Abstract
Microtubules give rise to intracellular structures with diverse morphologies and dynamics that are crucial for cell division, motility, and differentiation. They are decorated with abundant and chemically diverse posttranslational modifications that modulate their stability and interactions with cellular regulators. These modifications are important for the biogenesis and maintenance of complex microtubule arrays such as those found in spindles, cilia, neuronal processes, and platelets. Here we discuss the nature and subcellular distribution of these posttranslational marks whose patterns have been proposed to constitute a tubulin code that is interpreted by cellular effectors. We review the enzymes responsible for writing the tubulin code, explore their functional consequences, and identify outstanding challenges in deciphering the tubulin code.
Keywords: cytoskeleton, microtubule, microtubule-associated protein (MAP), post-translational modification (PTM), tubulin, TTLL, microtubule dynamics, microtubule motor, tubulin post-translational modifications, tubulin tyrosine ligase
Introduction
Microtubules are non-covalent cylindrical polymers formed by αβ-tubulin heterodimer building blocks. They possess two seemingly contradictory properties; they are highly dynamic, exhibiting rapid growth and shrinkage of their ends (1), but are also very rigid, with persistence lengths on the order of cellular dimensions (2). This duality is thought to underlie the versatile architectures of microtubule networks in cells (Fig. 1) and is tuned by a myriad of cellular effectors. These fall into two categories: effectors that bind to the microtubule and alter its properties non-covalently (motors and microtubule-associated proteins (MAPs))2 and effectors that chemically modify the tubulin subunits (tubulin posttranslational modification enzymes). Although the field has made tremendous progress in recent decades identifying a compendium of microtubule-interacting proteins and understanding their interplay and regulation in the cell, we are just now starting to unravel the basic mechanisms used by cells to chemically modify microtubules, despite the fact that tubulin posttranslational modifications have been known for over 40 years. A renaissance of interest into the roles of tubulin posttranslational modifications has been precipitated by the discovery in the last few years of the enzymes responsible for these modifications (3–8), methods for producing unmodified (9, 10), engineered, (11, 12) as well as chemically defined modified tubulin (13), and developments and refinements of in vitro microtubule-based assays using high-resolution microscopy and microfabricated substrates (14–16).
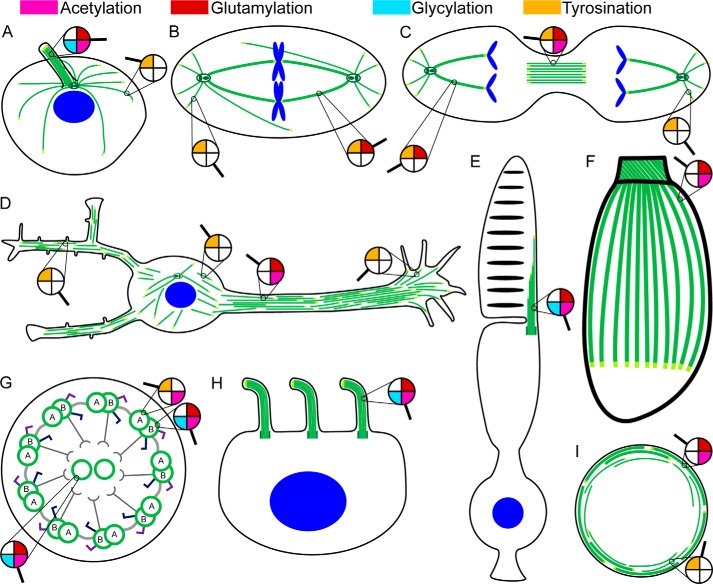
FIGURE 1.
Microtubules form complex arrays of spatio-temporally regulated supra-structures. A, radial interphase array. B, mitotic spindle array. C, midbody array. D, neuron with complex, parallel, and tiled array in the axon and mixed polarity array in dendrites. E, photoreceptor cells with a connecting cilium between their inner and outer segments. Individual microtubules extend to varying depths of the outer segment. F, protozoans contain a unique membrane-embedded array of subpellicular microtubules and an additional apical cylindrical structure termed the conoid that consists of unique comma-shaped open polymers formed from nine laterally associated tubulin protofilaments. G, cross-sectional view of the nine-fold symmetric axonemal array. Light gray, nexin linkers; dark gray, radial spokes; dark blue, inner arm dyneins; purple, outer arm dyneins. H, cell with multiple motile cilia. I, marginal microtubule band in platelets; stable microtubules are coiled in the peripheral edges, whereas dynamic, tyrosinated microtubules actively polymerize/depolymerize. Microtubules are shown in green; microtubule plus-ends are in light green, and nuclei are in blue. The distribution of tubulin posttranslational modifications in the various microtubule arrays is indicated by a magnifying glass (pink, acetylation; red, glutamylation; cyan, glycylation; orange, tyrosination).
Tubulin posttranslational modifications are chemically diverse, ranging from phosphorylation (17), acetylation (18, 19), palmitoylation (20), sumoylation (21), polyamination (22), and S-nitrosylation (23) to tyrosination (24), glutamylation (25, 26), and glycylation (27). Most of these modifications are reversible. Tubulin posttranslational modifications are evolutionarily conserved and abundantly represented in cellular microtubules. Most importantly, their distribution is stereotyped in cells. For example, interphase microtubules are enriched in tyrosination (28), whereas kinetochore fibers and midbody microtubules are enriched in detyrosination and glutamylation (Fig. 1, A–C) (29, 30). Axonal microtubules are enriched in detyrosination, acetylation, and glutamylation, whereas the dynamic growth cone microtubules are enriched in tyrosination (Fig. 1D) (31, 32). Microtubules in centrioles, cilia, and flagella are especially heavily glutamylated (Fig. 1, E, G, and H) (29, 33, 34). Glycylated microtubules are found predominantly in the axonemes of cilia and flagella (Fig. 1, A, G, and H) (35, 36); however, some cytoplasmic microtubules in paramecia are also glycylated (36). The more morphologically complex microtubule arrays exhibit the largest diversity and abundance of tubulin posttranslational modifications like the microtubule arrays found in neurons, cilia, or flagella or the highly specialized arrays found in some parasites such as toxoplasma and trypanosomes (Fig. 1F) (37). In some cases, even adjacent microtubules have completely different posttranslational modification signatures. This is beautifully illustrated in axonemes where the B tubule is highly glutamylated (38, 39), whereas the adjoining A tubule is not, but is enriched in tyrosination (Fig. 1G). Tubulin posttranslational modifications are also developmentally regulated. One striking example is during neuronal development that is accompanied by increases in glutamylation levels of both α-tubulin and β-tubulin, with β-tubulin glutamylation increasing mostly during the later stages of neuronal differentiation (40).
The enzymes that introduce these conserved modifications are essential to normal development (5, 41, 42). Underscoring their importance for normal cell physiology, increased levels of tubulin modifications are a hallmark of cancers and neurodegenerative disorders (reviewed in Ref. 43), and several neurodevelopmental disorders are linked to mutations in tubulin genes at sites that could interfere with modification enzyme function (reviewed in Ref. 44).
This microtubule chemical diversity was proposed to form the basis of a “tubulin code” that is read by cellular effectors (45). Despite the widespread appreciation for the ubiquity and functional importance of these modifications and their stereotyped distribution in organisms and cells, we do not currently understand how complex microtubule modification patterns are generated and what their functional consequences on cellular effectors are, i.e. we do not understand how the tubulin code is written and interpreted by cells.
