Summary
Most known disease-associated mutations are missense mutations involving changes of amino acids of proteins encoded by their genes. Given the plethora of genetic studies, sequenced exomes, and new protein structures determined each year, it is appropriate to revisit the role that structure plays in providing insights into the molecular basis of disease associated mutations. In that regard, a large-scale structural analysis on 6,025 disease-associated mutations as well as 4,536 neutral variations for comparison was performed. While buried amino acids are common among the disease-associated mutations as reported previously, more are statistically significantly enriched at observed or predicted functional sites. Interesting findings are that ligand-binding sites adjacent to protein-protein interfaces and residues involved in enzymatic function are especially vulnerable to disease-associated mutations. Finally, a compositional analysis of disease-associated mutations in comparison to variants identified in the 1000 Genomes Project provides a structural rationalization of the most disease-associated residue types.
Introduction
Since the sequencing of first human genome was completed a decade ago (Collins et al., 2004), tremendous efforts have been made to advance new sequencing methods that allow rapid, massively parallel sequencing at low cost (Metzker, 2010). These next-generation sequencing methods enable large-scale sequencing on thousands of individuals, generating a large amount of data available for comparative genome analysis (Abecasis et al., 2010; Abecasis et al., 2012). By identifying variations of DNA sequences, in particular, single-nucleotide polymorphisms (SNPs), we may begin to decipher the links between phenotypes and genotypes. Of particular interest are genetic mutations that cause various human, especially Mendelian, diseases. Statistical analyses of patients’ and normal people’s sequences often pinpoint mutations strongly associated with patients. Many such mutations are collected in databases such as the Online Database of Mendelian Inheritance in Man (OMIM) (McKusick, 2007) and the Human Gene Mutation Database (HGMD) (Stenson et al., 2008).
Since most cases in these databases are SNPs identified through statistical analysis, it is not clear whether a particular mutation is actually the cause of the implicated disease. As such, these mutations are usually referred to as being disease-associated. Most are non-synonymous SNPs (nsSNPs) that occur in the coding regions of genes and result in changes of amino acid type, i.e., missense mutations. It is therefore expected that the change of amino acid impairs the function of the involved protein. However, for the vast majority of cases, it is not clear how the mutation impacts the function of the protein. In this regard, studying the impact of mutations may provide a better understanding of the mechanisms of the corresponding diseases, and eventually may significantly increase the chance of finding a better treatment for patients with the disease.
Meanwhile, over the comparable period, the three dimensional structures of many proteins have been determined at high resolution (Berman et al., 2000). Often these proteins are co-crystalized with other biomolecules that are relevant to their functions, e.g., protein-protein complexes and protein-ligand complexes. Given these structural data, many questions regarding disease-causing mutations can be asked and addressed by a thorough inspection of these structures: Can we understand disease-associated mutations in the context of their locations in the protein’s structure? Where are disease-linked missense mutations located in the proteins? What are the functional and structural consequences of these mutations? Is there any location in the protein where variations are more likely to cause disease? What is the structural reason why certain types of mutations are more likely to be disease-associated? Answers to these questions not only deepen our understanding of the molecular mechanisms of diseases, but also have practical implications to the predictions of disease-association by automated computational tools and to personalized medicine.
Over a decade ago, Wang and Moult performed an early study on 262 disease-associated mutations from 26 proteins and found that about 80% of them destabilize proteins and about 5% involve ligand-binding (Wang and Moult, 2001). A subsequent study by Thornton and colleagues on 1,292 mutations from 63 proteins shows that about 28% are buried and related to protein stability, while 29% are involved in intermolecular interactions such as protein-protein interactions and ligand-binding (Steward et al., 2003). Similar results were also reported in another study (Sunyaev et al., 2000). However, mainly due to the paucity of solved protein structures, these studies were carried out on very few proteins. Since then, many new disease-causing mutations have been found, and the number of known protein structures has also grown exponentially, especially for those in complex with functional partners. The previous studies also lacked a comparison to neutral variations. The statistical significance of their discoveries, in particular, the value of structural analysis to the prediction of disease-association, is unclear.
Given this background, it is worthwhile to revisit the questions raised above. In this contribution, we performed a large scale analysis on 6,025 disease-associated mutations in 642 proteins with experimental structures, as well as 4,536 neutral mutations in 1,743 proteins for comparison. We take advantage of the fact that many of these proteins have multiple structures in complex with other proteins or ligands. Below, we first map these mutations to their locations onto protein structures. We then compare them with neutral variations and identify regions more likely to cause disease. We also analyze the likelihood of that different amino acid types are disease associated and provide structural explanations for some of the most deleterious mutations.
Results
We have collected 6,025 disease-associated missense mutations in 642 human genes and 4,536 neutral mutations in 1,743 human genes from the Aug 2013 release of the UNIPROT database (see Methods) (Wu et al., 2006). All these mutations have at least one experimentally determined protein structure containing the corresponding mutation. The locations of these mutations are subsequently analyzed.
Where are the disease-associated missense mutations in their protein structures?
Table 1 shows the statistics of disease-associated mutations. Among them, about 22% are buried in protein cores; 12% are found at protein-protein interfaces (PPIs); 36% are located in concave-shaped pockets; 12% and 2.9% are found in direct physical contact with small molecule ligands and metal ions, respectively; and a small fraction, 0.7%, are at either DNA-protein or RNA-protein interfaces. Since the experimental information is most likely incomplete, we further applied two computational tools, EFICAz for enzyme function prediction (Kumar and Skolnick, 2012), and FINDSITEcomb for ligand-binding site predictions to our data sets (Zhou and Skolnick, 2013). About 7% of disease-associated mutations are classified by EFICAz as being Functional Discriminating Residues (FDRs) for their corresponding enzymatic functions, and an additional 12% of mutations are at FINDSITEcomb predicted ligand-binding sites that did not have bound ligands in their PDB structures. It should be noted that these location classifications are not necessarily mutually exclusive, e.g., about 69% and 45% of observed small-molecule ligand binding sites are found in geometric pockets or predicted by FINDSITEcomb, respectively. Another interesting observation is that about 15% of disease-associated mutations of observed ligand-binding sites are also identified at the protein-protein interfaces. This is investigated in detail below.
