Background: Several synthetic glycosphingolipids have been tested to determine their capacity to activate type I NKT cells.
Results: Although the TCR binds with high affinity to all CD1d-presented glycolipids, only a few activate type I NKT cells in vivo.
Conclusion: TCR binding affinity does not necessarily predict antigenicity in vivo.
Significance: The prediction of the therapeutic efficacy of type I NKT cell antigens requires complementary assays.
Keywords: cytokine induction, glycolipid structure, immunology, protein crystallization, surface plasmon resonance (SPR), T cell receptor (TCR)
Abstract
The ability of different glycosphingolipids (GSLs) to activate type I natural killer T cells (NKT cells) has been known for 2 decades. The possible therapeutic use of these GSLs has been studied in many ways; however, studies are needed in which the efficacy of promising GSLs is compared under identical conditions. Here, we compare five unique GSLs structurally derived from α-galactosylceramide. We employed biophysical and biological assays, as well as x-ray crystallography to study the impact of the chemical modifications of the antigen on type I NKT cell activation. Although all glycolipids are bound by the T cell receptor of type I NKT cells in real time binding assays with high affinity, only a few activate type I NKT cells in in vivo or in vitro experiments. The differences in biological responses are likely a result of different pharmacokinetic properties of each lipid, which carry modifications at different parts of the molecule. Our results indicate a need to perform a variety of assays to ascertain the therapeutic potential of type I NKT cell GSL activators.
Introduction
Natural killer T (NKT)7 cells are a unique population of T lymphocytes with the capacity to impact a wide array of functions of the immune system, ranging from protection against infections to responses to tumors and the regulation of autoimmunity (1, 2). These cells accomplish this feat through their ability to secrete T helper type 1 (Th1) and T helper type 2 (Th2) cytokines, most notably IFN-γ and IL-4, respectively, and their ability to impact other white blood cells, such as natural killer cells, dendritic cells, and B cells (3–5). NKT cells, specifically type I NKT cells, have a semi-invariant T cell antigen receptor (TCR) that has an evolutionarily conserved α chain, formed by a Vα14/Jα18 joining in mice and a homologous Vα24/Jα18 in humans (3). The TCR α chain pairs with a less restricted β chain repertoire to impart a specificity for glycosphingolipids (GSLs) presented by CD1d. CD1d is a member of the CD1 family of antigen-presenting molecules (CD1a–e in humans, CD1d in mice) that is structurally similar to peptide-presenting MHC class I antigen-presenting molecules (6). The CD1d heavy chain is composed of three domains, α1, α2, and α3. Although the α3 domain noncovalently associates with β2-microglobulin, the α1-α2 superdomain forms a central hydrophobic antigen-binding groove (7). This groove further segregates into two connected pockets called A′ and F′. Each pocket binds one chain of a dual alkyl chain lipid antigen. In the case of GSLs, the sphingoid base binds in the F′ pocket, whereas the A′ pocket binds the fatty acid. This lipid binding orientation allows the sugar headgroup to be exposed in the center for recognition by the type I NKT cell TCR, with the α chain of the TCR providing the predominant binding energy (8, 9). Many GSLs have been analyzed for the activation of type I NKT cell TCRs, but the most commonly studied is α-galactosylceramide (αGalCer) (10, 11). This prototypical glycolipid activates type I NKT cells to secrete both IL-4 and IFN-γ in vivo within 90 min. Since this initial discovery, many glycolipids have been studied that sway the response of the immune system predominantly toward either a Th1 or a Th2 response (12). One of the earliest Th1 skewing lipids studied to date is C-glycoside, in which the O-glycosidic linkage of αGalCer is replaced with a methylene group, known to stabilize this lipid (13). Although this lipid causes a pronounced Th1 response in mice, it is unable to activate human type I NKT cells. The importance of Th1 skewing in a mammalian system is important for driving the system toward an inflammatory response essential for tumor clearance (14) and vaccine adjuvant activity (15). Thus, type I NKT cell lipids that stimulate IFN-γ have been studied in the context of possible therapeutic development. Although much work has been done to analyze these cytokine skewed responses, the exact mechanism of cytokine polarization is not completely elucidated. In this study, we selected five different lipids, previously demonstrated to activate type I NKT cells, and tested their ability to activate human and mouse type I NKT cells side-by-side. We have further assessed their TCR binding affinities and crystallized the mouse ternary CD1d-GSL-TCR complexes to analyze the molecular interactions that ultimately lead to TCR triggering.
