Background: The mechanism by which MutY discriminates against the anti-substrate oxoG:C remains unclear.
Results: MutY binds the C of the anti-substrate oxoG:C in an exo-site.
Conclusion: MutY uses a combination of exo-site stabilization and active site destabilization to prevent pro-mutagenic processing of oxoG:C.
Significance: This work explains how MutY avoids accelerating mutagenesis by processing of an inappropriate substrate.
Keywords: 8-oxoguanine (8-oxoG), base excision repair (BER), DNA repair, DNA-protein interaction, structural biology
Abstract
The highly mutagenic A:oxoG (8-oxoguanine) base pair in DNA most frequently arises by aberrant replication of the primary oxidative lesion C:oxoG. This lesion is particularly insidious because neither of its constituent nucleobases faithfully transmit genetic information from the original C:G base pair. Repair of A:oxoG is initiated by adenine DNA glycosylase, which catalyzes hydrolytic cleavage of the aberrant A nucleobase from the DNA backbone. These enzymes, MutY in bacteria and MUTYH in humans, scrupulously avoid processing of C:oxoG because cleavage of the C residue in C:oxoG would actually promote mutagenic conversion to A:oxoG. Here we analyze the structural basis for rejection of C:oxoG by MutY, using a synthetic crystallography approach to capture the enzyme in the process of inspecting the C:oxoG anti-substrate, with which it ordinarily binds only fleetingly. We find that MutY uses two distinct strategies to avoid presentation of C to the enzyme active site. Firstly, MutY possesses an exo-site that serves as a decoy for C, and secondly, repulsive forces with a key active site residue prevent stable insertion of C into the nucleobase recognition pocket within the enzyme active site.
Introduction
The genomic integrity of living organisms is threatened by a wide variety of chemical and biological agents that damage DNA. One of the most common and genotoxic forms of damage is 8-oxoguanine (oxoG),3 which arises through the attack of reactive oxygen species on guanine (see Fig. 1A) (1). OxoG preferentially mispairs with adenine during replicative DNA polymerization, thereby giving rise to G:C to T:A transversion mutations (see Fig. 1B). Cellular resistance to the genotoxic effects of guanine oxidation is conferred by the evolutionarily conserved GO system, which consists of several proteins that intercept oxoG and its misreplication products at multiple stages of the mutagenesis process (2). The orthologous 8-oxo-dGTPases MutT (eubacteria) and hMTH1 (humans) sanitize the nucleotide precursor pool by directly hydrolyzing 8-oxo-dGTP to 8-oxo-dGMP; the paralogous DNA glycosylases MutM and hOGG1 (bacteria and humans, respectively) selectively cleave the oxoG nucleobase from oxoG:C base pairs (3, 4). Failure to repair oxoG:C results in the production of oxoG:A (5), a lesion that is particularly insidious because neither nucleobase conveys faithful information with which to direct repair of the other. Although the oxoG moiety is constitutionally aberrant and therefore readily distinguishable from undamaged DNA, the A moiety is only contextually aberrant and otherwise comprises one of four nucleobases in DNA; the difference between faithful repair and wreaking havoc on the genome therefore depends critically on recognition of the base-pairing partner opposite A, being either oxoG (lesion) or T (undamaged). Repair of this lesion utilizes an oxoG:A-specific DNA glycosylase, bacterial MutY or human MUTYH, which leaves the oxoG unaffected and instead cleaves the contextually aberrant A, severing its glycosidic bond (see Fig. 1C) (3, 6) The resulting abasic site undergoes short-patch DNA repair resynthesis to furnish primarily oxoG:C, along with a lesser amount of oxoG:A. The latter is again acted upon by MutY/MUTYH, and the former is processed by MutM/hOGG1.
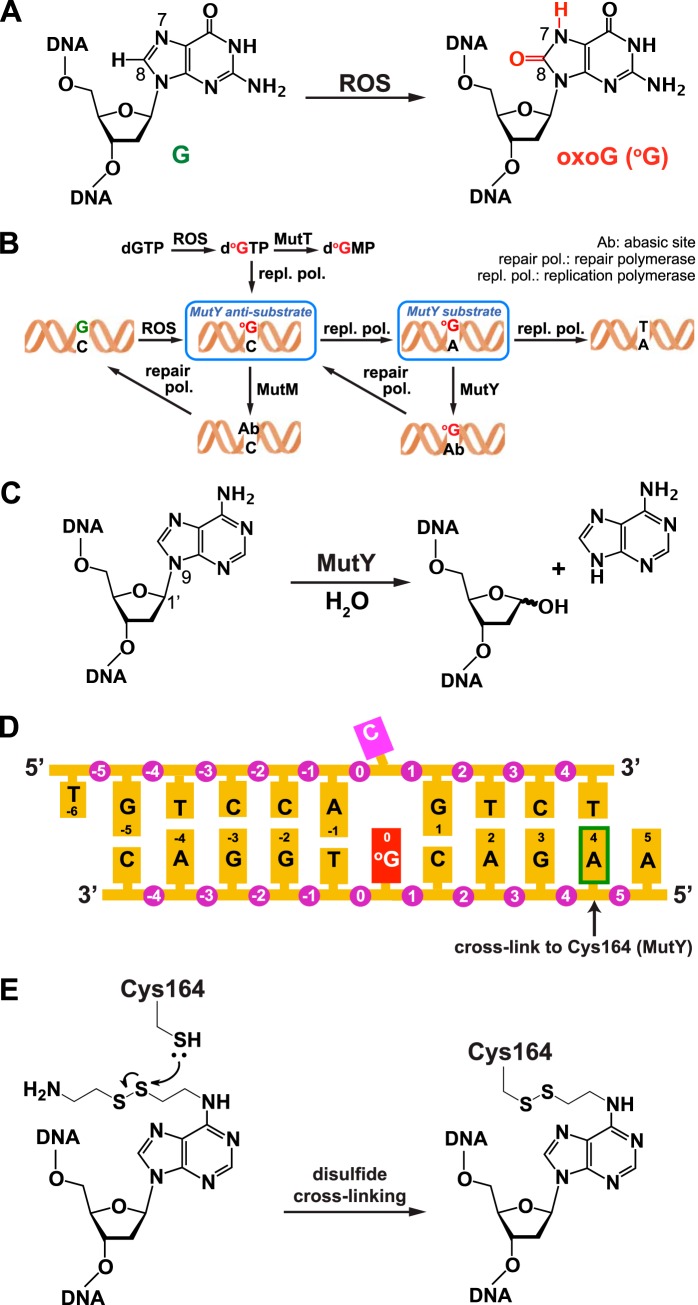
FIGURE 1.
