Abstract
The bacterial family Rickettsiaceae includes a group of well-known etiological agents of many human and vertebrate diseases, including epidemic typhus-causing pathogen Rickettsia prowazekii. Owing to their medical relevance, rickettsiae have attracted a great deal of attention and their host-pathogen interactions have been thoroughly investigated. All known members display obligate intracellular lifestyles, and the best-studied genera, Rickettsia and Orientia, include species that are hosted by terrestrial arthropods. Their obligate intracellular lifestyle and host adaptation is reflected in the small size of their genomes, a general feature shared with all other families of the Rickettsiales. Yet, despite that the Rickettsiaceae and other Rickettsiales families have been extensively studied for decades, many details of the origin and evolution of their obligate host-association remain elusive. Here we report the discovery and single-cell sequencing of ‘Candidatus Arcanobacter lacustris', a rare environmental alphaproteobacterium that was sampled from Damariscotta Lake that represents a deeply rooting sister lineage of the Rickettsiaceae. Intriguingly, phylogenomic and comparative analysis of the partial ‘Candidatus Arcanobacter lacustris' genome revealed the presence chemotaxis genes and vertically inherited flagellar genes, a novelty in sequenced Rickettsiaceae, as well as several host-associated features. This finding suggests that the ancestor of the Rickettsiaceae might have had a facultative intracellular lifestyle. Our study underlines the efficacy of single-cell genomics for studying microbial diversity and evolution in general, and for rare microbial cells in particular.
Introduction
The Rickettsiales are an order within the Alphaproteobacteria that comprise obligate intracellular endosymbionts of arthropods and mammals. They include the causative agents of many mild to severe diseases in humans and other animals, for example, epidemic and scrub typhus (Andersson et al., 1998; Cho et al., 2007), ehrlichiosis (Dunning Hotopp et al., 2006) heartwater (Collins et al., 2005) and anaplasmosis (Brayton et al., 2005). Their genomes are typically small (⩽2.1 Mb) due to reductive evolution and have an AT-rich composition (Darby et al., 2007). Among the four taxonomic families that are recognized within the Rickettsiales (Anaplasmataceae, Rickettsiaceae, Ca. Midichloriaceae and Holosporaceae), the most well known are the Rickettsiaceae, for they are human pathogens. They include Rickettsia prowazekii (epidemic typhus), Rickettsia rickettsia (Rocky Mountain spotted fever), Rickettsia typhi (murine typhus) and Orientia tsutsugamushi (scrub typhus) all of which are transmitted by hematophagous arthropods. Owing to the wide variety of diseases they are responsible for, genome sequencing efforts have so far focused on the pathogenic Rickettsia and Orientia genera, whereas other genera that are not carried by hematophagous hosts, also referred to as ‘Neglected Rickettsiaceae' (Schrallhammer et al., 2013) have largely been ignored (Merhej and Raoult, 2011). Currently, genomic sequences are available for 57 Rickettsia and 2 Orientia strains, but none for other genera of Rickettsiaceae even though 157 small subunit (SSU) rRNA sequences of uncultured members are available for them in the SILVA database ((Quast et al., 2012); release 119). Obviously, this apparent unbalance in taxon sampling is detrimental for our understanding of their evolution and emerging pathogenicity. With the advent of methodologies that allow the characterization of genomic sequences without the need for cultivation, the opportunity has arisen to explore the Rickettsiaceae diversity in an unbiased manner and gain a significantly better understanding of their evolution. In this work, we have used such a cultivation-independent approach and obtained and sequenced a single-cell amplified genome (SAG) from a novel alphaproteobacterium sampled from Damariscotta Lake (Martinez-Garcia et al., 2012). This alphaproteobacterium, for which we propose the name ‘Candidatus Arcanobacter lacustris' (from here on referred to as A. lacustris), represents a novel Rickettsiales lineage with a deep sister relationship to the Rickettsiaceae. By applying comparative and phylogenomic analyses, we provide a unique insight into the evolution of the Rickettsiaceae, and about the nature and abundance of this novel and rare alphaproteobacterial lineage.
Materials and methods
Single-cell sorting, lysis and whole-genome amplification
In an attempt to explore alphaproteobacterial diversity, a single-cell genomics pipeline was applied on an environmental freshwater sample obtained from Damariscotta Lake (USA; 28 April 2009) as described by (Martinez-Garcia et al., 2012). In brief, the single cell was obtained with fluorescent-activated cell sorting of the sample and subsequently lysed. Extracted DNA was subjected to real-time multiple displacement amplification (MDA). The MDA product was then verified by SSU rRNA qPCR. A second round of MDA (Swan et al., 2011) was required to obtain sufficient quantity of genomic DNA for Illumina sequencing. All above steps were carried out at the Bigelow Laboratory Single Cell Genomics Centre (East Boothbay, ME, USA).
Whole-genome sequencing, assembly and annotation
A short-insert paired-end TruSeq library (Illumina, San Diego, CA, USA) was prepared according to the manufacturer's prescriptions. The library, with insert size of ~400 bp, was sequenced on an Illumina HiSeq2000 instrument (Illumina), yielding a total of 9 171 854 read pairs, with each read being 100 bp long. Raw reads are deposited at NCBI's SRA under study number SRP055079. Before assembly, the quality of the sequence data were assessed with FastQC 0.10.1 (Andrews, 2012), and showed an average Phred Score per read of 38. The reads were assembled de novo using SPAdes 2.4 (Nurk et al., 2013) in single-cell mode (‘—sc') with default k-mer sizes (21, 33, 55), including read error correction and mismatch- and indel-correction steps. Resulting contigs were filtered in three steps. First, all contigs smaller than 200 bp or with an average k-mer coverage lower than 10 were removed. Second, ORFs were predicted with Prodigal 2.50 (Hyatt et al., 2010) and classified at Domain level by MEGAN 4.7 (Huson et al., 2011) based on a BLASTP search (Altschul et al., 1990) versus NCBI's nonredundant database (nr). All contigs in which less than a third of the predicted ORFs were classified as bacterial were removed. Finally, contigs were checked for contig edges that were inverted repeats relative to each other. These repeats are the result of ‘self-priming', a known MDA artifact (Lasken and Stockwell, 2007). In case such repeats were detected, the 5′ end of the contig was trimmed, accordingly. The remaining contigs were annotated using Prokka 1.11 (Seemann, 2014) using the ‘—rfam' flag and RNAmmer (Lagesen et al., 2007) as RNA predictor. In addition, Prokka was modified to allow prediction of partial ORFs on contig edges. Eukaryotic domains in predicted proteins were identified using the lists of eukaryotic-like domains available at effectors.org (Jehl et al., 2011). Putative type IV secretion system components and effectors were identified by a BLASTP search of the SecReT4 (Bi et al., 2013, 4) ‘T4SS components', ‘Experimentally verified effectors' and ‘T4SS effectors' databases. The genome has been deposited as a whole-genome shotgun project at DDBJ/EMBL/GenBank under the accession JYHA00000000. The version described in this paper is version JYHA01000000.
