They found that the κ-intimin allele had the highest clinical severity score compared with other alleles (56).
The third possibility is the variability of diagnostic tests. Barletta et al hypothesized that presence of symptoms in EPEC infections is related to the bacterial load (57). They analyzed stool samples from a passive surveillance diarrheal cohort study, including 1,034 Peruvian children. They isolated EPEC with a similar frequency from children with diarrhea and asymptomatic controls. However, quantitative real time PCR assay was applied to determine whether bacterial loads were related to diarrhea and it was found that bacterial load was significantly higher in the diarrhea group than in the control group among children with EPEC as the sole pathogen and among children less than 1 year old. However, it is evident that this detection method had some limitations due to the complexity in fecal samples.
Several other factors may also affect the results reported in different studies. For example, the control samples may be collected from pre- or post-symptomatic patients. Other issues include the sample size that may not be large enough or the fact that asymptomatic controls may transmit EPEC to other patients, which cannot be excluded (42). Finally, the environmental factors, such as poor hygiene and high fecal contamination, may also lead to the bacteria load in control groups (35).
In summary, cumulative data in recent years indicated that aEPEC are more prevalent than tEPEC in both developing and developed countries. However, tEPEC is still considered a bona fide pathogen due to their arsenal of virulence factors and association with severe, lethal disease. The recent emergence of aEPEC requires further epidemiological studies that can help elucidating whether certain serotypes are specifically linked to disease in humans. Further, more investigation is required to identify virulence/fitness factors of aEPEC that mediate the disease process or the ability to be maintained in patients and in healthy individuals. Future studies will answer whether aEPEC is a foe or innocent bystander in human disease.
Acknowledgments
This work was partially supported by NIH/NIAID grant AI079154 to A.G.T. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIAID or NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liu L, Johnson HL, Cousens S, Perin J, Scott S, Lawn JE, et al. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet. 2012;379:2151–61. doi: 10.1016/S0140-6736(12)60560-1. [DOI] [PubMed] [Google Scholar]
- 2.Bhutta ZA, Das JK. Global burden of childhood diarrhea and pneumonia: what can and should be done? Pediatrics. 2013;131:634–6. doi: 10.1542/peds.2012-3737. [DOI] [PubMed] [Google Scholar]
- 3.Neter E, Westphal O, Luderitz O, Gino RM, Gorzynski EA. Demonstration of antibodies against enteropathogenic Escherichia coli in sera of children of various ages. Pediatrics. 1955;16:801–8. [PubMed] [Google Scholar]
- 4.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochoa TJ, Barletta F, Contreras C, Mercado E. New insights into the epidemiology of enteropathogenic Escherichia coli infection. Trans R Soc Trop Med Hyg. 2008;102:852–6. doi: 10.1016/j.trstmh.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaper JB. Defining EPEC. Revista de Microbiologia. 1996;27:130–3. [Google Scholar]
- 7.Jensen C, Ethelberg S, Olesen B, Schiellerup P, Olsen KEP, Scheutz F, et al. Attaching and Effacing Escherichia coli strains isolated from Danish children: Clinical significance and microbiological characteristics. Clin Microbiol Infect. 2007;13:863–72. doi: 10.1111/j.1469-0691.2007.01773.x. [DOI] [PubMed] [Google Scholar]
- 8.Trabulsi LR, Keller R, Tardelli Gomes TA. Typical and atypical enteropathogenic Escherichia coli. Emerg Infect Dis. 2002;8:508–13. doi: 10.3201/eid0805.010385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bieber D, Ramer SW, Wu CY, Murray WJ, Tobe T, Fernandez R, et al. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–8. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 10.Frankel G, Phillips AD, Rosenshine I, Dougan G, Kaper JB, Knutton S. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol Microbiol. 1998;30:911–21. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- 11.Gomez-Duarte OG, Kaper JB. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect Immun. 1995;63:1767–76. doi: 10.1128/iai.63.5.1767-1776.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tozzoli R, Scheutz F. Diarrhoeagenic Escherichia coli Infections in Humans. In: Morabito S, editor. Pathogenic Escherichia coli. Caister Academic Press; Norfolk, UK: 2014. pp. 1–18. [Google Scholar]