Experimental Procedures
GSL Synthesis
The following GSLs used in this study have all been described previously: EF77 (16), GCK127 and -152 (17), NC-αGC (18), and 7DW8-5 (19–23).
In Vitro GSL Presentation Assays
The GSL cell-free presentation assay has been described previously (24). 96-Well plates were coated with 1 μg of CD1d and were incubated overnight with various concentrations of GSLs. The CD1d molecules were then co-cultured with type I NKT cell hybridomas overnight. The GSL cell-based assay also has been described (25). Briefly, antigen-presenting cells (1 × 105 per well) were pulsed with 100 ng of the indicated lipid and were incubated overnight. The cells were then combined with 5 × 104 Vα14Vβ8.2 NKT cell hybridomas for 24 h. The DN3A4-1.2 (1.2) type I NKT hybridoma cell line has been described previously (26). TCR engagement was measured using a sandwich ELISA for IL-2 cytokines in the supernatant of hybridoma cultures.
Human Type I NKT Cell Assay
The isolation and expansion of human Vα24+ NKT cell lines has been published previously (27). Human donor peripheral blood mononuclear cells (PBMCs, 1–1.5 × 106/ml) were isolated and cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% (v/v) FBS and 1% (v/v) Pen/Strep/glutamine (10,000 units of penicillin, 10,000 μg of streptomycin, 29.2 mg/ml l-glutamine; Invitrogen), and cultures were expanded by weekly re-stimulation with αGalCer-pulsed, irradiated PBMC and recombinant human IL-2. PBMCs (1 × 105 per well) were pulsed with GSLs, seeded in 96-well plates, and cultured in the presence of 5 × 104 Vα24+ human NKT cells for 24 h. GM-CSF release was evaluated in a sandwich ELISA following the manufacturer's instructions (R&D Systems).
Mice
C57BL/6 were purchased from The Jackson Laboratory. All mice were housed in specific pathogen-free conditions, and the experiments were approved by the Institutional Animal Care and Use Committee of the La Jolla Institute for Allergy and Immunology. Mice were injected with 1 μg of lipids intravenously, and the sera of immunized mice were subjected to sandwich ELISAs to measure mouse IFN-γ levels.
Mouse and Human CD1d and TCR Preparation
As reported previously (28), mouse CD1d-β2-microglobulin heterodimeric protein was expressed in a baculovirus expression system, and human CD1d-β2-microglobulin was prepared similarly to the mouse protein. The TCR was prepared by refolding as reported previously (29).
CD1d Loading
Each GSL was dissolved in DMSO at a concentration of 1 or 2 mg/ml. The individual GSLs were diluted in a vehicle solution (50 mm Tris-HCl, pH 7.0, 4.8 mg/ml sucrose, 0.5 mg/ml sodium deoxycholate, and 0.022% Tween 20) and were incubated at 80 °C for at least 20 min. The GSLs were then combined with CD1d protein in an approximate 3:1 molar ratio of lipid to protein overnight in the presence of 50 mm Tris-HCl, pH 7.0.
SPR Kinetic Analysis
Biotinylated CD1d was processed and purified, and studies were conducted using a BIAcore 3000 (GE Healthcare) as reported previously (29), and lipid loading was done as described above. Following overnight GSL loading, ∼300 response units of the CD1d-GSL complex were immobilized onto a streptavidin sensor chip surface by injecting the CD1d-GSL mixture at 5 μl/min using HBS (10 mm HEPES, 150 mm NaCl, 3.0 mm EDTA, pH 7.4) running buffer. A reference channel was bound with unloaded CD1d, and increasing concentrations of refolded TCR were passed over at a flow rate of 30 μl/min. Kinetic association and dissociation curves were calculated using BIAevaluation software version 4.1.
