Abstract
The microtubule (MT) cytoskeleton gives cells their shape, organizes the cellular interior, and segregates chromosomes. These functions rely on the precise arrangement of MTs, which is achieved by the coordinated action of MT-associated proteins (MAPs). We highlight the first and most important examples of how different MAP activities are combined in vitro to create an ensemble function that exceeds the simple addition of their individual activities, and how the Xenopus laevis egg extract system has been utilized as a powerful intermediate between cellular and purified systems to uncover the design principles of self-organized MT networks in the cell.
Keywords: cell division, cytoskeleton, microtubule, microtubule-associated protein (MAP), mitotic spindle
Introduction
The microtubule (MT)2 cytoskeleton forms the skeletal framework that gives eukaryotic cells their shape and organizes their cytoplasm by positioning organelles, providing tracks for transport, and establishing cell polarity. In an interphase cell, the MT cytoskeleton is also critical for cell motility and a key constituent of cilia and flagella. During cell division, the MT cytoskeleton gets remodeled into a spindle structure that segregates chromosomes. Each of these functions relies on a specific MT architecture, which must be capable of rapid and prolonged change followed by an eventual resumption of a steady state to respond to the cellular environment and morphology changes during growth and differentiation.
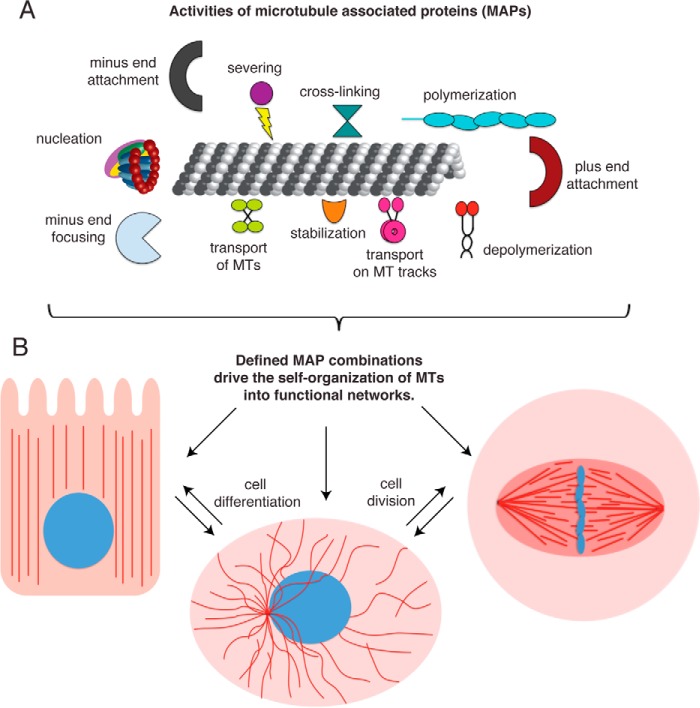
MTs are made of α/β-tubulin heterodimers, which assemble into a polar, cylindrical structure in the presence of GTP and above the so-called critical concentration in vitro. MT growth phases alternate with swift shrinkage phases (dynamic instability), and their transitions are referred to as catastrophe (switching from growth to shrinkage) and rescue (switching from shrinkage to growth) (1). In cells, a plethora of different MT-associated proteins (MAPs) regulate the MT-inherent abilities of MT nucleation and dynamics (Fig. 1A) (2). In addition, MT cross-linking proteins connect MTs into networks and molecular motors use MTs as tracks for cargo transport or transport MTs themselves (Fig. 1A). Altogether, different combinations of these four basic groups of MAP activities drive the self-organization of the MT cytoskeleton into discrete three-dimensional patterns (Fig. 1B) (3). Thus, they establish, maintain, and disassemble functional MT structures that are observed on the cellular level.
FIGURE 1.
Organization of the microtubule cytoskeleton by MAPs. A, representative classes of MAP activities are depicted schematically. B, different combinations of MAPs drive the self-organization of MTs into functional networks, which determine the organization of the cell, differ between cell cycles states, and change during cell differentiation.
Traditionally, individual MAPs were identified by loss-of-function experiments in cells followed by their detailed in vivo and in vitro characterization. During the past decade, high-throughput genomic and proteomic screens accelerated MAP discovery by cataloging RNAi phenotypes and identifying novel microtubule binders, resulting in comprehensive lists of candidates involved in organizing the MT cytoskeleton in various cell states (4–7). Now, the challenge is to understand how these MAPs work together to establish the physiological MT architecture of the cell. What specific MAP building blocks can generate the MT networks that shape a dendrite or a polarized epithelial cell (Fig. 1B)? More generally, what are the basic principles for constructing a functional MT structure?
A key step toward answering these questions is a detailed understanding of the mechanism of individual motor and non-motor MAPs, which has been emerging for many MAPs at the biochemical and structural level (8–11). Furthermore, pioneering in vitro reconstitution approaches demonstrated how individual MAPs, when mixed with static or dynamic MTs, act at the single MT level and contribute to simple microtubule patterns (3). However, the next critical step is to not only understand the mechanism of one MAP, but how combinations of MAPs together with MTs create physiological cytoskeletal structures in vitro. This will also address the more general question of how biological molecules, which act in the Å scale and in a transient fashion, generate self-organized assemblies in the μm scale that enable cell function.
In this review, we discuss recent findings from in vitro reconstitutions of more than one type of MAP with MTs, which provide details on how complexity is formed in multicomponent systems. A surprising outcome of this approach is that the combination of two MAP activities is not necessarily additive, but can be synergistic. We will focus on the three basic MAP activities of MT dynamics regulation, transport, and nucleation, with the goal of linking these activities and explaining their effects within the context of larger cytoskeletal structures. The fourth MAP activity of MT cross-linking was recently reviewed separately (11, 12). Last, we will highlight basic principles of MT self-organization derived from studies in cell extract, which is a powerful system that serves as an intermediate between cellular and purified systems.
In Vitro Reconstitution of Microtubule Dynamics
Microtubule Polymerases and Depolymerases Regulate Microtubule Dynamics
To organize MTs into functional networks, MAPs stimulate or reduce MT dynamics by affecting MT growth, shrinkage, and stabilization. MAPs achieve this by selectively binding to α/β-tubulin dimers in defined conformations that will favor the process to be catalyzed (13). A central point of regulation is the intrinsic curvature of the tubulin dimer, which it displays in isolation and which can be exploited to reduce MT stability (14). In contrast, straightening of the tubulin dimer seems to occur only within the MT lattice and promotes MT growth (15). Before describing some of the protein networks that affect MT dynamics and that have been reconstituted in vitro, we briefly introduce the individual molecular players.