- 13.Hernandes RT, Elias WP, Vieira MA, Gomes TA. An overview of atypical enteropathogenic Escherichia coli. FEMS Microbiol Lett. 2009;297:137–49. doi: 10.1111/j.1574-6968.2009.01664.x. [DOI] [PubMed] [Google Scholar]
- 14.Torres AG, Arenas-Hernández M, Martínez-Laguna Y. Overview of Escherichia coli. In: Torres AG, editor. Pathogenic Escherichia coli in Latin. Bentham Science Publishers Ltd; America: 2010. pp. 1–7. [Google Scholar]
- 15.Arenas-Hernandez MM, Martinez-Laguna Y, Torres AG. Clinical implications of enteroadherent Escherichia coli. Curr Gastroenterol Rep. 2012;14:386–94. doi: 10.1007/s11894-012-0277-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fagundes-Neto U, Scaletsky IC. The gut at war: the consequences of enteropathogenic Escherichia coli infection as a factor of diarrhea and malnutrition. Sao Paulo Med J. 2000;118:21–9. doi: 10.1590/S1516-31802000000100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine MM, Bergquist EJ, Nalin DR, Waterman DH, Hornick RB, Young CR, et al. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet. 1978;1:1119–22. doi: 10.1016/s0140-6736(78)90299-4. [DOI] [PubMed] [Google Scholar]
- 18.Gomes TAT, Gonzalez-Pedrajo B. Pathogenic Escherichia coli in Latin. Bentham Science Publishers Ltd; America: 2010. Enteropathogenic Escherichia coli (EPEC). In: Torres AG, editor; pp. 25–47. [Google Scholar]
- 19.Levine MM, Nataro JP, Karch H, Baldini MM, Kaper JB, Black RE, et al. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J Infect Dis. 1985;152:550–9. doi: 10.1093/infdis/152.3.550. [DOI] [PubMed] [Google Scholar]
- 20.Yatsuyanagi J, Saito S, Sato H, Miyajima Y, Amano K, Enomoto K. Characterization of enteropathogenic and enteroaggregative Escherichia coli isolated from diarrheal outbreaks. J Clin Microbiol. 2002;40:294–7. doi: 10.1128/JCM.40.1.294-297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viljanen MK, Peltola T, Junnila SY, Olkkonen L, Jarvinen H, Kuistila M, et al. Outbreak of diarrhoea due to Escherichia coli O111:B4 in schoolchildren and adults: association of Vi antigen-like reactivity. Lancet. 1990;336:831–4. doi: 10.1016/0140-6736(90)92337-h. [DOI] [PubMed] [Google Scholar]
- 22.Hedberg CW, Savarino SJ, Besser JM, Paulus CJ, Thelen VM, Myers LJ, et al. An outbreak of foodborne illness caused by Escherichia coli O39:NM, an agent not fitting into the existing scheme for classifying diarrheogenic E. coli. J Infect Dis. 1997;176:1625–8. doi: 10.1086/517342. [DOI] [PubMed] [Google Scholar]
- 23.Yatsuyanagi J, Saito S, Miyajima Y, Amano K, Enomoto K. Characterization of atypical enteropathogenic Escherichia coli strains harboring the astA gene that were associated with a waterborne outbreak of diarrhea in Japan. J Clin Microbiol. 2003;41:2033–9. doi: 10.1128/JCM.41.5.2033-2039.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao R, Qiu S, Wang Y, Yang G, Su W, Song L, et al. Quinolone-resistant Escherichia coli O127a:K63 serotype with an extended-spectrum-beta-lactamase phenotype from a food poisoning outbreak in China. J Clin Microbiol. 2012;50:2450–1. doi: 10.1128/JCM.00276-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Estrada-Garcia T, Lopez-Saucedo C, Thompson-Bonilla R, Abonce M, Lopez-Hernandez D, Santos JI, et al. Association of diarrheagenic Escherichia coli Pathotypes with infection and diarrhea among Mexican children and association of atypical Enteropathogenic E. coli with acute diarrhea. J Clin Microbiol. 2009;47:93–8. doi: 10.1128/JCM.01166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Estrada-Garcia T, Cerna JF, Paheco-Gil L, Velazquez RF, Ochoa TJ, Torres J, et al. Drug-resistant diarrheogenic Escherichia coli, Mexico. Emerg Infect Dis. 2005;11:1306–8. doi: 10.3201/eid1108.050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Afset JE, Bevanger L, Romundstad P, Bergh K. Association of atypical enteropathogenic Escherichia coli (EPEC) with prolonged diarrhoea. J Med Microbiol. 2004;53:1137–44. doi: 10.1099/jmm.0.45719-0. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen RN, Taylor LS, Tauschek M, Robins-Browne RM. Atypical enteropathogenic Escherichia coli infection and prolonged diarrhea in children. Emerg Infect Dis. 2006;12:597–603. doi: 10.3201/eid1204.051112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santona S, Diaz N, Fiori PL, Francisco M, Sidat M, Cappuccinelli P, et al. Genotypic and phenotypic features of enteropathogenic Escherichia coli isolated in industrialized and developing countries. J Infect Dev Ctries. 2013;7:214–9. doi: 10.3855/jidc.3054. [DOI] [PubMed] [Google Scholar]
