Abstract
The study of cytoskeletal polymers has been an active area of research for more than 70 years. However, despite decades of pioneering work by some of the brightest scientists in biochemistry, cell biology, and physiology, many central questions regarding the polymers themselves are only now starting to be answered. For example, although it has long been appreciated that the actin cytoskeleton provides contractility and couples biochemical responses with mechanical stresses in cells, only recently have we begun to understand how the actin polymer itself responds to mechanical loads. Likewise, although it has long been appreciated that the microtubule cytoskeleton can be post-translationally modified, only recently have the enzymes responsible for these modifications been characterized, so that we can now begin to understand how these modifications alter the polymerization and regulation of microtubule structures. Even the septins in eukaryotes and the cytoskeletal polymers of prokaryotes have yielded new insights due to recent advances in microscopy techniques. In this thematic series of minireviews, these topics are covered by some of the very same scientists who generated these recent insights, thereby providing us with an overview of the State of the Cytoskeleton in 2015.
Keywords: actin, cytoskeleton, intermediate filament, microtubule, tubulin, vimentin, bacterial cytoskeleton, polymer dynamics, polymer mechanics, septins
Introduction
The study of the protein polymers comprising the cytoskeleton is one of the more mature fields within biological chemistry and cell biology. Since the discovery of actin in the 1940s (1), tubulin in 1967 (2), intermediate filaments shortly thereafter (3, 4) (Fig. 1), and finally the septins in the 1970s (5), the cytoskeletal polymers have been the focus of intense study, from structural analysis at atomic resolution to physiological functions in mammals, producing some of the earliest seminal observations of any biological molecular components made across such a broad range of scales (e.g. Refs. 6 and 7). Even the prokaryotic cytoskeletal polymers, which are relative newcomers, have now been investigated for more than 25 years (8, 9). Given these long-standing intensive and extensive studies, one might presume that there are but minor details left to learn from the cytoskeletal polymer systems, and that all focus now is on higher order regulation of these and other biological machineries as complex systems. Indeed, many genomic and proteomic approaches have likewise been brought to bear on the regulation of the cytoskeleton polymers (e.g. Refs. 10–13). However, despite a nearly exhaustive library of regulatory factors and binding partners, there are many fundamental questions about the polymers themselves that are only now starting to be answered, many of which have broad repercussions for biology and human disease. In this minireview series, we highlight some of the important outstanding problems of and recent insights into each of the cytoskeletal systems, the sum of which provides a brief overview of a mature yet still thriving field of research.
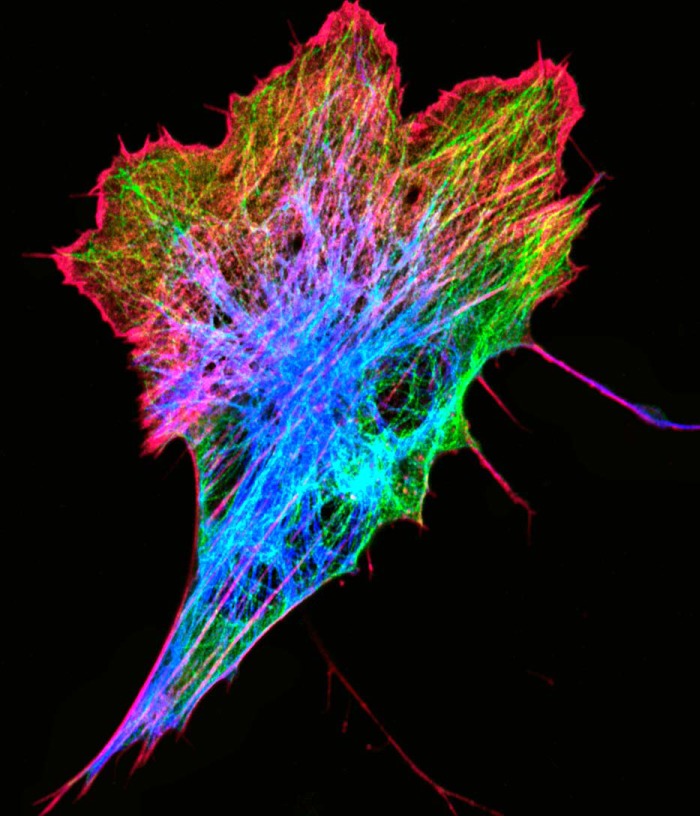
FIGURE 1.