Table 1.
Locations of disease-associated amino acid mutations in protein structures.
| Location | Count (Frequency) |
|---|---|
| Buried | 2211 (0.22) |
| PPI | 726 (0.12) |
| Ligand-binding | 714 (0.12) |
| Metal Ion-binding | 174 (0.029) |
| DNA/RNA-binding | 42(0.007) |
| Geometric Pocket | 2177 (0.36) |
| EFICAz | 420 (0.07) |
| FINDSITE | 738 (0.12) |
| Other (Exposed) | 1451 (0.30) |
| Total | 6025 |
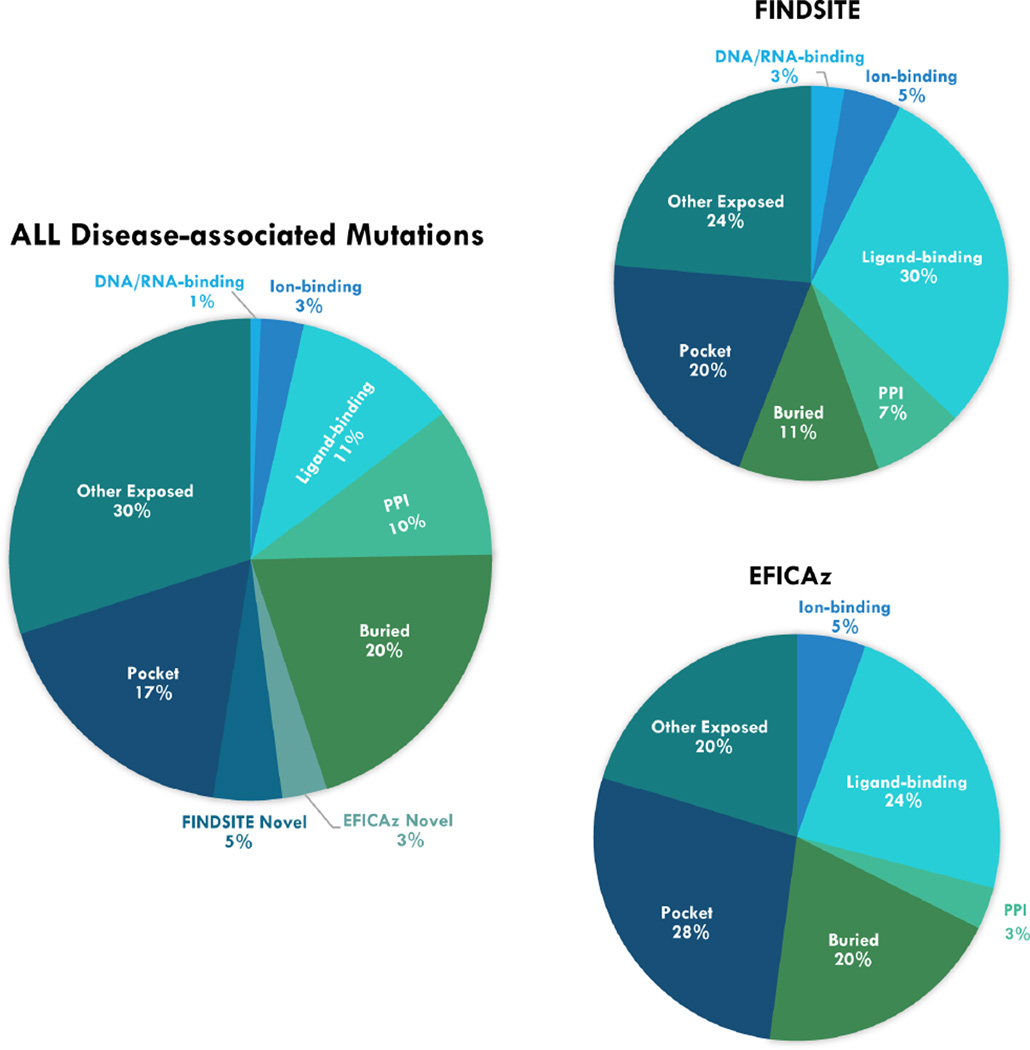
Fig. 1 shows a pie chart obtained by assigning a unique primary location to each mutation, using the following order: DNA/RNA-binding, ion-binding, small-molecule ligand-binding, protein-protein interface, buried, EFICAz, FINDSITEcomb, pocket, and other exposed. About 15% of disease-associated mutations are located at known functional sites, i.e., they involve binding to other molecules other than proteins; 10% are at protein-protein interfaces; and 20% of mutations are within protein cores and are likely important for maintaining stability. EFICAz and FINDSITEcomb additionally flagged 3% and 5% of mutations that are potentially important for the functions of the corresponding protein. After these classifications, 17% of mutations are found in pockets, which might engage in unknown interactions with some biomolecules. Overall, this classification scheme could assign a primary functional or structural role for 70% of disease-associated mutations. The remaining 30% of mutations involve exposed surface residues and are located at either a flat surface or small pocket. They are potentially candidate interaction sites for unknown protein-protein interactions or interactions with biomolecules that do not require large concave pockets.
Fig. 1.
Pie charts of disease-associated mutations according to their primary locations. On the left shows the overall distributions, on the right are the charts of positives hits by FINDSITE and EFICAz, respectively.