Generation and repair of oxoG:C and oxoG:A lesions in DNA. A, the primary oxoG lesion is formed by attack of reactive oxygen species (ROS) on a guanine residue. B, pathway for repair of oxoG lesions in DNA. C, hydrolytic cleavage of the target glycosidic bond catalyzed by MutY. D, duplex DNA sequence used in this work. The arrow indicates the adenine that is modified (as shown in panel E) to cross-link to an engineered Cys residue in MutY. E, structure of the modified adenine used in disulfide cross-linking and of the covalent linkage it forms with MutY.
The severe risk posed by oxoG:A mispairs, and correspondingly, the criticality of oxoG:A repair by MutY and MUTYH, are underscored by the extraordinarily high rate of G:C to T:A transversion mutations in MutY-deficient bacteria (7, 8) and by the established role of MUTYH as a protector against genomic instability in humans. Loss-of-function mutations in MUTYH are associated with a high rate of G:C to T:A transversions in colorectal cancer patients (9). Furthermore, inactivation of MUTYH leads to tumorigenic mutations in other genes, including the tumor suppressor APC (9) and the protooncogene KRAS (10). The oxoG:A mispair can also be generated in DNA via replicative misincorporation of 8-oxodGTP; recent studies have shown that inhibition of hMTH1 elevates genomic oxoG levels and is profoundly toxic to transformed human cells (11, 12). The realization that adenine DNA glycosylases are among the most important guardians of the genome has fueled interest in understanding the structure and function of MutY/MUTYH.
MutY and MUTYH belong to a large superfamily of structurally related DNA glycosylases having a signature helix-hairpin-helix motif followed by a Gly/Pro-rich loop and a key catalytic aspartic acid residue (HhH-GPD superfamily) (13, 14). The domain bearing the HhH-GPD motif lies at the N terminus of the protein and contains the catalytic pocket that recognizes and cleaves the substrate adenine (15, 16). MutY (and MUTYH) also contains a C-terminal domain that recognizes oxoG, thereby providing a means for discrimination of oxoG:A lesions versus undamaged T:A base pairs (17, 18). Equally important is avoidance of C excision from oxoG:C base pairs as this would accelerate transversion mutation by provoking repair-dependent formation of oxoG:A base pairs; indeed, MutY and mammalian MYH appear incapable of acting upon oxoG:C (19, 20). The structure of a lesion recognition complex having a catalytically incompetent but recognition-competent mutant MutY protein (D144N) bound to DNA bearing a central oxoG:A lesion has shed light on the mechanism of oxoG:A recognition (16). The substrate adenine is extruded from the DNA stack and inserted into an extrahelical active pocket, whereas the oxoG nucleobase is fully intrahelical, contacting primarily the N-terminal domain but also the C-terminal domain. In a related lesion recognition complex structure having a wild-type enzyme active site but an uncleavable “mutated” target nucleobase, 2′-fluoro-2′-deoxyadenosine, (FLRC, for fluorinated lesion recognition complex), the substrate adenine is plunged even more deeply into the active pocket, so as to reveal that adenine excision proceeds via general acid catalysis (21). The FLRC is therefore considered the structure that most accurately represents MutY poised to catalyze base excision. Although the molecular basis underlying recognition and excision of the oxoG:A substrate is well established both structurally and biochemically, the means by which MutY discriminates against oxoG:C as an anti-substrate remains a mystery. Here we have addressed this issue by analyzing at atomic resolution two complexes in which MutY has been captured in the act of interrogating a target oxoG:C base pair. These structures and the accompanying biochemical analysis suggest that MutY possesses a gatekeeper at the active site entrance that diverts C residues.
Experimental Procedures
Geobacillus stearothermophilus MutY Preparation
Point mutations (P164C, D144N) were introduced into the G. stearothermophilus MutY gene by QuikChange mutagenesis using a parent construct wherein the gene was cloned into the pET28 (Novagen) expression vector, resulting in an ORF with an N-terminal His tag and thrombin cleavage site. Protein was expressed and purified essentially as described before (16).
Synthesis of Cross-linking DNA
DNA oligomer 5′-AAGAC(oxoG)TGGAC-3′ (A, modified A with a disulfide tether for cross-linking; O6-Phenyl-dI-CE and oxoG phosphoramidite were purchased from Glen Research) was synthesized on an ABI 392 DNA synthesizer using standard reagents. To prepare the tether-containing DNA, the O6-Ph-dI-containing DNA was incubated with 1 ml of 1 m aqueous cystamine containing 0.25 mm β-mercaptoethanol at 55 °C for 18 h. 1 ml of NH4OH containing 0.25 mm β-mercaptoethanol was added to the resulting mixture and incubated at room temperature for 6 h to cleave off the DNA from the resin. The solution was desalted by NAP column chromatography (GE Healthcare), and concentrated by a SpeedVac. The residue was purified by urea PAGE, dissolved in 10 mm Tris, pH 8.0 buffer, and annealed with the complimentary strand (purchased from Eurofins MWG Operon).