Canonical cytoskeleton elements in a zebrafish fibroblast. The image shows a Zf4 cell expressing GFP-tubulin (green) and mCherry-vimentin (blue), which has been fixed with paraformaldehyde, permeabilized, and stained with Alexa Fluor 647 phalloidin (red). The image is a maximal intensity projection of a confocal z-series taken on a Zeiss 810 LSM equipped with an Airyscan detector.
It has long been appreciated that the actin cytoskeleton is one of the most obviously contractile structural elements, even within non-muscle cells (14). Indeed, mechanical forces required to perform many diverse cellular functions are mediated through the actin cytoskeleton (15). The importance of these diverse functions is illustrated by the evolution of a wide variety of actin regulators and nucleators, which combine to specify distinct actin structures throughout the cell (16). Over the past decade, it has become clear that several proteins associated with the actin cytoskeleton change their biochemical properties in response to mechanical stress (17–19). Given these observations, understanding the mechanical properties of the actin polymer itself has become increasingly important. In the current issue, in the article entitled “Actin Mechanics and Fragmentation,” De La Cruz and Gardel discuss how actin filaments, both locally and at the larger scale of filament networks, respond to mechanical inputs to alter local biochemical interactions. They discuss how the mechanical properties of the actin polymer contribute to the dynamics of the polymer itself and its interactions with filament regulators such as severing proteins, nucleators, and capping proteins. As we work toward understanding how cellular mechanics contribute to human diseases (20), a solid understanding of the mechanical properties of actin in its various architectures will be invaluable.
Perhaps even more important than actin for the mechanical properties of cells and tissues are the intermediate filaments, which can harden under strain (21) and stabilize organelles within the cytoplasm (22). In fact, the composite mechanical properties of intermediate filaments co-polymerized with associated actin filaments are not simply additive, but can produce differential stiffness based on the interactions of the two networks (23). Perhaps unsurprisingly, several human diseases can result from mutations in intermediate filament genes, which likely alter the mechanical properties of their encoded filaments. In this issue of the JBC, in the article entitled “Intermediate Filaments Play a Pivotal Role in Regulating Cell Architecture and Function,” Lowery et al. review how the most widely distributed of the intermediate filaments, vimentin, serves as an interface between biochemical and mechanical signals to influence development and disease. As these reviews make clear, the biophysical and biochemical properties of the cytoskeleton are inseparable and play profound roles in cellular physiology.
Much of our understanding of the cytoskeletal machinery can be summarized by the famous Feynman quote, “What I cannot create, I do not understand.” Indeed, the ability of biochemists to assemble various actin architectures in vitro using combinations of purified components has led to a formal and mechanistic understanding of how these structures can perform work in the cell (e.g. Ref. 24). Such an in vitro assembly approach has been elegantly employed to demonstrate the function of bacterial polymers as well (25). More recently, this approach has become a powerful part of the toolset for unraveling the complex and dynamic structures of the microtubule cytoskeleton as a more complete list of regulatory proteins is elucidated. In the current issue, in the article entitled “Building the Microtubule Cytoskeleton Piece by Piece,” Alfaro-Aco and Petry highlight some of the recent advances made using in vitro combinations of microtubule-associated proteins (MAPs)3 to reveal how their combined activities result in specific ensemble functions. In some cases, these combined activities result in complex functions and structures that are not simply the sum of the individual parts, but rather form a synergistic system with effectively new functions. In addition to the complexity of multicomponent MAP interactions, recent work has also begun to uncover the roles of the long-appreciated but poorly understood tubulin post-translational modifications. As discussed by Yu et al. in the current issue, in the article entitled “Writing and Reading the Tubulin Code,” these varied post-translational modifications appear to comprise a code that, when read by the MAPs and other proteins, specifies distinct microtubule architectures and dynamics in the cell. As an additional complexity, the enzymes responsible for these post-translational modifications may also be subject to spatiotemporal regulation. It is clear that, although we may now have the building blocks to generate a diverse array of microtubule structures, we are still only beginning to understand how the cell uses these building blocks for spatial control of physiological functions.