Tubulin Posttranslational Modifications: Variations upon a Dimer
The tubulin αβ-heterodimer is composed of a compact globular body and unstructured, negatively charged tubulin C-terminal tails (Fig. 2A). Although the tubulin body is highly conserved from Saccharomyces cerevisiae to humans, the C-terminal tails are the sites of largest sequence variation between organisms as well as among tubulin isoforms from the same organism. Despite their sequence variability, all tubulin tails are highly negatively charged, with glutamate residues being overrepresented (reviewed in Ref. 46). The overwhelming majority of tubulin posttranslational modifications concentrate on the C-terminal tails that serve as interaction sites for molecular motors and MAPs and thus can tune the activity of these effectors (reviewed in Refs. 43, 45, and 46).
FIGURE 2.

Posttranslational modifications map to both the body and the tails of the αβ-tubulin heterodimer. A, ribbon representation of the tubulin heterodimer (green, α-tubulin; blue, β-tubulin) with the disordered tubulin tails shown schematically using sequences for the α1A and βIVb tubulin isoforms. Sites of acetylation and polyamination are shown in stick representation (magenta and dark blue, respectively). The α-tubulin C-terminal tyrosine (orange) is subject to enzymatic removal/ligation (detyrosination/tyrosination cycle). Tail glutamates are subject to monoglutamylation and polyglutamylation (n denotes the number of glutamates in the elongated chain). Tail glutamates are also subject to monoglycylation and polyglycylation (m denotes the number of glycines in the elongated chain). B, zoomed-in view showing the acetylated β-tubulin Lys-252. C and D, view of the α-tubulin (C) and β-tubulin (D) longitudinal interfaces showing the position of mapped polyamination sites as dark blue sticks. α-Tubulin Lys-40 is shown in stick representation (magenta). Am, amination. Ac, acetylation.
Modifications on the C-terminal tails include detyrosination/tyrosination of α-tubulin (24), the removal of the penultimate glutamate of α-tubulin (forming Δ2-tubulin) (47), and glutamylation (25, 26) and glycylation (27) of α- and β-tubulin tails. The tubulin body is also subject to varied posttranslational modifications, such as palmitoylation (48), phosphorylation (17), S-nitrosylation (23), and polyamination (22). Acetylation of α-tubulin Lys-40 is the only modification known to occur within the microtubule lumen (49–51). With the exception of α-tubulin Lys-40 acetylation, posttranslational modifications on the tubulin body have not been intensively studied.
Detyrosination/Tyrosination
Most mammalian α-tubulin isoforms are synthesized with a genomically encoded C-terminal tyrosine that can then undergo enzymatic removal and re-addition as part of a detyrosination/tyrosination cycle (Fig. 2A). Tubulin tyrosination has long been used as a marker of microtubule stability in cells; tyrosinated microtubules persist 3–5 min, whereas long-lived microtubules are detyrosinated (lifetimes of 2–16 h) and resistant to cold- or nocodazole-induced depolymerization (28). For example, stable axonal microtubules are predominantly detyrosinated, whereas highly dynamic growth cone and dendritic microtubules are tyrosinated (52, 53). The modification per se does not seem to alter microtubule stability (54) but rather functions as a signal for the recruitment of cellular effectors to the microtubule (55) (reviewed in Ref. 43).
Δ2-Tubulin
Following detyrosination, the penultimate glutamate residue of the α-tubulin C-terminal tail can be further removed, producing Δ2-tubulin. This irreversible modification prevents the re-addition of the C-terminal tyrosine, thus removing this tubulin species from the tyrosination cycle (47). This modification also serves as a marker for stable microtubules in cells and is especially enriched on axonal microtubules (47).
Glutamylation
Microtubule glutamylation is the posttranslational addition of glutamate residues to the C-terminal tails of both α-tubulin and β-tubulin, targeting multiple internal sites in the glutamate-rich tails (56). The first glutamate is added through an isopeptide bond between the γ-carboxyl group of tubulin's encoded glutamate residue and the amino group of the incoming glutamate. The glutamates added beyond the initial branching point are linked through peptide bonds on α-carboxyl groups (57). Glutamylation is widely conserved across unicellular flagellates and multicellular organisms with the exception of higher-order plants (3). Glutamylation is enriched on neuronal microtubules, and also microtubules of the mitotic spindle (29), basal bodies, and axonemes of cilia and flagella (29) (Fig. 1). Glutamylation levels in cells are regulated through the opposing actions of both glutamylating and deglutamylating enzymes (5, 40, 58).
Glycylation
Microtubule glycylation is the addition of glycine residues to the side chains of glutamates on α- and β-tubulin C-terminal tails. Multiple glutamate residues in a tubulin tail can be glycylated, and subsequent additions of glycine extend this modification to form glycine chains (59). Glycylation is conserved among unicellular flagellates and multicellular organisms with ciliated tissues (60). Monoglycylation is ubiquitous in ciliated tissues, whereas only a subset contains polyglycylated microtubules (61). Glycylation is important for the stability, length, and function of motile cilia (7, 62, 63), the formation and maintenance of primary cilia, and control of cell proliferation (64).
Acetylation
α-Tubulin is acetylated on Lys-40 (65), a residue located within a short highly flexible loop projecting into the microtubule lumen (49, 51). Acetylation is enriched on microtubules in cilia and basal bodies as well as on a subset of stable, long-lived microtubules in the cytoplasm (lifetimes ∼2–16 h (66)). The direct effects of acetylation on microtubule dynamics and mechanical stability are not clear. Early studies showed no effects on brain tubulin polymerization (67); however, this tubulin contains a combination of other posttranslational modifications that could have masked the effects of acetylation. The answer to the question of stability will have to await in vitro microtubule dynamics as well as persistence length measurements with homogenous unmodified and acetylated tubulin. Interestingly, studies in Caenorhabditis elegans have revealed that Lys-40 acetylation is important for the formation and integrity of specialized 15-protofilament microtubules that are found in touch receptor neurons. In the absence of tubulin acetyltransferase (TAT), these microtubules appear radially compressed, and many are splayed open (68, 69).
In addition to acetylation on Lys-40, a second acetylation site has been reported more recently on β-tubulin Lys-252 in free heterodimers. This modification inhibits the incorporation of tubulin into microtubules (19), most likely due to its proximity to the nucleotide-binding site on α-tubulin at the interface between the α- and β-tubulin protomers (Fig. 2B). It has been proposed that acetylation at this site interferes with a conformational switch in the tubulin heterodimer needed for robust microtubule incorporation (19).
Polyamination
Polyamination has recently been discovered as an irreversible modification of tubulin that results in the covalent addition of polyamines, including putrescine, spermine, and spermidine, to various glutamines on α- and β-tubulin (22) (Fig. 2, A, C, and D). Polyamines are highly abundant in brain tissue. Polyamination sites map close to polymerization interfaces where they can impact tubulin polymerization and microtubule stability (Fig. 2, C and D), possibly helping to maintain cytoskeleton organization during neuronal development (22). Indeed, tubulin polyamination confers stability to microtubules, preventing cold- and Ca2+-mediated depolymerization (22). This might explain the persistence of a small fraction of microtubules that are resistant to cold- or Ca2+-induced depolymerization encountered during brain tubulin cycling and that consist of more positively charged tubulin isotypes (22, 70).
Palmitoylation
Palmitoylation, the modification of cysteine residues with a fatty acid group, is found on membrane-associated proteins, providing a posttranslational means of embedment. In mammals, tubulin palmitoylation was initially characterized in platelet microtubules. However, the significance of this modification remains unclear outside of S. cerevisiae where palmitoylation of α-tubulin Cys-377 affects nuclear positioning in anaphase (48).
S-Nitrosylation
S-Nitrosylation involves the addition of nitric oxide to various cysteine residues of both α-tubulin and β-tubulin. First observed in rat brain lysates (23), the in vivo function of this tubulin modification is unknown.