Glycosylase Assay of the Cross-linked Complex
To test the cleavage activity of the cross-linked complex, we first cross-linked the oxoG-containing strand to the protein by mixing 40 μm protein and 27 μm DNA in buffer containing 15 mm Tris, pH 7.6, and 100 mm NaCl at 4 °C for 4 h. The resulting mixture was purified by MonoQ ion-exchange chromatography, buffer-exchanged to 20 mm Tris, pH 8, 200 mm NaCl, and then diluted to 4 μm. The 5′ end of the complimentary strand (containing either A or C at the position that base pairs with oxoG) was labeled with 32P and diluted to 200 nm with buffer containing 10 mm Tris, pH 8. 1 μl of the single-strand-cross-linked complex and 1 μl of the complimentary strand solution were added to 8 μl of reaction buffer containing 50 mm Tris, pH 7.4, 1 mm EDTA, 100 mm NaCl, resulting in a 10-μl reaction. At each time point recorded, 2 μl was taken out from the reaction mixture, and 5 μl of 2 m NaOH was added to quench the reaction. The mixture was heated up to 90 °C for 15 min to facilitate strand scission. An equal amount of formamide was added into the quenched reaction, and the sample was stored at −20 °C until subjected to gel analysis. Each sample was heated up to 90 °C for 4 min before being electrophoresed by 15% urea gel for 2 h at 35 mA. The gel was developed for 2 h and imaged with a PhosphorImager.
Glycosylase Assay of the Catalytically Active MutY and the D144N Mutant MutY
To test the cleavage activity of MutY in the uncross-linked condition, we prepared MutY in two forms: the catalytically active protein (bearing a P164C mutation for cross-linking and a wild-type active pocket) and the D144N MutY (also bearing a P164C mutation, as well as a D144N mutation in the active pocket). The 30-mer oligonucleotides (5′-TTTTTTTTTTGTCCAXGTCTTTTTTTTTTT-3′; X is A in the oxoG:A DNA and C in the oxoG:C DNA) was end-labeled with 32P and annealed to the unlabeled complementary strand (5′-AAGAC°GTGGAC-3′; °G is 8-oxoguanine). The duplex was then incubated with catalytically active MutY or D144N MutY at room temperature for various amounts of time. Reactions were quenched by NaOH before they were analyzed by gel electrophoresis. The reaction conditions were similar to that used in the assay of the cross-linked complex.
Protein-DNA Cross-Linking Reaction and Crystallization
The protein-DNA complex was formed by incubating 40 μm protein and 27 μm DNA in buffer containing 15 mm Tris, pH 7.6, and 100 mm NaCl at 4 °C for 24 h. The complex was purified by MonoQ ion-exchange chromatography and dialyzed against buffer containing 10 mm Tris, pH 7.4, 90 mm NaCl, 10 μm β-mercaptoethanol at 4 °C overnight. The purified complex was concentrated to 175 μm and crystallized by the hanging-drop vapor diffusion method at room temperature. The anti-substrate complex (ASC) was crystallized in 100 mm Tris, pH 8, 250 mm NaOAc, 29% (w/v) PEG 4000. The D144N ASC was crystallized in 100 mm Tris, pH 8.6, 1.75 m (NH4)2SO4. Crystals appeared after several days and were allowed to grow for a couple of weeks. The ASC crystal was transferred to cryoprotectant solution containing 100 mm Tris, pH 8, 250 mm NaOAc, 29% (w/v) PEG 4000, and 10% glycerol; the D144N ASC crystal was transferred to cryoprotectant solution containing 100 mm Tris, pH 8.6, 1.75 m (NH4)2SO4, and 25% glycerol. To reduce the ASC crystal, we transferred the crystal from the original reservoir solution to a reducing drop that contained 100 mm Tris, pH 8, 250 mm NaOAc, 29% (w/v) PEG 4000, and 25 mm tris(carboxyethyl)phosphine (TCEP). The drop was equilibrated against a reservoir that had the same composition under room temperature for 20 h. The resulting crystals were transferred to cryoprotectant solution containing 100 mm Tris, pH 8, 250 mm NaOAc, 29% (w/v) PEG 4000, 25 mm TCEP, and 10% glycerol and frozen into liquid nitrogen for data collection.
Data Collection and Structure Refinement
Data were collected on beamline 24-ID at the Northeastern Collaborative Access Team (NE-CAT) at the Advanced Photon Source at the Argonne National Laboratory and processed using XDS (22). Resolution cut-off was chosen by examining several indicators of data quality, such as I/σ(I), completeness, and CC½ of the highest resolution shell. Crystal of the ASC belongs to the space group P212121, with unit cell dimensions of a = 38 Å, b = 85 Å, and c = 142 Å Crystal of the D144N ASC belongs to the space group P61, with unit cell dimensions of a = 105 Å, b = 105 Å, and c = 235 Å. Data collection and refinement statistics are summarized in Table 1. The coordinates of the protein from the structure of P164C/D144N G. stearothermophilus MutY bound to oxoG:A-containing DNA (Protein Data Bank (PDB) ID: 1RRQ) were used as the initial search model. Election density for double-stranded DNA was clearly evident in the σA-weighted Fo − Fc map right after molecular replacement. The model was improved by rigid body fit and subsequent simulated annealing, TLS (Translation/Libration/Screw), and individual B-factor refinement using Phenix (23). In both the ASC and the D144N ASC structures, the electron density of the extruded cytosine was clearly evident in their respective positions in the σA-weighted Fo − Fc map after the adjacent nucleotides were put into the model. Model building was performed in Coot (24) while constantly monitoring Rfree (25). Water molecules were added to the model using both automated methods in Phenix (23) and manual inspection of difference maps. Figures were prepared using PyMOL version 1.7 (26).
TABLE 1.