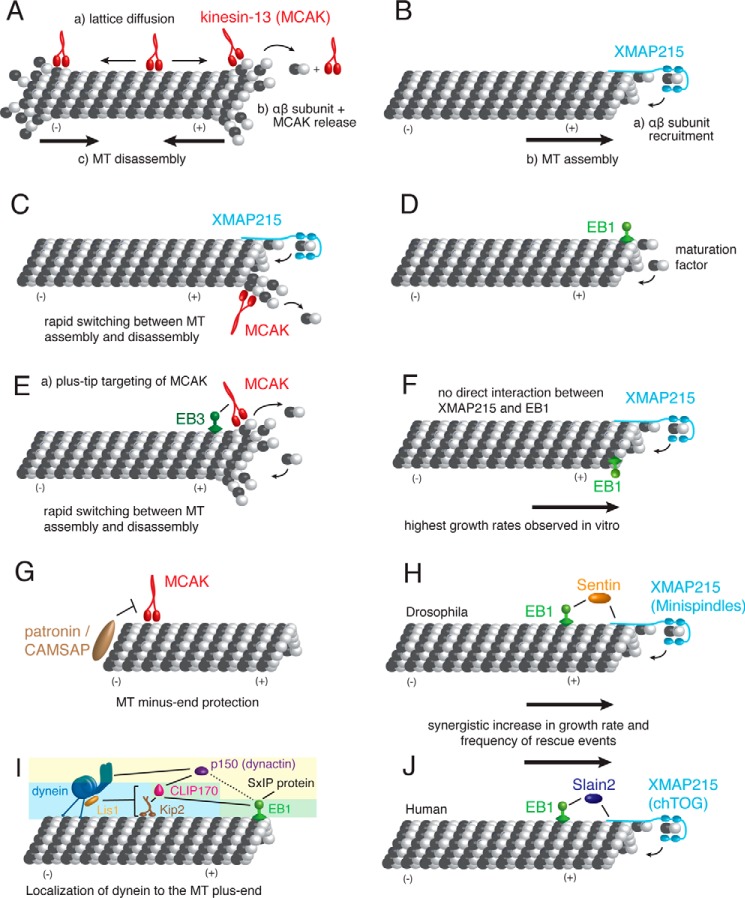
MT depolymerases destabilize MTs by promoting catastrophe to regulate MT stability and length (8, 13). Although many MT depolymerases exist in the cell, one of the best characterized ones in vitro is the kinesin-13 MCAK/XKCM1, which increases catastrophe rates in cells and regulates the size of MT structures (16, 17). During mitosis, it plays an important role for chromosome segregation at kinetochores (18) and during anaphase (19). In vitro, MCAK reaches both MT ends via a lattice diffusion process, where MCAK hydrolyzes ATP and distorts MT filaments by stabilizing tubulin dimer curvature to promote subunit release (20–23), facilitating MT disassembly and eliminating its dependence on the age of the MT (Fig. 2A) (24).
FIGURE 2.
In vitro reconstitutions of microtubules and more than one MAP. A, C, E, and G depict in vitro reconstitutions with the MT depolymerase MCAK. B, F, H, and J display in vitro studies with the MT polymerase XMAP215. D, E, F, H, I, and J show how EBs modulate MT dynamics by recruiting factors to the MT plus-end. Each of the in vitro studies (A–J) are discussed in the main text.
MT polymerases oppose depolymerases and promote growth or rescue depolymerizing MTs (8, 25). One of the best-studied MT stabilizing agents in vitro is the MT polymerase XMAP215/ch-TOG (26), which enhances MT growth rates up to 10-fold. Individual molecules remain bound to growing plus-ends during multiple rounds of tubulin dimer addition mediated by several so-called TOG domains (26, 27). TOG domains preferentially bind the curved conformation of tubulin dimers, which straighten when incorporated into the MT lattice and released by XMAP215 (Fig. 2B) (28).
In the cell, MT polymerases and depolymerases do not act individually on MTs, but work in a coordinated fashion to construct cell cycle-specific and local MT structures (29). This is apparent by the high polymerization rates and catastrophe frequencies of MTs in vivo (30–32) that are not displayed in MT dynamics from purified tubulin (33, 34) or with individual MAPs (see above). The first in vitro reconstitutions with combinations of the major MT polymerase, XMAP215, and the catastrophe factor, MCAK, approached physiological MT dynamics as observed in Xenopus extracts and demonstrated that a balance between an MT polymerase and an MT depolymerase is central to MT dynamics in cells (Fig. 2C) (35). However, MT dynamics and length distributions did not fully match in vivo parameters, indicating involvement of other MAPs.
Adding Complexity via + TIPs
The next level of complexity for regulating MT dynamics is added by special MT plus-end tracking proteins (+TIPs), which additionally recruit MT polymerases and depolymerases, as well as other MAPs (10, 36). The most central +TIPs, which recruit most other +TIPs to growing MT ends and are critical regulators of MT dynamics, are end-binding (EB) proteins (36–38). The canonical +TIP end-binding protein 1 (EB1) itself acts as a maturation factor by transitioning GTP to GDP-tubulin at the MT end in vitro (Fig. 2D) (39). +TIPs bind via their SxIP motif containing the amino acid sequence (S/T)X(I/L)P motif to EB homology (EBH) domains (40) or via CAP-Gly domains that recognize a EEY/F-COO− motif at the C terminus of EBs (10, 41). It has been demonstrated in vitro that the +TIP proteins CLIP-170, MACF, STIM1, and CLASP2 are directly recruited by EB1 to MT plus-ends (40, 42–45).
Interestingly, in cells, the MT depolymerase MCAK tracks growing MT plus-ends, rather than shortening ones, which relies on the interaction with EBs (21, 46). In vitro, plus-tip tracking of MCAK could be reconstituted in the presence of the EB family member EB3, which directly binds to MCAK via a SKIP motif and counteracts MCAK by stabilizing the MT growth phase (Fig. 2E) (47). EB3 increases the association rate of MCAK with MTs, thereby targeting MCAK to growing MT ends. This increases the catastrophe frequency and thus, MCAK and EB3 together induce rapid switching between MT growth and MCAK-induced depolymerization in vitro, providing an example of how EBs can regulate MT dynamics through direct recruitment of a modulator of MT dynamics (47). This plus-tip activity of MCAK is functionally important in mitosis, where MCAK regulates the length of MTs to promote robust attachments between spindle MTs and kinetochores (48) and at centromeres, where depolymerization activity is controlled by Aurora B and gets locally activated by the inner centromere KinI stimulator (ICIS) (49, 50). In cells, the ICIS homologue TIP150 was also shown to be an EB-dependent +TIP, which further enhances MT plus-end localization of MCAK (51), indicating yet another layer of MCAK targeting to the MT plus-end via a +TIP that is itself also recruited by EBs. In an analogous manner, MCAK interacts with the EB1-dependent plus-tip tracking kinesin 8 Kif18b, and this interaction is required for robust MT depolymerization in cells (52, 53). The specificity of MCAK as a plus-end depolymerase could be further regulated by the minus-end-binding proteins patronin/CAMSAP (54–56) and MCRS1 (57), which block MCAK activity at the MT minus-end to enhance MT stability (Fig. 2G). However, the mechanism(s) of how EB1, EB3, TIP150, Kif18b, patronin, and MCRS1 work in concert to affect the depolymerase activity of MCAK, and how this influences the interplay with XMAP215, remains to be determined.