How useful is structural/functional information for predicting disease-association?
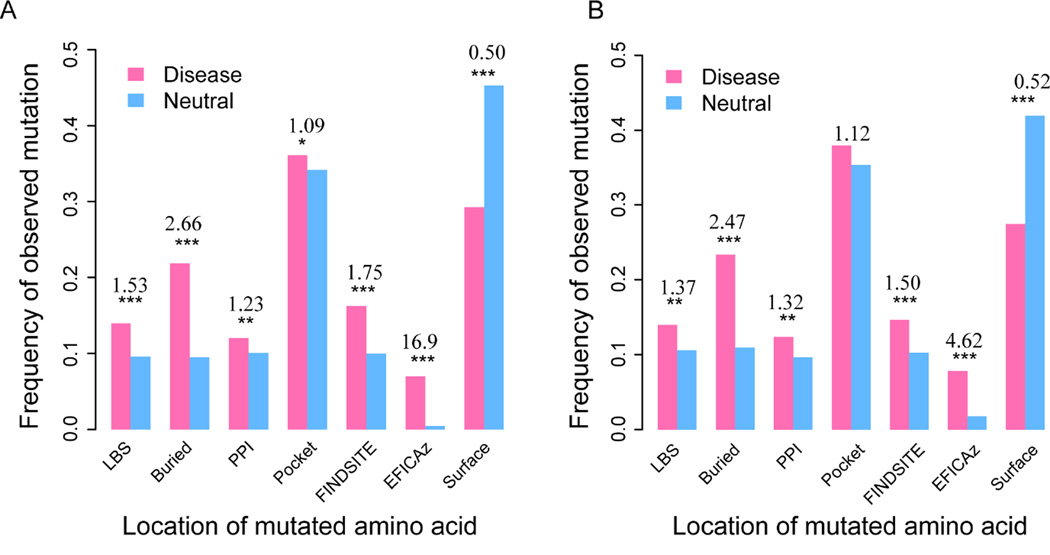
In order to answer this question, we compared the frequency of disease-associated mutations versus neutral mutations at different locations derived from our structural and functional analysis mentioned above. As shown in Fig. 2A, in all locations except for unannotated, exposed regions, disease mutations are more likely to appear than neutral mutations. In the protein interior, the frequency of disease mutations is over two times that of neutral mutations. This give an odds ratio (OR) of 2.66, which is the odds of disease versus neutral among buried mutations over the odds of disease versus neutral among all other mutations in our sets. The result is statistically highly significant with a p value 4.0×10−67 (Fisher’s exact test, two-tailed). Likewise, ligand-binding sites are about 50% more likely to be observed in the disease set than in the neutral set, which gives a significant OR of 1.53 (p = 6.8×10−12). At an OR of 1.75 (p = 4.4×10−21), the performance of FINDSITEcomb in terms of OR is slightly better than that obtained by counting observed ligand-binding sites in experimental structures. Within a protein-protein interface, or pocket region, disease mutations are slightly, but significantly, more frequent than neutral mutations, yielding ORs of 1.23 (p = 1.3×10−3) and 1.09 (p = 0.04), respectively. The latter is less significant, partly due to the fact that not all residues identified in a predicted, geometric protein pocket are important for protein function or stability; thus, there are more noisy signals in this region. Surprisingly, EFICAz predictions have the highest OR of about 17 (p = 1.6×10−78). Many of these predictions are at highly conserved active sites important for enzymatic function, though they only cover about 7% of disease mutations. On the other hand, among unannotated surface residues, neutral mutations have a high percentage at 45%, compared to about 30% of disease mutations. This is expected, as surface residues without a functional or structural role are less likely to cause disease.
Fig. 2.
Disease-associated mutations versus neutral ones assigned by their location in the protein’s structure. Each bar represents the fraction of mutations in either disease-associated or neutral data sets. The numbers above pairs of bars are the odds ratios. Stars denote statistical significance according to one-tail Fisher’s exact test (* − 0.01 < P < 0.05, ** − 0.001 < P < 0.01, *** − P < 0.001, no star – P > 0.05) (A) All mutations. (B) Subset of mutations from the same set of proteins that contain both disease-associated and neutral mutations.
Since neutral mutations are from a much more diverse set of proteins than disease associated mutations (1743 vs. 642 proteins), there is the concern that the data set might be biased. In order to eliminate such a concern, we further performed the same analysis on the subset of mutations that appear in the same set of proteins. Fig. 2B shows results that qualitatively are in agreement with the analysis on the full set shown above.