Data collection and refinement statistics
| ASC | Reduced ASC | D144N ASC | |
|---|---|---|---|
| Data collection statistics | |||
| Resolution rangea (Å) | 41.41–2.21 (2.29–2.21) | 41.37–2.32 (2.40–2.32) | 85.01–2.59 (2.68–2.59) |
| Space group | P212121 | P212121 | P61 |
| Unit cell | |||
| a, Å | 37.7 | 37.6 | 105.3 |
| b, Å | 84.6 | 83.8 | 105.3 |
| c, Å | 142.5 | 142.7 | 235.4 |
| α, ° | 90 | 90 | 90 |
| β, ° | 90 | 90 | 90 |
| γ, ° | 90 | 90 | 120 |
| Total reflectionsa | 166,023 (15,823) | 73,154 (7344) | 264,923 (26,696) |
| Unique reflectionsa | 23,611 (2286) | 20,028 (1992) | 45,769 (4610) |
| Completenessa (%) | 99.86 (99.87) | 98.45 (100.00) | 99.97 (99.85) |
| Multiplicitya | 7.0 (6.9) | 3.6 (3.7) | 5.8 (5.8) |
| Mean I/σ(I)a | 12.67 (1.31) | 9.11 (1.45) | 15.60 (1.66) |
| Rmergea,b | 0.1275 (1.477) | 0.118 (0.8846) | 0.1212 (1.209) |
| Refinement statistics | |||
| Rworka,c (%) | 18.44 (30.68) | 18.70 (26.78) | 16.94 (28.38) |
| Rfreea,d (%) | 24.96 (38.85) | 25.89 (30.43) | 22.80 (32.22) |
| Number of non-hydrogen atoms | 3333 | 3260 | 6575 |
| Macromolecules | 3191 | 3191 | 6530 |
| Ligands | 15 | 9 | 16 |
| Water | 127 | 60 | 29 |
| Protein residues | 366 | 366 | 749 |
| r.m.s.e deviation from ideality | |||
| Bond length (Å) | 0.01 | 0.011 | 0.012 |
| Bond angles (°) | 1.4 | 1.37 | 1.45 |
| Ramachandran plotf (%) | |||
| Favored | 96.2 | 95.6 | 92.1 |
| Allowed | 3.2 | 3.5 | 5.3 |
| Outliers | 0.6 | 0.9 | 2.6 |
| Average B-factor | 57.6 | 64.5 | 57.7 |
| Macromolecules | 58 | 64.8 | 57.8 |
| Ligands | 54.7 | 46.7 | 38.2 |
| Solvent | 48.3 | 53.7 | 44.6 |
| PDB ID | 4YOQ | 4YPH | 4YPR |
a Values in parenthesis refer to the highest resolution shell.
b Rmerge = Σ|I − 〈I〉 |/Σ〈I〉; where I is the observed intensity.
c Rwork = Σ|Fo − Fc |/Σ|Fo|; where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
d Rfree was calculated based on 5% data randomly selected and omitted throughout structure refinement (32).
e r.m.s., root mean square.
f Calculated using MOLPROBITY (33).
Results
Experimental System
Previous work has demonstrated that MutY does not readily crystallize with duplex DNA containing a target oxoG:A base pair, but that crystallization of this complex can be fostered through introduction of an intermolecular disulfide cross-link at a site several base pairs removed from the target base pair (Fig. 1, D and E) (16, 27). Anticipating the necessity to provide similarly for stabilization of MutY on an oxoG:C target base pair from which it would ordinarily dissociate, we again employed disulfide cross-linking in the studies described below. The previously established cross-linking system was used here (16); the protein component is MutY wild type apart from having an engineered Cys residue at position 164, and the DNA component comprises two complementary 11-mer paired to leave a one-nucleotide overhang at each end and with a C2 thiol attached to nucleobase A4 in the major groove. The resulting cross-linked complex furnished, upon optimization, high-quality crystals that enabled structure determination of the ASC via molecular replacement and model building with refinement to 2.2 Å resolution (Table 1).
General Features of MutY Interrogating an Anti-substrate oxoG:C Base Pair
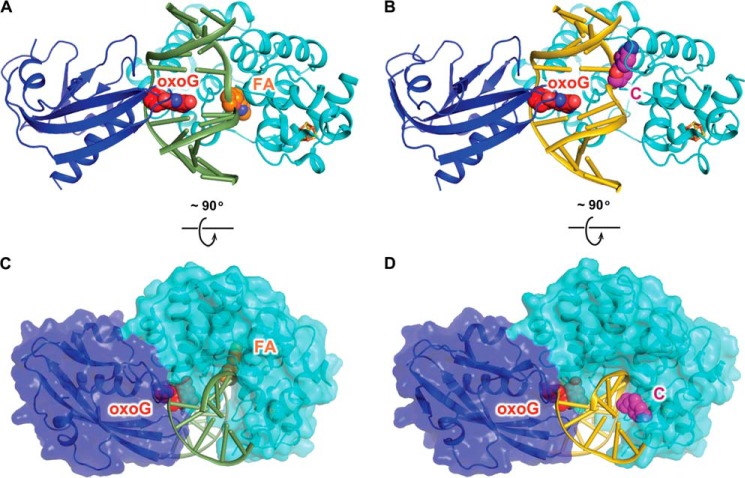
The overall architecture of the ASC is very similar to that of the FLRC (heavy atom root mean square deviation = 0.323 Å; Fig. 2, A and B), with the protein completely encircling a DNA duplex sharply bent at the site of the target oxoG:C base pair. The N-terminal domain penetrates the duplex from the minor groove side opposite the fully intrahelical oxoG nucleobase, engaging the Watson-Crick edge of the oxoG in hydrogen-bonding interactions, whereas the C-terminal domain engages oxoG from the major groove side (Figs. 2B and 3H). Indeed, nearly identical in the ASC and FLRC structures are the protein component plus the entire oxoG-containing strand and even the flanking regions of the strand containing the nucleobase targeted for excision. Notwithstanding these similarities, pronounced structural divergence is apparent in the immediate vicinity of the target nucleobase, A (FLRC) or C (ASC) (compare Fig. 2, A and C versus B and D, respectively; see also Fig. 3, A–D). Although the target nucleobase is fully extrahelical in both the FLRC and the ASC structures, the actual location of the target nucleobase within the complex and the local conformation of the DNA backbone at that site bear marked divergence. In FLRC, the substrate mimetic 2′-β-fluoroadenosine is deeply inserted into the active pocket, which lies at the junction between the two structural modules that comprise the catalytic domain (the six-helix barrel module and the [4Fe-4S] cluster module). By contrast, in the ASC, the anti-substrate C is not plunged into the active site pocket but instead is nestled into an exo-site groove formed by a helix-hairpin-helix motif (α8, α9, and the loop in between) belonging to the [4Fe-4S] module (Fig. 2, C and D).
FIGURE 2.