- 30.Goosney DL, Gruenheid S, Finlay BB. Gut feelings: enteropathogenic E. coli (EPEC) interactions with the host. Annu Rev Cell Dev Biol. 2000;16:173–89. doi: 10.1146/annurev.cellbio.16.1.173. [DOI] [PubMed] [Google Scholar]
- 31.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. 2013;26:822–80. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bielaszewska M, Prager R, Kock R, Mellmann A, Zhang W, Tschape H, et al. Shiga toxin gene loss and transfer in vitro and in vivo during enterohemorrhagic Escherichia coli O26 infection in humans. Appl Environ Microbiol. 2007;73:3144–50. doi: 10.1128/AEM.02937-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sekse C, Muniesa M, Wasteson Y. Conserved Stx2 phages from Escherichia coli O103:H25 isolated from patients suffering from hemolytic uremic syndrome. Foodborne Pathog Dis. 2008;5:801–10. doi: 10.1089/fpd.2008.0130. [DOI] [PubMed] [Google Scholar]
- 34.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–22. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 35.Ochoa TJ, Contreras CA. Enteropathogenic Escherichia coli infection in children. Curr Opin Infect Dis. 2011;24:478–83. doi: 10.1097/QCO.0b013e32834a8b8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wheeler JG, Sethi D, Cowden JM, Wall PG, Rodrigues LC, Tompkins DS, et al. Study of infectious intestinal disease in England: rates in the community, presenting to general practice, and reported to national surveillance. The Infectious Intestinal Disease Study Executive. BMJ. 1999;318:1046–50. doi: 10.1136/bmj.318.7190.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenkins C, Smith HR, Lawson AJ, Willshaw GA, Cheasty T, Wheeler JG, et al. Serotypes, intimin subtypes, and antimicrobial resistance patterns of atypical enteropathogenic Escherichia coli isolated in England from 1993 to 1996. Eur J Clin Microbiol Infect Dis. 1996;25:19–24. doi: 10.1007/s10096-005-0075-x. [DOI] [PubMed] [Google Scholar]
- 38.Staples M, Doyle CJ, Graham RM, Jennison AV. Molecular epidemiological typing of enteropathogenic Escherichia coli strains from Australian patients. Diagn Microbiol Infect Dis. 2013;75:320–4. doi: 10.1016/j.diagmicrobio.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 39.Moller-Stray J, Eriksen HM, Bruheim T, Kapperud G, Lindstedt BA, Skeie Å , et al. Two outbreaks of diarrhoea in nurseries in Norway after farm visits, April to May 2009. Euro Surveill. 2012:17. doi: 10.2807/ese.17.47.20321-en. [DOI] [PubMed] [Google Scholar]
- 40.Sakkejha H, Byrne L, Lawson AJ, Jenkins C. An update on the microbiology and epidemiology of enteropathogenic Escherichia coli in England 2010-2012. J Med Microbiol. 2013;62:1531–4. doi: 10.1099/jmm.0.062380-0. [DOI] [PubMed] [Google Scholar]
- 41.Nair GB, Ramamurthy T, Bhattacharya MK, Krishnan T, Ganguly S, Saha DR, et al. Emerging trends in the etiology of enteric pathogens as evidenced from an active surveillance of hospitalized diarrhoeal patients in Kolkata, India. Gut Pathog. 2010;2:4. doi: 10.1186/1757-4749-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Enserink R, Scholts R, Bruijning-Verhagen P, Duizer E, Vennema H, de Boer R, et al. High detection rates of enteropathogens in asymptomatic children attending day care. PLoS One. 2014;9:e89496. doi: 10.1371/journal.pone.0089496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ochoa TJ, Ecker L, Barletta F, Mispireta ML, Gil AI, Contreras C, et al. Age-related susceptibility to infection with diarrheagenic Escherichia coli among infants from Periurban areas in Lima, Peru. Clin Infect Dis. 2009;49:1694–702. doi: 10.1086/648069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Donnenberg MS, Finlay BB. Combating enteropathogenic Escherichia coli (EPEC) infections: the way forward. Trends Microbiol. 