Mutations adjacent to PPIs are more likely associated with disease than mutations within PPIs
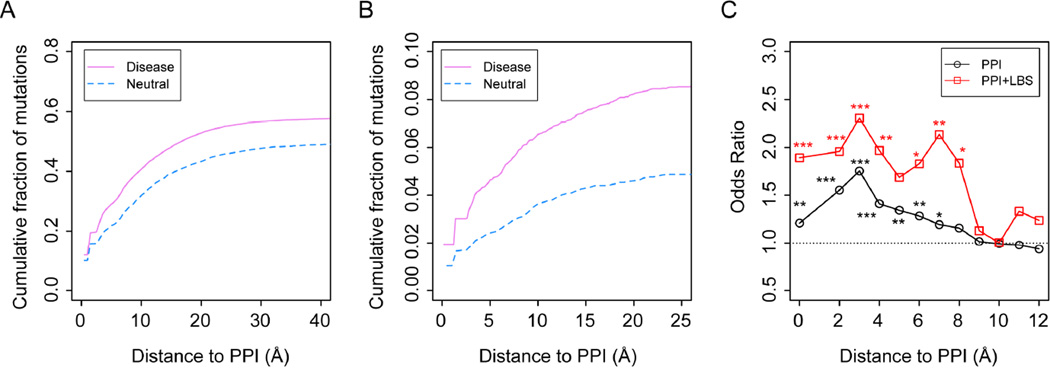
Overall, about 58% of disease associated mutations are found in at least one protein-protein complex structure versus 50% of neutral mutations are found in a protein-protein complex. We further analyzed the distribution of mutations in protein-protein complexes according to their distance from observed PPIs (Fig 3A). As shown above, about 12.0% of all disease mutations and 10.1% of neutral mutations in our data sets are located at the PPIs, i.e., 0 Å from the interface. This gives a small, but statistically significant, difference of 1.9%, corresponding to an increase of about 20% in OR to 1.23 for the disease associated mutations. The remaining difference in cumulative fraction comes mainly from the mutations close to but not at the PPIs. Within 3 Å from PPIs, the cumulative difference rapidly increases to 6.8%; this percentage further increases to 8.3% within 6 Å from PPIs. Thus, it appears that disease mutations are more likely to occur at and/or adjacent to PPIs than neutral mutations. Moreover, the biggest difference is attributed to regions immediately neighboring protein-protein interfaces, i.e., at a distance from 0 Å to 6 Å, rather than at the PPIs themselves. These difference leads to significant increase of OR values to as high as 1.75 at 3 Å from PPIs (Fig. 3C).
Fig. 3.
Distribution of mutations in the neighborhood of protein-protein interfaces. (A) Cumulative fractions of disease-associated mutations versus neutral ones according to their distance from known protein-protein interfaces. (B) Subsets of mutations that are also observed at small-molecule ligand binding sites, LBS. (C) Distributions of ORs according to distance from PPIs. A bin width of 2 Å is employed. Stars represent statistical significance as in Fig. 2.
Since disease-causing mutations are more likely to be buried than neutral mutations, as shown in Fig. 2, one natural explanation is that buried mutations close to PPIs are more likely disease-associated than neutral. The difference in cumulative fraction of buried mutations around PPIs between the disease and neutral sets is about 0.1% at 0 Å, 3.5% within 3 Å, and 5.1% within 6 Å from PPIs. Therefore, mutations of buried residues do make a major contribution to the observed phenomenon. However, this only explains about 61% of the observed difference.
A second main reason for this phenomenon is that adjacent to PPIs there are functionally relevant pockets involving ligand recognition. It has been shown that protein-protein association naturally creates geometric pockets (Gao and Skolnick, 2012). Some of these pockets could be selected by evolution for ligand-binding. As a result, functionally relevant ligand-binding pockets are enriched around PPIs. Mutations at these pockets likely lead to harmful effects. Indeed, if we focus on the subset of mutations that are also ligand-binding within protein-protein complexes, we found that disease-associated mutations are much more likely to be found in the neighborhood of PPIs versus neutral mutations. As shown in Fig. 3B, the difference in the cumulative fraction of mutations is 0.9% at 0 Å from PPIs, 2.0% within 3 Å from PPIs and 2.6% within 6 Å, which could explain about 31% of the corresponding 8.3% difference in overall cumulative fraction difference mentioned above. This also contributes to an overall “bump” in the OR shown in Fig. 3C. Notice that the risk of being disease-associated is about 30% higher in terms of OR when a mutation is observed in a ligand-binding site within 3 Å from a PPI.
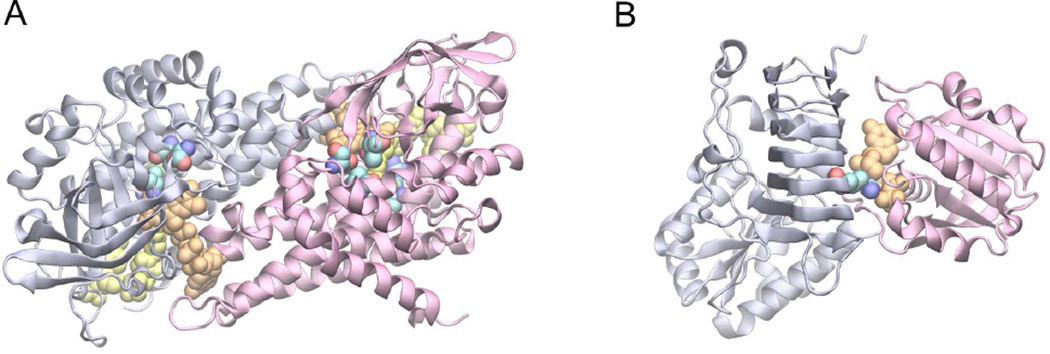
Fig. 4 shows two examples of disease-associated mutations located at/near PPIs. Medium-chain acyl-CoA dehydrogenase (ACADM) is one of four enzymes involved in fatty acid catabolism (Lee et al., 1996). The enzyme is responsible for the α,β-dehydrogenation of fatty acyl-CoA derivatives. The structure of this protein is a dimer of dimers. Fig. 4A shows the structure of the basic dimer unit, in which the co-factor FAD (Flavin-adenine dinucleotide) is sitting at the dimeric interface, whereas the fatty substrate is located close by. There are five known mutations located in the binding sites of these substrates within 5 Å. They are linked to ACADMD (Acyl-CoA dehydrogenase medium-chain deficiency), a disease that can cause sudden death of infants (OMIM access ID: 201450).
Fig. 4.
Examples of disease-associated mutations observed in the neighborhood of PPIs. (A) Dimeric medium-chain acyl-CoA_dehydrogenase. (B) Retinitis pigmentosa 2 and Arl3. In each snapshot, protein structures are shown in light blue and purple cartoon representations, small molecules are shown in orange and yellow Van der Waals, VDW, representations, and reference amino acids at the mutation sites are also shown in VdW representations where carbon, oxygen and nitrogen atoms are colored in cyan, red, and blue, respectively.