Overall structures of the ASC and the FLRC. A, structure of the FLRC (PDB ID: 3G0Q) showing 2′-β-fluoroadenosine (FA) in the active site. The N-terminal catalytic domain of MutY is colored in cyan, the C-terminal MutT domain is in blue, and the iron-sulfur cluster is shown as orange (iron) and yellow (sulfur) sticks. The DNA backbone is colored green, and the oxoG and 2′-β-fluoroadenosine are shown as red (oxoG) and orange (A) spheres. B, structure of the ASC (present work) showing C in the exo-site. The color scheme is the same as in A except that DNA backbone is colored gold and C is in magenta. C and D, the bottom panel shows the FLRC (C) and the ASC (D) structures rotated 90 degrees from the top panel view, with the protein shown in surface representation.
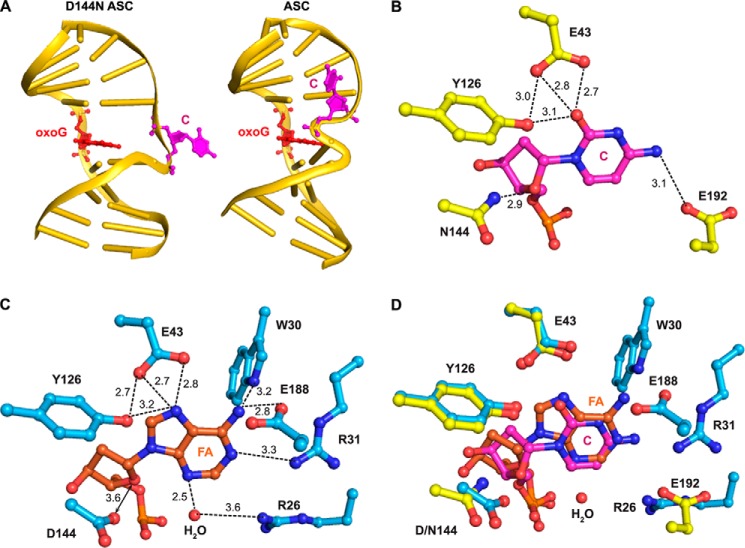
FIGURE 3.
Factors that determine the DNA conformation in the ASC. A, global changes in DNA structure. The protein components in the ASC and the FLRC are superimposed. The protein is not displayed, but the DNA that is bound to it is. B and C, close-up views of the target strand in the FLRC and the ASC. The dotted lines indicate the contacts between MutY and the DNA backbone. D, schematic diagram showing the protein-DNA interactions on the target strand in the FLRC and the ASC. E, exo-site contacts on cytosine in the ASC. F, the reversed Watson-Crick base pair observed in ASC. G, structure of a normal Watson-Crick base pair (from the ASC) The blue meshes in panels E and F are σA-weighted 2Fo − Fc map contoured at 1σ. Comparison of oxoG recognition by MutY in ASC and FLRC is shown in panels H–J. H, close-up view of oxoG recognition by MutY in the ASC. Important hydrogen-bonding interactions between oxoG and the protein are denoted by dotted lines. I, close-up view of oxoG recognition by MutY in the FLRC. J, protein superposition of panels A and B. The color coding scheme is the same as Fig. 2 except as specifically noted.
Although MutY bears a fully wild-type active site, the structure reveals continuous electron density throughout the 2′-deoxyribose and C nucleobase moieties, indicative of an intact glycosidic bond (Fig. 3E). Disulfide cross-linking has occasionally been observed to enable cleavage of an ordinarily dispreferred substrate,4 and the lack of any apparent cleavage in the ASC suggested that the MutY active site possesses extremely strong discrimination against the anti-substrate C. To rule out the possibility that disulfide cross-linking per se abrogates the catalytic activity of MutY, we turned to in vitro cleavage assays on disulfide cross-linked MutY complexes having either a substrate A or an anti-substrate C at the site of the target nucleobase. The reaction was initiated by adding the (anti) substrate-containing strand to a complex of MutY disulfide cross-linked to the oxoG-containing strand. As shown in Fig. 4, A and B, the substrate-containing strand underwent rapid cleavage on the time-scale of minutes (60% within 10 min), whereas the anti-substrate-containing strand was intact after 2 days. Thus, the powerful discrimination of A versus C is an intrinsic property of MutY and is observed even in the extreme situation in which the enzyme and DNA are physically linked; these results reinforce the notion of C as a true anti-substrate for MutY.
FIGURE 4.
Glycosylase assay of MutY. A, a representative urea gel showing cleavage of A opposite oxoG by MutY (left set of lanes) and lack of cleavage of C opposite oxoG under the same conditions. (right set of lanes) To test the glycosylase activity of the cross-linked complex used in crystallography, we cross-linked the strand containing oxoG to the protein and then added to it the 32P-labeled complimentary strand (containing either A or C to pair with oxoG). The reaction was quenched at various time points by adding NaOH into the mixture and heating at 90 °C for 15 min (see “Experimental Procedures” for detailed procedure). B, fraction of DNA cleaved in the two reactions in A plotted as a function of time. Each data point shows an average of three parallel experiments, and the error bar shows the standard deviation from the average. C, cleavage assays of catalytically active MutY (containing only the P164C mutation used in disulfide cross-linking) and the D144N mutant protein (also containing the P164C mutation) (neither of the two proteins is cross-linked to DNA in this experiment). During the course of the experiment, the active P164C MutY achieved almost 100% cleavage of the oxoG:A base pair, whereas no cleavage product was detected for the D144N mutant protein. Neither the catalytically active MutY nor the D144N mutant protein showed any detectable activity toward the anti-substrate oxoG:C.
