Background: Transmembrane domain interactions in gap junction channels are poorly understood.
Results: Tryptophan substitution experiments involving all four TM domains of Cx32 revealed tight packing.
Conclusion: After modeling, tight packing was found to occur in the midregion. Pore-facing residues were highly sensitive to substitution, whereas lipid-facing residues were variably tolerant.
Significance: Connexin-based channels are more densely packed than their innexin-based counterparts.
Keywords: connexin, gap junction, gating, mutagenesis, transmembrane domain, Cx32, TM domain interactions, comparative model, hemichannel, tryptophan scanning
Abstract
Tryptophan was substituted for residues in all four transmembrane domains of connexin32. Function was assayed using dual cell two-electrode voltage clamp after expression in Xenopus oocytes. Tryptophan substitution was poorly tolerated in all domains, with the greatest impact in TM1 and TM4. For instance, in TM1, 15 substitutions were made, six abolished coupling and five others significantly reduced function. Only TM2 and TM3 included a distinct helical face that lacked sensitivity to tryptophan substitution. Results were visualized on a comparative model of Cx32 hemichannel. In this model, a region midway through the membrane appears highly sensitive to tryptophan substitution and includes residues Arg-32, Ile-33, Met-34, and Val-35. In the modeled channel, pore-facing regions of TM1 and TM2 were highly sensitive to tryptophan substitution, whereas the lipid-facing regions of TM3 and TM4 were variably tolerant. Residues facing a putative intracellular water pocket (the IC pocket) were also highly sensitive to tryptophan substitution. Although future studies will be required to separate trafficking-defective mutants from those that alter channel function, a subset of interactions important for voltage gating was identified. Interactions important for voltage gating occurred mainly in the mid-region of the channel and focused on TM1. To determine whether results could be extrapolated to other connexins, TM1 of Cx43 was scanned revealing similar but not identical sensitivity to TM1 of Cx32.
Introduction
Gap junctions mediate direct intercellular communication between animal cells. A typical gap junction includes hundreds or thousands of gap junction channels localized to a region of cell contact. The channels permit the passage of ions, nutrients, and cellular metabolites up to about 1 kDa in size (1). The connexin protein family constitutes gap junction channels in mammalian tissues, and 21 different connexin proteins have been identified in humans (2). Connexins are named according to their molecular mass; for example, connexin32 (Cx32) has an estimated molecular mass of 32 kDa. Connexins are also classified in groups based on sequence and evolutionary origins (3). For example, Cx32 is a β-connexin, and it shows higher sequence identity with β-connexins (e.g. Cx26, Cx30, and Cx31) than α-connexins (e.g. Cx40, Cx43, and Cx50).
Connexins are expressed in specific and overlapping patterns, and Cx32 is expressed in liver, Schwann cells, and oligodendrocytes (2). Mutations in the human Cx32 gene (GJB1) are associated with a peripheral neuropathy known as Charcot-Marie-Tooth disease type X (CMTX)4 (4) with over 400 mutations identified in patients (5). In Schwann cells, Cx32 appears to form a critical pathway for the flow of cellular metabolites between layers of the myelin sheath (6), and mutations ranging from complete loss of function to fairly conservative missense mutations induce similar severity of disease (7). CMTX mutations have been characterized in a variety of experimental systems and have been shown to alter trafficking, voltage gating, and permeability (5). Because neuropathy is usually the only clinical symptom associated with CMTX mutations, it is suspected that other connexins compensate for the loss of Cx32 in tissue such as liver (5).
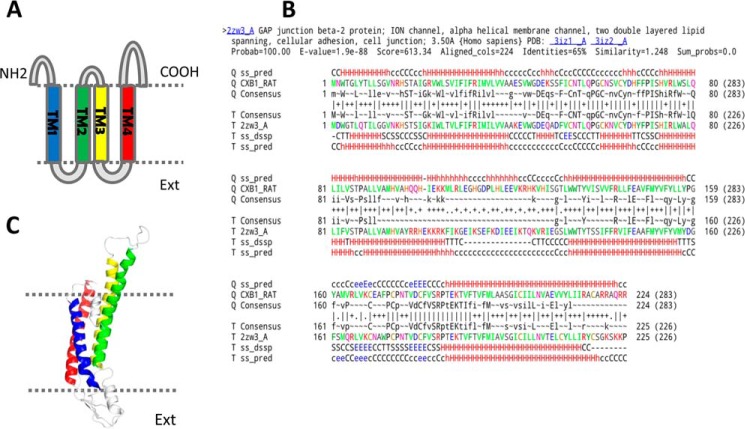
Connexins have four transmembrane domains (Fig. 1A, TM1–TM4) and cytoplasmic N and C termini (8) as indicated in Fig. 1. An intercellular gap junction channel is formed when six connexins oligomerize around a central pore, creating a connexon, also referred to as a hemichannel, which docks with a connexon in an adjacent cell (9). To date, only one atomic structure of a gap junction channel has been published (10). The structure of Cx26 obtained by x-ray crystallography represents a complex intercellular channel formed by head to head docking of two connexons. Each connexon is composed of six connexins arranged symmetrically around a central pore. As such, each pore is surrounded and stabilized by 24 membrane-spanning transmembrane (TM) domains. The TM domains maintain consistent α-helical secondary structure throughout the membrane (10).
FIGURE 1.
Topology and sequence of Cx32 shown together with one subunit of the modeled gap junction channel. A, membrane topology of Cx32 showing four transmembrane helices, cytoplasmic N and C termini, and two extracellular loops. Transmembrane helices are colored as follows: TM1 (blue), TM2 (green), TM3 (yellow), and TM4 (red). B, alignment of rCx26 and hCx32 obtained using HHpred, which was used to create the model of rCx32. C, one subunit of the modeled Cx32 channel viewed from the center of the pore. The model was created using MODELLER.
In the Cx26 structure, TM1 lies adjacent to the pore, interacting at the cytoplasmic end with an infolded N terminus. TM2 is kinked in the middle due to the presence of a conserved proline, and TM2 and TM3 extend beyond the membrane border at the cytoplasmic face. TM2 contributes to the pore at the cytoplasmic end, whereas TM3 and TM4 lie distal to the pore at the lipid-protein interface. Within the membrane-spanning region, the pore of the channel ranges from 15 Å in diameter at the extracellular end to 40 Å in diameter at the cytoplasmic end (10). Important elements of the pore include a hinged plug within the cytoplasmic vestibule of the channel, induced by an infolded N-terminal helix and a short 310 helix at the extracellular end, which is formed by EL1 (10). These important pore features are supported by three-dimensional projection structures (11, 12) and molecular dynamic refinements (13, 14).
Overall, the crystal structure (10) and MD simulated channel (13, 14) provide similar images of TM domain arrangement. However, TM domain interactions are dynamic, and each method provides only a snapshot of interactions in a channel conformation biased toward stability. In vivo, TM domain interactions occur soon after the co-translational process is initiated in the endoplasmic reticulum and play important roles in oligimerization and trafficking (15–17). In addition, TM domain interactions are important for dynamic processes, such as gating and signaling (18–20).
Tryptophan scanning analysis is a functional assay of TM domain interactions (21), based on the premise that the large bulky side chain of tryptophan will be poorly tolerated if inserted at a site of close interaction in a folded protein. It has been primarily applied to membrane proteins. For instance, the technique was first applied to the MotB protein, a bacterial integral membrane protein associated with flagellar motion (21), and it has since been applied to a range of membrane proteins, including the α-subunit of the nicotinic acetylcholine receptor (nAChR (22)), the mammalian hyperpolarization-activated cyclic nucleotide-gated channel (HCN (23)), and an innexin-based gap junction channel (20). Most commonly, tryptophan-scanning analysis leads to identification of sensitive sites positioned along a common face of a membrane-spanning helix (20–24), but in tightly packed regions of a membrane protein, broad sensitivity to tryptophan substitution is expected (21, 24).