One important benefit of an interfacial pocket formed adjacent to PPIs is that it could function as a molecular switch controlled by the association/dissociation of the protein complex. This is illustrated in an example involving two proteins, Retinitis pigmentosa 2 (RP2) and the small G protein Arf-like 3 (Arl3), which form a complex providing a GTP binding pocket at the PPI (Fig. 4B) (Veltel et al., 2008). It is thought that this is a molecular switch for regulating the ciliary process in photoreceptor cells. Mutations in RP2 that disrupt ligand-binding at the PPI, such as the Arginine finger R118, may cause X chromosome linked eye disease (OMIM ID: 312600).
Post-translational modifications and disease association
Post-translational modifications play essential roles in many biological processes. We examined how many disease-associated mutations are located at such a site compared to neutral mutations. From the UNIPROT knowledge base, we search for experimentally validated modification sites. We only found 27 cases in the disease mutation set and 11 cases in the neutral set, which gives a p value of 0.1 (Fisher’s exact test, two-tailed). The most common modifications are glycosylation, 9 in the disease mutations and 4 in the neutral mutations. If we drop the requirement of having solved structures, we are able to match 77 of 23,846 disease mutations and 68 of 37,687 neutral mutations, which lead to a significant p value of 5.9×10−4. As one would expect, mutations at post-translational modification sites are significantly more likely to be associated with disease. However, the cases that can be explained by such mutations are very small to the best of our knowledge.
Amino acid changes and disease association
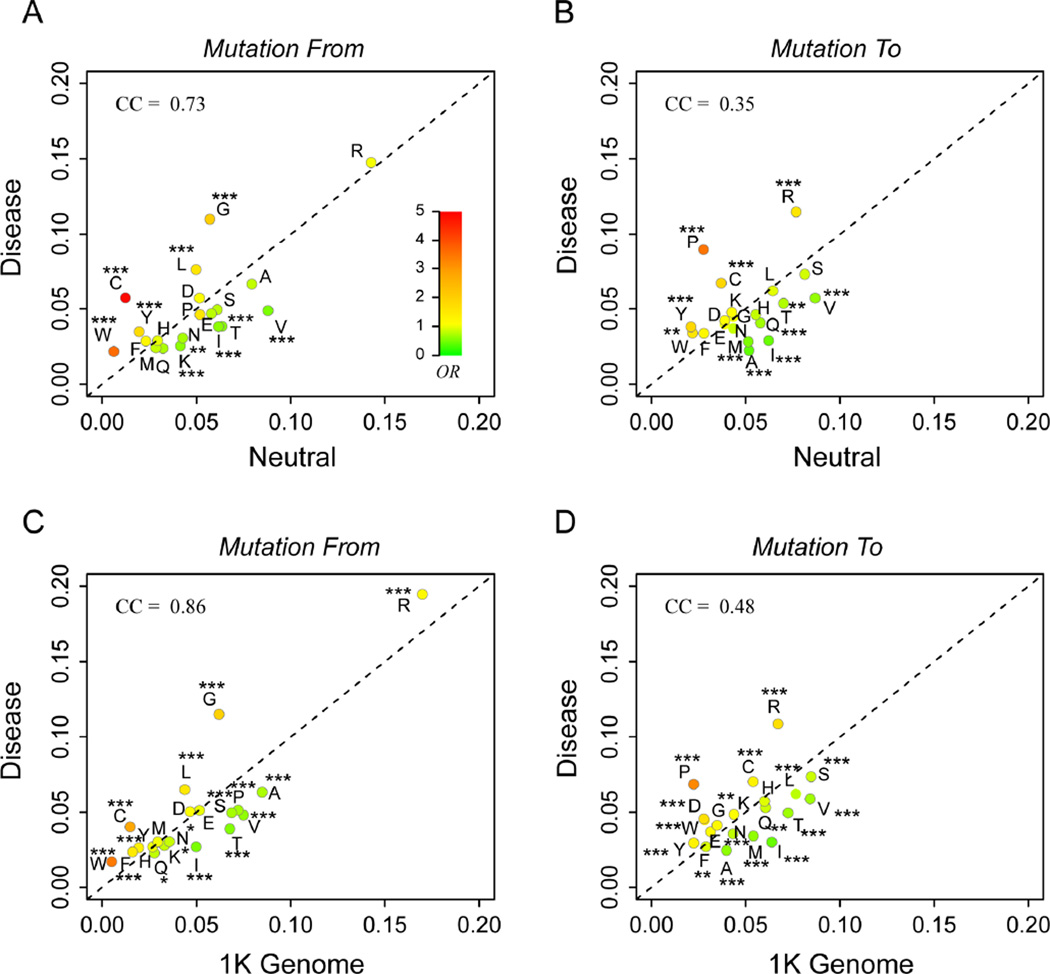
Finally, we ask the question whether there are certain types of amino acids changes that are more likely to be disease-associated. Fig. 5A shows the frequency of amino acid types from the original reference amino acid at the mutation sites. Arginine is most common mutation in both disease and neutral sets, due to the fact that four of the six Arginine codons contain the CpG dinucleotide that is relatively vulnerable to mutations (Cooper and Youssoufian, 1988). However, because the frequencies of Arginine mutations in these two sets are very close, 14.8% versus 14.3%, it is not a good indicator for predicting disease-association. In terms of OR, mutation from CYS is highest at 4.86, followed by TRP at 3.58, and Glycine at 2.04. On the other hand, mutations involving certain hydrophobic residues, such as Valine and Isoleucine are significantly underrepresented in disease mutations. One explanation is that Valine and Isoleucine have similar physico-chemical properties and are very close in the codon space, with only a single nucleotide difference (GUA, GUC, GUU of Valine and AUA, AUC, AUU of Isoleucine).