Factors That Determine the DNA Conformation in the ASC
As mentioned above, the target strand DNA conformation of the ASC diverges greatly from that in the FLRC. Although the FLRC shows the DNA backbone at the target site hyperextended out over the minor groove so as to enable A to reach the distal active site pocket, in the ASC, the backbone lies over the major groove and is considerably less extended from the duplex (Fig. 3A). This divergence of backbone conformation is localized nearly completely to the site of the target nucleobase, with the difference in the two complexes being 1.9 Å at p1, 11.2 Å at p0, and 0.6 Å at p−1 (where p indicates phosphate). Consistent with this profound but highly localized divergence, the amino acid contacts to p−1 and p1 are conserved in the FLRC and ASC, whereas the contacts to p0 are completely different (Fig. 3, B–D). Specifically, in both complexes, p−1 is contacted by the side-chain guanidinium of Arg-149 and p1 is contacted by the backbone and side-chain amides of Gln-48 and Gly-145 (Fig. 3, B–D). On the other hand, p0 is held in place in the ASC through engagement of the Lys-228 ammonium group in hydrogen-bonding interactions, whereas in the FLRC, p0 interacts with the Asn-146 side chain and Val-51 main chain. In addition to the backbone contacts that anchor the C nucleoside in the ASC, several amino acid residues, namely Lys-163, Cys-164, and Arg-167, make exquisitely matched hydrogen bonds to the C nucleobase, thereby giving the appearance of capturing the anti-substrate in an exo-site uniquely tailored to capture an extrahelical C (Fig. 3E). The DNA backbone contortions observed at the target site in the ASC are propagated locally to the A nucleobase (A−1) on its 5′ side, which flips over to form a reverse Watson-Crick hydrogen bond with the complementary T on the oxoG-containing strand (T−1) (Fig. 3F). To our knowledge, this is the first time that a reverse Watson-Crick A-T pair has been observed in DNA. When compared with the normal Watson-Crick base pair (Fig. 3G), both the base and the sugar of the adenine flip, allowing A−1 to maintain its anti glycosidic bond conformation; attempts to model A−1 as a syn-conformer resulted in worse fits to the electron density map and correspondingly worse structural statistics (data not shown).
To account for the influence of the disulfide cross-link on the structure of the complex derived thereby, we reduced treated crystal of the complex with a powerful disulfide-reducing agent, TCEP. Previous attempts in other systems to employ this strategy resulted in a loss of crystal integrity,5 but in the present case, no such loss was seen, and therefore we solved the structure of the complex produced by TCEP reduction to 2.3 Å resolution (Table 1). The resulting structure clearly exhibited a disappearance of the positive density seen in the ASC between the attachment points for the cross-link, Cys-164 and A−4. The local segment of protein structure in the vicinity of Cys-164 also differs slightly in the disulfide cross-linked and reduced complexes. Together, the loss of density for the disulfide, the adjustment of the loop bearing Cys-164, and the clear electron density for the loop in the TCEP-treated complex indicate that the disulfide cross-link had indeed undergone reductive cleavage to a dithiol (Fig. 5, A–C). In Fo − Fc maps of the reduced complex prior to modeling of the target cytosine, positive electron density was observed at the same exo-site position in which the extruded cytosine base had been modeled in the disulfide cross-linked complex, thereby confirming that the target cytosine remained bound in the exo-site (Fig. 5E). In both the non-reduced and the reduced complexes, the main-chain carbonyls of the cross-linked residue, Cys-164, and its N-terminal neighbor, Lys-163, appear to hydrogen-bond with the N4-H of the extruded cytosine, although the contact distance and geometry of the Cys-164 are rendered less favorable in the reduced complex because of the loop movement described above. Given that residue 164 appears to play such an important role in stabilizing the extrahelical Cys in the exo-site, we sought to account for the effect of mutating residue-164 from Pro to Cys. Thus, we superimposed the wild-type protein from the (uncross-linked) abasic site complex (PDB ID: 1RRS) with the protein in the ASC (having Pro-164 and Cys-164, respectively) and found that with the wild-type protein, the exo-site cytosine remains within hydrogen-bonding distance of the backbone carbonyls of Pro-164 and Lys-163 (Fig. 5, G and H). This analysis clearly indicates that wild-type MutY bears the same exo-site as P164C MutY and hence is capable of binding an extrahelical cytosine in analogous fashion to that seen in the ASC.
FIGURE 5.
Structure of the reduced ASC. A, close-up view of the cross-linking site in the ASC, colored as in Fig. 3C. B, close-up view of the cross-linking site in the reduced ASC. Protein and DNA are in pink. C, protein superposition of panels A and B. D, exo-site in the ASC, colored as in Fig. 2B. Important hydrogen-bonding interactions between the protein and the cytosine are denoted in dotted lines, with the associated numbers, showing distances in Å. E, exo-site in the reduced ASC, colored as in panel B. F, protein superposition of panels D and E. G, superposition of the wild-type protein in the abasic site complex (PDB ID: 1RRS) with the protein in the ASC. The wild-type protein (with Pro-164) and the DNA in ASC are shown. H, superposition of panels D and G. The σA-weighted 2Fo − Fc map is contoured at 1σ in panels A, B, and E. The Fo − Fc map is contoured at 3σ in panels A and B and at 2.5σ in panel E.
The Active Site D144N Mutation in the ASC (D144N ASC) Induces Translocation of the Anti-substrate C from the Exo-site to the Active Site
The active site residue Asp-144, which is known to be critical for catalysis by MutY (15, 28) (see also Fig. 4C), is believed to stabilize the oxocarbenium intermediate in adenine excision. Asp-144 also plays a crucial role in enabling full insertion and correct positioning of the substrate adenine within the active site (21); specifically, mutation of Asp-144 to Asn yielded a lesion recognition complex in which the substrate adenine is partially dislodged from the active site pocket. To assess the impact of the D144N mutation on the structure of the ASC, we cross-linked the D144N protein to DNA in the same fashion as described above and solved the structure of this complex (D144N ASC) to 2.6 Å resolution (Table 1). The asymmetric unit comprises two nearly identical complexes in the crystal structure of the D144N ASC (heavy atom root mean square deviation = 0.275 Å); we chose protomer B for our analysis. The D144N mutation has little effect on the disposition of the protein and most of the DNA duplex, as judged by comparison of the ASC structure with that of D144N ASC (Fig. 6A). Remarkably however, the D144N mutation alone is sufficient to cause the anti-substrate C to transition from the exo-site to the active site pocket, via a highly localized but substantial change in the conformation of the target DNA strand at the locus of the anti-substrate C (Fig. 6A). The residue corresponding to Asp-144 is a conserved feature of all HhH-GPD glycosylases and is absolutely essential for catalytic activity (13–15, 29); the side-chain carboxylate of this residue is believed to drive catalysis by stabilizing the incipient oxocarbenium ion that develops at O4′ and C1′ during glycosidic bond scission. Such transition state stabilization can occur only if the Asp-144 carboxylate is positioned in close proximity O4′. Indeed, in the FLRC, the two electronegative oxygen atoms in question (Asp-144 carboxylate-O− and O4′) are only 3.6 Å apart, close enough that the two probably experience a repulsive interaction in the ground state; this unfavorable interaction would also drive catalysis via bound state destabilization (30, 31). This putative repulsive interaction would explain the conformational effect of the D144N mutation as the change from a carboxylate to carboxamide side chain at position 144 would replace a repulsive interaction with an attractive one. This same train of logic suggests that the Asp-144/O4′ interaction is repulsive enough to disfavor stable binding of the anti-substrate C to the enzyme active site.