Not surprisingly, the MT polymerase XMAP215 also does not act autonomously on MT plus-ends. In Xenopus egg extracts, an interaction between EB1 and XMAP215 is important for physiological MT growth rates between 10 and 20 μm/min, proper spindle assembly, and chromosome segregation (58). This effect was investigated in vitro, where EB1 and XMAP215 combined had a much larger effect on MT growth rates than XMAP215 (10-fold increase) or EB1 (1.5-fold increase) alone with rates up to 20 μm/min, the highest rates ever observed outside cells (Fig. 2F) (59). This must be close to the maximum possible polymerization rate because the association rate constant for tubulin addition approached the maximum diffusion limited rate with up to 7.6 μm−1 s−1 per protofilament (59). The two proteins do not interact before binding to the MT end nor through canonical EB1 protein interactions, and they do not affect each other's localization, suggesting that the synergistic effect is allosteric. EB1 could be substituted by the lattice-straightening drug taxol without lowering the synergistic growth rates with XMAP215, providing further evidence that EB1 may affect the structure of the MT lattice (59). Besides direct recruitment via EBs and via +TIPs that also bind to EBs, allosteric interactions through the MT lattice are yet another mode of how MAPs can affect each other's activity.
If EB1 and XMAP215 do not interact on an MT in vitro, how then can the interaction of EB1 and XMAP215, revealed by pulldown experiments from extracts (58), be explained? In Drosophila S2 cells, the XMAP215 homologue Minispindles (Msps) requires EB1 to track MT plus-ends in mitosis and interphase. Curiously, Msps also requires an interaction with the +TIP Sentin to track plus-ends (60). Sentin is the dominant cargo for EB1 in S2 cells, and its depletion led to shorter spindles and less dynamic MTs, as did the EB1 and XMAP215 co-depletion. However, EB1-recruited Sentin displayed growth acceleration and catastrophe-promoting activity independent of XMAP215 in addition to recruiting this MAP to plus-ends. In vitro reconstitution with these three factors generated the most dynamic MTs, displaying a synergistic increase in growth rate and number of rescue events (Fig. 2H) (60). Similarly, the human EB1-binding protein SLAIN2 contributes to the localization of the human XMAP215 homologue ch-TOG to growing MT plus-ends in interphase and strongly stimulated processive MT polymerization (Fig. 2J) (61). Depletion or disruption of the SLAIN2-ch-TOG complex led to disorganization of MT arrays. Thus, XMAP215/ch-TOG and EB1 together modulate MT dynamics via allosteric interactions and via the +TIP SLAIN2. This in vitro reconstitution of three MAPs and dynamic MTs provides a valuable basis for future investigations of +TIP networks because SLAIN2 further binds to EB1, CLIPs, and CLIP-associated proteins.
Dynamic Competition at Crowded Microtubule Ends
The studies on Sentin and SLAIN2 exemplify how complex +TIP networks regulate dynamics at growing MT plus-ends. However, the variety and number of +TIPs exceed the number of binding sites at the growing MT plus-end, raising the question how a particular +TIP ensemble is assembled and regulated in the cell. A recent in vitro study addressed for the first time how a cargo complex is targeted to MT plus-ends under competitive conditions, in this case the minus-end-directed motor protein dynein (62). Dynein's adapter complex dynactin contains a p150Glued subunit, which contains a CAP-Gly domain and is targeted via EB1 to MT plus-ends. However, if a more competitive binder occupies this binding site, p150Glued can use the EB homology domain of CLIP-170 for its plus-end localization instead (Fig. 2I). This finding explains why both EB1 and CLIP-170 are necessary in vivo for this localization pathway. Besides this dynactin-dependent pathway, alternative pathways exist to enrich dynein at the MT plus-end, which does not depend on dynactin but the dynein regulator LIS1 (63–65). By reconstituting such an alterative pathway from yeast in vitro, Roberts et al. (66) revealed that the yeast LIS1 and a CLIP-170 homologue are sufficient to couple dynein to Kip2, a plus-end-directed kinesin (Fig. 2I). Dynein resists its plus-end-directed transport by Kip2, but this is overcome by both CLIP-170 and EB1, which strengthen the interaction between Kip2 and MTs. These alternative pathways may not be mutually exclusive, depending on cell type, in particular because transporting dynein to the MT plus-end is a critical first step to allow for essential dynein transport events toward the minus-end (65). These pioneering in vitro studies of more than three MAP types with MTs demonstrate that there is a binding hierarchy for limited and alternative binding sites, which guide dynamic assembly of +TIP ensembles. The in vitro reconstitutions further show that different MAP combinations regulate individual MT dynamics and thereby establish MT network architecture and function.
Coordinated Transport by Molecular Motors in Vitro
Besides regulating the intrinsic dynamics of MTs, the precise arrangement of MTs must be controlled to build the MT cytoskeleton of the cell. This construction role is carried out by molecular motors and MT-bundling proteins. Molecular motors are best known for transporting various forms of cargo along MTs throughout the cell. The mechanism of individual motor types has been well studied, in particular the plus-end-directed kinesin motor family (9). Although many functionally distinct kinesin motor proteins in the kinesin superfamily exist in a cell and provide a diversity of cargo binding domains (67), there is only one cytoplasmic dynein 1 that acts as the major processive minus-end-directed motor in most eukaryotic cells (68). Instead, multiple adapter proteins recruit the soluble pool of cytoplasmic dynein to specifically transport a variety of cargoes, such as organelles, viruses, and mRNAs.
Recently, two in vitro reconstitution studies with motor proteins and multiple components provided novel insight into how their transport is coordinated. In contrast to yeast dynein (69), purified mammalian dynein is not processive (70), although cytoplasmic dynein transports its cargoes over long intracellular distances. This mystery could be resolved by the addition of the accessory dynactin complex and the dynein binding region of the cargo adapter BICD2, or other cargo-specific adapter proteins, to purified dynein, which surprisingly stimulated the processivity of human dynein in vitro (71, 72). Thus, dynein becomes specifically activated for long intracellular transport only when the motor is bound to its cargo.