Although reduced or abolished function is the most common reporter that tryptophan has been inserted at a site of interaction, a range of other consequences has also been described, including altered agonist sensitivity in the GABAA receptor (24), slow activation of HERG channels (25), and altered response to transjunctional voltage of innexin-based gap junctions (20). This suggests that tryptophan scanning can be used as a reporter of TM domain interactions that are critical for particular protein functions, as well as an overall reporter of protein structure.
In this study, tryptophan scanning mutagenesis was applied to all four transmembrane domains (TM1–TM4) of the gap junction protein Cx32 to broadly identify sites of transmembrane domain interaction that are essential for channel function. Scanning all four domains allowed the results to be interpreted from the broadest possible perspective. Sensitive sites were mapped onto a molecular model of Cx32 channel, created by comparative modeling using as a template the Cx26 atomic model. The broad map of TM domain interactions will serve as the basis for studies aimed at identifying interactions essential for protein trafficking and localization, docking, and gating.
Materials and Methods
Site-directed Mutagenesis
The genes encoding rat connexin32 (rCx32) and rat connexin Cx43 (rCx43) were cloned into the EcoRI site of PGem7zf (+) and were a generous gift from Dr. Bruce Nicholson (University of Texas, Health Science Center at San Antonio). The Stratagene QuikChange® or QuikChange Lightning® mutagenesis method (Agilent Technologies-Stratagene Products, Santa Clara, CA) was used to create tryptophan substitution mutants. Primers were designed using the QuikChange® Primer Design Program (Agilent Technologies-Stratagene Products, Santa Clara, CA) and custom-synthesized by Integrated DNA Technologies (Coralville, IA) in 25-nmol quantities with standard desalting. Mutations were confirmed by sequencing through the coding region (Roswell Park Cancer Institute DNA Sequencing Facility, Buffalo, NY).
In Vitro Transcription
Prior to in vitro transcription, plasmid DNA was linearized with XbaI, which cut downstream of the Cx32 coding region or KpnI which cut downstream of the Cx43 coding region. Linearized DNA was cleaned and concentrated according to the GENECLEAN protocol (MP Biomedicals, Santa Ana, CA), and RNA was prepared using either an SP6 (Cx32) or T7 (Cx32) mMessage mMachine RNA kit (Applied Biosystems/Ambion, Austin, TX). RNA was purified with lithium chloride and quantified using gel electrophoresis and ethidium bromide staining by comparison with an RNA 250 control (Applied Biosystems/Ambion, Austin, TX). Cx32 RNA was diluted to ∼50 ng/μl prior to injection of 41 nl per oocyte (total RNA ∼ 2 ng/oocyte) and Cx43 RNA was diluted to ∼12 ng/μl prior to injection of 41 nl per oocyte (total RNA ∼0.5 ng/oocyte).
Oocyte Expression
The technique of recording intercellular currents from paired Xenopus oocytes was carried out as described previously (26). Oocytes were removed from ovulating Xenopus laevis females. They were cleaned and digested in Oocyte Ringers 2 (OR2: 82.5 mm NaCl, 2 mm KCl, 1 mm MgCl2, 5 mm HEPES, pH 7.4) and maintained in modified Barth's (MB) solution (88 mm NaCl, 1 mm KCl, 0.41 mm CaCl2, 0.82 mm MgSO4, 1 mm MgCl2, 0.33 mm Ca(NO3)2, 20 mm HEPES, pH 7.4) for injection, pairing, and recording. Oocytes clumps were cut into small sections and incubated in OR2 supplemented with collagenase (type 1A, Sigma) for 10–30 min, and the follicular layer was subsequently removed using fine forceps. The following day oocytes were pre-injected with 0.05 ng of morpholino antisense oligonucleotide directed against Xenopus Cx38 (Gene Tools LLC, Philomath, OR). Approximately 24 h after pre-injection, cRNA was injected, and oocytes were incubated at 18 °C for 12–24 h, stripped of their vitelline membranes, and paired overnight in agar wells. Agar wells were prepared with 1% agar dissolved in OR2, whereas the bathing media consisted of MB1. All mutants were tested in heterotypic pairings (e.g. mutant/Cx32 or mutant/Cx43) to avoid additive effects of mutations on function.
Functional Analysis
To assess junctional conductance, paired oocytes were clamped at −20 mV using two Geneclamp amplifiers (Molecular Devices, Sunnyvale, CA). One cell was then pulsed to +80 and −120 mV eliciting a 2-s transjunctional current (Ij) in the partnered oocyte. Currents were measured at their maximal level, which occurred within the first 100 ms of the voltage pulse for most wild type channels and most mutants. For a few mutants, current activated rather than inactivated in response to transjunctional voltage (Vj). For these “reverse-gating” mutants, the current was measured at the end of a 2-s voltage pulse. Functional mutants were further assessed with a set of longer voltage steps applied in 10-mV increments to maximum Vj values of ±100 mV.
We calculated transjunctional conductance (Gj) for each pair (mutant/Cx32). Between three and 20 pairs were tested for each mutant. Results were normalized to the conductance of oocyte pairs expressing wild type connexins on the same day. For Cx32-associated mutants, pairs were only included for analysis when positive control pairs displayed high junctional conductance (Gj >25 μS for Cx32/Cx32) and negative controls were uncoupled (Gj <0.5 μS for Cx32/oligonucleotide). For Cx43-associated mutants, experiments were considered when the mean conductance of wild type controls was greater than 10 μS, and the mean conductance of negative controls (Cx43/oligonucleotide) was less than 2 μS.
Data Management
Pairings between wild type connexins served as a positive control and provided the baseline conductance for each batch of cells. For the tryptophan scan of Cx32, the primary focus of this study, very high conductance positive controls were obligatory as they allowed clear identification of nonfunctional mutants and aided with discrimination between nonfunctional mutants and those with reduced function. A typical Cx32/Cx32-positive control resulted in a mean Gj ∼50 μS (n = 8). Working within this range of saturating conductance also served to negate the effect of small variations in RNA quantification or injection on overall coupling levels (20).
To determine whether tryptophan substitution significantly altered function, the conductance induced by each mutant was averaged. This average was then compared with the average conductance induced by wild type Cx32 on the same day. For Cx32, statistical significance was determined at a level of p < 0.05 after a Student's t test was applied to the raw data. Also for Cx43, a Student's t test was used to compare each mutant to wild type Cx43 pairs on the same day. Conductance was considered to be abolished when the mean Gj was zero, and reduced when the mean Gj was significantly smaller than wild type (p < 0.05).
Model of rCx32 Hemichannel
The program MODELLER 9.13 (27) was used to develop a comparative molecular model of the rCx32 hemichannel using the atomic model structure of hCx26 as a template (10) (Protein Data Bank code 2zw3). To improve the alignment, HHpred (28) was employed to match the secondary structure in the hCx26 model with the secondary structure predicted for rCx32 (Fig. 1). Symmetry restraints obtained from the hCx26 crystal structure were then applied to the Cx32 hemichannel molecular model, identifying the structure with the lowest objective function (molpdf). In doing so, we selected as representative structure the model with the highest fitness to structural restraints obtained from hCx26. Further stereochemical quality check of the model was assessed by evaluating the selected model by both PROCHECK (29) and WHAT_CHECK (30).