FIGURE 6.
Structure of the D144N ASC. A, global structure of D144N ASC and ASC. The protein portion is not displayed, but DNA that is bound to it is. DNA in both structures is shown in gold, oxoG is shown in red, and C is shown in magenta. The two structures are viewed from the same orientation. B, close-up view of the active site of the D144N ASC. C, close-up view of the active site of the FLRC. D, an overlay of B and C. Protein in FLRC is shown in cyan, 2′-β-fluoroadenosine (FA) is shown in orange, and ordered water is shown as red spheres. Protein in D144N ASC is shown in yellow, and cytosine is shown in magenta. Hydrogen-bonding interactions are denoted in dotted lines, with the associated numbers, showing distances in Å.
Whether the target nucleoside prefers to bind in the exo-site or active site is determined by the balance of forces originating not only from the attractive versus repulsive interactions between Asp-144 and the substrate sugar moiety, but also the interactions established between the target nucleobase and its binding pocket within the enzyme active site. This nucleobase-specific interaction repertoire provides an explanation as to why C in the anti-substrate complex prefers an exo-site disposition, whereas A in the physiologic substrate (as represented by the FLRC) prefers the active site pocket. Comparison of the contacts made by A versus C in the active site pocket (compare Fig. 6, B and C) reveals that A makes more extensive contacts with the active site pocket than does C.
Discussion
To perform faithfully their role as guardians of the genome, DNA glycosylases must not only cleave their cognate substrate, but must also avoid cleaving or even binding stably to non-substrates. The stakes are particularly high with MutY and C:oxoG as cleavage of the C in this base pair would provoke error-prone repair replication and hence accelerate the mutagenic process. For this reason, we refer to C:oxoG as an anti-substrate for all adenine DNA glycosylases. The results presented here provide a structural rationale for the mechanism by which MutY avoids pro-mutagenic repair of C:oxoG. We propose that MutY uses at least two distinct mechanisms to prevent accidental cleavage of a bound C:oxoG that has undergone extrusion of the target C from the helical stack. Firstly, the exo-site serves as a decoy that stabilizes the extrahelical cytosine at a location remote from the enzyme active site. Secondly, even if C manages to shake off the grasp of the exo-site, the enzyme active site rejects the non-cognate nucleoside because the balance of attractive and repulsive forces between the 2′-deoxyribose moiety and Asp-144, and between C and the active site pocket, disfavors engagement of C in the active site pocket. Finally, the observation that even when forced to remain bound to each other for 2 days, the anti-substrate fails to undergo detectable cleavage by MutY suggests the likelihood that there exists yet a third proofreading mechanism, namely the inability of the enzyme to catalyze cleavage of C should it perchance enter the active site.
Author Contributions
L.W. and G.V. designed the study and wrote the manuscript. L.W. performed the experiments. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank the entire staff of beamline 24-ID of the Advance Photon Source at Argonne National Laboratory for expert assistance with x-ray data collection and processing.
This research was supported by National Institutes of Health Grant CA100742. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 4YOQ, 4YPH, and 4YPR) have been deposited in the Protein Data Bank (http://wwpdb.org/).
M. Zhou and G. Verdine, unpublished data.
L. Wang and G. Verdine, unpublished data.
- oxoG
- 8-oxoguanine
- 8-oxo-dGTP
- 8-oxo-deoxyguanosine triphosphate
- 8-oxo-dGMP
- 8-oxo-deoxyguanosine monophosphate
- ASC
- anti-substrate
- FLRC
- fluorinated lesion recognition complex
- TCEP
- tris(carboxyethyl)phosphine
- h
- human.