The sorting of cargoes in the cell is regulated, yet cargoes in a cell exhibit bidirectional movement because multiple copies of dynein and kinesin simultaneously bind to them (73). How directed transport can be achieved by a mixed motor ensemble was unclear. Derr et al. (74) developed a unique in vitro tool in the form of a programmable, synthesized cargo using three-dimensional DNA origami to study this question; specifically, how do motor type, number, spacing, and orientation affect cargo transport? In ensembles of one to seven identical-polarity motors, motor number had a minimal affect on directional velocity, whereas ensembles of opposite-polarity motors engaged in a tug-of-war resolvable by disengaging one motor species. Although this pioneering study provided new insight into how molecular motors coordinate intracellular transport, more research will be required to understand how motor ensembles move cargo. MTs themselves can also be cargoes, and many MAPs transport, slide, or push MTs to arrange them into functional cytoskeletal structures, which has recently been reviewed (11, 12).
Microtubule Nucleation
Another key step in generating a specific MT architecture is to regulate when, where, and how MTs are made. In a cell, MTs are mostly observed to originate from MT organizing centers (MTOCs). Although MTOCs were originally synonymous with centrosomes, many other MTOCs have been identified in the meantime, such as chromosomes, the nuclear envelope, the Golgi apparatus, the plasma membrane, and MTs themselves (75). MT nucleation from MTOCs depends on the nucleation factor γ-tubulin (γ-TB), which associates with additional γ-TB complex (GCP) proteins. In higher eukaryotes, γ-TB and GCPs 2–6 form a distinctive ring-shaped structure, hence its name γ-TB ring complex (γ-TuRC, Fig. 1A). Although in vitro polymerized MTs contain variable protofilament numbers (76), it contains predominantly 13 in vivo. This defined number is templated by a ring of 13 γ-TB molecules in γ-TuRC that then binds tubulin dimers via the α subunit interface (77).
The nucleation activity of purified γ-TB complexes is surprisingly low and led to the suggestion that γ-TB complexes must be activated besides being localized to their respective MTOC. Genetic screens in Saccharomyces cerevisiae, in which GCPs 4–6 are absent and two γ-TB molecules form a tetrameric γ-TB small complex (γ-TuSC) with GCPs 2 and 3, and Schizosaccharomyces pombe, in which GCPs 4–6 are not essential, identified many γ-TuSC-interacting proteins. Similarly, in higher eukaryotes, γ-TuRC-interacting proteins were recently uncovered by immunoprecipitations and co-purifications (78, 79). In parallel, cell-free Xenopus laevis egg extract has been a powerful system to identify new MTOCs and MT nucleation effectors, such as centrosomes, chromatin beads (80), RCC1 (Ran guanine nucleotide exchange factor) beads (81), Aurora A beads (82), and MTs themselves during branching MT nucleation (83). However, only a few of the binding partners and effectors were tested for direct and individual effect in a purified in vitro experiment. Specifically, the kinase NME7 and the centrosomal protein CDK5RAP2 enhanced γ-TuRC MT nucleation activity by 2.5- and 7-fold, respectively, in vitro (78, 84). Thus, evidence in yeast and higher eukaryotic cells suggests that an assembly of proteins is required to recruit, transport, and tether γ-TB complexes to MTOCs, where they are specifically activated, but how they achieve this remains to be determined.
Assembly and Scaling of Self-organized Microtubule Structures
The ultimate challenge is to synthesize a model of how individual MAP activities, which regulate MT dynamics, transport, nucleation, and cross-linking, establish cellular MT structures. Because mitotic and meiotic spindles are easily built and altered in Xenopus egg extract and we known many of the biochemical parameters of spindle MAPs, they have been favored structures for both theoretical and experimental examination of the principles of self-organization and scaling. Theoretical modeling has been instrumental for our understanding of how larger scale structures such as the spindle form (85, 86). Recently, it was discovered that bipolar structures with antiparallel fluxing MTs as in a spindle could be formed in silico with dynamic MTs, an MT cross-linking force, and antiparallel sliding activity, and pole formation was achieved by the addition of a NuMA-like minus-end cross-linker and directed transport of MT depolymerization activity toward minus-ends (87). Realistic MT lifetimes and MT length distributions required dynamic instability and minus-end depolymerization activities, yet meiotic spindle assembly could only be modeled by simulating MT nucleation specifically throughout the spindle and not only from the chromatin zone. The hypothesis that MT nucleation and transport drive spindle assembly, whereas MT dynamics are constant throughout the cytoplasm, has since been experimentally confirmed by novel measurements of spindle dynamics (88) and direct observations of γ-TuRC within the mitotic spindle (89).
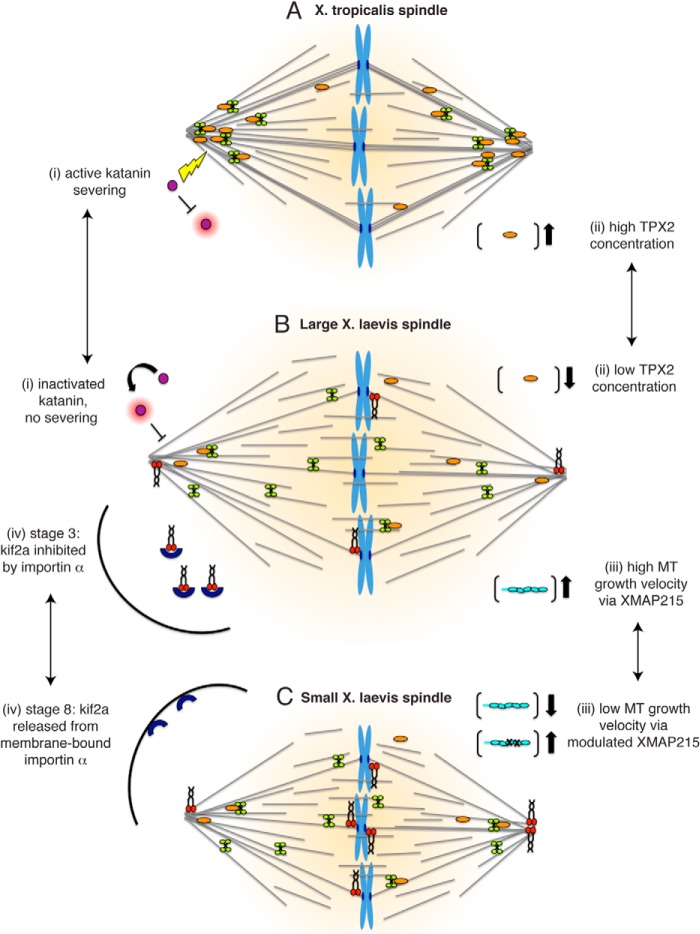
One of the major structural parameters of the spindle that experimentalists have tried to tackle in vitro is size scaling. Xenopus tropicalis extract spindles are smaller than their X. laevis counterparts and serve as an optimal system to identify the molecular basis of spindle size difference (90). Surprisingly, MT depolymerization activity via the MT-severing enzyme katanin was much higher in X. tropicalis egg cytoplasm than in X. laevis and was more concentrated at X. tropicalis spindle poles (91). Katanin inhibition increased spindle length to a greater degree in X. tropicalis than X. laevis, suggesting that it acts as a scaling factor. Although the concentration of katanin was similar in both egg extracts, the catalytic p60 subunit of X. tropicalis katanin lacks an inhibitory Aurora B kinase phosphorylation site, which could explain its higher activity and implied that it is differentially regulated (Fig. 3, A and B).