Crystallization and Structure Determination
Following overnight GSL loading, the CD1d-GSL complexes were incubated with refolded TCR at room temperature for at least 30 min, followed by isolation of the complexes using Superdex S200 10/300 GL (GE Healthcare) equilibrated in buffer (50 mm HEPES, pH 7.4, 150 mm NaCl). The isolated fractions containing the ternary complexes were concentrated, and crystals were grown at 22.3 °C by sitting drop vapor diffusion while mixing 0.5 μl of protein solution with 0.5 μl of precipitate. Precipitates were 0.2 m ammonium citrate dibasic, pH 5.1, 20% PEG 4000 for EF77, 17% polyethylene glycol, 8% tacsimate, pH 4, for 7DW8-5, 20% polyethylene glycol, 0.2 m ammonium citrate dibasic for NC-αGC, 17% polyethylene glycol, 0.2 m potassium acetate for GCK127, and 16% polyethylene glycol, 0.2 m ammonium citrate dibasic for GCK152. Single crystals were harvested and flash-cooled in mother liquor containing 20% glycerol. Crystals were collected at the Stanford Synchrotron Radiation Lightsource and the Advanced Light Source and processed with the software Mosflm (30) or HKL2000 (31). The structures were solved by molecular replacement in CCP4 (Collaborative Computational Project, Number 4) (32). First, the coordinates for the CD1d protein of PDB code 2Q7Y were used, followed by the TCR coordinates of PDB code 3QUZ. Models were rebuilt into σA-weighted 2Fo − Fc and Fo − Fc difference electron density maps using COOT (33). The GSLs were built into 2Fo − Fc map and refined using REFMAC (34). Refmac geometric libraries for the glycolipids were obtained using the PRODRG server (35). Data collection and refinement statistics are summarized in Table 1.
TABLE 1.
Refinement statistics for the CD1d-GSL-TCR complexes
NA means not available.
| Data collection statistics | CD1d-GCK127-TCR | CD1d-GCK152-TCR | CD1d-NC-αGC-TCR | CD1d-EF77-TCR | CD1d-7DW8–5-TCR |
|---|---|---|---|---|---|
| PDB ID | 4Y4F | 4Y4H | 4Y16 | 4Y4K | 4Y2D |
| Space group | P21 | P21 | C2221 | C2221 | P21 |
| Cell dimension | |||||
| a, b, c (Å) | 78.6, 149.7, 101.4 | 79.4, 150.4, 102.5 | 79.6, 191.9, 151.9 | 79.1, 191.4, 151.3 | 79.4, 150.3, 100.8 |
| α, β, γ (°) | 90, 96.5,90 | 90, 96.4, 90 | 90, 90, 90 | 90, 90, 90 | 90, 96.2, 90 |
| Resolution range (Å) (outer shell) | 40–3.2 (3.31–3.2) | 40–3.1 (3.15–3.1) | 66.21–2.60 (2.71–2.60) | 95.7–2.9 (3.06–2.9) | 500–3.05 (3.12–3.05) |
| No. of reflections | 38,286 | 42,694 | 34,681 | 25,092 | 44,418 |
| Rmerge (%) | 13 (63.9) | 11.3 (59.9) | 10.3 (47.8) | 17.2 (59.4) | 9.3 (53.6) |
| Rpim (%) | NA | NA | 7.0 (32.6) | 11.1 (38.4) | NA |
| Multiplicity | 3.3 (3.3) | 3.8 (4.0) | 2.9 (2.9) | 3.2 (3.4) | 3.0 (3.0) |
| Average I/sI | 13.7 (2.1) | 21.5 (2.7) | 6.8 (2.3) | 5.9 (2.2) | 13.2 (2.0) |
| Completeness (%) | 98.2 (91.1) | 99.4 (100) | 96.2 (95.7) | 97.4 (99.1) | 99.3 (99.8) |
| Refinement statistics | |||||
| No. of atoms | 12,599 | 12,469 | 6,532 | 6,305 | 11,943 |