Estimation of genome completeness and genome size
Genome completeness and consequently genome size were estimated as follows. HMM profiles of the 129 Rickettsiales panorthologous marker genes (see Supplementary Methods) were constructed with HMMER3 (Eddy, 1998) and used to search the predicted proteome. In earlier work (Rinke et al., 2013), an ‘unweighted' completeness estimate would be obtained by dividing the number of present marker genes by the total number of marker genes. Thus, each present marker gene would contribute equally to the completeness estimate, thereby assuming that all marker genes are spread evenly in the genome. A better, ‘weighted' estimate is obtained when weighing each marker gene with the median genomic distance to the closest marker gene in the data set (unpublished observation). For example, ribosomal protein genes, which are generally organized in operons, have a lesser individual weight than other genes that are generally more evenly spread out in the genome. Here for each of the 20 Rickettsiales genomes (see Figure 1; except for draft genomes Holospora undulata and Ca. Odyssella thessalonicensis) we measured the distance of all present 129 Rickettsiales panorthologous markers in one genome to its closest upstream and downstream neighbors, normalized distances by the genome size and used the normalized median distance recorded for each marker as a weight to estimate how clustered this marker generally is. Thereby, marker genes that tend to be located near other marker genes get a lower weight than marker genes that are relatively isolated. The weights were normalized again by dividing them by the sum of all weights, so that a genome containing all markers would have a completeness estimate of 1. Finally, the completeness estimate for A. lacustris was obtained by taking the sum of all normalized weights of marker genes found in this genome. The genome size was then estimated by dividing the total assembly length by the completeness estimate.
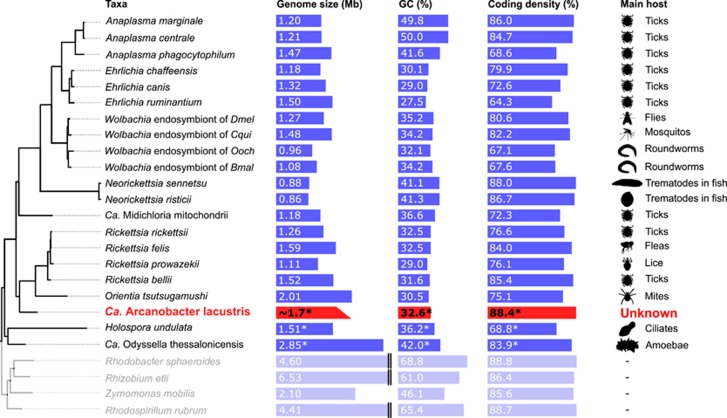
Figure 1.
A comparison of genome characteristics and host species of Rickettsiales for which genomic sequences are available. The widths of the bars that represent the genome size, GC% and coding density are scaled. Asterisks (*) indicate draft genomes. The tree shown on the left is the same as the PhyloBayes genome tree (see Figure 2).
Phylogenetic analyses
Unless otherwise stated, all alignments were made with MAFFT 7.050b (Katoh and Standley, 2013) using the local pair option (mafft-linsi) and trimmed with trimAl 1.4 (Capella-Gutiérrez et al., 2009), removing sites for which more than half of the taxa contained a gap. Maximum likelihood (ML) phylogenies were inferred with RAxML 7.2.8 (Stamatakis, 2006), using 100 rapid bootstraps under the Γ model for rate heterogeneity among sites with the GTR substitution matrix for nucleotide alignments and the LG substitution matrix (Le and Gascuel, 2008) for protein alignments. Bayesian phylogenies (BI) were inferred with PhyloBayes MPI 1.4f (Lartillot et al., 2013) using four MCMC chains under the CAT-Poisson model. The log likelihood, total tree length, α-parameter and number of categories of all trees per chain were traced and visually inspected to choose the burn-in cutoff. Whenever convergence (maxdiff<0.1) between chains was not fully achieved, the topology of individual chain trees were examined to ensure that they were congruent for critical nodes. Trees were drawn with FigTree 1.4 (Andrew Rambaut, http://tree.bio.ed.ac.uk/software/figtree/). Additional details regarding the phylogenetic analyses that were carried out in the present study are available in the Supplementary Methods.
Genome content comparison
The occurrence of KEGG Pathways (Kanehisa et al., 2014) in A. lacustris and members of the Rickettsiaceae were compared as follows: first, the prodigal predicted (partial) protein sequences from A. lacustris and the protein RefSeq accessions from Rickettsia prowazekii Madrid E, Rickettsia belli RML369 and Orientia tsutsugamushi Ikeda were used in a BLASTP search (E-value cutoff: 10−5) of NCBI nr. The BLASTP results were then imported into MEGAN-5.1.5 and the occurrence of all KEGG Pathways were visually compared.
The Rickettsiales-specific clusters of orthologous groups (rickCOGs; see Supplementary Methods) served as a basis to compare the genome content of A. lacustris with the Anaplasmataceae, Rickettsiaceae, Holosporaceae and Ca. Midichloria mitochondrii; see Figure 2 for family attribution). A Venn diagram was constructed using the R package VennDiagram (Chen and Boutros, 2011) to visualize how many rickCOGs were unique for A. lacustris or each of the Rickettsiales family and how many were common between two or more families. Each rickCOG was considered as unique to a given family if it contained a sequence from at least one taxon that belonged to that family and contained no sequences from other families. A rickCOG was considered as common between two or more families if it contained sequences from at least one taxon of each of those families and contained no sequences from the other remaining families. Outgroup taxa (see Figure 2) were not considered in this analysis.
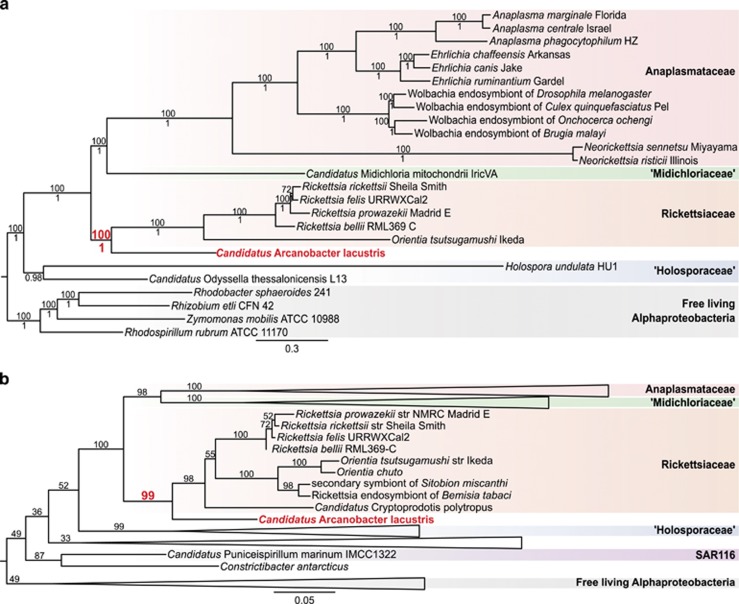
Figure 2.
Phylogenetic placement of A. lacustris. A. lacustris and branch support values on branches of interest have been marked with bold red. (a) Phylogenetic tree based on a concatenated alignment of 64 Rickettsiales panorthologs. Tree topology and posterior probability branch support values (shown below branches) were inferred using PhyloBayes (model: CAT-Poisson). Bootstrap branch support values (shown above branches) were calculated with RAxML (model: Γ+LG). Separate full RAxML and PhyloBayes phylogenetic trees are shown in Supplementary Figure 1. (b) Phylogenetic tree based on SSU rRNA gene. Tree topology and bootstrap support values (shown above branches) were inferred with RAxML. Major taxonomic groups have been collapsed to improve readability. Full phylogenetic tree is shown in Supplementary Figure 2.