Results
Effects of Tryptophan Substitution on Junctional Conductance
Tryptophan substitutions were created at about 15 sites in each TM domain with targeted regions determined by hydropathy analysis (TMHMM 2.0; Ref. 31). All substitutions fell within the membrane-spanning region defined during early biochemical experiments of connexins (8) and also within the membrane-spanning regions of the Cx26 atomic model (10). Tryptophan substitutions were avoided at predicted membrane boundaries as large aromatic side chains can influence positioning of helices within the membrane (32). In the Cx26 crystal structure, helices TM2 and TM3 extend beyond the membrane at the cytosolic face (10), and these cytoplasmic regions were not analyzed in this study.
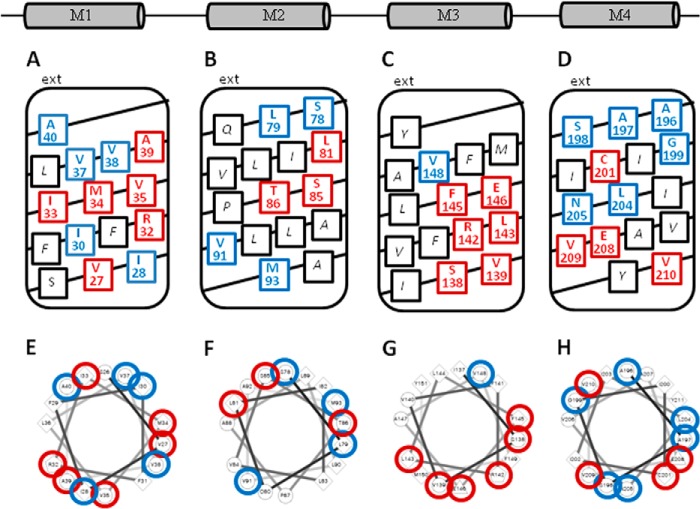
The effects are summarized in Fig. 2. Results are organized according to TM domain, and the conductance for each mutant is normalized to the conductance of oocyte pairs expressing wtCx32 on the same day (Fig. 2, bottom of each histogram). The numbers beside each bar in Fig. 2 represent the number of pairs with measurable conductance versus the number of pairs tested. Color-coding helps distinguish mutants with reduced function (Fig. 2, blue text) and abolished function (red text). The distribution of sensitive sites in each transmembrane helix is shown in helical net format and helical wheel format in Fig. 3.
FIGURE 2.
Results of tryptophan substitution summarized in histogram format. Domains sketched adjacent to each graph show the orientation of TM domains (NH2, cytoplasmic N terminus; EL1, first extracellular loop; CL, cytoplasmic loop; EL2, second extracellular loop; COOH, C terminus). Numbers beside each bar indicate the number of pairs with measurable conductance along with the number of pairs tested (e.g. number functional/number tested). Each bar represents the mean junctional conductance between oocytes paired heterotypically (Cx32/mutant) and normalized to Cx32/Cx32 pairs of the same batch on the same day. Conductance was reduced (blue), abolished (red), or unaffected (black). NF, nonfunctional, *, statistically different from wild type at p ≤ 0.05 as determined by Student's t test.
FIGURE 3.
Helical wheel and helical net plots highlighting sites where tryptophan substitution either reduced (blue), abolished (red), or did not affect (black) gap junction intercellular conductance. A–D, helical net plots. E–H, helical wheel plots generated using wheel.pl, Version 1.4.
Tryptophan Substitution in TM1
InTM1, tryptophan was substituted at sites Ser-26 through Ala-40 and completely abolished function at six locations (V27W, R32W, I33W, M34W, L35W, and A39W). Five other substitutions in TM1 significantly reduced function (I28W, I30W, V37W, V38W, and A40W) totaling 11 out of 15 sites (∼76%) that were sensitive to tryptophan substitution. On the helical net plot (Fig. 3A), a sensitive region defined by a ring around the middle of TM1 is apparent, including Arg-32, Ile-33, Met-34, and Val-35. Even outside the mid-region, there is only a narrow helical face (Fig. 3E) that remains tolerant to tryptophan substitution, including Ser-26, Phe-29, Ile-30, Leu-36, Val-37, and Ala-40. This is consistent with a packing arrangement involving a critical region of interaction in middle of the TM1 helix, along with widespread interactions in other regions.
Tryptophan Substitution in TM2
In TM2, tryptophan was substituted at sites Ser-78 through Met-92. Gap junction function was completely abolished with mutations L81W, S85W, and T86W, although conductance was significantly reduced for four other mutants (S78W, L79W, V91W, and M93W), totaling seven out of 16 sites (44%) that were sensitive to tryptophan substitution. The helical net plot (Fig. 3B) and helical wheel plot (Fig. 3F) demonstrate that sensitive sites are clustered along a broad face of the helix. Interestingly, replacing Pro-87 with tryptophan did not significantly reduce function. A similar observation was made for the P87C substitution during cysteine scanning studies (33) in contrast to results for Cx26 where removal of the conserved proline in M2 induced a reverse-gating phenotype (34). The importance of the conserved M2 proline has been demonstrated in other connexins (35), although its structural importance has yet to be determined. It is likely that the bend in TM2 is important for maintaining the open state of the channel and that other interactions help maintain the kink in Cx32.
Tryptophan Substitution in TM3
In TM3, tryptophan was substituted at sites Ile-137 through Tyr-151. Function was abolished at six sites (S138W, V139W, R142W, L143W, F145W, and E146W) and significantly reduced at one (V148W), totaling seven out of 15 sites (∼47%) that were sensitive to tryptophan substitution. The helical net plot (Fig. 3C) highlights a sensitive face of TM3, which covers three rotations of the helix and includes three conserved hydrophilic residues Ser-138, Arg-142, and Glu-146. Three other hydrophobic residues along this face of the helix (Val-139, Leu-143, and Phe-145) were also sensitive to tryptophan substitution implying that the effects are not limited to changes in hydrophobicity or charge. However, this face of the helix lacks sensitivity to tryptophan substitution toward the extracellular end (F149W and M150W are functional), suggesting that the packing of TM3 may be relaxed toward the extracellular surface. On the helical wheel plot (Fig. 3G), sensitive sites cluster along one side of the helix, and it is apparent that in a fairly broad helical face, about half of the helix is tolerant to tryptophan substitution.
Although our study identified many mutants with reduced and abolished function, only in rare cases did tryptophan substitution increase conductance. Substitutions at either end of TM3 increased conductance with mean Gj values of I137W and Y151W greater than double that of wtCx32 (Fig. 2C). Although the difference was only significant for Y151W (p < 0.05, Student's t test), the results are consistent with reports that large aromatic residues play an important role in positioning helices within the membrane (32) and suggest that positioning aromatic residues close to the TM border could influence structure and function of the channel.
Tryptophan Substitution in TM4
Tryptophan was substituted at sites Ala-196 through Tyr-211 in TM4. Function was completely abolished at four sites (Cys-201, Glu-208, Val-209, and Val-210) and significantly reduced at six (Ala-196, Ala-197, Ser-198, Gly-199, Leu-204, and Asn-205), totaling 10 out of 15 sites (∼67%) that were sensitive to tryptophan substitution. The distribution of sensitive sites is shown in helical net format (Fig. 3D) and helical wheel format (Fig. 3H) where it appears that sensitive sites are distributed broadly around and along the TM4 helix.
Cx32 Hemichannel Model
A comparative molecular model of rCx32 hemichannel was created to display the location of tryptophan-sensitive sites in three dimensions. The model is based on the crystal structure of human Cx26 (10), which, like Cx32, is a β-type connexin. rCx32 and hCx26 share about 65% sequence identity (Fig. 1B); therefore, the architecture of gap junction channels composed on rCx32 and hCx26 is expected to be similar.