FIGURE 3.
Spindle scaling. A and B, different regulation of katanin activity has been traced to make X. tropicalis spindles smaller than X. laevis spindles. TPX2 concentrations modulate the spindle architecture and lead to a concentration of MTs the poles of the X. tropicalis spindle. B and C, spindle length of the X. laevis spindle is determined by MT growth velocity, which can be modulated via XMAP215 activity. X. laevis spindles become smaller throughout development and between stage 3 and stage 8; the observed size difference can be contributed to differential regulation of kif2a.
The differences between X. laevis and X. tropicalis extract spindles were further dissected based on evidence that X. tropicalis spindles resist inhibition of two factors essential for assembly of the larger X. laevis spindles: RanGTP and Eg5 (92). The factor that is regulated by RanGTP and binds to Eg5 is TPX2, which is 3-fold more abundant in X. tropicalis extracts. Increased TPX2 level in X. laevis reduced spindle length and sensitivity to Ran and Eg5 inhibition. Curiously, the increased TPX2 led to heightened recruitment of Eg5 at the spindle poles, which consequently increased local MT density, suggesting that the balance of TPX2 and Eg5 modulates spindle architecture (Fig. 3, A and B).
In parallel, further progress has been made in understanding how spindle geometry is controlled within one organism, and systematic, genome-wide screens for mitotic proteins in Caenorhabditis elegans, Drosophila S2 cells, and vertebrate cultured cells identified proteins that influence spindle length (93). Within X. laevis egg extract spindles, MT polymerization is important for regulating spindle length. By engineering versions of XMAP215 with gradually increasing enzymatic activity, spindle length can be increased linearly with MT growth velocity, and thus XMAP215 controls the total mass of spindle MTs (Fig. 3, B and C). This occurs without changing MT density, lifetime, and spindle shape, suggesting that spindle size is determined separately and by mass balance (94). Similarly, the factors that determine spindle shape are not well established, and recent studies in human cells and C. elegans have shown that spindle shape scales anisotropically with spindle length and chromosome number (95, 96).
If one considers the reduction in cell volume from a fertilized egg to a 1000-cell embryo, it is obvious why spindle size needs to be scaled (97). The kinesin 13 kif2a was identified as a driver of this developmental spindle scaling. The MT-destabilizing activity of Kif2a is inhibited in stage 3 spindles by the transport receptor importin α and activated in stage 8 when importin α partitions to a membrane pool (Fig. 3, B and C) (98). Changing spindle size in developing embryos had no effect on chromosome segregation, but interfered with spindle orientation, suggesting that it is coupled to cell size through a ratiometric mechanism controlling microtubule destabilization. This idea was further explored via an innovative system, in which Xenopus egg extracts were encapsulated using microfluidic technology (99, 100). Both studies beautifully demonstrated that reductions in cytoplasmic volume, rather than developmental cues or changes in cell shape, were sufficient to recapitulate spindle scaling observed in Xenopus embryos. Thus, the amount of cytoplasmic material provides a mechanism for regulating the size of intracellular structures.
Concluding Remarks
With a near complete list of MAPs that build the MT cytoskeleton, a rich resource has been made available to now determine how MAP activities are coordinated in time and space to establish functional MT structures. Biochemical and structural studies of individual MAPs and their characterization at a mechanistic level will remain essentials that will benefit from recent advances in cryo-electron microscopy to achieve near atomic resolution of macromolecular assemblies. In vitro reconstitutions of combinations of MAPs will help explain how to build complexity, aided by innovations to precisely position individual molecules, to control the geometry of the sample chamber, and to routinely observe assemblies at the single molecule level and in a parallel manner using microfluidics. Ultimately, this will help uncover how MAPs can create macroscopic structures that form the MT cytoskeleton and narrow the gap that currently exists between purified in vitro systems and extract systems as well as in silico systems. This will simultaneously address an important, general question that the biochemistry of this century is facing: After having learned how individual proteins and protein complexes work, how do numerous factors and protein complexes act in concert to generate self-organized assemblies that enable cell function?
Acknowledgments
We apologize that because of space constraints, many important contributions could not be incorporated. We thank Eric Griffis and Julie Welburn for critical reading of the manuscript and helpful comments.
This work was supported by National Institutes of Health/NIGMS Grant 4R00GM100013, the Pew Scholars Program in the Biomedical Sciences, the Sidney Kimmel Foundation, and the David and Lucile Packard Foundation (all to S. P.). This is the third article in the Thematic Minireview series “The State of the Cytoskeleton in 2015.” The authors declare that they have no conflicts of interest with the contents of this article.
- MT
- microtubule
- MAP
- MT-associated protein
- MTOC
- MT organizing center
- MCAK
- mitotic centromere-associated kinase
- +TIP
- plus-end tracking protein
- EB
- end-binding protein
- CLIP
- class II-associated invariant chain peptide
- γ-TB
- γ-tubulin
- GCP
- γ-TB complex protein.