To annotate the rickCOGs that were unique for A. lacustris and uniquely shared with specific Rickettsiales families, a BLAST analysis was performed. rickCOGs were aligned with MAFFT and resulting alignments were used as query to search nr with PSIBLAST (E-value cutoff: 10−10; Altschul et al., 1997). If a rickCOG only contained a single sequence, it was used directly as query to search nr with BLASTP (E-value cutoff: 10−5). The protein identity of the rickCOG was then called based on the best PSI-BLAST or BLASTP hit.
Estimation of environmental abundance
The metagenomic data sets of the freshwater lakes Damariscotta (spring and summer), Mendota (spring and summer), Sparkling (spring and summer; Martinez-Garcia et al., 2012), Ekoln, Erken and Vattern (Zaremba-Niedzwiedzka et al., 2013) were used to estimate the relative abundance of A. lacustris and 10 LD12 SAGs in freshwater lakes (Zaremba-Niedzwiedzka et al., 2013) by recruiting the metagenomic reads to their contigs with NUCmer (Kurtz et al., 2004). Detected reads were filtered based on identity (⩾80%), alignment length (⩾100 bp and ⩾90% of read length). Obtained read counts were corrected for genome completeness estimates (calculated for the LD12 SAGs as described above by using 139 well conserved bacterial marker genes (Rinke et al., 2013)), and subsequently used to calculate abundance relative to total metagenome size. Abundances of the LD12 SAGs relative to A. lacustris were calculated per metagenome by dividing their completeness-corrected read counts.
Small subunit rRNA amplicon data sets were screened using an approach described by (Lagkouvardos et al., 2014). In brief, all raw SSU rRNA amplicon sequence data from environmental samples in the databases SRA (June 2013) (Kodama et al., 2012) and VAMPS (‘Not Normalized' September 2014; Huse et al., 2010); http://vamps.mbl.edu/) were extracted and organized by sample in independent data sets, retaining sample-associated metadata. The databases were searched using BLAST and the full-length 16S rRNA gene sequence of A. lacustris and LD12 (accession no. Z99997.1) as query. The detected amplicon reads were filtered with respect to size (⩾200 nucleotides), alignment length (⩾80% of read length) and identity (⩾95%). For those data sets that contained A. lacustris hits, abundances relative to amplicon data set size were calculated. For detected data sets that contained hits for both organisms, the relative abundance of LD12 compared with A. lacustris was calculated by dividing the LD12 hit count with the A. lacustris hit count.
Results
General features of the single-cell amplified genome
With the aim of exploring genetic diversity of environmental Alphaproteobacteria, we applied a single-cell genomics pipeline on an environmental freshwater sample obtained from Damariscotta Lake (USA; sampled in April 2009). Because single-cell sequence data are difficult to assemble owing to extreme sequence depth bias, contamination sensitivity, potential chimeric sequences and other MDA artifacts, we employed SPAdes (Nurk et al., 2013), an assembler that is specifically designed to handle such data and checked the resulting assembly thoroughly for contamination and MDA artifacts.
The resulting assembly of the SAG consists of 151 contigs comprising a total size of 822 563 bp with an average GC content of 32.6% and coding density of 88.4% (Table 1). A total of 882 protein-coding genes were identified, of which 45 were annotated as repeat-containing proteins (transposases, ankyrin repeat proteins, integrases and so on). On the basis of a weighted single-copy gene count, we estimate a completeness of 48% and consequently predict a genome size of 1.7 Mb. SAGs are generally incomplete and a completeness estimate of 50% is considered typical. Compared with other members of the Rickettsiales, A. lacustris has a relatively large genome, an average GC content and high coding density (Figure 1).
Table 1. A. lacustris draft genome features.
| Feature | Value |
|---|---|
| Number of contigs | 151 |
| N50 (bp) | 12 448 |
| Total size (bp) | 822 563 |
| GC (%) | 32.6 |
| ORFs | 882 |
| rRNA | 1a |
| tRNA | 10 |
| Coding density (%) | 88.4 |
| Average intergenic space length (bp) | 99 |
| Completeness estimate (%) | 48 |
| Genome size estimate (Mb) | 1.7 |
| Transposases | 26 |
| Integrases | 9 |
| Ankyrin repeat proteins | 5 |
| Other repeat type proteins | 5 |
The SSU rRNA gene
Phylogenetic relationship to other Rickettsiales
To assess the phylogenetic relationships relative to other Rickettsiales, phylogenomic analyses based on Rickettsiales panorthologs and on SSU rDNA were performed. For the phylogenomic analysis, 12 197 rickCOGs were constructed from all proteins encoded in 20 representative Rickettsiales and four outgroup Alphaproteobacteria. From these, 129 panorthologs were extracted that were present in exactly one copy per genome in all genomes excluding A. lacustris. Out of these, 64 were present in A. lacustris (Supplementary Table 1). To assess the effect of missing data and tree reconstruction method, ML and BI phylogenies were inferred from the ‘64-panortholog' data set and the ‘129-panortholog' data set. A deep sister relationship to the Rickettsiaceae was retrieved with strong support (bootstrap support (BS):⩾94, posterior probability (PP):⩾0.99), robust to tree reconstruction methods and missing data (Figure 2a; Supplementary Figure 1). In addition, a phylogenetic analysis of 64 SSU rRNA sequences from Rickettsiales and outgroup Alphaproteobacteria was performed. A deep sister relationship to the Rickettsiaceae was also retrieved in this analysis (BS: 99; Figure 2b; Supplementary Figure 2). Despite the inclusion of more Rickettsiales taxa in the analysis, no significant affiliation with any of the established families was observed, indicating that A. lacustris represents a novel clade.
Genome content comparisons
The unique phylogenetic placement of A. lacustris as a sister clade to Rickettsiaceae allowed us to investigate the evolution of the latter family in more detail. To this end, we inferred candidate genes lost by the last Rickettsiaceae common ancestor by searching for KEGG pathways (Kanehisa et al., 2014) for which component genes were present in A. lacustris, but completely absent in Rickettsiaceae members R. prowazekii, R. bellii and O. tsutsugamushi. Pathways with the majority of the genes missing in Rickettsiaceae were the flagellar assembly (18 genes) and bacterial chemotaxis (9 genes). Other identified pathways and protein complexes include threonine metabolism (three genes; thrA, thrB and thrC), glycolysis (two genes; pgi and eno), panthothenate and CoA biosynthesis (two genes: coaD and coaX), terpenoid backbone synthesis (thiB, thiP and thiQ), the thiamine ABC transporter (three genes: tbpA, thiP and thiQ) and an antibiotics ABC-2 transporter (two genes: yadH and yadG).