Phosphorylation
β-Tubulin phosphorylation was originally documented in rat brain tubulin (17). Tubulin phosphorylation is a poorly characterized modification whose functional significance is unclear. α-Tubulin is phosphorylated on an unidentified tyrosine residue near its C terminus by the kinase Syk (71), whereas β-tubulin is phosphorylated on Ser-172 (72). Ser-172 phosphorylation inhibits polymerization, likely a consequence of decreased nucleotide binding due to this residue's proximity to the exchangeable nucleotide-binding site on β-tubulin.
Writers of the Tubulin Code: Who Are They?
The staggering chemical complexity of tubulin is produced by diverse protein families ranging from kinases and acetyltransferases to ATP-dependent ligases and carboxypeptidases. Many of these enzymes were not identified until the last decade. The first tubulin modification enzyme isolated and cloned was tubulin tyrosine ligase (TTL), the enzyme responsible for the ATP-dependent re-addition of the genomically encoded tyrosine residue to the C terminus of α-tubulin (73). TTL loss has drastic effects for the viability of the organism as TTL knock-out mice die shortly after birth due to disorganized neuronal arrays (41). TTL suppression is also strongly linked to tumorigenesis as well as tumor aggressiveness (74).
The most abundant and variable components of the tubulin code, glutamylation and glycylation, are products of enzymes that belong to the tubulin tyrosine ligase-like (TTLL) family. Enzymes of this family share a core domain structurally homologous to TTL and an ATP-dependent amino acid ligation mechanism, which is also shared with more distantly related amino acid ligases such as glutathione S-transferase or d-Ala:d-Ala ligase (3, 6, 75). All TTLLs preferentially modify microtubules (6, 76), unlike TTL, which modifies soluble tubulin (75).
Mammals encode 13 TTLLs (Table 1). TTLL1, -4, -5, -6, -7, -9, -11, and -13 are glutamylases (3, 6, 39, 77, 78), whereas TTLL3, -8, and -10 are glycylases ((4, 7, 79); reviewed in Ref. 43). TTLL2 appears to be a glutamylase based on homology, but has yet to be biochemically characterized. TTLL12 is inactive as both a glutamylase and a glycylase but is proposed to function as a pseudoenzyme that alters tubulin tyrosination and DNA methylation levels indirectly (80).
TABLE 1.
Biochemical characteristics of TTL and TTLL family enzymes
MT, microtubule.
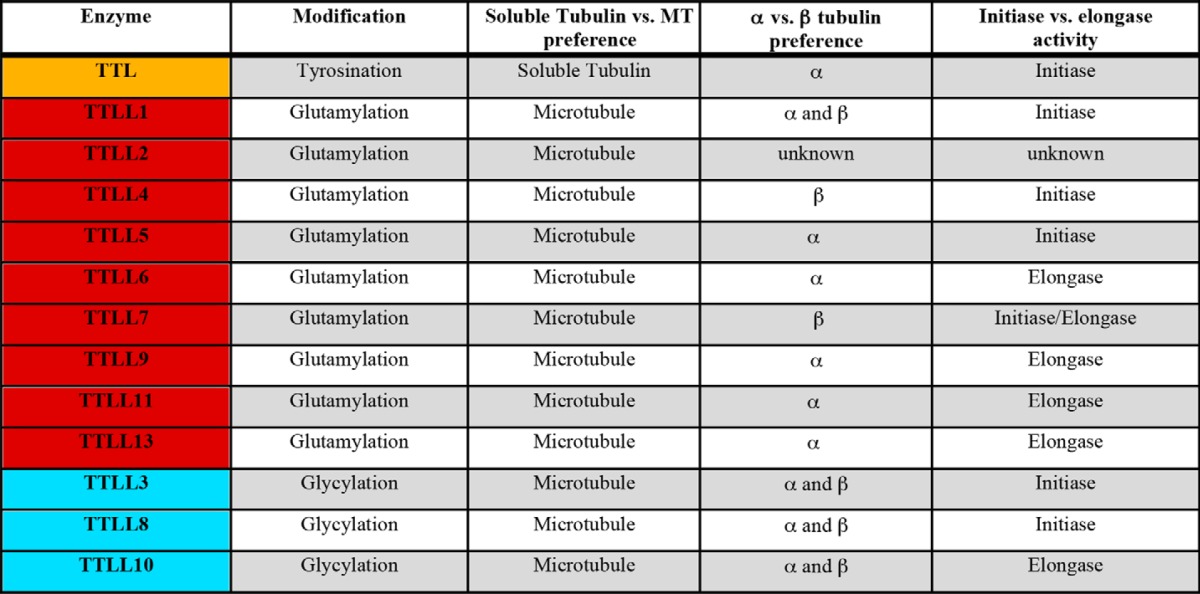
Beyond the amino acid they specifically incorporate, TTLLs are distinguished by their preferences for α- or β-tubulin and whether they are initiases or elongases (Table 1). Initiases add the first glutamate or glycine to a tubulin tail internal glutamate, whereas elongases extend these modifications with additional glutamates or glycines, respectively. TTLL4, an initiase, shows a preference for β-tubulin, whereas TTLL5, also an initiase, prefers α-tubulin (6). TTLL6, -11, and -13 elongate glutamate chains preferentially on α-tubulin (6). TTLL3 seems to initiate glycylation with equal preference for α- and β-tubulin (4), whereas TTLL8 shows a preference for initiating a glycine chain on β-tubulin (4).
The different nature of the substrates involved, a γ-carboxyl group for initiation and an α-carboxyl group for elongation, makes the delineation of the TTLL family enzymes into initiases and elongases quite attractive. However, exceptions seem to exist to this framework. For example, the β-tubulin glutamylase TTLL7, the most abundant glutamylase in neuronal tissue, catalyzes both initiation and elongation (76). Even more surprising, as initiating and elongating glycines are added to markedly different substrates (glutamate and glycine, respectively), the two TTLL3 glycylase homologs in Drosophila melanogaster are capable of both initiating and elongating glycine chains (4). It is important to note that most investigations into the substrate specificity of the TTLL enzymes involve cellular overexpression. The elevated concentrations encountered in these types of experiments can lead to increased promiscuity that can confound true physiological specificity.
Most TTLL enzymes are much larger than the conserved ∼370-residue TTL core that defines the family. TTLL enzymes typically range in size from ∼400 amino acids (TTLL1 and -9) to ∼1200 amino acids and larger (TTLL4 and -5). Most TTLL enzymes with autonomous activity (TTLL3, -4, -5, -6, -7, -8, -10, -11, and -13) are larger than 800 residues, containing conserved sequences both N-terminal and C-terminal to the TTL core (4, 6, 76, 79). Although detailed structure-function studies for TTLL enzymes are lacking so far, in the cases where examined, sequences outside the TTL core domain are required for proper subcellular targeting. For example, both TTLL6 and the longer isoform of TTLL7 require their C-terminal domains for ciliary localization (6).
TTLL1, the first tubulin glutamylase to be isolated, is part of an ∼360-kDa five-protein complex that preferentially initiates α-tubulin glutamylation (3). Because this is the only enzyme of the family that was isolated biochemically, whereas the others were identified from sequence searches based on their common TTL core, it is not clear whether other TTLLs function as part of larger complexes in vivo. What is notable in the case of TTLL1 is that the subunit that contains the enzyme active site (polyglutamylase subunit 3 (PGs3)) has no glutamylation activity when expressed in isolation and requires the other four subunits for microtubule binding and modification activity (3). The Chlamydomonas reinhardtii glutamylase TTLL9 also has a binding partner that targets it to modify ciliary microtubules (81). TTLL2 may also need to be part of a larger complex for proper targeting and microtubule binding (6).