| Protein | 12,389 | 12,247 | 6,333 | 6,183 | 11,627 |
| Ligand | 126 | 96 | 73 | 56 | 208 |
| Carbohydrate | 84 | 126 | 80 | 66 | 108 |
| Water | 0 | 0 | 46 | 0 | 0 |
| R/Rfree (%) | 21.8/25.7 | 24.2/28.3 | 21.1/24.4 | 21.9/26.5 | 21.7/25.7 |
| Ramachandran plot (%) | |||||
| Favored | 96.91 | 95.52 | 96.36 | 96.11 | 95.93 |
| Allowed | 99.75 | 99.68 | 100 | 99.75 | 99.87 |
| Root mean square deviations | |||||
| Bonds (Å) | 0.006 | 0.01 | 0.01 | 0.005 | 0.005 |
| Angles (°) | 0.879 | 1.226 | 1.248 | 1.076 | 0.994 |
| B-factors (Å2) | |||||
| Protein | 89.25 | 75.42 | 41.75 | 37.58 | 75.66 |
| Ligand | 67.87 | 58.88 | 33.77 | 28 | 50.79 |
| Carbohydrate | 83.86 | 87.85 | 54.56 | 44.54 | 96.54 |
Results
Modifications of the αGalCer Structure
We have compared the structure and antigenicity of αGalCer analogs modified in three different parts as follows: the galactose headgroup, fatty acyl chain, and the sphingoid base (Fig. 1). In addition, GCK127 and GCK152 have their O-glycosidic linkages replaced with an E-alkene linker between the sugar and lipid moieties, reminiscent of C-glycoside (α-C-GalCer), whereas GCK152 also has a truncated fatty acid with a terminal phenyl group (17). Naphthylcarba-αGalCer (NC-αGC) has an aromatic 6″-OH galactose modification, similar to the previously crystallized naphthylurea (NU)-αGC. However, unlike NU-αGC where the naphthyl is linked via a urea group, NC-αGC has a carbamate linker, which provides more flexibility to the naphthyl moiety (18). The rationale of introducing this flexible linker was to assess whether the rigidity of the urea linker or the aromatic nature and size of the naphthyl group cause the reported structural change in the A′ roof of CD1d (36). EF77 is a plakoside A-like glycolipid, a GSL isolated from the marine sponge, Plakortis simplex (37), and is related to the previously crystallized SMC124 lipid (16). The sugar headgroup and O-glycosidic linkage are identical to αGalCer, but the acyl chain has been modified to mimic plakoside A, with a cyclopropyl group and double bond between carbons 4 and 5 (16). Like EF77, 7DW8-5 also has a modification in the acyl group. The length of the chain has been shortened to 11 carbons and a parafluor-phenyl group has been added (19, 20). The rationale behind those different modifications is that changes in the lipid moiety would be expected to mainly influence the lipid interaction with CD1d, whereas changes in the galactose moiety and linkage can affect interaction with CD1d; the TCR and they could confer resistance to endoglycosidases upon injection into mice and thus could increase the half-life in serum.
FIGURE 1.
Chemical structures of glycosphingolipids. Green, carbohydrate galactose sugar moiety; red, fatty acid chain; blue, sphingoid base of αGalCer; yellow, structural modifications of each GSL compared with the αGalCer template structure.
Real Time TCR Binding Kinetics
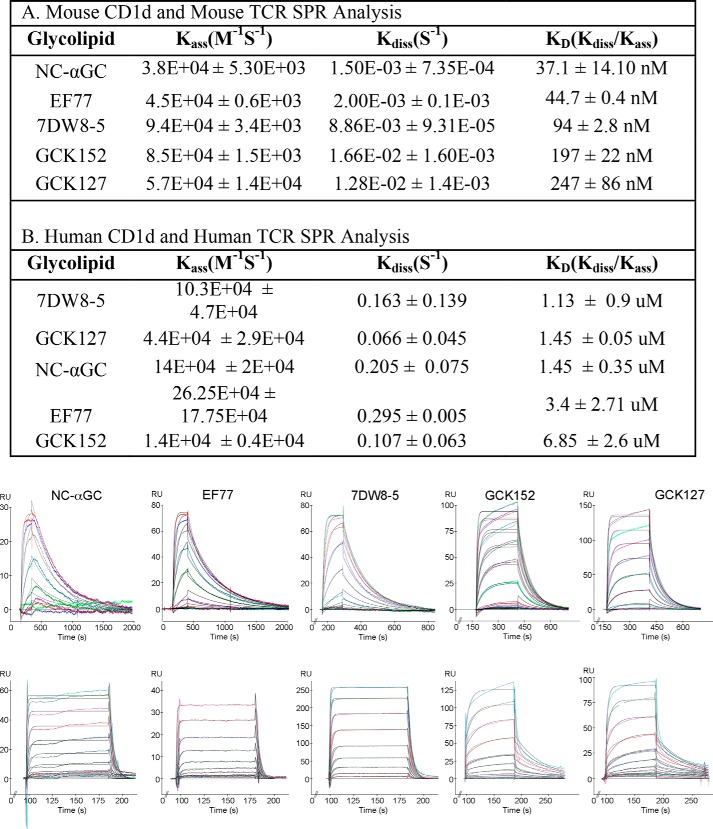