Clearly, one of the most important roles the cytoskeleton plays in biology is to spatially organize the cell into specific domains and shapes designed to enable specific functions. In most animal cells, the cell cortex not only defines the cell shape, but also provides forces needed for migration (26), provides polarity cues to development (27), and defines membrane domains (28). Emerging evidence suggests that, of the various components of the cortex, the septins are critically important for integrating the plasma membrane with the various cytoskeletal networks in spatially defined regions and domains. In the current issue of the JBC, in the article entitled “Septin Form and Function at the Cell Cortex,” Bridges and Gladfelter discuss the current state of the art in our understanding of how the septin polymers may recognize and alter local membrane composition and cooperate with other cytoskeletal polymers to alter local cell shape, and how these functions may be perturbed in human disease.
For many years, the consensus view in biology was that only the eukaryotes required cytoskeletal systems to spatially order their internal cellular structures and segregate their chromosomes, but biologists have been disabused of this notion by the demonstration of dynamic protein polymers in bacteria that perform these same functions (9, 29). Although the first bacterial polymers discovered have relatively clear counterparts in the eukaryotic cytoskeleton (30), the variety of the prokaryotic cytoskeletons reflects the diversity of the prokaryotic kingdoms themselves (31). Furthermore, although the core function of subunit polymerization dependent on nucleotide binding may be conserved, the way in which these cytoskeletons carry out their functions has distinct differences. In this issue, in the article entitled “Bacterial Filament Systems: Toward Understanding Their Emergent Behavior and Cellular Functions,” Eun et al. discuss these diverse prokaryotic filament systems and how comparing their similarities and differences is starting to provide insights into the general rules governing polymer assembly and dynamics, and how these affect prokaryotic cellular functions.
It is clear from the spectrum of minireviews gathered in this issue of the JBC that the cytoskeletal polymers still have much to teach us about how cells organize themselves in space and time, how they respond to and integrate both physical and chemical cues, and how these mechanisms have evolved to effectively carry out these diverse functions using combinations of conserved building blocks. Indeed, these minireviews also highlight the notion that, although we have a nearly complete set of building blocks in this post-genomic era, it is how nature combines these building blocks that ultimately creates emergent properties and unexpected functions. Given the complexities of these combinations and their broad impact on human health and biology, the State of the Cytoskeleton in 2015 appears to be quite strong for years to come.
This work was authored, in whole or in part, by National Institutes of Health staff. The authors declare that they have no conflicts of interest with the contents of this article.
- MAP
- microtubule-associated protein.
References
- 1. Straub F. B. (1942) Actin. in Studies from the Institute of Medical Chemistry University Szeged, (Szent-Györgyi A., ed) Vol. II, pp. 3–15, S. Karger, Basel, New York [Google Scholar]
- 2. Borisy G. G., Taylor E. W. (1967) The mechanism of action of colchicine: binding of colchincine-3H to cellular protein. J. Cell Biol. 34, 525–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ishikawa H., Bischoff R., Holtzer H. (1968) Mitosis and intermediate-sized filaments in developing skeletal muscle. J. Cell Biol. 38, 538–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huneeus F. C., Davison P. F. (1970) Fibrillar proteins from squid axons. I. Neurofilament protein. J. Mol. Biol. 52, 415–428 [DOI] [PubMed] [Google Scholar]
- 5. Byers B., Goetsch L. (1976) A highly ordered ring of membrane-associated filaments in budding yeast. J. Cell Biol. 69, 717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Suck D., Kabsch W., Mannherz HG. (1981) Three-dimensional structure of the complex of skeletal muscle actin and bovine pancreatic DNase I at 6-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 78, 4319–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Witke W., Sharpe A. H., Hartwig J. H., Azuma T., Stossel T. P., Kwiatkowski D. J. (1995) Hemostatic, inflammatory, and fibroblast responses are blunted in mice lacking gelsolin. Cell 81, 41–51 [DOI] [PubMed] [Google Scholar]