Tubulin glutamylation levels are established by the balance between modification enzymes of the TTLL family and the enzymes that remove these modifications. Deglutamylation is carried out by a novel family of carboxypeptidases (CCPs) from the M14D subfamily of metallocarboxypeptidases (5, 58, 82). CCP enzymes also show functional diversification. Mammals typically produce six different CCP enzymes (CCP1–6) with unique substrate specificities. CCP1, -4, and -6 shorten polyglutamate chains and also produce Δ2-tubulin, whereas CCP5 removes branching point glutamates (5). Interestingly, CCP1 is capable of removing both elongated and branching point glutamates when they are added by TTLL6, but cannot remove branching point glutamates added by TTLL4 (5), suggesting that both location and length of the polyglutamate chain attached to the tubulin tail are determining factors of CCP enzyme specificity. The specificities of CCP2 and CCP3 were originally unclear, fueling speculation that either might function as a deglycylase or a detyrosinase. However, recent characterization revealed that both function solely as deglutamylases that also generate Δ2-tubulin (83). Despite decades of effort, the identity of the tubulin detyrosinase remains unknown. To date, no enzyme with tubulin deglycylating activity has been identified either. Identification of these enzymes would complete the compendium of enzymes responsible for generating the most prevalent posttranslational modifications of tubulin and would finally allow functional and mechanistic studies into maintenance and control of the levels and patterns of these modifications.
Tubulin acetyltransferase is responsible for acetylating α-tubulin on Lys-40. It was initially identified in C. elegans and is conserved across a wide range of species from flagellates to humans (8, 84). The acetyltransferase San modifies β-tubulin on Lys-252 (19), but has multiple substrates in addition to tubulin. Tubulin deacetylases (HDAC6, SIRT2) have been extensively studied, but insights into their precise mechanism of action have been complicated by the fact that they have multiple substrates other than tubulin (reviewed in Ref. 43).
Polyamination is catalyzed by transglutaminases, a family of enzymes capable of cross-linking the side chain of glutamine residues to various primary amines via non-standard isopeptide bonds. Transglutaminases catalyze the polyamination of both free tubulin and tubulin already incorporated into microtubules (22). Prior to establishing a connection with microtubules, transglutaminases and polyamines were known to be important in neuronal differentiation and degeneration (85).
Tubulin phosphorylation is poorly characterized, and kinases linked to tubulin phosphorylation so far have a range of other substrates. α-Tubulin is phosphorylated by the Syk tyrosine kinase (71). Cyclin-dependent kinase 1 phosphorylates Ser-172 on β-tubulin in addition to modifying several other MAPs (72). The physiological significance of these phosphorylation events is currently not understood.
Writers of the Tubulin Code: Specificity and Combinatorial Use
The complex microtubule modification patterns observed in cells are a function of the tissue distribution, developmental regulation, and biochemical properties of tubulin posttranslational modification enzymes (i.e. substrate specificity and kinetic parameters) in addition to the tissue-specific enrichment of certain tubulin isoforms. In addition to these first order factors, pre-existing modifications and their patterns may influence the further addition and removal of modifications. Furthermore, the preceding factors are convoluted with the dynamics of the microtubules themselves (which can potentially be also influenced by modifications) and the effects of tubulin and microtubule-binding proteins. Faced with this multilevel regulatory complexity, analysis requires the ability to generate chemically defined tubulin and microtubule substrates for in vitro reconstitution experiments. Such substrates can then be used to characterize the basic biochemical properties of the modifying enzymes. These defined substrates and enzymes will then allow quantitative investigation of the tubulin code ranging from the dynamics of the modified microtubules themselves to generation of temporal and spatial microtubule modification patterns, to the effects on microtubule effectors.
To date, the overwhelming majority of in vitro studies of microtubules and their regulators have employed tubulin purified from brain tissue. This tubulin is highly heterogeneous as it contains multiple posttranslational modifications (phosphorylation, acetylation, detyrosination, glutamylation) as well as multiple isoforms (eight α-tubulin and seven β-tubulin) that give rise to tens of different variants (86). Although microtubules in cells show topographically defined modification patterns, the isolation procedure of microtubules from brain tissue results in complete scrambling of all the tubulin modifications and isoforms and thus makes the task of deciphering a tubulin code impossible. Recent advances now allow the purification of naive, unmodified tubulin from various sources (9, 10) as well as recombinant tubulin (11, 12) in which posttranslational modification sites can be mutated. Using unmodified human tubulin, we have shown how to generate defined posttranslationally modified tubulin and microtubules that are tyrosinated, glutamylated, and acetylated (13). Variable levels of glutamylation can be achieved and quantitatively measured using mass spectrometry (13).
TTL was the first tubulin modification enzyme to be structurally characterized (75). The enzyme recognizes tubulin via a bipartite recognition strategy involving low-affinity, high-specificity recognition of the flexible α-tubulin tail and moderate-affinity interactions with the tubulin body at interfaces that prevent the incorporation of soluble tubulin into microtubules (75, 87). TTL competes with stathmin for tubulin binding (88), raising the interesting question of how other tubulin-binding proteins in the cell can positively or negatively regulate tyrosination.
Although these structural and functional studies have shed light on the mechanism used by TTL to distinguish between soluble and polymeric tubulin, how members of the larger TTLL family specifically recognize the microtubule polymer is still unknown due to the lack of any structural information. Moreover, the molecular basis for the preference for either α-tubulin or β-tubulin tails is unknown, but is central to understanding how the tubulin code is generated.
A systematic biochemical and structural dissection of the enzymes that modify tubulin constitutes a first step toward understanding the molecular requirements for generating the large tubulin chemical diversity observed in cells. However, we might be quickly approaching a situation where the resolution of in vitro reconstitution assays will exceed the resolution of detecting microtubule modification patterns in cells, so far limited by tools to specifically recognize modifications in vivo and the resolution of conventional light microscopy. The former problem promises to benefit from in vitro studies, which bring with them the hope of generating recombinant antibodies with higher specificity and resolution for various modifications or fluorescent amino acid analogs compatible with engineered TTLL enzyme active sites that could potentially be used for live cell imaging. Recent years have seen a revolution in high-resolution microscopy techniques that in conjunction with such labeling tools promise to get us closer to a high-resolution dynamic map of tubulin posttranslational modifications in cells.
Pattern Formation in Vivo: Spatial and Kinetic Control
Tubulin posttranslational modification enzymes display tissue specificity as well as distinct subcellular localization (62, 66, 77). This spatial regulation can give rise to diverse modification patterns in cells and tissues. However, at a more local level, the dynamic behavior of the microtubules themselves intersects with the kinetic properties of the enzymes, giving rise to spatial and temporal patterns. For example, TTL is specific for soluble tubulin and does not tyrosinate microtubules (73, 75). Its kinetic parameters allow it to rapidly tyrosinate the tubulin cytoplasmic pool. Conversely, the detyrosination reaction concentrates on the microtubule polymer. As a result, the newly growing end of a microtubule would be enriched in tyrosinated tubulin, whereas older segments can have a lower density of tyrosinated tubulin and thus differentially recruit factors that are sensitive to the tubulin tyrosination status (75). Early experiments using anti-tyrosinated tubulin antibodies hint at such gradients in cells (89).