First, we determined the TCR interactions of these lipids presented by both mouse and human CD1d using SPR (Fig. 2). As has been seen previously, the affinity of the mouse TCR to the GSL-CD1d complexes is higher (in the nanomolar range), whereas the affinity of the human TCR for the GSL-CD1d complexes is weaker (in the micromolar range). According to the analysis using mouse CD1d and TCR (Fig. 2A), the lipids cluster into a lower affinity and a higher affinity group. GCK127 (KD = 247 ± 86 nm) and GCK152 (KD = 197 ± 22 nm) show the lowest Vα14Vβ8.2 TCR affinity. This is similar to the affinity reported for the parent α-C-GalCer (KD = 247 nm) (36) but is ∼10-fold weaker than αGalCer, which in our hands ranges in affinity from 11 to 25 nm (18, 29). Of note, the binding affinity is still high compared with mouse TCR affinities for MHC-presented peptides, which most often are in the micromolar range (39, 40). The higher affinity group is composed of the NC-αGC (KD = 37.1 ± 14.10 nm), similar to NU-αGC (36), EF77 (44.7 ± 0.4 nm), and 7DW8-5 (94 ± 2.8 nm). The division into lower and higher affinity groups was not maintained in the SPR analysis using the human Vα24Vβ11 TCR and human CD1d (Fig. 2B). GCK152 and EF77 were the weakest binders with KD values of 6.85 ± 2.6 and 3.4 ± 2.71 μm, respectively. The other lipids had similar affinity to αGalCer, which in our hands ranges from 1 to 3 μm. GCK127 (1.45 ± 0.05 μm) and NC-αGC (1.45 ± 0.35 μm) were very similar, and 7DW8-5 resulted in the highest TCR affinity (1.13 ± 0.9 μm). We noted that in the mouse studies the off-rate for the type I NKT cell TCR for both GCK127 and GCK152 (kd = 1.28 ± 0.0014 × 10−2 and 1.66 ± 0.0016 × 10−2 s−1, respectively) is 10 times faster than the other ligands, including αGalCer (kd = 2.2 ± 0.52 × 10−3 s−1) (data not shown), but is similar to α-C-GalCer (36). Therefore, we assume that the GCK glycolipids were not able to induce the closure of the roof over the CD1d F′ pocket. As reported previously, some GSLs like αGalCer induce the formation of the F′ roof closure prior to TCR docking by orienting CD1d side chains at Leu-84, Val-149, and Leu-150 to an optimal conformation for engagement by the TCR CDR3α residue Leu-99. The pre-formed F′ roof closure has been correlated with a slower off-rate of the type I NKT cell TCR (41). In the human SPR studies, we noted that the off-rates for all the GSLs were similar, likely due to the inability of human CD1d to pre-form the closed F′ roof, as the Leu-84 of mouse CD1d is altered to Phe-84 in human CD1d, and a fully closed F′ roof has not been observed in the hCD1d-αGalCer structure (42).
FIGURE 2.
Real time TCR binding kinetics. Binding of refolded mouse Vα14Vβ8.2 TCR (A) or human Vα24Vβ11 TCR (B) to the indicated glycolipids presented by mouse and human CD1d, respectively. One representative sensorgram is depicted at the bottom showing the binding response of increasing concentrations of TCR (colored curves) and the calculated fit (black curves).