Fig. 5.
Composition analysis of disease-associated mutations versus neutral mutations from the (A&B) UNIPROT database and the (C&D) 1000 genomes database. Amino acids are labeled by their one-letter names. The color of each type of amino acid represent ORs of disease mutations versus neutral ones. The same color scale of OR shown as the insert in (A) is adopted throughout all other panels. Statistical significance of association is indicated by stars in the same way as in Fig 2. CC is the Pearson correlation coefficient. The dashed line represents an OR of 1.
Fig. 5B shows the frequencies of amino acids to different mutated residues. Statistical analysis suggests that mutation to PRO is most likely damaging with an OR of 3.46, followed by CYS at 1.87. It is interesting to note that the frequencies of disease causing mutations show a much lower correlation to frequencies of neutral mutations when one compares mutated-to AAs versus mutation-from AAs. The corresponding Pearson correlation coefficients are 0.35 versus 0.73. This suggests that the mutation-from frequencies are largely determined by similar reasons, for example, distribution of codons and distances in the codon space (i.e., those with one nucleotide difference are of high possibilities), whereas mutation-to frequencies are largely determined by reasons very different between the disease and neutral sets.
The 1000 Genomes Project has revealed a large set of variants in about 1000 healthy individuals (Abecasis et al., 2012). One could argue that these variants are a good neutral background for the purpose of our analysis. We repeated the same composition analysis using the data from the 1000 Genomes Project. As shown in Fig. 5C&D, we obtained very similar results as those using the UNIPROT neutral set. Mutations from and to CYS give an OR of 2.79 and 1.33 respectively; mutations from TRP have an OR of 3.58; and the OR of mutations to PRO is 3.21. These are also mutations that are most likely associated with diseases than neutral mutations.
We then seek structural explanation for the top three most deleterious types of mutations involving CYS, PRO, and TRP amino acids. First, we ask why mutations from and to a CYS residue are harmful? Since it is known that CYS often form disulfide bonds, it is hypothesized that a CYS mutation either disrupts useful disulfide bonds in the case of mutation from or introduces an unwanted disulfide bond in case of mutation to. To test this hypothesis, we examined if there are CYS disulfide bonds in the corresponding protein’s structure. In the mutation from set, about 2/3 of CYS mutations are likely to form a disulfide bond with another CYS residue nearby within 4.5 Å, whereas only about 1/3 of CYS mutations in the neutral set have another CYS nearby. This gives a highly significant p value of 3.2×10−6. Therefore, disease-associated CYS mutations are more likely to disrupt an original disulfide bond important for protein stability or function. On the other hand, in the mutation-to data sets, we found that after the mutation to CYS, there are 45 cases where there is another CYS nearby in the disease set, versus only 7 cases in the neutral set (p = 0.007). In these cases, the CYS mutation may introduce an unwanted disulfide bond that could lead to protein malfunction.
Second, why is a mutation to PRO likely to be harmful? PRO is a unique amino acid that does not have an amine hydrogen for backbone hydrogen bond formation. In addition, it has a more restricted dihedral angle space than a typical amino acid. This restriction often creates a kink in helical structures at the position of a PRO. As a result, mutations to PRO often disrupt helical structures. We performed an analysis of the secondary structure where a mutation to PRO occurs, with the result shown in Table 2. The most common PRO mutation in the disease set lies within a 310 helix, about 36% percent, in contrast to 23% cases in the neutral set (p = 0.006). The second common secondary structural element where a PRO mutation occurs is a turn; 26% in disease versus 18% in neutral (p = 0.1) sites, where a PRO mutation might disrupt hydrogen bonding. By contrast, in the coil region where there is no ordered secondary structure, a PRO mutation is much less likely present at 20%, in comparison to 32% in the neutral set (p = 0.009). The result supports the conclusion that a PRO mutation’s disruptive presence in the secondary structure is often disease-associated.
Table 2.
Analysis of secondary structure location of mutations to Proline.
| Type | Disease | Neutral | Pa |
|---|---|---|---|
| 310 helix | 195 (0.36) | 29 (0.23) | 0.003 |
| α-helix | 21 (0.039) | 7 (0.056) | – |
| Turn | 139 (0.26) | 23 (0.18) | 0.049 |
| β-sheet | 38 (0.071) | 11 (0.088) | – |
| Bend | 35 (0.065) | 15 (0.12) | – |
| Coil | 110 (0.20) | 40 (0.32) | 0.005 |
| Total | 538 | 125 |
Note:
Insignificant P-value is indicated by –.
Lastly, we investigate mutation from TRP that is often linked to disease. TRP is the largest amino acid that plays an important role in protein folding and stability. On average, a TRP residue makes 8.5 side chain contacts versus 6.0 side chain contacts of other types of amino acids. Further calculation estimates that a TRP mutation in the disease set leads to an mean increase of the free energy of protein folding ΔΔG of 3.4 kcal/mol (126 cases, SD = 1.61 kcal/mol), versus 2.9 kcal/mol (27 cases, SD = 1.36 kcal/mol) of TRP mutations in the neutral set (p = 0.076, t-test, two-tailed). This analysis suggests that mutations from TRP likely destabilize the protein’s structure and result in less fitness. In the vast majority of cases analyzed, this destabilization likely has a significant impact on the function of protein, thus leading to various disease conditions.