Several tubulin posttranslational modifications such as acetylation and glutamylation are correlated with stable, long-lived microtubules. Although we still do not understand the causality between modifications and microtubule stability, recent mechanistic work on tubulin acetyltransferase revealed that its slow catalytic rate, coupled with its exploration of the microtubule length, allows it to preferentially mark long-lived microtubules at enzyme concentrations that are substoichiometric to tubulin (51). An understanding of the differential kinetic parameters of TTLL family members as well as other classes of tubulin modification enzymes is likely to illuminate how their molecular properties generate complex temporal and spatial modification patterns.
Reading the Tubulin Code
It has been known for more than two decades that tubulin C-terminal tails can regulate the interaction of motors and MAPs with the microtubule as well as influence the polymerization properties of tubulin. Early experiments demonstrated a reduction in the processivity of both kinesin and dynein on partially proteolyzed microtubules missing their C-terminal tails (90). Removal of tubulin C-terminal tails also inhibits spastin- and katanin-mediated microtubule severing (91, 92). Early blot overlay assays revealed that the microtubule binding affinities of Tau, MAP1A, MAP1B, and MAP2 are influenced by polyglutamylation (93, 94). Single molecule tracking experiments coupled with antibody labeling to identify the posttranslational status of microtubules in cells revealed a specialization of several kinesins for modified microtubules (95). Furthermore, tail deletions and point mutations of glutamylation and glycylation sites in Tetrahymena α- and β-tubulin revealed their importance for the viability of the organism (96, 97).
Subtilisin-treated tubulin missing both α-tubulin and β-tubulin C-terminal tails has a critical concentration 50-fold lower than tubulin and forms, in addition to microtubules, other polymeric species such as sheets, rings, and aggregates (98). These and other early experiments indicate that tubulin tails and their modifications can tune both the basic properties of the microtubule polymer and its interaction with cellular effectors. However, further mechanistic investigations into the effects of the tails and their posttranslational modifications on motor and MAP activity were hampered by the unknown identity of most tubulin modification enzymes as well as the inability to generate distinctly modified tubulin or tubulin that can be engineered for in vitro experiments.
Several microtubule plus-end tracking proteins, including CLIP-170 and p150Glued, contain cytoskeleton-associated protein glycine-rich (CAP-Gly) domains that track the growing microtubule plus-end by specifically recognizing the α-tubulin C-terminal tyrosine (99). More recent studies using TTL KO fibroblasts show that the depolymerizing kinesin-13 mitotic centromere-associated kinesin (MCAK) is more active on tyrosinated than detyrosinated microtubules, thus providing a possible mechanistic explanation for the increased stability of detyrosinated microtubules in cells (55). Experiments using KO mice for one of the TTLL1 subunits show that decreased glutamylation on axonal microtubules lowers the affinity of kinesin-3 and reduces synaptic vesicle trafficking (42). Glutamylation also regulates sliding velocities of axonemal microtubules, likely by modulating the microtubule binding affinity of inner arm dynein (38, 39). Recent experiments using engineered S. cerevisiae tubulin revealed the differential regulation of several kinesins and cytoplasmic dynein by different α- and β-tubulin isoforms as well as detyrosination (100).
Despite significant progress in the last few years, the effects of tubulin posttranslational modifications on the recruitment and activity of most motors and MAPs are still largely unknown and are just beginning to be investigated. For many modifications, such as polyamination, phosphorylation, and glycylation, the effects are completely unknown and will no doubt be the focus of future experiments once methods to produce well characterized microtubules carrying these modifications are developed.
How Does the Cell Interpret the Tubulin Code?
Although the tubulin code is gradually yielding its secrets, what is not known is how the cell ultimately integrates the information encoded in tubulin posttranslational modifications. However, what is clear is that the organism invests a large amount of coding capacity for modification enzymes, that their loss can be deleterious to the organism, and that nontrivial amounts of energy are expended to modify tubulin. Moreover, tubulin modification enzymes appear to be under strong evolutionary selection. The power of the bottom-up reconstitution approach has been amply demonstrated in the last five decades in the study of basic cell biological processes. The analysis of tubulin posttranslational modifications is rapidly entering this stage. We see several major challenges: to characterize the dynamics and mechanical properties of modified microtubules; to understand the basic principles that give rise to the differential specificities of tubulin modification enzymes; to understand how motors and microtubule-associated proteins are influenced by modifications and how their action in turn modulates the behavior of this dynamic polymer; and to generate modification patterns that mimic those found in cells and build complex microtubule array geometries. These basic first steps should get us closer to understanding how the cell interprets the tubulin code.
This work was supported, in whole or in part, by National Institutes of Health Grant 1ZIANS003122-05 (to A. R. M.). This is the fourth article in the Thematic Minireview series “The State of the Cytoskeleton in 2015.” The authors declare that they have no conflicts of interest with the contents of this article.
- MAP
- microtubule-associated protein
- TTL
- tubulin tyrosine ligase
- TTLL
- tubulin tyrosine ligase-like
- CCP
- cytoplasmic carboxypeptidase.
References
- 1. Mitchison T., Kirschner M. (1984) Microtubule assembly nucleated by isolated centrosomes. Nature 312, 232–237 [DOI] [PubMed] [Google Scholar]
- 2. Gittes F., Mickey B., Nettleton J., Howard J. (1993) Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J. Cell Biol. 120, 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Janke C., Rogowski K., Wloga D., Regnard C., Kajava A. V., Strub J. M., Temurak N., van Dijk J., Boucher D., van Dorsselaer A., Suryavanshi S., Gaertig J., Eddé B. (2005) Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science 308, 1758–1762 [DOI] [PubMed] [Google Scholar]
- 4. Rogowski K., Juge F., van Dijk J., Wloga D., Strub J. M., Levilliers N., Thomas D., Bré M. H., Van Dorsselaer A., Gaertig J., Janke C. (2009) Evolutionary divergence of enzymatic mechanisms for posttranslational polyglycylation. Cell 137, 1076–1087 [DOI] [PubMed] [Google Scholar]
- 5. Rogowski K., van Dijk J., Magiera M. M., Bosc C., Deloulme J. C., Bosson A., Peris L., Gold N. D., Lacroix B., Bosch Grau M., Bec N., Larroque C., Desagher S., Holzer M., Andrieux A., Moutin M. J., Janke C. (2010) A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell 143, 564–578 [DOI] [PubMed] [Google Scholar]
- 6. van Dijk J., Rogowski K., Miro J., Lacroix B., Eddé B., Janke C. (2007) A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol. Cell 26, 437–448 [DOI] [PubMed] [Google Scholar]