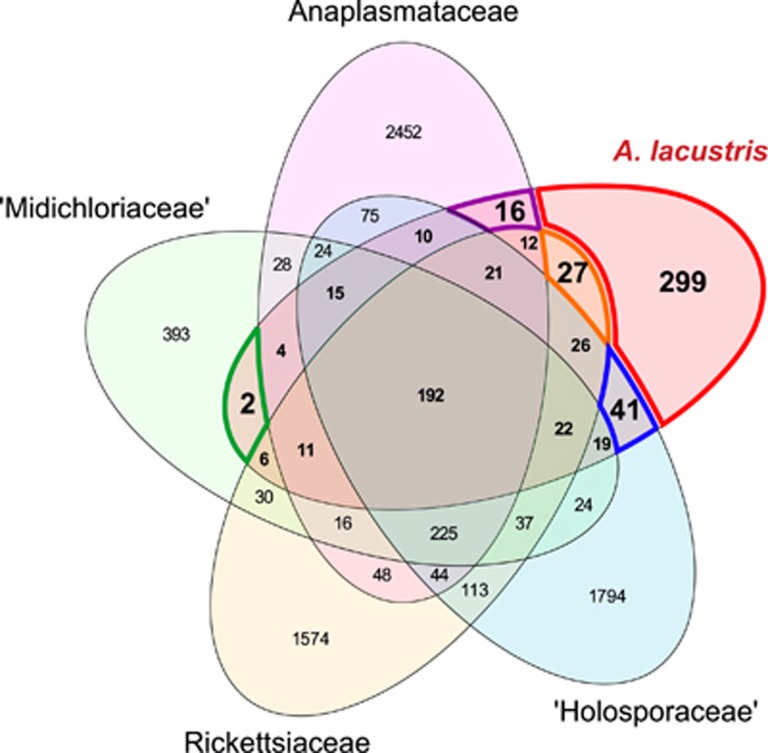
We then compared the genome content with other Rickettsiales families by utilizing the rickCOGs that were also used for the phylogenomic analyses. All A. lacustris predicted proteins (882) belonged to a total of 723 ortholog clusters. Among these, 299 (41%) were unique to A. lacustris (Figure 3), including 143 (20%) that did not have detectable homologs (E-value⩽10−6) in the NCBI nr database. Those for which homologs could be detected included 8 ortholog clusters putatively involved in heme metabolism and 14 putatively mobile elements (Supplementary Table 2). Twenty-seven ortholog clusters were uniquely shared with the Rickettsiaceae and included six putatively involved in toxin-antitoxin systems that were encoded by three gene pairs that each putatively contained one toxin and one antitoxin gene. A total of 16 ortholog clusters were uniquely shared with the Anaplasmataceae and include a DNA primase and a putative phage-related ATPase. The flagellar hook protein FlgK and a PAP2 superfamily protein were uniquely shared with ‘Midichloriaceae'. A remarkable high number (41) of ortholog clusters were shared with the ‘Holosporaceae'. These included seven chemotaxis proteins.
Figure 3.
Five-set Venn diagram depicting gene content comparison between A. lacustris and other Rickettsiales families based on Rickettsiales-specific clusters of orthologous groups. The borders of intersections of interest are marked and colored: A. lacustris unique genes (red), genes uniquely shared between A. lacustris and either Anaplasmataceae (purple), Rickettsiaceae (orange), ‘Holosporaceae' (blue) or ‘Midichloriaceae (green).
Evolutionary origins of flagellar and chemotaxis proteins
To assess whether the flagellar and chemotaxis genes were lost in the last Rickettsiaceae common ancestor or independently acquired, we sought to investigate their evolutionary origins.
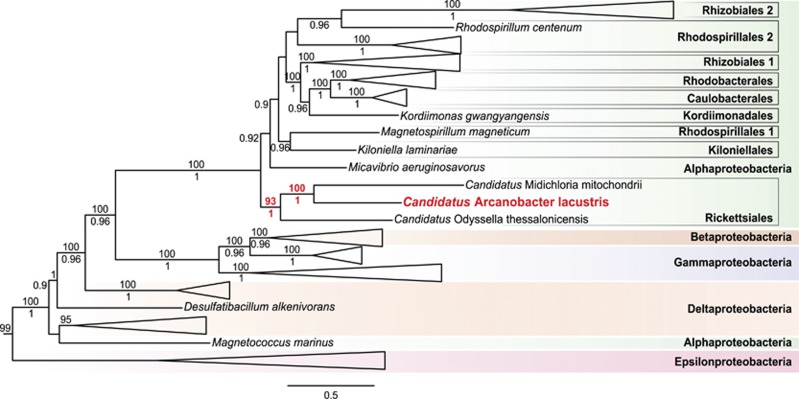
In addition to the 18 flagellar genes identified above, 4 more were identified. The flagellar genes are often located in pairs or on their own, with exception of two clusters: one containing fliW, flgL, flgK, flgJ, flgI, motA and motB and one containing fliC, fliD, fliS, fliL and fliM. All key components of the flagellum (hook, filament, basal body and motor) are represented. Phylogenetic analyses were performed on a set of 14 conserved flagellar genes (Liu and Ochman, 2007; Sassera et al., 2011) of which 8 were present in A. lacustris. The data set was expanded with newly sequenced alphaproteobacterial representatives and ML and BI phylogenies were inferred for both concatenated ‘14 core flagella' and ‘8 core flagella' data sets. Resulting trees were congruent with the expected species phylogeny in which A. lacustris groups with Rickettsiales at or near the root of the Alphaproteobacteria (Figure 4; Supplementary Figure 3). This strongly suggests that the flagellar genes were vertically inherited. Two chains of the ‘8-core flagella' Bayesian phylogenies did not display full species tree congruence, but A. lacustris clustered with Rickettsiales with high confidence, supporting the idea that these genes were vertically inherited.
Figure 4.
Phylogenetic analysis based on concatenated alignment of 14 conserved flagellar genes. A. lacustris and branch support values on branches of interest have been marked with bold red. Tree topology and posterior probability branch support values (shown below branches) were inferred with PhyloBayes (model: CAT-Poisson). Posterior probabilty support values <0.70 were omitted. Bootstrap branch support values (shown above branches) were calculated with RAxML (model: Γ+LG). Separate full PhyloBayes and RAxML trees are shown in Supplementary Figure 3. Major taxonomic groups have been collapsed and outgroup has been trimmed to improve readability.
Chemotaxis genes are very rare in sequenced Rickettsiales and have only been detected in one species whose host is a protist, Ca. Odyssella thessalonicensis. In A. lacustris, the genes are distributed between a cheAWYBR cluster, a cheZY cluster and two genes encoding for methyl-accepting chemotaxis proteins (mcp, and pctC). Proteins encoded by these genes are most similar to homologs in the Rhodospirillales, except for pctC that shows more similarity to homologs in Caulobacterales and Rhizobiales.
Genes indicating a host-associated or intracellular lifestyle
As A. lacustris is a member of the Rickettsiales, which comprises obligate intracellular bacteria, we screened its genome content for any indication of host-associated lifestyle. Indeed, we identified several gene families that are indicative of such a lifestyle:
First, an ATP/ADP translocase homolog was identified. This transporter allows host-adapted bacteria to parasitize their hosts energy by exchanging their cytoplasmic ADP with host ATP (Krause et al., 1985). Although not all homologs of this enzyme necessarily transport ATP (Audia and Winkler, 2006), all are only found in intracellular bacteria and chloroplasts and represent a reliable marker for intracellular lifestyle. We sought to predict the substrate of this homolog with a phylogenetic analysis including all functionally characterized homologs (Linka et al., 2003; Audia and Winkler, 2006; Schmitz-Esser et al., 2008; Vahling et al., 2010). In the resulting tree, the A. lacustris homolog branched at the base of three duplication events (PP: 0.77) in the Rickettsiaceae and was not closely related to any of the other homologs, including those that were characterized (Supplementary Figure 4) leaving us unable to predict its function with confidence.