Discussion
Since the vast majority of disease-associated mutations are assigned based on statistical analysis, it is not clear whether a mutation is actually the cause of the implicated disease. To provide a better understanding of mutations from a structural prospective, we performed a large-scale analysis of 6,025 disease-associated mutations in 642 proteins. We found that about 20% of mutations are buried in protein cores and that these mutations may destabilize protein structures; whereas about 11% of mutations are involved in ligand-binding, and another 10% are involved in protein-protein interactions. About 17% are in pockets, 3% involve ion binding and 1% of DNA/RNA-binding. These numbers are consistent with previous studies conducted on smaller data sets (Steward et al., 2003; Sunyaev et al., 2000). Using computational approaches, we further predict about another 8% of mutations are likely ligand-binding or essential for enzyme function. Together, we find a structural/functional annotation for about 70% of mutations. As more experimental structures are determined for functional complexes of proteins, e.g., in complex with other ligand or proteins for their function, we expect to see more mutations explained by the functional roles of the mutated residues.
In order to demonstrate the usefulness of the structural/functional data, one needs to compare disease-associated mutations to neutral mutations. This is largely ignored or not thoroughly pursued in previous studies. Here, we show a statistically significant difference between disease-associated and neutral mutations. As pointed out previously (Ng and Henikoff, 2006), one of most effective descriptors is the degree of surface exposure of the protein residue at the mutation site. Buried residues in the disease set have the second highest OR at 2.66 compared to neutral ones, whereas exposed residues not involving any interactions or pocket-like feature are more likely to be neutral at an OR of 0.50. Functional sites, such as ligand-binding sites and protein-protein interface residues, all have significant ORs at 1.53 and 1.23. However, they are not as effective as one naïvely expects. One reason might be due to purification selections, which removed many deleterious, fatal mutations at these functional sites. It is also worth to mention that computational predictions achieved good performance. Predictions by FINDSITEcomb and EFICAz yield OR values at 1.75 and 16.9, respectively, which is comparable or better than results obtained by counting known ligand-binding sites from experimental structures. Most interestingly, it is the predicted loss of enzymatic function that is most strongly disease associated. These are encouraging results that demonstrate the potential of computational, knowledge-based methods.
One novel observation is that mutations adjacent to protein-protein interfaces are more likely associated with disease than mutations at the PPIs themselves. At 1.75, the OR is highest about 3Å from PPIs, which is about 40% higher than the OR at PPIs. There are two contributing factors. One is that the mutations at buried sites within each monomer but close to the PPIs are more likely to be disease-causing, presumably by destabilizing the protein complex. The second is due to the presence interfacial pockets, which are ligand-binding pockets at or adjacent to PPIs (Gao and Skolnick, 2012). Mutations within interfacial pockets could disrupt functionally important ligand-protein interactions. This is supported by subsequent analysis, which shows that mutations within interfacial pockets have more than double the OR of mutations at PPIs.
Although amino acid type change is one main factor for assessing pathogenicity of human mutations in many automatic annotation tools (Adzhubei et al., 2010; Kumar et al., 2009), in-depth analysis of why it works is somewhat lacking, though there have been studies about the frequencies of disease-associated mutations (de Beer et al., 2013; Vitkup et al., 2003). Obviously, if the mutation involves very different amino acid types, e.g., from hydrophobic to charged residues, it is more likely to cause an issue than mutations among hydrophobic residues. Ideally, one would like to study all 380 pairs of non-synonymous mutations. However, the number of known disease-causing mutations is currently too limited for such a study. We opt instead to focus on each type of amino acids, grouped by mutations from and mutation to a given amino acid type. We found that certain types of mutations, such as mutations involving CYS, TRP, or PRO, are more likely to be disease-associated. Through structural analysis, CYS mutations often involve breaking disulfide bridges or forming unwanted disulfide bonds; TRP mutations usually significantly destabilize structures, and mutations to PRO tend to disrupt helical structures. These analyses provide some examples of how structural analysis could provide further insights into the mechanisms of disease.
Methods
Data set
From the Aug 2013 release of UNIPROT knowledge base (Wu et al., 2006), we collected 23,846 disease-associated variants and 37,782 polymorphisms in human, the latter we assume are neutral mutations. The classification was manually assigned according to the literature and a previously curated database such as OMIM (McKusick, 2007). These variants are all missense mutations with changes of amino acid type. Unclassified variants were ignored. From this collection, we further selected those with at least one experimentally determined structure deposited in the PDB (Berman et al., 2000). We verified that the protein structure contains the same amino acid at the position indicated in the UNIPROT mutation data. To avoid possible bias, if the gene contains more than 50 variants, we randomly selected no more than 50 variants in each gene. If multiple variants are found corresponding to the same residue position in a gene, we also randomly selected only one variant. This procedure yielded 6,025 disease-associated mutations in 642 proteins, and 4,536 neutral mutations in 1,743 proteins. These two sets are the main data set used in our study. They share 355 common proteins with 4,209 disease mutations and 1,113 neutral mutations.
Data of the 1000 Genomes Project variants are from reference (de Beer et al., 2013). It should be noted that the counts of mutations from OMIM are mislabeled in that study. The counts of mutations from were mistaken as mutation to, and vice versa. The mistake in (de Beer et al., 2013) gives incorrect amino acid mutation frequencies which are inconsistent with a previous study (Vitkup et al., 2003), whereas our results are in agreement with (Vitkup et al., 2003).