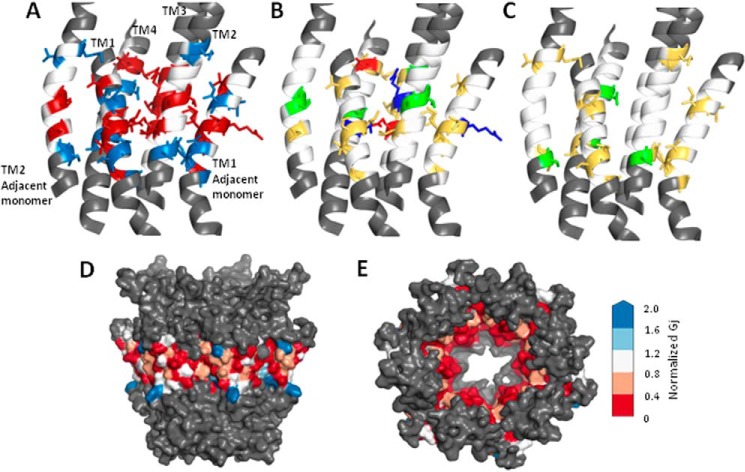
In the modeled Cx32 hemichannel, tryptophan-sensitive sites are widely distributed within the TM domains and appear to cluster in regions that face the pore or other TM domains. The three-dimensional location of all residues where tryptophan scanning produced altered function is presented in Fig. 4A. Because it is difficult to interpret the consequences of substitutions involving removal of charged and polar amino acids, the location of tryptophan-sensitive sites in the Cx32 model was further investigated by considering side-chain properties. Fig. 4, B and C, show the results of tryptophan scanning according to residue type. Fig. 4B focuses on sites where tryptophan completely abolished function and shows that a significant proportion of these are hydrophobic. In fact, many are hydrophobic sites distributed at interfaces between helices. There also appears to be a clustering of tryptophan-sensitive hydrophobic residues in the mid-region of the membrane, including the sensitive mid-region of TM1 (Arg-32 to Val-35, Fig. 3A).
FIGURE 4.
Location of tryptophan-sensitive residues on a Cx32 model. A–C, schematic representation of four TM domains of one Cx32 subunit, including TM1 from the clockwise adjacent monomer and TM2 from the counterclockwise adjacent monomer. The N-terminal helix of the central monomer, which lies adjacent to TM1, has been omitted for clarity. The region assessed by tryptophan scanning is shown in white when no effect was produced and using an appropriate color for altered function (see below). Residues outside the targeted tryptophan scan are colored gray. A, location of all residues where tryptophan mutations produced altered function: abolished function (red side chains), significantly reduced function (blue side chains). B, location of all residues where tryptophan mutation produced nonfunctional channels with side chains colored by residue type: red, denoting negatively charged side chains; blue representing positively charged side chains; green representing polar side chains; and yellow representing hydrophobic (apolar or aromatic) side chains. C, reduced function sites with side chains colored by residue type, with red denoting negatively charged side chains, blue representing positively charged side chains, green representing polar side chains, and yellow representing hydrophobic (apolar or aromatic) side chains. D and E, results of tryptophan scanning displayed on a surface rendered Cx32 model with residues color-coded according to normalized conductance values as indicated in the spectrum on the right. Colors range from red (abolished function) and white (unaffected) to blue (increased function) D, side view of the hemichannel with cytoplasmic end at top. E, view through the pore from the cytoplasmic mouth, with the N terminus removed to show color coding of tryptophan-sensitive sites.
Sites where tryptophan significantly reduced but did not abolish function are highlighted in Fig. 4C. The native amino acid at these mildly sensitive sites tended to be nonpolar/aromatic (yellow) or polar (green). There were no charged residues (red/blue) in the group of substitutions, and they tended to occur toward the ends of transmembrane domains. Sensitive sites are concentrated just above (extracellular to) and below (cytoplasmic to) the sensitive mid-region apparent in Fig. 4B.
Fig. 4, D and E, demonstrates the sensitivity of lipid- and pore-facing regions to tryptophan substitution. Using a surface-rendering version of the Cx32 hemichannel model, tryptophan sensitivity was graded based on mean conductance change. The correspondence between normalized Gj (from Fig. 2) and color assigned in the model is provided to the right. All tested residues are colored, although only those at the surface are visible in the surface-rendering model.
When membrane-spanning regions are highlighted (Fig. 4D), it is apparent that they comprise a relatively narrow portion of the external surface of the hemichannel. The colored region represents tested sites, including about four rotations of the TM helices (15–16 residues). Residues at the predicted TM borders were excluded from analysis based on previous reports that large aromatic side chains can influence positioning of TM helices (32). The side view (Fig. 4D) clearly shows membrane-facing contours splayed outward at the predicted cytoplasmic and extracellular boundaries of the membrane, supporting the premise that aromatic side chains are involved in positioning TM helices. Blue residues in Fig. 4D represent sites where tryptophan induced a gain of function. Specifically, the substitutions were I137W and Y151W, which fall at opposite ends of TM3.
In general, tryptophan substitution at the lipid interface had variable effects with some sites unaffected by tryptophan substitution (Fig. 4D, white), others reduced (Fig. 4D, pink/red) or increased (Fig. 4D, blue). A small number of lipid-facing sites showed high sensitivity to tryptophan substitution ((Fig. 4D, red). These residues could mediate interactions with lipids, adjacent proteins, or be involved in TM domain interactions in channel conformations that differ from the conformation of the model. The highly sensitive ((Fig. 4D, red) residues facing the lipid bilayer occur within a narrow strip along the TM4 helix (Ala-196, Ile-203, and Val-210) suggesting that a slight helical rotation could alter their position relative to other TM domains.
Fig. 4E highlights the sensitivity of pore-facing residues to tryptophan substitution. The pore is viewed from the cytoplasmic mouth, with the N terminus omitted for clarity. Only residues considered to be within the membrane-spanning region are colored, representing their sensitivity to tryptophan substitution. As one might predict, pore-facing residues appear to be highly sensitive with most of the pore contours appearing red (Fig. 4E). In the Cx32 model, the pore lining is contributed by TM1 (Ile-30, Ile-33, Met-34, and Val-37) and TM2 (Leu-81, Ile-82, Ser-85, Thr-86, Leu-89, and Met-93).
Intracellular Water Pocket
Molecular dynamic simulations of an hCx26 hemichannel revealed a water pocket positioned between the intracellular ends of the transmembrane domains (14). This water pocket, named th IC pocket, extends about one-third of the way through the membrane and is stabilized by hydrophilic residues in TM1, TM3, and TM4. As expected from the high sequence identity between hCx26 and rCx32, a similar environment exists in our modeled Cx32 hemichannel. The location of the IC pocket in hCx26 is shown in Fig. 5A (adapted from 14) and in the Cx32 hemichannel model in Fig. 5B. In Cx32, the IC pocket is formed partially by hydrophilic residues in TM1 (Arg-32), TM2 (Thr-86), TM3 (Ser-138, Arg-142, and Glu-146), and TM4 (Glu-208). Tryptophan substitution at these sites rendered channels nonfunctional.
FIGURE 5.