Second, eight genes putatively encoding components of a virB type IV secretion system (T4SS) were identified, distributed over three loci. The loci consist of (i) virB8, virB9, virB10, virB11 and virD4, (ii) virB4 and virB2 and (iii) a second virB8. virB type T4SSs in Rickettsiales are thought to be of the ‘effector translocator' type involved in host interaction and this hypothesis was recently experimentally enforced in Ehrlichia chaffeensis and Anaplasma marginale (Lockwood et al., 2011; Liu et al., 2012). We investigated whether the T4SS genes in A. lacustris were related to these systems or horizontally acquired with a phylogenetic analysis. The resulting tree, which was based on a concatenation of five well-conserved genes (virB4, virB8, virB9, virB10, virB11), suggests that the T4SS genes were vertically inherited and thus suggests that the T4SS in A. lacustris is likely to be of the ‘effector translocator' type as well (Supplementary Figure 5).
Last, we were able to predict a number of candidate effector proteins. By searching the SecReT4 (experimentally verified) effectors database (Bi et al., 2013, 4), we found 18 putative T4SS effectors (Supplementary Table 3). These included several ankyrin repeat proteins, phosphoglucomutases and Fic-family proteins. Because many effectors have been shown to contain domains that are typically found in eukaryotes, we searched for additional putative effectors with eukaryotic-like domains by using the Effective database (Jehl et al., 2011). Hits (Z-score⩾4) included three proteins containing a glycosyltransferase domain (pfam: ‘Gly_transf_sug', acc: PF04488), one containing a PhoPQ-activated pathogenicity-related protein domain (pfam: ‘PhoPQ_related' acc: PF10142) and one containing a galactosyl tranferase domain (pfam: ‘Glyco_transf_34', acc: PF05637).
Environmental abundance and diversity
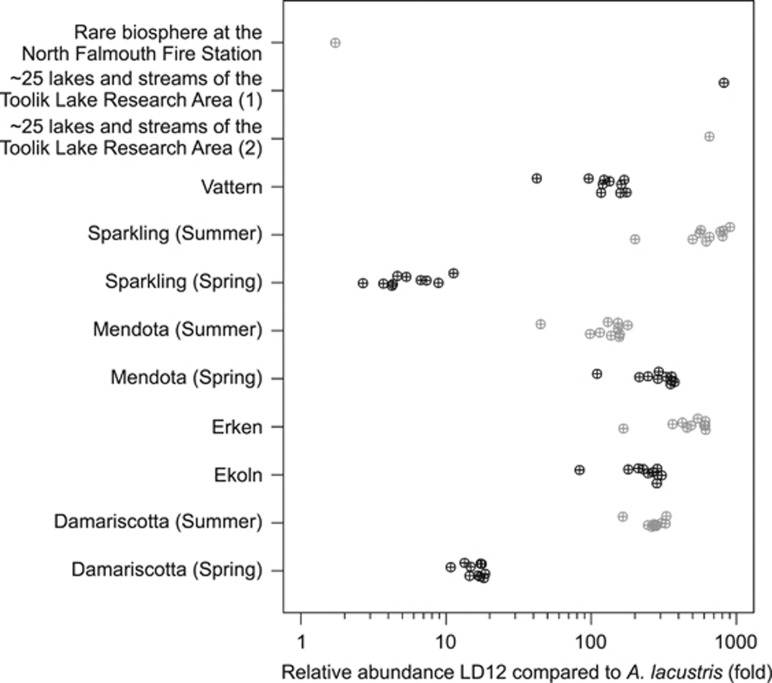
To estimate the abundance of this novel Rickettsiales lineage in freshwater environment, reads from metagenomic data sets of six lakes, including the spring metagenome of Damariscotta Lake from which the single cell was sampled (Martinez-Garcia et al., 2012), were recruited to A. lacustris contigs. Only 0.004–0.014% of the metagenomic reads could be recruited (Table 2). Interestingly, it was not the metagenome of lake Damariscotta (spring), but that of lake Erken that had the highest relative abundance of recruited reads. Compared with LD12, another freshwater alphaproteobacterium for which SAGs are available, A. lacustris is generally 100–1000 times less abundant in these metagenomic data sets (Figure 5).
Table 2. Relative abundance of A. lacustris related reads in metagenomic and SSU amplicon data sets.
| Data set | Type | Size (n) | Recruited reads (n) | Correct for completeness (n) | Relative abundance (%) |
|---|---|---|---|---|---|
| Damariscotta (Spring) | 454 metagenome | 343 495 | 14 | 29 | 0.008 |
| Damariscotta (Summer) | 454 metagenome | 399 994 | 15 | 31 | 0.008 |
| Mendota (Spring) | 454 metagenome | 484 350 | 17 | 35 | 0.007 |
| Mendota (Summer) | 454 metagenome | 562 100 | 11 | 23 | 0.004 |
| Sparkling (Spring) | 454 metagenome | 137 643 | 4 | 8 | 0.006 |
| Sparkling (Summer) | 454 metagenome | 53 174 | 1 | 2 | 0.004 |
| Ekoln | 454 metagenome | 324 764 | 18 | 37 | 0.011 |
| Erken | 454 metagenome | 665 775 | 46 | 95 | 0.014 |
| Vattern | 454 metagenome | 332 583 | 18 | 37 | 0.011 |
| SRR305966a | SSU amplicon (SRA) | 528 037 | 13 | — | 0.002 |
| SRR305967a | SSU amplicon (SRA) | 435 378 | 8 | — | 0.002 |
| ERR204530a | SSU amplicon (SRA) | 1998 | 4 | — | 0.2 |
| ERR204548a | SSU amplicon (SRA) | 1154 | 1 | — | 0.087 |
| ERR204555a | SSU amplicon (SRA) | 866 | 1 | — | 0.115 |
| ERR204613a | SSU amplicon (SRA) | 604 | 1 | — | 0.166 |
| ERR204649a | SSU amplicon (SRA) | 2139 | 1 | — | 0.047 |
| ERR204651a | SSU amplicon (SRA) | 673 | 1 | — | 0.149 |
| ERR204690a | SSU amplicon (SRA) | 3946 | 1 | — | 0.025 |
| ERR204702a | SSU amplicon (SRA) | 919 | 2 | — | 0.218 |
| ERR204743a | SSU amplicon (SRA) | 3074 | 1 | — | 0.033 |
| RARE_MSF_Bv6v4a | SSU amplicon (VAMPS) | 216 537 | 7 | — | 0.003 |
| RARE_NFF_Bv6v4a | SSU amplicon (VAMPS) | 307 126 | 22 | — | 0.007 |
| RARE_WHF_Bv6v4a | SSU amplicon (VAMPS) | 440 607 | 2 | — | 0.0005 |
Study titles and environment type can be found in Supplementary Table 5.