References
- 1. Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature 362, 709–715 [DOI] [PubMed] [Google Scholar]
- 2. David S. S., O'Shea V. L., Kundu S. (2007) Base-excision repair of oxidative DNA damage. Nature 447, 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Michaels M. L., Cruz C., Grollman A. P., Miller J. H. (1992) Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl. Acad. Sci. U.S.A. 89, 7022–7025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Michaels M. L., Miller J. H. (1992) The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174, 6321–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shibutani S., Takeshita M., Grollman A. P. (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349, 431–434 [DOI] [PubMed] [Google Scholar]
- 6. Slupska M. M., Baikalov C., Luther W. M., Chiang J. H., Wei Y. F., Miller J. H. (1996) Cloning and sequencing a human homolog (hMYH) of the Escherichia coli mutY gene whose function is required for the repair of oxidative DNA damage. J. Bacteriol. 178, 3885–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moriya M., Grollman A. P. (1993) Mutations in the mutY gene of Escherichia coli enhance the frequency of targeted G:C → T:A transversions induced by a single 8-oxoguanine residue in single-stranded DNA. Molec. Gen. Genet. 239, 72–76 [DOI] [PubMed] [Google Scholar]
- 8. Nghiem Y., Cabrera M., Cupples C. G., Miller J. H. (1988) The mutY gene: a mutator locus in Escherichia coli that generates G·C → T·A transversions. Proc. Natl. Acad. Sci. U.S.A. 85, 2709–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Al-Tassan N., Chmiel N. H., Maynard J., Fleming N., Livingston A. L., Williams G. T., Hodges A. K., Davies D. R., David S. S., Sampson J. R., Cheadle J. P. (2002) Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat. Genet. 30, 227–232 [DOI] [PubMed] [Google Scholar]
- 10. Lipton L., Halford S. E., Johnson V., Novelli M. R., Jones A., Cummings C., Barclay E., Sieber O., Sadat A., Bisgaard M.-L., Hodgson S. V., Aaltonen L. A., Thomas H. J. W., Tomlinson I. P. M. (2003) Carcinogenesis in MYH-associated polyposis follows a distinct genetic pathway. Cancer Res. 63, 7595–7599 [PubMed] [Google Scholar]
- 11. Gad H., Koolmeister T., Jemth A.-S., Eshtad S., Jacques S. A., Ström C. E., Svensson L. M., Schultz N., Lundbäck T., Einarsdottir B. O., Saleh A., Göktürk C., Baranczewski P., Svensson R., Berntsson R. P.-A., Gustafsson R., Strömberg K., Sanjiv K., Jacques-Cordonnier M.-C., Desroses M., Gustavsson A.-L., Olofsson R., Johansson F., Homan E. J., Loseva O., Bräutigam L., Johansson L., Höglund A., Hagenkort A., Pham T., Altun M., Gaugaz F. Z., Vikingsson S., Evers B., Henriksson M., Vallin K. S. A., Wallner O. A., Hammarström L. G. J., Wiita E., Almlöf I., Kalderén C., Axelsson H., Djureinovic T., Puigvert J. C., Häggblad M., Jeppsson F., Martens U., Lundin C., Lundgren B., Granelli I., Jensen A. J., Artursson P., Nilsson J. A., Stenmark P., Scobie M., Berglund U. W., Helleday T. (2014) MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature 508, 215–221 [DOI] [PubMed] [Google Scholar]
- 12. Huber K. V. M., Salah E., Radic B., Gridling M., Elkins J. M., Stukalov A., Jemth A.-S., Göktürk C., Sanjiv K., Strömberg K., Pham T., Berglund U. W., Colinge J., Bennett K. L., Loizou J. I., Helleday T., Knapp S., Superti-Furga G. (2014) Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature 508, 222–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thayer M. M., Ahern H., Xing D., Cunningham R. P., Tainer J. A. (1995) Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 14, 4108–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nash H. M., Bruner S. D., Schärer O. D., Kawate T., Addona T. A., Spooner E., Lane W. S., Verdine G. L. (1996) Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol. 6, 968–980 [DOI] [PubMed] [Google Scholar]
- 15. Guan Y., Manuel R. C., Arvai A. S., Parikh S. S., Mol C. D., Miller J. H., Lloyd S., Tainer J. A. (1998) MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat. Struct Biol. 5, 1058–1064 [DOI] [PubMed] [Google Scholar]
- 16. Fromme J. C., Banerjee A., Huang S. J., Verdine G. L. (2004) Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature 427, 652–656 [DOI] [PubMed] [Google Scholar]
- 17. Noll D. M., Gogos A., Granek J. A., Clarke N. D. (1999) The C-terminal domain of the adenine-DNA glycosylase MutY confers specificity for 8-oxoguanine·adenine mispairs and may have evolved from MutT, an 8-oxo-dTPase. Biochemistry 38, 6374–6379 [DOI] [PubMed] [Google Scholar]
- 18. Volk D. E., House P. G., Thiviyanathan V., Luxon B. A., Zhang S., Lloyd R. S., Gorenstein D. G. (2000) Structural similarities between MutT and the C-terminal domain of MutY. Biochemistry 39, 7331–7336 [DOI] [PubMed] [Google Scholar]
- 19. Lu A. L., Fawcett W. P.(1998) Characterization of the recombinant MutY homolog, an adenine DNA glycosylase, from yeast Schizosaccharomyces pombe. J. Biol. Chem. 273, 25098–25105 [DOI] [PubMed] [Google Scholar]
- 20. Parker A., Gu Y., Lu A. L. (2000) Purification and characterization of a mammalian homolog of Escherichia coli MutY mismatch repair protein from calf liver mitochondria. Nucleic Acids Res. 28, 3206–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee S., Verdine G. L. (2009) Atomic substitution reveals the structural basis for substrate adenine recognition and removal by adenine DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 106, 18497–18502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kabsch W. (2010) Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. Brünger A. T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 26. Delano W. L. (2014) The PyMOL Molecular Graphics System, version 1.7, Schrödinger, LLC, New York [Google Scholar]
- 27. Verdine G. L., Norman D. P. G. (2003) Covalent trapping of protein-DNA complexes. Annu. Rev. Biochem. 72, 337–366 [DOI] [PubMed] [Google Scholar]
- 28. Werner R. M., Stivers J. T. (2000) Kinetic isotope effect studies of the reaction catalyzed by uracil DNA glycosylase: evidence for an oxocarbenium ion-uracil anion intermediate. Biochemistry 39, 14054–14064 [DOI] [PubMed] [Google Scholar]
- 29. Labahn J., Schärer O. D., Long A., Ezaz-Nikpay K., Verdine G. L., Ellenberger T. E. (1996) Structural basis for the excision repair of alkylation-damaged DNA. Cell 86, 321–329 [DOI] [PubMed] [Google Scholar]
- 30. Parikh S. S., Walcher G., Jones G. D., Slupphaug G., Krokan H. E., Blackburn G. M., Tainer J. A. (2000) Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc. Natl. Acad. Sci. U.S.A. 97, 5083–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barrett T. E., Schärer O. D., Savva R., Brown T., Jiricny J., Verdine G. L., Pearl L. H. (1999) Crystal structure of a thwarted mismatch glycosylase DNA repair complex. EMBO J. 18, 6599–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brünger A. T. (1993) Assessment of phase accuracy by cross validation: the free R value. Methods and applications. Acta Crystallogr. D Biol. Crystallogr. 49, 24–36 [DOI] [PubMed] [Google Scholar]
- 33. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375-W383 [DOI] [PMC free article] [PubMed] [Google Scholar]