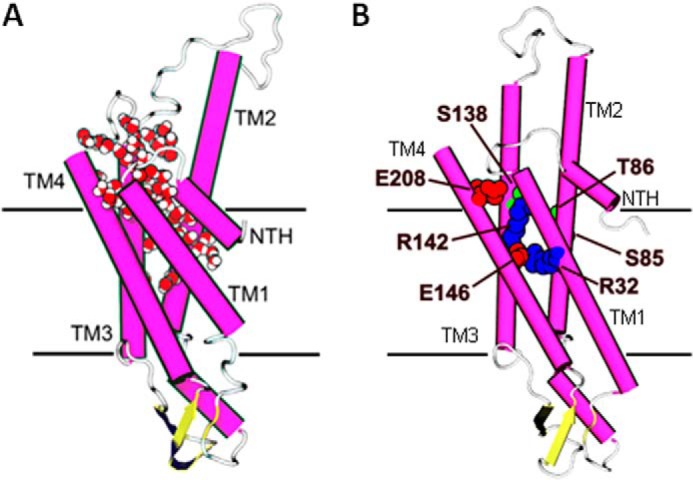
Several sites sensitive to tryptophan substitution face a putative water pocket identified during MD simulations of a Cx26 hemichannel (14). A, one monomer of the Cx26 MD-simulated model showing location of a water pocket accessible from the intracellular face of the membrane. Protein has been colored by secondary structure (magenta, α-helix; yellow, β-sheet; white, loop). Water molecules filling the IC pocket are shown in van der Waals representation colored by element (red, oxygen; white, hydrogen) (Ref. 14 reprinted with permission from Biophys. J.). B, Cx32 model showing only the nonfunctional related residues with hydrophilic properties, red representing negatively charged side chains, blue representing positively charged side chains, and green representing polar side chains.
TM Domain Interactions and Vj Gating
The broad map of tryptophan-sensitive sites provides a framework for further investigations aimed at defining specific interactions important for oligomerization, trafficking docking, gating, and other channel functions. Future studies may focus on specific residues or on specific aspects of channel function to tease out this information.
In this study, a subset of interactions important for channel gating was identified using techniques of cellular electrophysiology complemented by molecular modeling approaches. This group of mutants was identified based on an altered response to application of transjunctional voltage (Vj) involving activation rather than inactivation. Cx32 is mildly sensitive to Vj, displaying half-maximal inactivation at Vj values around ±70 mV and a maximal conductance decrease of about 40% (39). In contrast, a group of tryptophan mutants produced junctional currents that were small at rest but activated in a time- and voltage-dependent manner in response to Vj. This phenotype is a fairly common consequence of amino acid substitutions within the TM domains of connexins and innexins (20, 33–38), and it is known to result from reduced open probability at Vj = 0 mV (36). For these channels, there is a shift in the conductance versus voltage relationship such that Gjmax is offset from 0 mV (37). Because reverse-gating channels have a reduced open probability at Vj = 0 mV, they often fail to induce coupling when paired homotypically. In this study, all mutants were paired with wild type Cx32 for the functional assay, and the phenotype was readily observed.
Voltage-dependent gating of gap junction channels is known to involve at least two distinct mechanisms, a loop gate and a Vj gate, which are likely to have independent sensors and gates (39). It is not known whether the reverse-gating phenotype results from disruption of the loop gate, the Vj gate, or another as yet unidentified gating mechanism. The reverse-gating sites are highlighted in Fig. 6A, which shows the amino acid sequence of each TM domain in Cx32 with tryptophan-sensitive sites color-coded as in previous figures with red representing abolished function and blue representing reduced function. Below the individual amino acids, letters are used to indicate substitutions that induced a reverse-gating phenotype, including tryptophan substitution mutants identified in this study (W), cysteine substitution mutants (C (33, 38)), and CMTX mutants (36).
FIGURE 6.
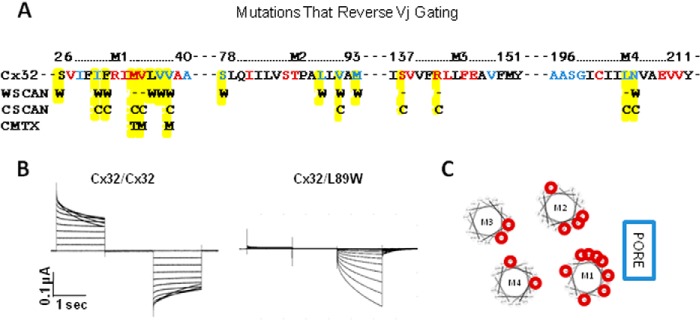
Sites where mutations “reverse” voltage gating in Cx32, possibly through a mechanism that involves disruption of TM domain interactions. A, amino acid sequence of TM domains highlighting tryptophan mutants with abolished (red) or reduced (blue) function. Letters below individual amino acids indicate the substitutions that reverse voltage gating (W, tryptophan mutant; C, cysteine mutant; other, CMTX mutant). Data from Skerrett et al. (33), Toloue et al. (38), and Oh et al. (36). B, junctional currents recorded from oocytes expressing Cx32/Cx32 (left) and Cx32/L89W (right). When recording intercellular currents, both oocytes were clamped at −20 mV, and currents were recorded from a continuously clamped oocyte although its partner was pulsed to +80 and −120 mV in 10-mV steps. C, helical wheel plots arranged to represent the relative positions of TM domains with respect to the channel pore. Sites where point mutations are reverse voltage gating are circled in red.
Fig. 6B shows examples of intercellular currents mediated by wtCx32 (left) and a reverse-gating mutant Cx32L89W (right). Note that Cx32/Cx32 currents inactivate symmetrically in a time- and voltage-dependent manner, whereas Cx32/L89W currents activate in response to Vj of only one polarity. All reverse-gating mutants activated in response to relatively positive Vj but displayed different levels of voltage sensitivity and kinetics of activation. Three of the tryptophan substitution mutants tested in this study displayed the reverse-gating phenotype but no significant change in conductance. They include S26W, F31W, and L36W (Fig. 6A). This is possible because normalized conductance (displayed in histograms in Fig. 1) is based on maximal junctional currents. For most channels, maximal conductance occurred at the start of the voltage pulse, but for reverse-gating mutants, maximum conductance was reached at the end of the voltage pulse (Fig. 6B).
All gating-sensitive sites are displayed in helical wheel format in Fig. 6C with helices arranged as described in the atomic model of Cx26 (10) and in our Cx32 hemichannel model. Most of the sensitive sites lie in TM1, with four interactions apparent in TM2 and two each in TM3 and TM4. The distribution of gating-sensitive TM domain interactions suggests a model where gating occurs via global conformational changes centered around TM1. Because TM1 faces the pore and interacts with the infolded N terminus (10), it makes sense that disrupting or rearranging TM domain interactions could cause the channel to “fall” into a closed state. It is likely that the collapsed state involves displacement of the N-terminal helix caused by disruption of interactions with TM1. This explanation is supported by molecular dynamic simulations of Cx26M34T where changes in hydrophobic interactions between Met-34 with the N-terminal tryptophan (Trp-2) destabilize the pore funnel (40).
TM1 Interactions in Cx43
To determine whether the results of tryptophan scanning could be extrapolated to other gap junction channels, TM1 of Cx43 was scanned. Thirteen tryptophan substitution mutants were created in TM1, and the mutants were expressed in Xenopus oocytes, and conductance was assessed as described for Cx32. The results are summarized in histogram format in Fig. 7A. The first TM domain of Cx43 was highly sensitive to tryptophan substitution with eight of 13 sites displaying a significant reduction in coupling. Because Cx43 interacts with endogenous XeCx38, a minimal level of coupling, about 3% of the normalized conductance was observed in the negative control (Fig. 7A, oligo). This made it impossible to differentiate between reduced and abolished coupling. Hence, all sensitive sites are indicated with red coloring (Fig. 7, A and B). Despite this complication, the results were similar to those observed for TM1 of Cx32, including high sensitivity in the mid-region of the helix and broad sensitivity throughout. The helical face, including Arg-33 (Arg-32 in Cx32), is sensitive below and above the conserved arginine, similar to the results in Cx32. Differences are obvious toward the extracellular end of TM1, where the sequences differ significantly (Cx32 Leu-36, Val-37, and Val-38 versus Cx43 Leu-37, Gly-38, and Thr-39), and the scan of Cx32 included additional residues (Ala-39 and Ala-40). In general, however, the results were quite consistent. This clearly contrasts the results of tryptophan scanning of an innexin-based gap junction channel (ShakBL) where sensitivity was limited to a narrow helical face (Fig. 7C) (20). The scan of Cx43 further confirms that TM1 is more tightly packed in connexons than innexons.