Figure 5.
Relative abundance of alphaproteobacterium LD12 compared with A. lacustris. The relative abundances were assessed for public lake metagenomes Vattern, Sparkling (Summer), Sparkling (Spring), Mendota (Summer), Mendota (Spring), Erken, Ekoln, Damariscotta (Summer) and Damariscotta (Spring) and public SSU amplicon data sets Rare biosphere at the North Falmouth Fire Station (VAMPS accession: RARE_NFF_Bv6v4), ~25 lakes and streams of the Toolik Lake Research Area sequencing run 1 and 2 (SRA accessions: SRR305966, SRR305967). Each point represents the relative abundance of a LD12 SAG (described in (Zaremba-Niedzwiedzka et al., 2013)) for lake metagenomes and the relative abundance of the LD12 SSU sequence (accession no. Z99997.1) for the amplicon data sets. Points are jittered and colored alternating black and dark gray per data set to improve readability.
Broadening the scope of the analysis, public available SSU amplicon databases of SRA (Kodama et al., 2012) and VAMPS (Huse et al., 2010) were screened for reads with high sequence similarity to A. lacustris SSU. Hits were identified in 11 SRA data sets, where they constituted between ~0.002% and ~0.2% of the total data set and in three VAMPS data sets, where hits constituted between ~0.0005% and ~0.007% of the total data set (Table 2). In contrast, when the same screen was performed with LD12 SSU, 115 SRA data sets and seven VAMPS data sets were hit. In amplicon data sets where both A. lacustris and LD12 hits were found, LD12 was approximately between 100 and 1000 times more abundant, with exception of ‘Rare biosphere at the North Falmouth Fire Station', Damariscotta (spring) and Sparkling (spring) data sets (Figure 5). Yet, the lower fold difference in these data sets can be explained by a lower LD12 hit count and not a higher A. lacustris count.
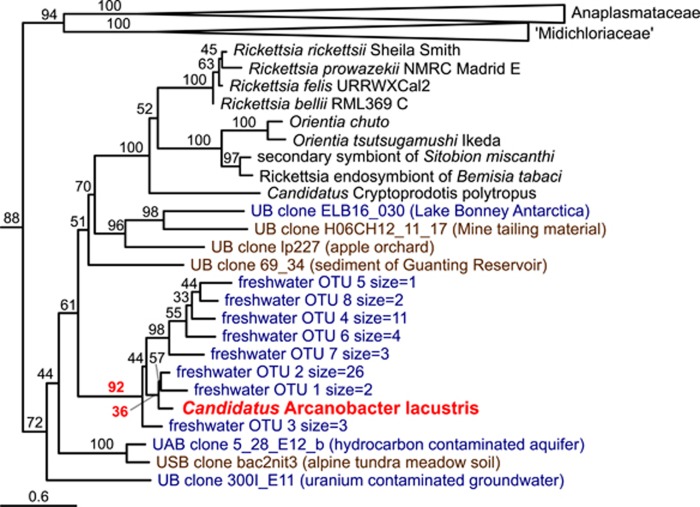
To get a rough idea of the phylogenetic diversity that can be found in the novel lineage that A. lacustris represents, reads that were identified in the screens above and additional (partial) SSU sequences with high similarity found in the NCBI-nt database were incorporated in the SSU phylogeny used earlier in this study. In the resulting tree, A. lacustris formed a monophyletic group with eight OTUs (BS: 92) containing only reads originating from freshwater environments (Figure 6; Supplementary Figure 6). Interestingly, a clade of 28 reads originating from the North Falmouth water distribution system were the closest relatives of A. lacustris with moderate support (BS: 36). In addition, other closely related reads originated were associated with freshwater environments as well (Supplementary Table 4).
Figure 6.
Phylogenetic tree based on SSU rRNA gene including A. lacustris related reads identified SRA and VAMPS SSU amplicon data sets and related sequences in NCBI-nt. The A. lacustris leaf and branch support values on branches of interest have been marked with bold red. For NCBI-nt sequences, their environmental origin is stated between parentheses. Names of reads and NCBI-nt sequences have been colored based on the type of environment where the sample was taken (freshwater: blue, soil: brown). Major taxonomic groups have been collapsed. Outgroup has been omitted to improve readability. Full phylogenetic tree is shown in Supplementary Figure 6.
Discussion
In this work we sought to obtain novel insights into the evolution of Rickettsiaceae by using a methodology that is cultivation independent and by targeting environmental Alphaproteobacteria as opposed to targeting those that are medically or agriculturally relevant. We have identified A. lacustris, an environmental alphaproteobacterium isolated from the freshwater Damariscotta Lake. Below we discuss its inferred lifestyle, its implications for our understanding of Rickettsiaceae evolution and its apparent extreme rarity in the sampled biosphere.
Lifestyle
We were able to isolate and identify A. lacustris from a freshwater sample because it was a ‘free' single cell within the sample at the time of sorting. Even though this could indicate a free-living lifestyle, it does not necessarily have to be the case. One can think of several other possible scenarios that lead to a ‘free' single cell at the time of sorting: (i) A. lacustris is facultative intracellular, and the microbe was captured during a free-living stadium of its lifecycle, (ii) it is obligate intracellular but was released after it caused its host cell to lyse, similar to other Rickettsiales (Hackstadt, 1996) or (iii) it was inside a host cell at the time of sampling, but was released when the host cell lysed owing to mechanical processing of the sample (that is, filtration or cell sorting). In summary, from this knowledge alone we cannot distinguish between an obligate free-living, facultative intracellular or obligate intracellular lifestyle.
We therefore inspected its genomic content to get more insights. Several factors were found that are supportive of an intracellular lifestyle. The presence of an ATP/ADP translocase homolog is the strongest indicator, as homologs of this protein have been found in intracellular bacteria only (Schmitz-Esser et al., 2004). In addition, the presence of several components of a virB-type T4SS and a number of putative effectors suggest a host-associated lifestyle. We suggest that the T4SS is of the effector secretion type, as it shares a recent common ancestor with the vir-type T4SS of E. chaffeensis and A. marginale for which it has been recently shown that they are capable of secreting effector proteins into host cells (Lockwood et al., 2011; Liu et al., 2012). Conversely, we found an array of flagellar and chemotaxis genes that could indicate a (partially) free-living lifestyle. For example, both systems could support A. lacustris in locating and targeting nutrients and/or new host cells. However, as these systems are also encoded by the obligate intracellular symbiont Ca. Odyssella thessalonicensis, these systems do not necessarily indicate a free-living lifestyle.
Taken together, we suggest either a facultative or obligate intracellular lifestyle for A. lacustris. The identity of the host remains obscure, but the relative high amount of uniquely shared protein families with Ca. Odyssella thessalonicensis (hosted by the amoebae Acanthamoeba) and Holospora undulata (hosted by the ciliate Paramecium), hints toward a protist host.