References
- 1. Mitchison T., Kirschner M. (1984) Dynamic instability of microtubule growth. Nature 312, 237–242 [DOI] [PubMed] [Google Scholar]
- 2. Desai A., Mitchison T. J. (1997) Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83–117 [DOI] [PubMed] [Google Scholar]
- 3. Nédélec F., Surrey T., Karsenti E. (2003) Self-organisation and forces in the microtubule cytoskeleton. Curr. Opin. Cell Biol. 15, 118–124 [DOI] [PubMed] [Google Scholar]
- 4. Goshima G., Wollman R., Goodwin S. S., Zhang N., Scholey J. M., Vale R. D., Stuurman N. (2007) Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316, 417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hughes J. R., Meireles A. M., Fisher K. H., Garcia A., Antrobus P. R., Wainman A., Zitzmann N., Deane C., Ohkura H., Wakefield J. G. (2008) A microtubule interactome: complexes with roles in cell cycle and mitosis. PLoS Biol. 6, e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hutchins J. R. A., Toyoda Y., Hegemann B., Poser I., Hériché J. K., Sykora M. M., Augsburg M., Hudecz O., Buschhorn B. A., Bulkescher J., Conrad C., Comartin D., Schleiffer A., Sarov M., Pozniakovsky A., Slabicki M. M., Schloissnig S., Steinmacher I., Leuschner M., Ssykor A., Lawo S., Pelletier L., Stark H., Nasmyth K., Ellenberg J., Durbin R., Buchholz F., Mechtler K., Hyman A. A., Peters J. M. (2010) Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328, 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neumann B., Walter T., Hériché J. K., Bulkescher J., Erfle H., Conrad C., Rogers P., Poser I., Held M., Liebel U., Cetin C., Sieckmann F., Pau G., Kabbe R., Wünsche A., Satagopam V., Schmitz M. H., Chapuis C., Gerlich D. W., Schneider R., Eils R., Huber W., Peters J. M., Hyman A. A., Durbin R., Pepperkok R., Ellenberg J. (2010) Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464, 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Howard J., Hyman A. A. (2007) Microtubule polymerases and depolymerases. Curr. Opin. Cell Biol. 19, 31–35 [DOI] [PubMed] [Google Scholar]
- 9. Gennerich A., Vale R. D. (2009) Walking the walk: how kinesin and dynein coordinate their steps. Curr. Opin. Cell Biol. 21, 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Akhmanova A., Steinmetz M. O. (2008) Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9, 309–322 [DOI] [PubMed] [Google Scholar]
- 11. Dogterom M., Surrey T. (2013) Microtubule organization in vitro. Curr. Opin. Cell Biol. 25, 23–29 [DOI] [PubMed] [Google Scholar]
- 12. Subramanian R., Kapoor T. M. (2012) Building complexity: insights into self-organized assembly of microtubule-based architectures. Dev. Cell 23, 874–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brouhard G. J., Rice L. M. (2014) The contribution of αβ-tubulin curvature to microtubule dynamics. J. Cell Biol. 207, 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nogales E., Wang H. W. (2006) Structural mechanisms underlying nucleotide-dependent self-assembly of tubulin and its relatives. Curr. Opin. Struct. Biol. 16, 221–229 [DOI] [PubMed] [Google Scholar]
- 15. Alushin G. M., Lander G. C., Kellogg E. H., Zhang R., Baker D., Nogales E. (2014) High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell 157, 1117–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kline-Smith S. L., Walczak C. E. (2002) The microtubule-destabilizing kinesin XKCM1 regulates microtubule dynamic instability in cells. Mol. Biol. Cell 13, 2718–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wordeman L. (2005) Microtubule-depolymerizing kinesins. Curr. Opin. Cell Biol. 17, 82–88 [DOI] [PubMed] [Google Scholar]
- 18. Rogers G. C., Rogers S. L., Schwimmer T. A., Ems-McClung S. C., Walczak C. E., Vale R. D., Scholey J. M., Sharp D. J. (2004) Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature 427, 364–370 [DOI] [PubMed] [Google Scholar]
- 19. Maney T., Wagenbach M., Wordeman L. (2001) Molecular dissection of the microtubule depolymerizing activity of mitotic centromere-associated kinesin. J. Biol. Chem. 276, 34753–34758 [DOI] [PubMed] [Google Scholar]
- 20. Desai A., Verma S., Mitchison T. J., Walczak C. E. (1999) Kin I kinesins are microtubule-destabilizing enzymes. Cell 96, 69–78 [DOI] [PubMed] [Google Scholar]
- 21. Moores C. A., Yu M., Guo J., Beraud C., Sakowicz R., Milligan R. A. (2002) A mechanism for microtubule depolymerization by KinI kinesins. Mol. Cell 9, 903–909 [DOI] [PubMed] [Google Scholar]
- 22. Helenius J., Brouhard G., Kalaidzidis Y., Diez S., Howard J. (2006) The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature 441, 115–119 [DOI] [PubMed] [Google Scholar]
- 23. Cooper J. R., Wagenbach M., Asbury C. L., Wordeman L. (2010) Catalysis of the microtubule on-rate is the major parameter regulating the depolymerase activity of MCAK. Nat. Struct. Mol. Biol. 17, 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gardner M. K., Zanic M., Gell C., Bormuth V., Howard J. (2011) Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell 147, 1092–1103 [DOI] [PubMed] [Google Scholar]
- 25. Al-Bassam J., Chang F. (2011) Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 21, 604–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brouhard G. J., Stear J. H., Noetzel T. L., Al-Bassam J., Kinoshita K., Harrison S. C., Howard J., Hyman A. A. (2008) XMAP215 is a processive microtubule polymerase. Cell 132, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Widlund P. O., Stear J. H., Pozniakovsky A., Zanic M., Reber S., Brouhard G. J., Hyman A. A., Howard J. (2011) XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc. Natl. Acad. Sci. U.S.A. 108, 2741–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ayaz P., Munyoki S., Geyer E. A., Piedra F. A., Vu E. S., Bromberg R., Otwinowski Z., Grishin N. V., Brautigam C. A., Rice L. M. (2014) A tethered delivery mechanism explains the catalytic action of a microtubule polymerase. eLife 3, e03069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Niethammer P., Kronja I., Kandels-Lewis S., Rybina S., Bastiaens P., Karsenti E. (2007) Discrete states of a protein interaction network govern interphase and mitotic microtubule dynamics. PLoS Biol. 5, e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cassimeris L., Pryer N. K., Salmon E. D. (1988) Real-time observations of microtubule dynamic instability in living cells. J. Cell Biol. 107, 2223–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Verde F., Dogterom M., Stelzer E., Karsenti E., Leibler S. (1992) Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J. Cell Biol. 118, 1097–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rusan N. M., Fagerstrom C. J., Yvon A. M., Wadsworth P. (2001) Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-α tubulin. Mol. Biol. Cell 12, 971–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Horio T., Hotani H. (1986) Visualization of the dynamic instability of individual microtubules by dark-field microscopy. Nature 321, 605–607 [DOI] [PubMed] [Google Scholar]