FIGURE 7.
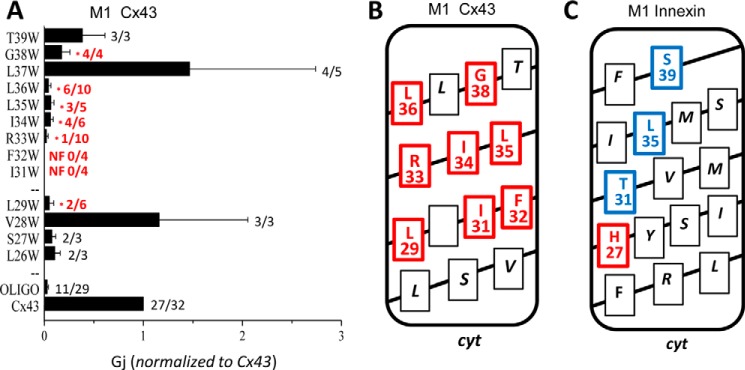
Tryptophan scanning reveals similarities and differences in TM domain packing of connexins and innexins. A, tryptophan scan of Cx43 TM1. Histogram shows mean junctional conductance recorded for tryptophan substitution mutants expressed in Xenopus oocytes. Mutants were paired heterotypically with wild type Cx32, and conductance was normalized to that of wild type Cx43 pairs of the same batch on the same day. A baseline level of conductance is noted in the Cx43/oligonucleotide control because Cx43 pairs with XeCx38 minimally after knockdown with the antisense oligonucleotide. This prevented discrimination between substitutions that abolished and reduced function. The numbers beside each bar represent the number with measurable conductance versus the number tested (e.g. number of functional/number of tested). * = statistically reduced compared with wtCx43 at p ≤ 0.05 as determined Student's t test. B, helical net plot highlighting location of sites in Cx43 where tryptophan substitution abolished (red) or significantly reduced (blue) conductance. C, for comparison, tryptophan scanning results for TM1 of an innexin (ShakBL) are shown in helical net format (redrawn from Ref. 20). TM1 of the innexins is loosely packed compared with Cx32 and Cx43.
Discussion
The technique of tryptophan scanning was applied to Cx32 to determine the location and importance of TM domain interactions in gap junction channels. Our approach involved a primary screen of sites in all TM domain, and our assay was the formation of functional intercellular channels between paired oocytes. A surprisingly high percentage of mutants displayed reduced or abolished function revealing that interactions between transmembrane helices are extensive and critical.
The high sensitivity to tryptophan substitution seems atypical of channels and transporters. Published outcomes of tryptophan scanning reveal tolerance ranges from very high, in the case of the S4 domain of HERG K+ channels where all substitutions were tolerated (25), to tolerance across a broad helical face (24) or within a specific region of the helix (19, 21). The extent of sensitivity in Cx32 was particularly surprising in TM1 considering our previous study of a gap junction channel composed on an innexin revealed a very narrow tryptophan-sensitive helical face (20).
Model of Cx32 Hemichannel
Results were displayed and interpreted using a modeled Cx32 hemichannel. The model was developed based on the atomic model (crystal structure) of a Cx26 gap junction channel (10), and due to the high level of sequence similarity between the two connexins, the modeled hemichannel of Cx32 is nearly identical to the Cx26 structure on which it is based. The model is expected to represent an open state of the channel, as is the Cx26 atomic model, based on pore diameter and crystallization conditions. Molecular dynamic simulations have been applied to refine the atomic model of Cx26 and suggest that a widened pore and very minor rearrangements of the transmembrane helices are more consistent with measurements of channel conductance in the open state (13).
In terms of transmembrane domain architecture, there is relatively little difference between our Cx32 model, the Cx26 atomic model (10), and previous MD simulations (13, 14). Important features revealed on our model include dense packing of TM domains, concentrated sensitivity in the mid-region, and a clustering of voltage-sensitive sites within and around TM1. Pore-facing sites were found to be highly sensitive to tryptophan substitution in contrast to lipid-facing regions, which displayed variable tolerance. One important difference between the modeled structures that was critical to interpretation of our results was the IC pocket apparent in the MD simulations of Araya-Secchi et al. (14). We chose to include the IC pocket in our model because it helps to explain the high sensitivity of a number of hydrophilic residues.
Importance of Mid-region
Sites where tryptophan abolished function cluster in the mid-region of the membrane include a complete rotation of TM1 (Arg-32, Ile-33, Met-34, and Val-35), two hydrophilic residues in TM2 (Ser-85 and Thr-86), several sites in TM3 (Arg-142, Leu-143, Phe-145, and Glu-146), and one residue in TM4 (Cys-201). This suggests that an integral structural component(s) of TM domain assembly or function is/are centered in this region. Previous structural analyses support the existence of critical interactions between TM domains in the mid-region of the channel, where side-chain properties such as length or branching have been reported to influence channel function. For instance at position Met-34, shorter side chains (Ala, Thr, and Cys) cause a reduction in function; side-chains similar in length (Leu and Gln) produce channels with normal function, and very large side chains (Trp) completely abolish function (38). Similar observations occur with respect to branching at position Val-91 (38).
Voltage Gating
It has been proposed that voltage-dependent gating occurs when charged residues in the N terminus cause the short N-terminal helix to be displaced, resulting in closure of the channel (10, 40). In this study, 11 tryptophan mutants were identified that altered Vj gating. In conjunction with analysis of cysteine mutants (37) and CMTX mutants (36), 16 sites have been identified in Cx32 where TM domain interactions are likely to influence channel gating. About half of these occur in TM1, although the other eight appear to be located in other helices facing TM1 (Fig. 6).
The distribution of sites where amino acid substitutions induce the reverse-gating phenotype is consistent with a model where interactions between the infolded N terminus and TM1 influence the gated state of the channel (10, 40). Most of the reverse-gating sites occur in the mid-region of the channel, which appears to involve many critical interactions. Those located outside this central region may result in global conformational adjustments that ultimately influence packing in this region. A plausible scenario is that reverse gating represents an intermediate step between functional and nonfunctional channels that occurs when minor perturbations alter the orientation of TM1 with respect to the pore. Disruption of interactions, if minor, cause the channel to collapse into a closed state that can be influenced to open by application of Vj, although larger perturbations completely abolish function. This is substantiated by studies showing that “size matters” at residue Met-34, and “branching matters” for residue Val-91 (38). In both of these instances, minor modifications to side chain properties induce reverse gating, and major modifications abolish function.
There is relevance in further identifying conformational changes associated with reverse gating because it appears to be a common result of missense mutations associated with connexin diseases and therefore a pathological state of the channel. For instance, the reverse-gating mutations in Cx26 M34T cause deafness (41), and the mutations M34T, V35M, and V38M in Cx32 cause CMTX (36). Voltage gating in gap junction channels is complex and involves gates that act in response to Vm and Vj (39). The reverse-gating phenotype may involve a shift in Vj sensitivity (36, 37), a modified response of the Vm gate to transjunctional voltage, or a conformation unrelated to either of the previously characterized voltage-gating mechanisms. One observation argues against the involvement of typical Vj-gating mechanisms. Cx26 and Cx32 have Vj gates of opposite polarity (38, 39) yet reverse-gating mutants of both connexins open in response to relatively positive Vj.