Evolution
From our phylogenomic and phylogenetic analyses, we observed that A. lacustris was not closely affiliated with any of the currently established Rickettsiales families. Rather, it represented a previously unexplored branch, basal to the Rickettsiaceae. Although this topology has been observed before for Ca. Midichloria mitochondrii (Sassera et al., 2011), our analyses strongly showed that it affiliated with the Anaplasmataceae instead (Figure 2). This affiliation has been observed by other studies as well (Driscoll et al., 2013; Montagna et al., 2013). The phylogenetic novelty of A. lacustris is further underlined by the large fraction (143 out of 299) of A. lacustris unique protein families that, not only have no detected homologs in other Rickettsiales families, but also none in NCBI's nonredundant database. This phylogenetic placement provided us with a unique opportunity to obtain novel insights into the evolution of the Rickettsiaceae. The most significant insight is the presence of an array of flagellar and chemotaxis genes, which are completely absent in all current Rickettsiaceae genomes. As we showed that flagellar genes were vertically inherited, this implies that they were lost in the last common ancestor of the Rickettsiaceae, likely owing to the heavy genome reduction forces that are characteristic of all Rickettsiaceae. In line with this hypothesis A. lacustris is estimated to have a relatively large genome size compared with most Rickettsiales (~1.7 Mb vs ~1.0–1.5 Mb; Figure 1).
The presence of flagellar genes follows the current trend in which an increasing amount of Rickettsiales members are found to encode flagellar genes or have been observed to synthesize actual flagella: Ca. Midichloria mitochondrii (Sassera et al., 2011; Mariconti et al., 2012), Ca. Odyssella thessalonicensis, (Georgiades et al., 2011), Lyticum, (Boscaro et al., 2013) and Trichorickettsia and Gigarickettsia (Vannini et al., 2014) are examples that contradict the previous assumption that flagella were lost in all Rickettsiales and fortifies the suggestion that flagella were present in the last common Rickettsiales ancestor.
The Rickettsiales have traditionally been put forward as candidates for the closest relatives of mitochondria (Fitzpatrick et al., 2006; Williams et al., 2007). However, there is still no consensus about exact phylogenetic placement of mitochondria. This is mainly caused by the tree reconstruction artifacts long branch attraction and compositional bias: the Rickettsiales, SAR11 and mitochondria all share a fast evolutionary rate and have AT-rich genomes (Grote et al., 2012; Wang and Wu, 2015). Indeed, the affiliation of SAR11 with mitochondria was convincingly shown to be a compositional bias artifact (Brindefalk et al., 2011; Rodríguez-Ezpeleta and Embley, 2012; Viklund et al., 2012). The availability of A. lacustris, which breaks the relatively long branch leading to the Rickettsiaceae, reduces the long branch attraction problem and may help future studies to identify the closest extant relatives of mitochondria with more confidence.
Rarity
After exploring the diversity and abundance of A. lacustris in metagenomic data sets, it seemed that this bacterium and relatives are rare in the sampled biosphere. Compared with the free-living freshwater alphaproteobacterium LD12, it is about 100–1000 times less abundant. Even in both available metagenomes of Damariscotta Lake, the source of this bacterium, only up to 15 reads (0.008%) could be identified as an A. lacustris relative. However, this apparent rarity might be an artifact: many environmental samples go through a size filtration step before DNA extraction and follow-up metagenomic sequencing, effectively filtering out a large fraction of eukaryotic cells and thus putative A. lacustris host cells. Because of this and the inferred intracellular lifestyle, we cannot rule out the possibility that A. lacustris is more abundant in the biosphere than we can observe in current metagenomic and SSU amplicon data sets.
Conclusion
In conclusion, we present here the existence of a novel Rickettsiales member that represents a lineage that is a deep sister relative to the Rickettsiaceae. This lineage appears to be ultrarare in the sampled biosphere and occupies a freshwater niche. We predict a facultative or obligate intracellular lifestyle, perhaps in association with a protist and finally, observe the presence of chemotaxis genes and vertically inherited flagellar genes. This study has highlighted the power of single-cell genomics with regard to exploring the biological dark matter and its implications for understanding microbial evolution. It would very likely have not been possible to discover A. lacustris and characterize its genome with standard cultivation-based and metagenomics approaches, because of its seemingly extreme rarity and symbiotic lifestyle. Single-cell genomics and other cultivation-independent techniques will be of great value for future studies that aim to gain insight into the evolution of the Rickettsiaceae and other uncultivable or hard to cultivate microbial lineages.
Acknowledgments
All sequencing was performed by the SNP and SEQ Technology Platform, Science for Life Laboratory at Uppsala University, a national infrastructure supported by the Swedish Research Council (VR-RFI) and the Knut and Alice Wallenberg Foundation. We thank the Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) at Uppsala University for providing computational resources. We thank Ilias Lagkouvardos for help in the analysis of the environmental distribution of A. lacustris and Davide Sassera for providing the flagella data set. This work was supported by grants to TJGE and SGEA of the Swedish Research Council (621-2009-4813; 621-2011-4669) and to TJGE and MH of the European Research Council (ERC Starting grant 310039-PUZZLE_CELL and 281633-EvoChlamy). FS is a recipient of the DOC fellowship of the Austrian Academy of Sciences at the Division of Microbial Ecology, University of Vienna, Austria.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SGE, Zomorodipour A, Andersson JO, Sicheritz-Pontén T, Alsmark UCM, Podowski RM et al. (1998). The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396: 133–140. [DOI] [PubMed] [Google Scholar]
- Andrews S. (2012). FastQC A Quality Control tool for High Throughput Sequence Data http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Audia JP, Winkler HH. (2006). Study of the five Rickettsia prowazekii proteins annotated as ATP/ADP translocases (Tlc): only Tlc1 transports ATP/ADP, while Tlc4 and Tlc5 transport other ribonucleotides. J Bacteriol 188: 6261–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi D, Liu L, Tai C, Deng Z, Rajakumar K, Ou H-Y. (2013). SecReT4: a web-based bacterial type IV secretion system resource. Nucleic Acids Res 41: D660–D665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscaro V, Schrallhammer M, Benken KA, Krenek S, Szokoli F, Berendonk TU et al. (2013). Rediscovering the genus Lyticum, multiflagellated symbionts of the order Rickettsiales. Sci Rep 3: 3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brayton KA, Kappmeyer LS, Herndon DR, Dark MJ, Tibbals DL, Palmer GH et al. (2005). Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc Natl Acad Sci USA 102: 844–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindefalk B, Ettema TJG, Viklund J, Thollesson M, Andersson SGE. (2011). A phylometagenomic exploration of oceanic alphaproteobacteria reveals mitochondrial relatives unrelated to the SAR11 clade. PLoS One 6: e24457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Boutros PC. (2011). VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho N-H, Kim H-R, Lee J-H, Kim S-Y, Kim J, Cha S et al. (2007). The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host–cell interaction genes. Proc Natl Acad Sci USA 104: 7981–7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NE, Liebenberg J, Villiers EP, de Brayton KA, Louw E, Pretorius A et al. (2005). The genome of the heartwater agent Ehrlichia ruminantium contains multiple tandem repeats of actively variable copy number. Proc Natl Acad Sci USA 102: 838–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby AC, Cho N-H, Fuxelius H-H, Westberg J, Andersson SGE. (2007). Intracellular pathogens go extreme: genome evolution in the Rickettsiales. Trends Genet 23: 511–520. [DOI] [PubMed] [Google Scholar]