Structural and Functional Analysis
Using the experimental structures, we map the location of each mutation. For each PDB record, we analyzed the structure given by the basic asymmetric unit, as well as author assigned biounit if it contains protein-protein complexes. The HET code in each PDB header was used to determine the types of ligand, i.e., metal-ion, small-molecule ligand, or DNA/RNA. Note that this analysis does not guarantee that a ligand has biologically relevant interaction with its co-crystalized protein. Nevertheless, in many cases, biologically relevant molecules recognize the same binding sites on proteins as other non-biological molecules. Atomic contacts between protein and ligand were determined by the program LPC (Sobolev et al., 1999). If the original amino acid at the mutation site contains at least one heavy atom making physical contact with a ligand, it is assigned as ligand-binding. Similarly, protein-protein interface residues are defined by a heavy-atom distance of 4.5 Å. The distance between a variant residue and PPI is defined as the shortest heavy-atom distance between the residue and any PPI residue. If a protein has multiple complex structures, we chose the minimal distance among all these complexes. Solvent Accessible Surface Area (SASA) was calculated for each original residue of the mutations using the program NACCESS (Hubbard and Thornton, 1993). If a residue has less than a 1% relative SASA percentage, it is defined as a buried residue. Otherwise, the residue is an exposed surface residue. Again, if the residue is observed in multiple structures, we employed the lowest relative SASA value. We also ran FINDSITEcomb (Zhou and Skolnick, 2013) to annotate computationally ligand-binding sites, and EFICAz (Kumar and Skolnick, 2012) to predict Functional Discriminating Residues for predicted enzymes. Calculations of the free energy differences, ΔΔG, of TRP mutations were conducted using the program DMutant, which is based on statistical potentials (Zhou and Zhou, 2002) and ranked among the top performers in an independent assessment (Khan and Vihinen, 2010). Secondary structure analysis was carried out using DSSP (Kabsch and Sander, 1983).
Statistical Analysis
With a few noted exceptions, a Fisher’s exact test was conducted on the contingency table given in Table 3. A positive case is a variant containing a certain feature, such as ligand binding, or a mutation from a CYS residue. A negative case is a variant without such a feature. Outcomes are either disease-associated or neutral. The counts of positive/negative cases with disease are denoted as dpos/dneg, respectively, and similarly, npos/nneg, for positive/negative cases for neutral ones. Odds are calculated for positive and negative cases separately, and their ratio yields the OR.
Table 3.
Contingency table for the statistical test.
| Disease | Neutral | Odds | |
|---|---|---|---|
| Positive | dpos | npos | dpos/npos |
| Negative | dneg | nneg | dneg/nneg |
| Total | 6,025 | 4,325 | – |
Supplementary Material
Highlights.
A comparative structural analysis of disease-associated and neutral mutations
Assessment of structural regions most vulnerable to disease mutations
Mutations adjacent to protein-protein interfaces are strongly disease associated
Compositional analysis in comparison to mutations from the 1K Genomes Project
Acknowledgements
This work is supported by grant No GM-37408 of the Division of General Medical Sciences of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Availability. The data sets are available as supplementary material and also online at http://cssb.biology.gatech.edu/nssnp.
References
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I, Schmidt S, Peshkin L, Ramensky V, Gerasimova A, Bork P, Kondrashov A, Sunyaev S. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Lander ES, Rogers J, Waterston RH International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet. 1988;78:151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- de Beer TA, Laskowski RA, Parks SL, Sipos B, Goldman N, Thornton JM. Amino acid changes in disease-associated variants differ radically from variants observed in the 1000 genomes project dataset. PLoS Comput Biol. 2013;9:e1003382. doi: 10.1371/journal.pcbi.1003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Skolnick J. The distribution of ligand-binding pockets around protein-protein interfaces suggests a general mechanism for pocket formation. Proc Natl Acad Sci USA. 2012;109:3784–3789. doi: 10.1073/pnas.1117768109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SJ, Thornton JM. 'NACCESS', Computer Program, Department of Biochemistry and Molecular Biology. University College London: 1993. [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure - pattern-recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Khan S, Vihinen M. Performance of protein stability predictors. Hum Mutat. 2010;31:675–684. doi: 10.1002/humu.21242. [DOI] [PubMed] [Google Scholar]
- Kumar N, Skolnick J. EFICAz2.5: application of a high-precision enzyme function predictor to 396 proteomes. Bioinformatics. 2012;28:2687–2688. doi: 10.1093/bioinformatics/bts510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng P. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Wang M, Paschke R, Nandy A, Ghisla S, Kim JJ. Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity. Biochemistry. 1996;35:12412–12420. doi: 10.1021/bi9607867. [DOI] [PubMed] [Google Scholar]
- McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. Am J Hum Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies - the next generation. Nature reviews Genetics. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annual review of genomics and human genetics. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Ball E, Howells K, Phillips A, Mort M, Cooper DN. Human Gene Mutation Database: towards a comprehensive central mutation database. J Med Genet. 2008;45:124–126. doi: 10.1136/jmg.2007.055210. [DOI] [PubMed] [Google Scholar]
- Steward RE, MacArthur MW, Laskowski RA, Thornton JM. Molecular basis of inherited diseases: a structural perspective. Trends in genetics : TIG. 2003;19:505–513. doi: 10.1016/S0168-9525(03)00195-1. [DOI] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends in genetics : TIG. 2000;16:198–200. doi: 10.1016/s0168-9525(00)01988-0. [DOI] [PubMed] [Google Scholar]
- Veltel S, Gasper R, Eisenacher E, Wittinghofer A. The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3. Nat Struct Mol Biol. 2008;15:373–380. doi: 10.1038/nsmb.1396. [DOI] [PubMed] [Google Scholar]
- Vitkup D, Sander C, Church GM. The amino-acid mutational spectrum of human genetic disease. Genome biology. 2003;4:R72. doi: 10.1186/gb-2003-4-11-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Moult J. SNPs, protein structure, and disease. Hum Mutat. 2001;17:263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang HZ, Lopez R, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Skolnick J. FINDSITEcomb: a threading/structure-based, proteomic-scale virtual ligand screening approach. J Chem Inf Model. 2013;53:230–240. doi: 10.1021/ci300510n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HY, Zhou YQ. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.