Crystal Structures of the Trimolecular Complexes
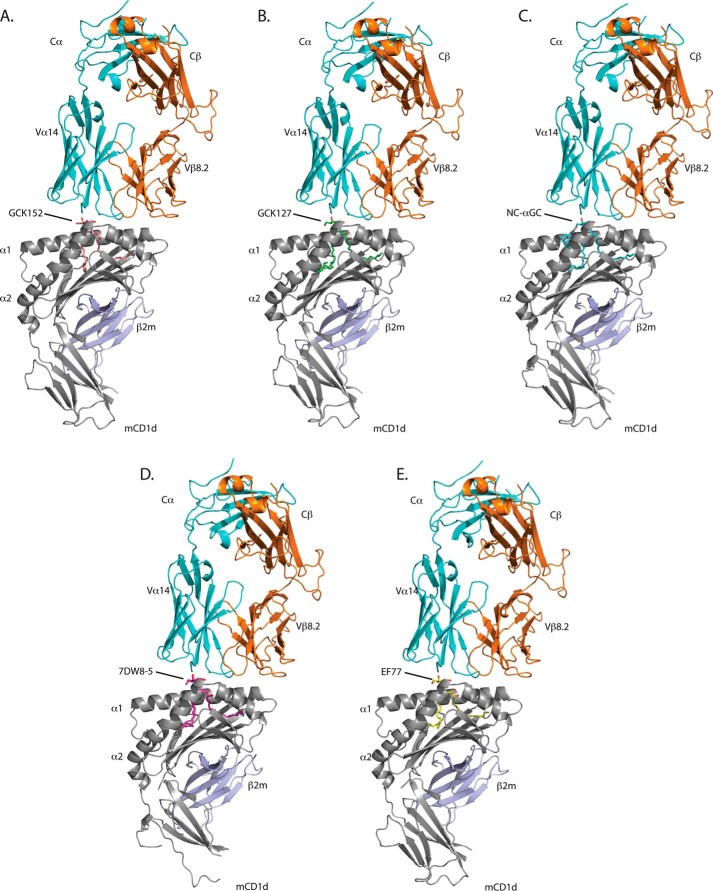
To determine how each of these lipids is recognized by the TCR of type I NKT cells, we crystallized each lipid in complex with mouse CD1d and the mouse Vα 14Vβ8.2 TCR. Crystallographic parameters are listed in Table 1. The overall structures of all complexes were consistent with the previously crystallized CD1d-glycolipid-TCR complexes, with the TCR docked offset over the F′ pocket of CD1d in a parallel orientation to the two α-helices (Fig. 3), notably different from the diagonal footprint generated in MHC-peptide-TCR complexes. All GSLs bind with the fatty acid chain in the A′ pocket of CD1d; the sphingoid base nestles inside the F′ pocket, and the galactose moiety was exposed at the CD1d-binding groove portal for TCR recognition. Electron density for all of the lipids is well defined except for the acyl chain of 7DW8-5 (Fig. 4). In this case, the electron density implies that the acyl chain can orient itself both clockwise and counterclockwise around cysteine 12 within the A′ binding pocket. Thus, we have modeled both acyl chain orientations of 7DW8-5 with 50% occupancy (occurrence) (Fig. 4F).
FIGURE 3.
Ternary crystal structures of mouse CD1d-GSL-Vα14Vβ8.2 NKT cell TCR complexes. A, GCK152; B, GCK127; C, NC-αGC; D, 7DW8-5; E, EF77. CD1d is shown in gray and β2M in blue. TCRα chain is shown in cyan and TCRβ chain in orange.
FIGURE 4.
Electron density of GSLs shown with CD1d. Note: α2 helix was removed for clarity. A, NC-αGC; B, EF77; C, 7DW8-5; D, GCK152; E, GCK127. F, dual binding motif for acyl chain of 7DW8-5. 2FoFc electron density is drawn as a blue mesh around the glycolipid (colored sticks) and contoured at 1σ. CD1d is shown in gray.