- 7. Wloga D., Webster D. M., Rogowski K., Bré M. H., Levilliers N., Jerka-Dziadosz M., Janke C., Dougan S. T., Gaertig J. (2009) TTLL3 is a tubulin glycine ligase that regulates the assembly of cilia. Dev. Cell 16, 867–876 [DOI] [PubMed] [Google Scholar]
- 8. Akella J. S., Wloga D., Kim J., Starostina N. G., Lyons-Abbott S., Morrissette N. S., Dougan S. T., Kipreos E. T., Gaertig J. (2010) MEC-17 is an α-tubulin acetyltransferase. Nature 467, 218–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Drummond D. R., Kain S., Newcombe A., Hoey C., Katsuki M., Cross R. A. (2011) Purification of tubulin from the fission yeast Schizosaccharomyces pombe. Methods Mol. Biol. 777, 29–55 [DOI] [PubMed] [Google Scholar]
- 10. Widlund P. O., Podolski M., Reber S., Alper J., Storch M., Hyman A. A., Howard J., Drechsel D. N. (2012) One-step purification of assembly-competent tubulin from diverse eukaryotic sources. Mol. Biol. Cell 23, 4393–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnson V., Ayaz P., Huddleston P., Rice L. M. (2011) Design, overexpression, and purification of polymerization-blocked yeast αβ-tubulin mutants. Biochemistry 50, 8636–8644 [DOI] [PubMed] [Google Scholar]
- 12. Minoura I., Hachikubo Y., Yamakita Y., Takazaki H., Ayukawa R., Uchimura S., Muto E. (2013) Overexpression, purification, and functional analysis of recombinant human tubulin dimer. FEBS Lett. 587, 3450–3455 [DOI] [PubMed] [Google Scholar]
- 13. Vemu A., Garnham C. P., Lee D. Y., Roll-Mecak A. (2014) Generation of differentially modified microtubules using in vitro enzymatic approaches. Methods Enzymol. 540, 149–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bieling P., Kandels-Lewis S., Telley I. A., van Dijk J., Janke C., Surrey T. (2008) CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J. Cell Biol. 183, 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gell C., Bormuth V., Brouhard G. J., Cohen D. N., Diez S., Friel C. T., Helenius J., Nitzsche B., Petzold H., Ribbe J., Schäffer E., Stear J. H., Trushko A., Varga V., Widlund P. O., Zanic M., Howard J. (2010) Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol. 95, 221–245 [DOI] [PubMed] [Google Scholar]
- 16. Laan L., Dogterom M. (2010) In vitro assays to study force generation at dynamic microtubule ends. Methods Cell Biol. 95, 617–639 [DOI] [PubMed] [Google Scholar]
- 17. Eipper B. A. (1972) Rat brain microtubule protein: purification and determination of covalently bound phosphate and carbohydrate. Proc. Natl. Acad. Sci. U.S.A. 69, 2283–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. L'Hernault S. W., Rosenbaum J. L. (1985) Chlamydomonas α-tubulin is posttranslationally modified by acetylation on the ϵ-amino group of a lysine. Biochemistry 24, 473–478 [DOI] [PubMed] [Google Scholar]
- 19. Chu C. W., Hou F., Zhang J., Phu L., Loktev A. V., Kirkpatrick D. S., Jackson P. K., Zhao Y., Zou H. (2011) A novel acetylation of β-tubulin by San modulates microtubule polymerization via down-regulating tubulin incorporation. Mol. Biol. Cell 22, 448–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Caron J. M. (1997) Posttranslational modification of tubulin by palmitoylation: I. In vivo and cell-free studies. Mol. Biol. Cell 8, 621–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rosas-Acosta G., Russell W. K., Deyrieux A., Russell D. H., Wilson V. G. (2005) A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol. Cell Proteomics 4, 56–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Song Y., Kirkpatrick L. L., Schilling A. B., Helseth D. L., Chabot N., Keillor J. W., Johnson G. V., Brady S. T. (2013) Transglutaminase and polyamination of tubulin: posttranslational modification for stabilizing axonal microtubules. Neuron 78, 109–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jaffrey S. R., Erdjument-Bromage H., Ferris C. D., Tempst P., Snyder S. H. (2001) Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell Biol. 3, 193–197 [DOI] [PubMed] [Google Scholar]
- 24. Barra H. S., Rodriguez J. A., Arce C. A., Caputto R. (1973) A soluble preparation from rat brain that incorporates into its own proteins [14C]arginine by a ribonuclease-sensitive system and [14C]tyrosine by a ribonuclease-insensitive system. J. Neurochem. 20, 97–108 [DOI] [PubMed] [Google Scholar]
- 25. Eddé B., Rossier J., Le Caer J. P., Desbruyères E., Gros F., Denoulet P. (1990) Posttranslational glutamylation of α-tubulin. Science 247, 83–85 [DOI] [PubMed] [Google Scholar]
- 26. Redeker V., Melki R., Promé D., Le Caer J. P., Rossier J. (1992) Structure of tubulin C-terminal domain obtained by subtilisin treatment: the major α- and β-tubulin isotypes from pig brain are glutamylated. FEBS Lett. 313, 185–192 [DOI] [PubMed] [Google Scholar]
- 27. Redeker V., Levilliers N., Schmitter J. M., Le Caer J. P., Rossier J., Adoutte A., Bré M. H. (1994) Polyglycylation of tubulin: a posttranslational modification in axonemal microtubules. Science 266, 1688–1691 [DOI] [PubMed] [Google Scholar]
- 28. Webster D. R., Gundersen G. G., Bulinski J. C., Borisy G. G. (1987) Differential turnover of tyrosinated and detyrosinated microtubules. Proc. Natl. Acad. Sci. U.S.A. 84, 9040–9044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bobinnec Y., Moudjou M., Fouquet J. P., Desbruyères E., Eddé B., Bornens M. (1998) Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil. Cytoskeleton 39, 223–232 [DOI] [PubMed] [Google Scholar]
- 30. Lacroix B., van Dijk J., Gold N. D., Guizetti J., Aldrian-Herrada G., Rogowski K., Gerlich D. W., Janke C. (2010) Tubulin polyglutamylation stimulates spastin-mediated microtubule severing. J. Cell Biol. 189, 945–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Konishi Y., Setou M. (2009) Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat. Neurosci. 12, 559–567 [DOI] [PubMed] [Google Scholar]
- 32. Liao G., Gundersen G. G. (1998) Kinesin is a candidate for cross-bridging microtubules and intermediate filaments. Selective binding of kinesin to detyrosinated tubulin and vimentin. J. Biol. Chem. 273, 9797–9803 [DOI] [PubMed] [Google Scholar]
- 33. Mary J., Redeker V., Le Caer J. P., Rossier J., Schmitter J. M. (1996) Posttranslational modifications in the C-terminal tail of axonemal tubulin from sea urchin sperm. J. Biol. Chem. 271, 9928–9933 [DOI] [PubMed] [Google Scholar]
- 34. Mary J., Redeker V., Le Caer J. P., Rossier J., Schmitter J. M. (1997) Posttranslational modifications of axonemal tubulin. J. Protein Chem. 16, 403–407 [DOI] [PubMed] [Google Scholar]
- 35. Levilliers N., Fleury A., Hill A. M. (1995) Monoclonal and polyclonal antibodies detect a new type of post-translational modification of axonemal tubulin. J. Cell Sci. 108, 3013–3028 [DOI] [PubMed] [Google Scholar]
- 36. Bré M. H., Redeker V., Quibell M., Darmanaden-Delorme J., Bressac C., Cosson J., Huitorel P., Schmitter J. M., Rossler J., Johnson T., Adoutte A., Levilliers N. (1996) Axonemal tubulin polyglycylation probed with two monoclonal antibodies: widespread evolutionary distribution, appearance during spermatozoan maturation and possible function in motility. J. Cell Sci. 109, 727–738 [DOI] [PubMed] [Google Scholar]
- 37. Schneider A., Plessmann U., Weber K. (1997) Subpellicular and flagellar microtubules of Trypanosoma brucei are extensively glutamylated. J. Cell Sci. 110, 431–437 [DOI] [PubMed] [Google Scholar]
- 38. Kubo T., Yanagisawa H. A., Yagi T., Hirono M., Kamiya R. (2010) Tubulin polyglutamylation regulates axonemal motility by modulating activities of inner-arm dyneins. Curr. Biol. 20, 441–445 [DOI] [PubMed] [Google Scholar]