Residues Facing the Pore
Our Cx32 hemichannel model, consistent with currently accepted models of connexin-based gap junction structure, places TM1 and TM2 adjacent to the pore (10, 13, 14). Tryptophan substitutions were poorly tolerated at TM domain sites associated with pore-lining positions (Fig. 4E), which might be expected for a couple of reasons. First, at the cytoplasmic end of the channel, inserting a large bulky side chain is likely to disrupt positioning of the N terminus. Second, high sensitivity to tryptophan insertion is expected at the extracellular end of the pore where it narrows.
Previous cysteine accessibility studies presented TM3 as the main pore-lining domain due to extensive accessibility along the helix; however, many residues in TM1 were accessible to sulfhydryl reagents. These correlate well with pore-lining residues in the atomic model (10) and the Cx32 hemichannel model developed for this study. Five cysteine mutants in TM1 (I30C, F31C, I33C, M34C, and V35C) were accessible to sulfhydryl reagent (33). Three of these are sites where tryptophan substitutions rendered the channels nonfunctional (I33W, M34W, and V35W) and one exhibited reduced function (I30W). Only Phe-31 was functional as a tryptophan mutant (F31W), which might be expected given the nature of the substitution. Interpretation of the results of cysteine scanning was complicated by the fact that four of the cysteine mutants (I30C, F31C, M34C, and V35C) displayed the reverse-gating phenotype, and one was accessible in nonperfused cells (I33C) where sulfhydryl reagents should not have been able to access the pore. However, the results of cysteine accessibility are largely consistent with the position of these TM1 residues in our Cx32 hemichannel model.
Residues Facing the Lipid Bilayer
It was generally expected that substitutions would be well tolerated at the protein-lipid interface where the bulky hydrophobic side chain of tryptophan would be easily accommodated by the flexible environment of the fatty acid tails (21). Surprisingly, tryptophan substitutions on the external surface of the channel had a wide range of effects when viewed on the Cx32 hemichannel model (Fig. 4D, membrane-spanning regions colored). The colored region represents about four rotations of a TM helix, corresponding to about 15 residues that were subjected to tryptophan scanning. The membrane-spanning domains appear as a relatively narrow region on the external surface of the hemichannel, and the channel appears to splay outward at the membrane borders. Several substitutions at the TM domain borders caused an increase in conductance (blue residues in Fig. 3A). Several substitutions at the TM domain borders caused an increase in conductance (Fig. 4D, blue). Other residues at the protein-lipid interface range from highly sensitive (Fig. 4D, red) to slightly sensitive (Fig. 4D, pink) and tolerant (Fig. 4D, white). High sensitivity was not expected at the lipid interface. The presence of Trp-sensitive sites in this region may suggest that the lipid-facing region of the channel plays important functions that have not yet been identified or that sensitive residues are sites of transmembrane domain interaction in a channel conformation not apparent in the modeled channel. Future studies should be aimed at clarifying the role of TM3 residues.
Comparing Connexin- and Innexin-based Channels
One of the earliest and most significant applications of tryptophan scanning involved a comparison of related proteins. Sharp et al. (21) identified structural differences between flagellar motility proteins MotA and MotB based on differing sensitivities to tryptophan. More recently De Feo et al. (43) have identified structural similarities between distantly related copper transporters using tryptophan scanning. DePriest et al. (20) previously demonstrated that TM1 of an innexin was quite tolerant to tryptophan substitution. The results of tryptophan scanning in Cx32 suggest that connexin-based channels are more densely packed than their innexin-based counterparts. To determine whether other connexins are also densely packed, TM1 of Cx43 was scanned with results comparable with those obtained for Cx32. Both Cx32 and Cx43 displayed a highly sensitive region in the middle of TM1, where tryptophan substitution was very poorly tolerated over at least an entire rotation of the helix. This is very different from the innexin, which showed limited sensitivity along one face of the TM1 helix (20).
Connexins and innexins reportedly evolved convergently (44) but are surprisingly similar in their topology and function (1, 12). Innexin channels generally have a larger outer diameter (12, 42), consistent with looser packing of TM domains. Advances in the biology of innexin channels are likely to provide exciting information about their similarities and differences.
Acknowledgments
We thank the students in Advanced Cell Biology at SUNY Buffalo State for technical assistance.
This work was supported in part by an American Heart Association National Affiliate Scientist Development grant and the State University of New York Buffalo State Incentive Funding Program. The authors declare that they have no conflicts of interest with the contents of this article.
- CMTX
- Charcot-Marie-Tooth disease type X
- TM
- transmembrane
- MD
- molecular dynamics
- μS
- microsiemens
- h
- human
- r
- rat.
References
- 1. Goodenough D. A., Paul D. L. (2009) Gap junctions. Cold Spring Harb. Perspect. Biol. 1, a002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Willecke K., Eiberger J., Degen J., Eckardt D., Romualdi A., Güldenagel M., Deutsch U., Söhl G. (2002) Structural and functional diversity of connexin genes in the mouse and human genome. Biol. Chem. 383, 725–737 [DOI] [PubMed] [Google Scholar]
- 3. Kumar N. M., Gilula N. B. (1996) The gap junction communication channel. Cell 84, 381–388 [DOI] [PubMed] [Google Scholar]
- 4. Bergoffen J., Scherer S. S., Wang S., Scott M. O., Bone L. J., Paul D. L., Chen K., Lensch M. W., Chance P. F., Fischbeck K. H. (1993) Connexin mutations in X-linked Charcot-Marie-Tooth disease. Science 262, 2039–2042 [DOI] [PubMed] [Google Scholar]
- 5. Kleopa K. A., Abrams C. K., Scherer S. S. (2012) How do mutations in GJB1 cause X-linked Charcot-Marie-Tooth disease? Brain Res. 1487, 198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balice-Gordon R. J., Bone L. J., Scherer S. S. (1998) Functional gap junctions in the Schwann cell myelin sheath. J. Cell Biol. 142, 1095–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shy M. E., Siskind C., Swan E. R., Krajewski K. M., Doherty T., Fuerst D. R., Ainsworth P. J., Lewis R. A., Scherer S. S., Hahn A. F. (2007) CMT1X phenotypes represent loss of GJB1 gene function. Neurology 68, 849–855 [DOI] [PubMed] [Google Scholar]
- 8. Milks L. C., Kumar N. M., Houghten R., Unwin N., Gilula N. B. (1988) Topology of the 32-kd liver gap junction protein determined by site-directed antibody localizations. EMBO J. 7, 2967–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bruzzone R., White T. W., Paul D. L. (1996) Connections with connexins: the molecular basis of direct intercellular signaling. Eur. J. Biochem. 238, 1–27 [DOI] [PubMed] [Google Scholar]
- 10. Maeda S., Nakagawa S., Suga M., Yamashita E., Oshima A., Fujiyoshi Y., Tsukihara T. (2009) Structure of the connexin 26 gap junction channel at 3.5 Å resolution. Nature 458, 597–602 [DOI] [PubMed] [Google Scholar]