- 34. Walker R. A., O'Brien E. T., Pryer N. K., Soboeiro M. F., Voter W. A., Erickson H. P., Salmon E. D. (1988) Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J. Cell Biol. 107, 1437–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kinoshita K., Arnal I., Desai A., Drechsel D. N., Hyman A. A. (2001) Reconstitution of physiological microtubule dynamics using purified components. Science 294, 1340–1343 [DOI] [PubMed] [Google Scholar]
- 36. Mimori-Kiyosue Y., Shiina N., Tsukita S. (2000) The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 10, 865–868 [DOI] [PubMed] [Google Scholar]
- 37. Rogers S. L., Rogers G. C., Sharp D. J., Vale R. D. (2002) Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 158, 873–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tirnauer J. S., O'Toole E., Berrueta L., Bierer B. E., Pellman D. (1999) Yeast Bim1p promotes the G1-specific dynamics of microtubules. J. Cell Biol. 145, 993–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maurer S. P., Cade N. I., Bohner G., Gustafsson N., Boutant E., Surrey T. (2014) EB1 accelerates two conformational transitions important for microtubule maturation and dynamics. Curr. Biol. 24, 372–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Honnappa S., Gouveia S. M., Weisbrich A., Damberger F. F., Bhavesh N. S., Jawhari H., Grigoriev I., van Rijssel F. J., Buey R. M., Lawera A., Jelesarov I., Winkler F. K., Wüthrich K., Akhmanova A., Steinmetz M. O. (2009) An EB1-binding motif acts as a microtubule tip localization signal. Cell 138, 366–376 [DOI] [PubMed] [Google Scholar]
- 41. Slep K. C. (2010) Structural and mechanistic insights into microtubule end-binding proteins. Curr. Opin. Cell Biol. 22, 88–95 [DOI] [PubMed] [Google Scholar]
- 42. Bieling P., Kandels-Lewis S., Telley I. A., van Dijk J., Janke C., Surrey T. (2008) CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J. Cell Biol. 183, 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dixit R., Barnett B., Lazarus J. E., Tokito M., Goldman Y. E., Holzbaur E. L. (2009) Microtubule plus-end tracking by CLIP-170 requires EB1. Proc. Natl. Acad. Sci. U.S.A. 106, 492–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gouveia S. M., Akhmanova A. (2010) Cell and molecular biology of microtubule plus end tracking proteins: end binding proteins and their partners. Int. Rev. Cell Mol. Biol. 285, 1–74 [DOI] [PubMed] [Google Scholar]
- 45. Kumar P., Wittmann T. (2012) +TIPs: SxIPping along microtubule ends. Trends Cell Biol. 22, 418–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mennella V., Rogers G. C., Rogers S. L., Buster D. W., Vale R. D., Sharp D. J. (2005) Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat. Cell Biol. 7, 235–245 [DOI] [PubMed] [Google Scholar]
- 47. Montenegro Gouveia S., Leslie K., Kapitein L. C., Buey R. M., Grigoriev I., Wagenbach M., Smal I., Meijering E., Hoogenraad C. C., Wordeman L., Steinmetz M. O., Akhmanova A. (2010) In vitro reconstitution of the functional interplay between MCAK and EB3 at microtubule plus ends. Curr. Biol. 20, 1717–1722 [DOI] [PubMed] [Google Scholar]
- 48. Domnitz S. B., Wagenbach M., Decarreau J., Wordeman L. (2012) MCAK activity at microtubule tips regulates spindle microtubule length to promote robust kinetochore attachment. J. Cell Biol. 197, 231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ohi R., Coughlin M. L., Lane W. S., Mitchison T. J. (2003) An inner centromere protein that stimulates the microtubule depolymerizing activity of a KinI kinesin. Dev. Cell 5, 309–321 [DOI] [PubMed] [Google Scholar]
- 50. Ohi R., Sapra T., Howard J., Mitchison T. J. (2004) Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol. Biol. Cell 15, 2895–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jiang K., Wang J., Liu J., Ward T., Wordeman L., Davidson A., Wang F., Yao X. (2009) TIP150 interacts with and targets MCAK at the microtubule plus ends. EMBO Rep. 10, 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tanenbaum M. E., Macurek L., van der Vaart B., Galli M., Akhmanova A., Medema R. H. (2011) A complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by Aurora kinases. Curr. Biol. 21, 1356–1365 [DOI] [PubMed] [Google Scholar]
- 53. Stout J. R., Yount A. L., Powers J. A., Leblanc C., Ems-McClung S. C., Walczak C. E. (2011) Kif18B interacts with EB1 and controls astral microtubule length during mitosis. Mol. Biol. Cell 22, 3070–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goodwin S. S., Vale R. D. (2010) Patronin regulates the microtubule network by protecting microtubule minus ends. Cell 143, 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jiang K., Hua S., Mohan R., Grigoriev I., Yau K. W., Liu Q., Katrukha E. A., Altelaar A. F., Heck A. J., Hoogenraad C. C., Akhmanova A. (2014) Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev. Cell 28, 295–309 [DOI] [PubMed] [Google Scholar]
- 56. Hendershott M. C., Vale R. D. (2014) Regulation of microtubule minus-end dynamics by CAMSAPs and Patronin. Proc. Natl. Acad. Sci. U.S.A. 111, 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Meunier S., Vernos I. (2011) K-fibre minus ends are stabilized by a RanGTP-dependent mechanism essential for functional spindle assembly. Nat. Cell Biol. 13, 1406–1414 [DOI] [PubMed] [Google Scholar]
- 58. Kronja I., Kruljac-Letunic A., Caudron-Herger M., Bieling P., Karsenti E. (2009) XMAP215-EB1 interaction is required for proper spindle assembly and chromosome segregation in Xenopus egg extract. Mol. Biol. Cell 20, 2684–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zanic M., Widlund P. O., Hyman A. A., Howard J. (2013) Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nat. Cell Biol. 15, 688–693 [DOI] [PubMed] [Google Scholar]
- 60. Li W., Moriwaki T., Tani T., Watanabe T., Kaibuchi K., Goshima G. (2012) Reconstitution of dynamic microtubules with Drosophila XMAP215, EB1, and Sentin. J. Cell Biol. 199, 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. van der Vaart B., Franker M. A., Kuijpers M., Hua S., Bouchet B. P., Jiang K., Grigoriev I., Hoogenraad C. C., Akhmanova A. (2012) Microtubule plus-end tracking proteins SLAIN1/2 and ch-TOG promote axonal development. J. Neurosci. 32, 14722–14728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Duellberg C., Trokter M., Jha R., Sen I., Steinmetz M. O., Surrey T. (2014) Reconstitution of a hierarchical +TIP interaction network controlling microtubule end tracking of dynein. Nat. Cell Biol. 16, 804–811 [DOI] [PubMed] [Google Scholar]
- 63. Caudron F., Andrieux A., Job D., Boscheron C. (2008) A new role for kinesin-directed transport of Bik1p (CLIP-170) in Saccharomyces cerevisiae. J. Cell Sci. 121, 1506–1513 [DOI] [PubMed] [Google Scholar]