- 39. Suryavanshi S., Eddé B., Fox L. A., Guerrero S., Hard R., Hennessey T., Kabi A., Malison D., Pennock D., Sale W. S., Wloga D., Gaertig J. (2010) Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity. Curr. Biol. 20, 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Audebert S., Desbruyères E., Gruszczynski C., Koulakoff A., Gros F., Denoulet P., Eddé B. (1993) Reversible polyglutamylation of α- and β-tubulin and microtubule dynamics in mouse brain neurons. Mol. Biol. Cell 4, 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Erck C., Peris L., Andrieux A., Meissirel C., Gruber A. D., Vernet M., Schweitzer A., Saoudi Y., Pointu H., Bosc C., Salin P. A., Job D., Wehland J. (2005) A vital role of tubulin-tyrosine-ligase for neuronal organization. Proc. Natl. Acad. Sci. U.S.A. 102, 7853–7858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ikegami K., Heier R. L., Taruishi M., Takagi H., Mukai M., Shimma S., Taira S., Hatanaka K., Morone N., Yao I., Campbell P. K., Yuasa S., Janke C., Macgregor G. R., Setou M. (2007) Loss of α-tubulin polyglutamylation in ROSA22 mice is associated with abnormal targeting of KIF1A and modulated synaptic function. Proc. Natl. Acad. Sci. U.S.A. 104, 3213–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garnham C. P., Roll-Mecak A. (2012) The chemical complexity of cellular microtubules: tubulin post-translational modification enzymes and their roles in tuning microtubule functions. Cytoskeleton (Hoboken) 69, 442–463, 10.1002/cm.21027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tischfield M. A., Cederquist G. Y., Gupta M. L., Jr., Engle E. C. (2011) Phenotypic spectrum of the tubulin-related disorders and functional implications of disease-causing mutations. Curr. Opin. Genet. Dev. 21, 286–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Verhey K. J., Gaertig J. (2007) The tubulin code. Cell Cycle 6, 2152–2160 [DOI] [PubMed] [Google Scholar]
- 46. Roll-Mecak A. (2015) Intrinsically disordered tubulin tails: complex tuners of microtubule functions? Semin. Cell Dev. Biol. 37, 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Paturle-Lafanechère L., Manier M., Trigault N., Pirollet F., Mazarguil H., Job D. (1994) Accumulation of Δ2-tubulin, a major tubulin variant that cannot be tyrosinated, in neuronal tissues and in stable microtubule assemblies. J. Cell Sci. 107, 1529–1543 [DOI] [PubMed] [Google Scholar]
- 48. Caron J. M., Vega L. R., Fleming J., Bishop R., Solomon F. (2001) Single site α-tubulin mutation affects astral microtubules and nuclear positioning during anaphase in Saccharomyces cerevisiae: possible role for palmitoylation of α-tubulin. Mol. Biol. Cell 12, 2672–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nogales E., Wolf S. G., Downing K. H. (1998) Structure of the αβ-tubulin dimer by electron crystallography. Nature 391, 199–203 [DOI] [PubMed] [Google Scholar]
- 50. Soppina V., Herbstman J. F., Skiniotis G., Verhey K. J. (2012) Luminal localization of α-tubulin K40 acetylation by cryo-EM analysis of fab-labeled microtubules. PLoS One 7, e48204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Szyk A., Deaconescu A. M., Spector J., Goodman B., Valenstein M. L., Ziolkowska N. E., Kormendi V., Grigorieff N., Roll-Mecak A. (2014) Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase. Cell 157, 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marcos S., Moreau J., Backer S., Job D., Andrieux A., Bloch-Gallego E. (2009) Tubulin tyrosination is required for the proper organization and pathfinding of the growth cone. PLoS One 4, e5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Baas P. W., Black M. M. (1990) Individual microtubules in the axon consist of domains that differ in both composition and stability. J. Cell Biol. 111, 495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Webster D. R., Wehland J., Weber K., Borisy G. G. (1990) Detyrosination of α-tubulin does not stabilize microtubules in vivo. J. Cell Biol. 111, 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Peris L., Wagenbach M., Lafanechère L., Brocard J., Moore A. T., Kozielski F., Job D., Wordeman L., Andrieux A. (2009) Motor-dependent microtubule disassembly driven by tubulin tyrosination. J. Cell Biol. 185, 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Redeker V., Rossier J., Frankfurter A. (1998) Posttranslational modifications of the C-terminus of α-tubulin in adult rat brain: α4 is glutamylated at two residues. Biochemistry 37, 14838–14844 [DOI] [PubMed] [Google Scholar]
- 57. Redeker V., Le Caer J. P., Rossier J., Promé J. C. (1991) Structure of the polyglutamyl side chain posttranslationally added to α-tubulin. J. Biol. Chem. 266, 23461–23466 [PubMed] [Google Scholar]
- 58. Kimura Y., Kurabe N., Ikegami K., Tsutsumi K., Konishi Y., Kaplan O. I., Kunitomo H., Iino Y., Blacque O. E., Setou M. (2010) Identification of tubulin deglutamylase among Caenorhabditis elegans and mammalian cytosolic carboxypeptidases (CCPs). J. Biol. Chem. 285, 22936–22941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vinh J., Loyaux D., Redeker V., Rossier J. (1997) Sequencing branched peptides with CID/PSD MALDI-TOF in the low-picomole range: application to the structural study of the posttranslational polyglycylation of tubulin. Anal. Chem. 69, 3979–3985 [DOI] [PubMed] [Google Scholar]
- 60. Bré M. H., Redeker V., Vinh J., Rossier J., Levilliers N. (1998) Tubulin polyglycylation: differential posttranslational modification of dynamic cytoplasmic and stable axonemal microtubules in paramecium. Mol. Biol. Cell 9, 2655–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dossou S. J., Bré M. H., Hallworth R. (2007) Mammalian cilia function is independent of the polymeric state of tubulin glycylation. Cell Motil. Cytoskeleton 64, 847–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bosch Grau M., Gonzalez Curto G., Rocha C., Magiera M. M., Marques Sousa P., Giordano T., Spassky N., Janke C. (2013) Tubulin glycylases and glutamylases have distinct functions in stabilization and motility of ependymal cilia. J. Cell Biol. 202, 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pathak N., Austin C. A., Drummond I. A. (2011) Tubulin tyrosine ligase-like genes ttll3 and ttll6 maintain zebrafish cilia structure and motility. J. Biol. Chem. 286, 11685–11695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rocha C., Papon L., Cacheux W., Marques Sousa P., Lascano V., Tort O., Giordano T., Vacher S., Lemmers B., Mariani P., Meseure D., Medema J. P., Bièche I., Hahne M., Janke C. (2014) Tubulin glycylases are required for primary cilia, control of cell proliferation and tumor development in colon. EMBO J. 33, 2247–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. LeDizet M., Piperno G. (1987) Identification of an acetylation site of Chlamydomonas α-tubulin. Proc. Natl. Acad. Sci. U.S.A. 84, 5720–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bulinski J. C., Richards J. E., Piperno G. (1988) Posttranslational modifications of α-tubulin: detyrosination and acetylation differentiate populations of interphase microtubules in cultured cells. J. Cell Biol. 106, 1213–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Maruta H., Greer K., Rosenbaum J. L. (1986) The acetylation of α-tubulin and its relationship to the assembly and disassembly of microtubules. J. Cell Biol. 103, 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cueva J. G., Hsin J., Huang K. C., Goodman M. B. (2012) Posttranslational acetylation of α-tubulin constrains protofilament number in native microtubules. Curr. Biol. 22, 1066–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Topalidou I., Keller C., Kalebic N., Nguyen K. C., Somhegyi H., Politi K. A., Heppenstall P., Hall D. H., Chalfie M. (2012) Genetically separable functions of the MEC-17 tubulin acetyltransferase affect microtubule organization. Curr. Biol. 22, 1057–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Brady S. T., Tytell M., Lasek R. J. (1984) Axonal tubulin and axonal microtubules: biochemical evidence for cold stability. J. Cell Biol. 99, 1716–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Peters J. D., Furlong M. T., Asai D. J., Harrison M. L., Geahlen R. L. (1996) Syk, activated by cross-linking the B-cell antigen receptor, localizes to the cytosol where it interacts with and phosphorylates α-tubulin on tyrosine. J. Biol. Chem. 271, 4755–4762 [DOI] [PubMed] [Google Scholar]