2013;21:317–9. doi: 10.1016/j.tim.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levine MM, Robins-Browne RM. Factors that explain excretion of enteric pathogens by persons without diarrhea. Clin Infect Dis. 2012;55:S303–11. doi: 10.1093/cid/cis789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kotloff KL, Nataro JP, Losonsky GA, Wasserman SS, Hale TL, Taylor DN, et al. A modified Shigella volunteer challenge model in which the inoculum is administered with bicarbonate buffer: clinical experience and implications for Shigella infectivity. Vaccine. 1995;13:1488–94. doi: 10.1016/0264-410x(95)00102-7. [DOI] [PubMed] [Google Scholar]
- 47.Manthey CF, Autran CA, Eckmann L, Bode L. Human milk oligosaccharides protect against enteropathogenic Escherichia coli attachment in vitro and EPEC colonization in suckling mice. J Pediatr Gastroenterol Nutr. 2014;58:165–8. doi: 10.1097/MPG.0000000000000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parissi-Crivelli A, Parissi-Crivelli JM, Giron JA. Recognition of enteropathogenic Escherichia coli virulence determinants by human colostrum and serum antibodies. J Clin Microbiol. 2000;38:2696–700. doi: 10.1128/jcm.38.7.2696-2700.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loureiro I, Frankel G, Adu-Bobie J, Dougan G, Trabulsi LR, Carneiro-Sampaio MM. Human colostrum contains IgA antibodies reactive to enteropathogenic Escherichia coli virulence-associated proteins: intimin, BfpA, EspA, and EspB. J Pediatr Gastroenterol Nutr. 1998;27:166–71. doi: 10.1097/00005176-199808000-00007. [DOI] [PubMed] [Google Scholar]
- 50.Durand D, Ochoa TJ, Bellomo SM, Contreras CA, Bustamante VH, Ruiz J, et al. Detection of secretory immunoglobulin A in human colostrum as mucosal immune response against proteins of the type III secretion system of Salmonella, Shigella and enteropathogenic Escherichia coli. Pediatr Infect Dis J. 2013;32:1122–6. doi: 10.1097/INF.0b013e318293306c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ochoa TJ, Mercado EH, Durand D, Rivera FP, Mosquito S, Contreras C, et al. Frequency and pathotypes of diarrheagenic Escherichia coli in Peruvian children with and without diarrhea. Rev Peru Med Exp Salud Publica. 2011;28:13–20. doi: 10.1590/s1726-46342011000100003. [DOI] [PubMed] [Google Scholar]
- 52.Opintan JA, Bishar RA, Newman MJ, Okeke IN. Carriage of diarrhoeagenic Escherichia coli by older children and adults in Accra, Ghana. Trans R Soc Trop Med Hyg. 2010;104:504–6. doi: 10.1016/j.trstmh.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 53.Afset JE, Bruant G, Brousseau R, Harel J, Anderssen E, Bevanger L, et al. Identification of virulence genes linked with diarrhea due to atypical enteropathogenic Escherichia coli by DNA microarray analysis and PCR. J Clin Microbiol. 2006;44:3703–11. doi: 10.1128/JCM.00429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Afset JE, Anderssen E, Bruant G, Harel J, Wieler L, Bergh K. Phylogenetic backgrounds and virulence profiles of atypical enteropathogenic Escherichia coli strains from a case-control study using multilocus sequence typing and DNA microarray analysis. J Clin Microbiol. 2008;46:2280–90. doi: 10.1128/JCM.01752-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Wakushima M, Aota T, Yoshida Y, Kita T, Maehara T, et al. Specific properties of enteropathogenic Escherichia coli isolates from diarrheal patients and comparison to strains from foods and fecal specimens from cattle, swine, and healthy carriers in Osaka City, Japan. Appl Environ Microbiol. 2013;79:1232–40. doi: 10.1128/AEM.03380-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Contreras CA, Ochoa TJ, Lacher DW, DebRoy C, Navarro A, Talledo M, et al. Allelic variability of critical virulence genes (eae, bfpA and perA) in typical and atypical enteropathogenic Escherichia coli in Peruvian children. J Med Microbiol. 2010;59:25–31. doi: 10.1099/jmm.0.013706-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barletta F, Ochoa TJ, Mercado E, Ruiz J, Ecker L, Lopez G, et al. Quantitative real-time polymerase chain reaction for enteropathogenic Escherichia coli: a tool for investigation of asymptomatic versus symptomatic infections. Clin Infect Dis. 2011;53:1223–9. doi: 10.1093/cid/cir730. [DOI] [PMC free article] [PubMed] [Google Scholar]