- 11. Oshima A., Tani K., Hiroaki Y., Fujiyoshi Y., Sosinsky G. E. (2007) Three-dimensional structure of a human connexin26 gap junction channel reveals a plug in the vestibule. Proc. Natl. Acad. Sci. U.S.A. 104, 10034–10039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oshima A., Matsuzawa T., Nishikawa K., Fujiyoshi Y. (2013) Oligomeric structure and functional characterization of Caenorhabditis elegans Innexin-6 gap junction protein. J. Biol. Chem. 288, 10513–10521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kwon T., Harris A. L., Rossi A., Bargiello T. A. (2011) Molecular dynamics simulations of the Cx26 hemichannel: evaluation of structural models with Brownian dynamics. J. Gen Physiol. 138, 475–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Araya-Secchi R., Perez-Acle T., Kang S. G., Huynh T., Bernardin A., Escalona Y., Garate J. A., Martínez A. D., García I. E., Sáez J. C., Zhou R. (2014) Characterization of a novel water pocket inside the human Cx26 hemichannel structure. Biophys. J. 107, 599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee S. P., Xie Z., Varghese G., Nguyen T., O'Dowd B. F., George S. R. (2000) Oligomerization of dopamine and serotonin receptors. Neuropsychopharmacology 23, S32–S40 [DOI] [PubMed] [Google Scholar]
- 16. Laird D. W. (2006) Life cycle of connexins in health and disease. Biochem. J. 394, 527–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith T. D., Mohankumar A., Minogue P. J., Beyer E. C., Berthoud V. M., Koval M. (2012) Cytoplasmic amino acids within the membrane interface region influence connexin oligomerization. J. Membr. Biol. 245, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matthews E. E., Zoonens M., Engelman D. M. (2006) Dynamic helix interactions in transmembrane signaling. Cell 127, 447–450 [DOI] [PubMed] [Google Scholar]
- 19. Otero-Cruz J. D., Báez-Pagán C. A., Caraballo-González I. M., Lasalde-Dominicci J. A. (2007) Tryptophan-scanning mutagenesis in the αM3 transmembrane domain of the muscle-type acetylcholine receptor–a spring model revealed. J. Biol. Chem. 282, 9162–9171 [DOI] [PubMed] [Google Scholar]
- 20. Depriest A., Phelan P., Martha Skerrett I. (2011) Tryptophan-scanning mutagenesis of the first transmembrane domain of the innexin Shaking-B(Lethal). Biophys. J. 101, 2408–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sharp L. L., Zhou J., Blair D. F. (1995) Tryptophan-scanning mutagenesis of MotB, an integral membrane protein essential for flagellar rotation in Escherichia coli. Biochemistry 34, 9166–9171 [DOI] [PubMed] [Google Scholar]
- 22. Guzmán G. R., Santiago J., Ricardo A., Martí-Arbona R., Rojas L. V., Lasalde-Dominicci J. A. (2003) Tryptophan scanning mutagenesis in the αM3 transmembrane domain of the Torpedo californica acetylcholine receptor: functional and structural implications. Biochemistry 42, 12243–12250 [DOI] [PubMed] [Google Scholar]
- 23. Ishii T. M., Nakashima N., Ohmori H. (2007) Tryptophan-scanning mutagenesis in the S1 domain of mammalian HCN channel reveals residues critical for voltage-gated activation. J. Physiol. 579, 291–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ueno S., Lin A., Nikolaeva N., Trudell J. R., Mihic S. J., Harris R. A., Harrison N. L. (2000) Tryptophan scanning mutagenesis in TM2 of the GABAA receptor α subunit: effects on channel gating and regulation by ethanol. Br. J. Pharmacol. 131, 296–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Subbiah R. N., Kondo M., Campbell T. J., Vandenberg J. I. (2005) Tryptophan scanning mutagenesis of the HERG K+ channel: the S4 domain is loosely packed and likely to be lipid-exposed. J. Physiol. 569, 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Skerrett I. M., Merritt M., Zhou L., Zhu H., Cao F.-L., Smith J. F., Nicholson B. J. (2001) Applying the Xenopus oocyte expression system to the analysis of gap junction proteins. Methods Mol. Biol. 154, 225–249 [DOI] [PubMed] [Google Scholar]
- 27. Sali A., Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 28. Söding J., Biegert A., Lupas A. N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK–a program to check the stereochemical quality of protein structures. J. App. Crystallogr. 26, 283–291 [Google Scholar]
- 30. Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Errors in protein structures. Nature 381, 272–272 [DOI] [PubMed] [Google Scholar]
- 31. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 32. Mall S., Broadbridge R., Sharma R. P., Lee A. G., East J. M. (2000) Effects of aromatic residues at the end of transmembrane α-helices on helix interactions with lipid bilayers. Biochemistry 39, 2071–2078 [DOI] [PubMed] [Google Scholar]
- 33. Skerrett I. M., Aronowitz J., Shin J. H., Cymes G., Kasperek E., Cao F.-L., Nicholson B. J. (2002) Identification of amino acid residues lining the pore of a gap junction channel. J. Cell Biol. 159, 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Suchyna T. M., Xu L. X., Gao F., Fourtner C. R., Nicholson B. J. (1993) Identification of a proline residue as a transduction element involved in voltage gating of gap junctions. Nature 365, 847–849 [DOI] [PubMed] [Google Scholar]
- 35. Arora A., Minogue P. J., Liu X., Reddy M. A., Ainsworth J. R., Bhattacharya S. S., Webster A. R., Hunt D. M., Ebihara L., Moore A. T., Beyer E. C., Berthoud V. M. (2006) A novel GJA8 mutation is associated with autosomal dominant lamellar pulverulent cataract: further evidence for gap junction dysfunction in human cataract. J. Med. Genet. 43, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oh S., Ri Y., Bennett M. V., Trexler E. B., Verselis V. K., Bargiello T. A. (1997) Changes in permeability caused by connexin 32 mutations underlie X-linked Charcot-Marie-Tooth disease. Neuron 19, 927–938 [DOI] [PubMed] [Google Scholar]
- 37. Skerrett I. M., Smith J. F., Nicholson B. J. (1999) Mechanistic differences between chemical and electrical gating of gap junctions. Curr. Top. Membr. 49, 249–269 [Google Scholar]
- 38. Toloue M., Woolwine Y., Karcz J., Kasperek E., Nicholson B. J., Skerrett I. M. (2008) Site-directed mutagenesis reveals putative regions of protein interaction within the transmembrane domains of connexins. Cell Commun. Adhes. 15, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Harris A. L. (2001) Emerging issues of connexin channels: biophysics fills the gap. Q. Rev. Biophys. 34, 325–472 [DOI] [PubMed] [Google Scholar]
- 40. Zonta F., Buratto D., Cassini C., Bortolozzi M., Mammano F. (2014) Molecular dynamics simulations highlight structural and functional alterations in deafness-related M34T mutation of connexin 26. Front. Physiol. 5, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Skerrett I. M., Di W. L., Kasperek E. M., Kelsell D. P., Nicholson B. J. (2004) Aberrant gating, but a normal expression pattern, underlies the recessive phenotype of the deafness mutant Connexin26M34T. FASEB J. 18, 860–862 [DOI] [PubMed] [Google Scholar]
- 42. Peracchia C. (1973) Low resistance junctions in crayfish. II. Two arrays of globules in junctional membranes. J. Cell Biol. 57, 66–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. De Feo C. J., Mootien S., Unger V. (2010) Tryptophan scanning analysis of the membrane domain of CTR-copper transporters. J. Membr. Biol. 234, 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Alexopoulos H., Böttger A., Fischer S., Levin A., Wolf A., Fujisawa T., Hayakawa S., Gojobori T., Davies J. A., David C. N., Bacon J. P. (2004) Evolution of gap junctions: the missing link? Curr. Biol. 14, R879–R880 [DOI] [PubMed] [Google Scholar]