- 72. Fourest-Lieuvin A., Peris L., Gache V., Garcia-Saez I., Juillan-Binard C., Lantez V., Job D. (2006) Microtubule regulation in mitosis: tubulin phosphorylation by the cyclin-dependent kinase Cdk1. Mol. Biol. Cell 17, 1041–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ersfeld K., Wehland J., Plessmann U., Dodemont H., Gerke V., Weber K. (1993) Characterization of the tubulin-tyrosine ligase. J. Cell Biol. 120, 725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mialhe A., Lafanechère L., Treilleux I., Peloux N., Dumontet C., Brémond A., Panh M. H., Payan R., Wehland J., Margolis R. L., Job D. (2001) Tubulin detyrosination is a frequent occurrence in breast cancers of poor prognosis. Cancer Res. 61, 5024–5027 [PubMed] [Google Scholar]
- 75. Szyk A., Deaconescu A. M., Piszczek G., Roll-Mecak A. (2011) Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nat. Struct. Mol. Biol. 18, 1250–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mukai M., Ikegami K., Sugiura Y., Takeshita K., Nakagawa A., Setou M. (2009) Recombinant mammalian tubulin polyglutamylase TTLL7 performs both initiation and elongation of polyglutamylation on β-tubulin through a random sequential pathway. Biochemistry 48, 1084–1093 [DOI] [PubMed] [Google Scholar]
- 77. Ikegami K., Mukai M., Tsuchida J., Heier R. L., Macgregor G. R., Setou M. (2006) TTLL7 is a mammalian β-tubulin polyglutamylase required for growth of MAP2-positive neurites. J. Biol. Chem. 281, 30707–30716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wloga D., Dave D., Meagley J., Rogowski K., Jerka-Dziadosz M., Gaertig J. (2010) Hyperglutamylation of tubulin can either stabilize or destabilize microtubules in the same cell. Eukaryot. Cell 9, 184–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ikegami K., Setou M. (2009) TTLL10 can perform tubulin glycylation when co-expressed with TTLL8. FEBS Lett. 583, 1957–1963 [DOI] [PubMed] [Google Scholar]
- 80. Brants J., Semenchenko K., Wasylyk C., Robert A., Carles A., Zambrano A., Pradeau-Aubreton K., Birck C., Schalken J. A., Poch O., de Mey J., Wasylyk B. (2012) Tubulin tyrosine ligase like 12, a TTLL family member with SET- and TTL-like domains and roles in histone and tubulin modifications and mitosis. PLoS One 7, e51258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kubo T., Yanagisawa H. A., Liu Z., Shibuya R., Hirono M., Kamiya R. (2014) A conserved flagella-associated protein in Chlamydomonas, FAP234, is essential for axonemal localization of tubulin polyglutamylase TTLL9. Mol. Biol. Cell 25, 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kalinina E., Biswas R., Berezniuk I., Hermoso A., Aviles F. X., Fricker L. D. (2007) A novel subfamily of mouse cytosolic carboxypeptidases. FASEB J. 21, 836–850 [DOI] [PubMed] [Google Scholar]
- 83. Tort O., Tanco S., Rocha C., Bièche I., Seixas C., Bosc C., Andrieux A., Moutin M. J., Avilés F. X., Lorenzo J., Janke C. (2014) The cytosolic carboxypeptidases CCP2 and CCP3 catalyze posttranslational removal of acidic amino acids. Mol. Biol. Cell 25, 3017–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Shida T., Cueva J. G., Xu Z., Goodman M. B., Nachury M. V. (2010) The major α-tubulin K40 acetyltransferase αTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl. Acad. Sci. U.S.A. 107, 21517–21522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Basso M., Berlin J., Xia L., Sleiman S. F., Ko B., Haskew-Layton R., Kim E., Antonyak M. A., Cerione R. A., Iismaa S. E., Willis D., Cho S., Ratan R. R. (2012) Transglutaminase inhibition protects against oxidative stress-induced neuronal death downstream of pathological ERK activation. J. Neurosci. 32, 6561–6569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wolff A., Denoulet P., Jeantet C. (1982) High level of tubulin microheterogeneity in the mouse brain. Neurosci. Lett. 31, 323–328 [DOI] [PubMed] [Google Scholar]
- 87. Prota A. E., Magiera M. M., Kuijpers M., Bargsten K., Frey D., Wieser M., Jaussi R., Hoogenraad C. C., Kammerer R. A., Janke C., Steinmetz M. O. (2013) Structural basis of tubulin tyrosination by tubulin tyrosine ligase. J. Cell Biol. 200, 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Szyk A., Piszczek G., Roll-Mecak A. (2013) Tubulin tyrosine ligase and stathmin compete for tubulin binding in vitro. J. Mol. Biol. 425, 2412–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ahmad F. J., Pienkowski T. P., Baas P. W. (1993) Regional differences in microtubule dynamics in the axon. J. Neurosci. 13, 856–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang Z., Sheetz M. P. (2000) The C-terminus of tubulin increases cytoplasmic dynein and kinesin processivity. Biophys. J. 78, 1955–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. McNally F. J., Vale R. D. (1993) Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell 75, 419–429 [DOI] [PubMed] [Google Scholar]
- 92. Roll-Mecak A., Vale R. D. (2005) The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr. Biol. 15, 650–655 [DOI] [PubMed] [Google Scholar]
- 93. Bonnet C., Boucher D., Lazereg S., Pedrotti B., Islam K., Denoulet P., Larcher J. C. (2001) Differential binding regulation of microtubule-associated proteins MAP1A, MAP1B, and MAP2 by tubulin polyglutamylation. J. Biol. Chem. 276, 12839–12848 [DOI] [PubMed] [Google Scholar]
- 94. Boucher D., Larcher J. C., Gros F., Denoulet P. (1994) Polyglutamylation of tubulin as a progressive regulator of in vitro interactions between the microtubule-associated protein Tau and tubulin. Biochemistry 33, 12471–12477 [DOI] [PubMed] [Google Scholar]
- 95. Cai D., McEwen D. P., Martens J. R., Meyhofer E., Verhey K. J. (2009). Single molecule imaging reveals differences in microtubule track selection between kinesin motors. PLoS Biol. 7, e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Duan J., Gorovsky M. A. (2002) Both carboxy-terminal tails of α- and β-tubulin are essential, but either one will suffice. Curr. Biol. 12, 313–316 [DOI] [PubMed] [Google Scholar]
- 97. Thazhath R., Liu C., Gaertig J. (2002) Polyglycylation domain of β-tubulin maintains axonemal architecture and affects cytokinesis in Tetrahymena. Nat. Cell Biol. 4, 256–259 [DOI] [PubMed] [Google Scholar]
- 98. Bhattacharyya B., Sackett D. L., Wolff J. (1985) Tubulin, hybrid dimers, and tubulin S: stepwise charge reduction and polymerization. J. Biol. Chem. 260, 10208–10216 [PubMed] [Google Scholar]
- 99. Akhmanova A., Steinmetz M. O. (2010) Microtubule +TIPs at a glance. J. Cell Sci. 123, 3415–3419 [DOI] [PubMed] [Google Scholar]
- 100. Sirajuddin M., Rice L. M., Vale R. D. (2014) Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat. Cell Biol. 16, 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]