- Driscoll T, Gillespie JJ, Nordberg EK, Azad AF, Sobral BW. (2013). Bacterial DNA sifted from the trichoplax adhaerens (animalia: placozoa) genome project reveals a putative rickettsial endosymbiont. Genome Biol Evol 5: 621–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen J et al. (2006). Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet 2: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. (1998). Profile hidden Markov models. Bioinformatics 14: 755–763. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick DA, Creevey CJ, McInerney JO. (2006). Genome phylogenies indicate a meaningful α-proteobacterial phylogeny and support a grouping of the mitochondria with the Rickettsiales. Mol Biol Evol 23: 74–85. [DOI] [PubMed] [Google Scholar]
- Georgiades K, Madoui M-A, Le P, Robert C, Raoult D. (2011). Phylogenomic analysis of odyssella thessalonicensis fortifies the common origin of Rickettsiales, Pelagibacter ubique and Reclimonas americana mitochondrion. PLoS One 6: e24857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote J, Thrash JC, Huggett MJ, Landry ZC, Carini P, Giovannoni SJ et al (2012) Streamlining and core genome conservation among highly divergent members of the SAR11 clade. Mbio 3: e00252–e002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T. (1996). The biology of Rickettsiae. Infect Agents Dis 5: 127–143. [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. (2010). Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12: 1889–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. (2011). Integrative analysis of environmental sequences using MEGAN4. Genome Res 21: 1552–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehl M-A, Arnold R, Rattei T. (2011). Effective—a database of predicted secreted bacterial proteins. Nucleic Acids Res 39: D591–D595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. (2014). Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42: D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol Biol Evol 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y, Shumway M, Leinonen R, International Nucleotide Sequence Database Collaboration.. (2012). The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res 40: D54–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause DC, Winkler HH, Wood DO. (1985). Cloning and expression of the Rickettsia prowazekii ADP/ATP translocator in Escherichia coli. Proc Natl Acad Sci USA 82: 3015–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C et al. (2004). Versatile and open software for comparing large genomes. Genome Biol 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rødland EA, Stærfeldt H-H, Rognes T, Ussery DW. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35: 3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagkouvardos I, Weinmaier T, Lauro FM, Cavicchioli R, Rattei T, Horn M. (2014). Integrating metagenomic and amplicon databases to resolve the phylogenetic and ecological diversity of the Chlamydiae. ISME J 8: 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J. (2013). PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol 62: 611–615. [DOI] [PubMed] [Google Scholar]
- Lasken RS, Stockwell TB. (2007). Mechanism of chimera formation during the Multiple Displacement Amplification reaction. BMC Biotechnol 7: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. (2008). An improved general amino acid replacement matrix. Mol Biol Evol 25: 1307–1320. [DOI] [PubMed] [Google Scholar]
- Linka N, Hurka H, Lang BF, Burger G, Winkler HH, Stamme C et al. (2003). Phylogenetic relationships of non-mitochondrial nucleotide transport proteins in bacteria and eukaryotes. Gene 306: 27–35. [DOI] [PubMed] [Google Scholar]
- Liu H, Bao W, Lin M, Niu H, Rikihisa Y. (2012). Ehrlichia type IV secretion effector ECH0825 is translocated to mitochondria and curbs ROS and apoptosis by upregulating host MnSOD. Cell Microbiol 14: 1037–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Ochman H. (2007). Stepwise formation of the bacterial flagellar system. Proc Natl Acad Sci 104: 7116–7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockwood S, Voth DE, Brayton KA, Beare PA, Brown WC, Heinzen RA et al. (2011). Identification of anaplasma marginale type iv secretion system effector proteins. PLoS One 6: e27724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariconti M, Epis S, Sacchi L, Biggiogera M, Sassera D, Genchi M et al. (2012). A study on the presence of flagella in the order Rickettsiales: the case of ‘Candidatus Midichloria mitochondrii'. Microbiology 158: 1677–1683. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia M, Swan BK, Poulton NJ, Gomez ML, Masland D, Sieracki ME et al. (2012). High-throughput single-cell sequencing identifies photoheterotrophs and chemoautotrophs in freshwater bacterioplankton. ISME J 6: 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhej V, Raoult D. (2011). Rickettsial evolution in the light of comparative genomics. Biol Rev 86: 379–405. [DOI] [PubMed] [Google Scholar]
- Montagna M, Sassera D, Epis S, Bazzocchi C, Vannini C, Lo N et al. (2013). ‘Candidatus Midichloriaceae' fam. nov. (Rickettsiales), an ecologically widespread clade of intracellular Alphaproteobacteria. Appl Environ Microbiol 79: 3241–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A et al. (2013). Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20: 714–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F et al (2013) Insights into the phylogeny and coding potential of microbial dark matter. Nature,. (2013) 499: 431–437. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Embley TM. (2012). The SAR11 group of alpha-proteobacteria is not related to the origin of mitochondria. PLoS One 7: e30520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassera D, Lo N, Epis S, D'Auria G, Montagna M, Comandatore F et al. (2011). Phylogenomic evidence for the presence of a flagellum and cbb3 oxidase in the free-living mitochondrial ancestor. Mol Biol Evol 28: 3285–3296. [DOI] [PubMed] [Google Scholar]
- Schmitz-Esser S, Haferkamp I, Knab S, Penz T, Ast M, Kohl C et al. (2008). Lawsonia intracellularis contains a gene encoding a functional Rickettsia-Like ATP/ADP translocase for host exploitation. J Bacteriol 190: 5746–5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Esser S, Linka N, Collingro A, Beier CL, Neuhaus HE, Wagner M et al. (2004). ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to Chlamydiae and Rickettsiae. J Bacteriol 186: 683–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrallhammer M, Ferrantini F, Vannini C, Galati S, Schweikert M, Görtz H-D et al. (2013). ‘Candidatus megaira polyxenophila' gen. nov., sp. nov.: considerations on evolutionary history, host range and shift of early divergent Rickettsiae. PLoS One 8: e72581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30: 2068–2069. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the Dark Ocean. Science 333: 1296–1300. [DOI] [PubMed] [Google Scholar]
- Vahling CM, Duan Y, Lin H. (2010). Characterization of an ATP Translocase Identified in the Destructive Plant Pathogen ‘Candidatus Liberibacter asiaticus'. J Bacteriol 192: 834–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini C, Boscaro V, Ferrantini F, Benken KA, Mironov TI, Schweikert M et al. (2014). Flagellar movement in two bacteria of the family Rickettsiaceae: a re-evaluation of motility in an evolutionary perspective. PLoS One 9: e87718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viklund J, Ettema TJG, Andersson SGE. (2012). Independent Genome Reduction and Phylogenetic Reclassification of the Oceanic SAR11 Clade. Mol Biol Evol 29: 599–615. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wu M. (2015). An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci Rep 5: 7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KP, Sobral BW, Dickerman AW. (2007). A robust species tree for the Alphaproteobacteria. J Bacteriol 189: 4578–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba-Niedzwiedzka K, Viklund J, Zhao W, Ast J, Sczyrba A, Woyke T et al. (2013). Single-cell genomics reveal low recombination frequencies in freshwater bacteria of the SAR11 clade. Genome Biol 14: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.