- 8. Bi E. F., Lutkenhaus J. (1991) FtsZ ring structure associated with division in Escherichia coli. Nature 354, 161–164 [DOI] [PubMed] [Google Scholar]
- 9. Jones L. J., Carballido-López R., Errington J. (2001) Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104, 913–922 [DOI] [PubMed] [Google Scholar]
- 10. Klemke R. L., Jiang X., Choi S., Kelber J. A. (2013) Proteomic and biochemical methods to study the cytoskeletome. Methods Mol. Biol. 1046, 203–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Radulovic M., Godovac-Zimmermann J. (2011) Proteomic approaches to understanding the role of the cytoskeleton in host-defense mechanisms. Expert Rev. Proteomics 8, 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Calligaris D., Verdier-Pinard P., Devred F., Villard C., Braguer D., Lafitte D. (2010) Microtubule targeting agents: from biophysics to proteomics. Cell Mol. Life Sci. 67, 1089–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sobczyk G. J., Wang J., Weijer C. J. (2014) SILAC-based proteomic quantification of chemoattractant-induced cytoskeleton dynamics on a second to minute timescale. Nat. Commun. 5, 3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Korn E. D. (1978) Biochemistry of actomyosin-dependent cell motility (a review). Proc. Natl. Acad. Sci. U.S.A. 75, 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fletcher D. A., Mullins R. D. (2010) Cell mechanics and the cytoskeleton. Nature 463, 485–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Waterman C. M., Skau C. (2015) Specification of architecture and function of actin structures by actin nucleation factors. Annu. Rev. Biophys. 44, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. del Rio A., Perez-Jimenez R., Liu R., Roca-Cusachs P., Fernandez J. M., Sheetz M. P. (2009) Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim T.-J., Zheng S., Sun J., Muhamed I., Wu J., Lei L., Kong X., Leckband D. E., Wang Y. (2015) Dynamic visualization of α-catenin reveals rapid, reversible conformation switching between tension states. Curr. Biol. 25, 218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoffman L. M., Jensen C. C., Chaturvedi A., Yoshigi M., Beckerle M. C. (2012) Stretch-induced actin remodeling requires targeting of zyxin to stress fibers and recruitment of actin regulators. Mol. Biol. Cell 23, 1846–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. DuFort C. C., Paszek M. J., Weaver V. M. (2011) Balancing forces: architectural control of mechanotransduction. Nat. Rev. Mol. Cell Biol. 12, 308–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma L., Xu J., Coulombe P. A., Wirtz D. (1999) Keratin filament suspensions show unique micromechanical properties. J. Biol. Chem. 274, 19145–19151 [DOI] [PubMed] [Google Scholar]
- 22. Guo M., Ehrlicher A. J., Mahammad S., Fabich H., Jensen M. H., Moore J. R., Fredberg J. J., Goldman R. D., Weitz D. A. (2013) The role of vimentin intermediate filaments in cortical and cytoplasmic mechanics. Biophys J. 105, 1562–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jensen M. H., Morris E. J., Goldman R. D., Weitz D. A. (2014) Emergent properties of composite semiflexible biopolymer networks. Bioarchitecture 4, 138–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pantaloni D., Le Clainche C., Carlier M. F. (2001) Mechanism of actin-based motility. Science 292, 1502–1506 [DOI] [PubMed] [Google Scholar]
- 25. Garner E. C., Campbell C. S., Weibel D. B., Mullins R. D. (2007) Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science 315, 1270–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bergert M., Erzberger A., Desai R. A., Aspalter I. M., Oates A. C., Charras G, Salbreux G., Paluch E. K. (2015) Force transmission during adhesion-independent migration. Nat. Cell Biol. 17, 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goehring N. W., Trong P. K., Bois J. S., Chowdhury D., Nicola E. M., Hyman A. A., Grill S. W. (2011) Polarization of PAR proteins by advective triggering of a pattern-forming system. Science 334, 1137–1141 [DOI] [PubMed] [Google Scholar]
- 28. Dobbelaere J., Barral Y. (2004) Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science 305, 393–396 [DOI] [PubMed] [Google Scholar]
- 29. Erickson H. P. (1995) FtsZ, a prokaryotic homolog of tubulin? Cell 80, 367–370 [DOI] [PubMed] [Google Scholar]
- 30. Erickson H. P. (2007) Evolution of the cytoskeleton. Bioessays 29, 668–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cabeen M. T., Jacobs-Wagner C. (2010) The bacterial cytoskeleton. Annu. Rev. Genet. 44, 365–392 [DOI] [PubMed] [Google Scholar]