TCR Interactions
Previous studies established that the TCR of type I NKT cells binds CD1d with a largely conserved binding footprint (43–46). Not surprisingly, the contact surfaces between CD1d and the TCR are very similar throughout the different ternary complexes (Table 2). All the TCR interactions with the antigen are formed with the conserved TCR α chain, although both TCR chains contact CD1d. TCR interactions with CD1d are dominated by the TCR α chain (818–870 Å2 total buried surface area), but there is substantial involvement of the TCR β chain (582–650 Å2 buried surface area) (Table 2). Contacts between TCRα and the ligand vary depending on the ligand modification, resulting in subtle differences in the presentation of the sugar to the TCR. Crystal structure analysis shows hydrogen bonding is maintained similar to αGalCer in the cases of 7DW8-5, GCK127, and EF77, with contacts between the galactose and residues Asn-30, Arg-95, and Gly-96 of the type I NKT TCR α chain (Fig. 4). GCK152 shows the most deviation from this binding. It fails to maintain the hydrogen bonds between the 4″ hydroxyl group and Asn-30 and the 3″ hydroxyl group and Arg-95 of the TCR. Furthermore, it also fails to make a hydrogen bond between the 3′-sphingoid base hydroxyl group and Asp-80 of the CD1d molecule. These differences likely partially account for its reduced TCR affinity. NC-αGC loses the hydrogen bond between the 4″ hydroxyl group of the sugar and Asn-30 of the TCR (Fig. 5C). It should be noted that due to the moderate resolution of the different complexes, a precise hydrogen bond distance is not given, and generally a cutoff of 3.5Å distance is applied. Similarly to the previously crystallized NU-αGC, the naphthyl anchor binds within a newly formed pocket in the A′ roof of CD1d, slightly pulling the galactose away from the TCR. Although NC-αGC was predicted to be more flexible than NU-αGC, when the two structures are overlaid (Fig. 5F), there are no discernable differences between them at the resolution of this study, suggesting that the size and nature of the aromatic modification, and not the flexibility of the linker region, drives the induced fit observed in CD1d. We noted that the total buried surface area between the TCRα and GSL was highest in the group of three ligands with the highest TCR affinity, NC-αGC, EF77, and 7DW8-5 (Table 2). However, considering NC-αGC, the number of H-bonds between TCR and antigen did not always correlate with the affinity, suggesting that van der Waals interactions can contribute greatly to the TCR binding affinity and can compensate for the loss of individual hydrogen bond interactions (Fig. 5).
TABLE 2.
Total buried surface areas between mouse TCR-CD1d and TCRα-glycolipid (in Å3)
| Contact surfaces | CD1d-ligand-TCR complex |
||||
|---|---|---|---|---|---|
| GCK127 | GCK152 | NC-αGC | EF77 | 7DW8–5 | |
| TCRα-ligand | 298 | 290 | 298 | 303 | 241 |
| TCRα-CD1d | 844 | 818 | 867 | 854 | 870 |
| TCRβ-CD1d | 610 | 647 | 633 | 637 | 582 |
FIGURE 5.
Mouse Vα14Vβ8.2 NKT cell TCR hydrogen bond interactions with CD1d and GSLs. A, GCK127; B, GCK152; C, NC-αGC; D, 7DW8-5; E, EF77; F, overlay of NC-αGC (cyan) with NU-αGC (purple) ribbon crystal structures. Hydrogen bonds between TCRα (cyan) and the various glycolipids (colored sticks) are indicated by blue dashed lines. Individual OH groups of the galactose sugar that participate in hydrogen bonding are labeled with position. CD1d α1-helix as a reference is shown in gray.
Biological Activity of the Glycolipids
As all glycolipid antigens were bound by the TCR with moderate to high affinity, we investigated their activity in vitro and in vivo. Different biological assays for each of the glycolipids have been reported previously, EF77 (16), GCK127 and -152 (17), NC-αGC (18), and 7DW8-5 (19–23); however, here we assessed their potency side-by-side using the same assays. First, we performed a cell-based antigen presentation assay, in which a mouse B cell lymphoma cell line (A20) stably expressing CD1d was pulsed with various lipids, and their ability to activate the mouse Vα14Vβ8.2 NKT cell hybridoma cell line 1.2 was analyzed. TCR engagement and subsequent hybridoma activation were measured by IL-2 secretion. All the GSLs stimulated the hybridoma compared with the mock control, with 7DW8-5, EF77, and GCK152 giving the most potent response (Fig. 6A).
FIGURE 6.
Biological assays with GSLs. A, cell-based assay in which an A20 B cell lymphoma cell line transfected with CD1d was pulsed with 100 ng of the indicated GSL and co-cultured with a Vα14Vβ8.2 NKT cell hybridoma (1.2). TCR engagement was measured after 24 h through IL-2 production in serum measured with a sandwich ELISA. Data represent samples in triplicate and are representative of three independent experiments. B, human type I NKT cells were activated by human PBMCs pulsed with the indicated glycolipid concentrations for 24 h. Cytokine levels for human GM-CSF were measured in the supernatant using a sandwich ELISA. Representative data from one of two experiments performed in triplicate wells using multiple human cell lines are shown. C, C57Bl/6 mice were injected with 1 μg of the indicated GSL, and sera were analyzed at 24 h. Serum samples were measured for IFN-γ cytokine levels by ELISA. Data are representative of two independent experiments from three mice per group. Error bars represent ± S.E. D. Cell-free antigen presentation assay. 96-Well plates were coated with recombinant CD1d and incubated overnight with the indicated concentration of glycolipids. After the initial incubation, Vα14Vβ8.2 NKT cell hybridoma 1.2 was added overnight. Serum IL-2 was measured using a sandwich ELISA. Representative data from 1 of 2 different hybridoma lines are shown.