- 64. Markus S. M., Punch J. J., Lee W. L. (2009) Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr. Biol. 19, 196–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yamada M., Toba S., Yoshida Y., Haratani K., Mori D., Yano Y., Mimori-Kiyosue Y., Nakamura T., Itoh K., Fushiki S., Setou M., Wynshaw-Boris A., Torisawa T., Toyoshima Y. Y., Hirotsune S. (2008) LIS1 and NDEL1 coordinate the plus-end-directed transport of cytoplasmic dynein. EMBO J. 27, 2471–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Roberts A. J., Goodman B. S., Reck-Peterson S. L. (2014) Reconstitution of dynein transport to the microtubule plus end by kinesin. eLife 3, e02641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Welburn J. P. (2013) The molecular basis for kinesin functional specificity during mitosis. Cytoskeleton 70, 476–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Allan V. J. (2011) Cytoplasmic dynein. Biochem. Soc. Trans. 39, 1169–1178 [DOI] [PubMed] [Google Scholar]
- 69. Reck-Peterson S. L., Yildiz A., Carter A. P., Gennerich A., Zhang N., Vale R. D. (2006) Single-molecule analysis of dynein processivity and stepping behavior. Cell 126, 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Trokter M., Mücke N., Surrey T. (2012) Reconstitution of the human cytoplasmic dynein complex. Proc. Natl. Acad. Sci. U.S.A. 109, 20895–20900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schlager M. A., Hoang H. T., Urnavicius L., Bullock S. L., Carter A. P. (2014) In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 33, 1855–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. McKenney R. J., Huynh W., Tanenbaum M. E., Bhabha G., Vale R. D. (2014) Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345, 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hirokawa N., Niwa S., Tanaka Y. (2010) Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68, 610–638 [DOI] [PubMed] [Google Scholar]
- 74. Derr N. D., Goodman B. S., Jungmann R., Leschziner A. E., Shih W. M., Reck-Peterson S. L. (2012) Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science 338, 662–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lüders J., Stearns T. (2007) Microtubule-organizing centres: a re-evaluation. Nat. Rev. Mol. Cell Biol. 8, 161–167 [DOI] [PubMed] [Google Scholar]
- 76. Evans L., Mitchison T., Kirschner M. (1985) Influence of the centrosome on the structure of nucleated microtubules. J. Cell Biol. 100, 1185–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kollman J. M., Merdes A., Mourey L., Agard D. A. (2011) Microtubule nucleation by γ-tubulin complexes. Nat. Rev. Mol. Cell Biol. 12, 709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Choi Y. K., Liu P., Sze S. K., Dai C., Qi R. Z. (2010) CDK5RAP2 stimulates microtubule nucleation by the γ-tubulin ring complex. J. Cell Biol. 191, 1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Teixidó-Travesa N., Villén J., Lacasa C., Bertran M. T., Archinti M., Gygi S. P., Caelles C., Roig J., Lüders J. (2010) The γTuRC revisited: a comparative analysis of interphase and mitotic human γTuRC redefines the set of core components and identifies the novel subunit GCP8. Mol. Biol. Cell 21, 3963–3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., Hyman A., Karsenti E. (1996) Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382, 420–425 [DOI] [PubMed] [Google Scholar]
- 81. Halpin D., Kalab P., Wang J., Weis K., Heald R. (2011) Mitotic spindle assembly around RCC1-coated beads in Xenopus egg extracts. PLoS Biol. 9, e1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ishihara K., Nguyen P. A., Groen A. C., Field C. M., Mitchison T. J. (2014) Microtubule nucleation remote from centrosomes may explain how asters span large cells. Proc. Natl. Acad. Sci. U.S.A. 111, 17715–17722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Petry S., Groen A. C., Ishihara K., Mitchison T. J., Vale R. D. (2013) Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 152, 768–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Liu P., Choi Y. K., Qi R. Z. (2014) NME7 is a functional component of the γ-tubulin ring complex. Mol. Biol. Cell 25, 2017–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Burbank K. S., Mitchison T. J., Fisher D. S. (2007) Slide-and-cluster models for spindle assembly. Curr. Biol. 17, 1373–1383 [DOI] [PubMed] [Google Scholar]
- 86. Wollman R., Civelekoglu-Scholey G., Scholey J. M., Mogilner A. (2008) Reverse engineering of force integration during mitosis in the Drosophila embryo. Mol. Syst. Biol. 4, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Loughlin R., Heald R., Nédélec F. (2010) A computational model predicts Xenopus meiotic spindle organization. J. Cell Biol. 191, 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Brugués J., Nuzzo V., Mazur E., Needleman D. J. (2012) Nucleation and transport organize microtubules in metaphase spindles. Cell 149, 554–564 [DOI] [PubMed] [Google Scholar]
- 89. Lecland N., Lüders J. (2014) The dynamics of microtubule minus ends in the human mitotic spindle. Nat. Cell Biol. 16, 770–778 [DOI] [PubMed] [Google Scholar]
- 90. Brown K. S., Blower M. D., Maresca T. J., Grammer T. C., Harland R. M., Heald R. (2007) Xenopus tropicalis egg extracts provide insight into scaling of the mitotic spindle. J. Cell Biol. 176, 765–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Loughlin R., Wilbur J. D., McNally F. J., Nédélec F. J., Heald R. (2011) Katanin contributes to interspecies spindle length scaling in Xenopus. Cell 147, 1397–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Helmke K. J., Heald R. (2014) TPX2 levels modulate meiotic spindle size and architecture in Xenopus egg extracts. J. Cell Biol. 206, 385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Goshima G., Scholey J. M. (2010) Control of mitotic spindle length. Annu. Rev. Cell Dev. Biol. 26, 21–57 [DOI] [PubMed] [Google Scholar]
- 94. Reber S. B., Baumgart J., Widlund P. O., Pozniakovsky A., Howard J., Hyman A. A., Jülicher F. (2013) XMAP215 activity sets spindle length by controlling the total mass of spindle microtubules. Nat. Cell Biol. 15, 1116–1122 [DOI] [PubMed] [Google Scholar]
- 95. Hara Y., Kimura A. (2013) An allometric relationship between mitotic spindle width, spindle length, and ploidy in Caenorhabditis elegans embryos. Mol. Biol. Cell 24, 1411–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Young S., Besson S., Welburn J. P. (2014) Length-dependent anisotropic scaling of spindle shape. Biol. Open 3, 1217–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Levy D. L., Heald R. (2012) Mechanisms of intracellular scaling. Annu. Rev. Cell Dev. Biol. 28, 113–135 [DOI] [PubMed] [Google Scholar]
- 98. Wilbur J. D., Heald R. (2013) Mitotic spindle scaling during Xenopus development by kif2a and importin α. eLife 2, e00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hazel J., Krutkramelis K., Mooney P., Tomschik M., Gerow K., Oakey J., Gatlin J. C. (2013) Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science 342, 853–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Good M. C., Vahey M. D., Skandarajah A., Fletcher D. A., Heald R. (2013) Cytoplasmic volume modulates spindle size during embryogenesis. Science 342, 856–860 [DOI] [PMC free article] [PubMed] [Google Scholar]