The ultimate goal for the synthesis of novel GSLs is to develop a possible therapeutic agent for human use. To address whether these lipids could activate human type I NKT cells, we used a model whereby antigen-presenting cells in the form of PBMCs and type I NKT cells were isolated from normal human donor blood samples. Various concentrations of lipids were added to PBMCs, and activation of human type I NKT cells was measured by GM-CSF cytokine secretion in the supernatant (one representative graph is shown, Fig. 6B). 7DW8-5, NC-αGC, and EF77 are strong activators, capable of stimulating human type I NKT cells in a dose-dependent manner. However, high concentrations of NC-αGC seemed to inhibit type I NKT cell activation. In comparison, both alkene versions of the parental C-glycoside compound, GCK127 and GCK152, failed to activate human type I NKT cells, even at high concentrations.
We further tested the lipids in vivo by i.v. injection of 1 μg of each lipid into mice. 24 h post-injection, we analyzed serum IFN-γ cytokine levels using a sandwich ELISA. NC-αGC and EF77 were able to generate a pronounced, systemic IFN-γ burst at this time point (Fig. 6C). To test whether the inability of the GCK compounds to show a strong response in any biological assay is due to unexpected degradation of the glycolipids during storage, we performed a cell-free antigen presentation assay. Recombinant CD1d was coated in 96-well plates, incubated overnight with the indicated concentration of glycolipids, and cultured with the Vα14Vβ8.2 NKT cell hybridoma 1.2. We expected that glycolipids that result in a strong TCR binding affinity should also induce robust T cell hybridoma activation, because TCR triggering and subsequent T cell activation is independent of T cell co-stimulation. As such, we expected a correlation between TCR binding strength and cytokine production. Both GCK127 and GCK152 induced a robust IL-2 secretion, demonstrating that both lipids were still fully intact throughout the course of this study (Fig. 6D). The same pattern was observed using a different type I NKT cell hybridoma (data not shown). GCK127 and GCK152 induced a lower IL-2 production, compared with 7DW8-5 and EF77, in correlating with their lower TCR affinity. However, surprisingly, NC-αGC, which had the highest TCR affinity, gave the lowest response.
In summary, we measured the formation of the trimolecular complex of CD1d, GSL antigen and the TCR by SPR and analyzed the nature of the molecular interactions by x-ray crystallography. This analysis provided insights into the ways these antigens can be bound and recognized. Differences in TCR affinity and binding kinetics were not highly predictive even of the ability of the compounds to stimulate type I NKT cell hybridomas in a cell-free antigen presentation assay. They were less predictive of the ability to stimulate type I NKT cell cytokine responses in vivo. Therefore, other properties, including solubility, interactions with lipid-binding proteins, uptake by different cell types, and chemical stability of each lipid appear to affect stimulation of type I NKT cells. Although NC-αGC is not able to activate type I NKT cells when pulsed into A20 cells, it strongly activates human type I NKT cells using pulsed PBMCs and murine type I NKT cells when injected intravenously into mice. In contrast, 7DW8-5 potently activates human type I NKT cells and murine type I NKT cell hybridomas using pulsed A20 cells but not when injected intravenously. EF77 was the only lipid that gave strong responses in all three assays, whereas the GCK series failed to give a robust response in vivo or in cell-based assays, even though the TCR binding kinetics were among the strongest for human type I NKT cells (Fig. 2).
Discussion
Many type I NKT cell-activating GSLs have been assessed to determine how they stimulate the human and mouse immune systems (12). Analysis of GSLs reported to be Th1-skewing clearly indicate that a variety of parameters are in play that will determine the in vivo immune response in mice and humans, and potency cannot be predicted by a purely biochemical approach or by any one type of assay. As mouse and human type I NKT cells have different fine specificities for CD1d presented antigens, structural studies of the human complexes are important to dissect the underlying structural differences (47). However, we were unsuccessful obtaining the required resolution for robust structure determination using the human molecules. Instead, biochemical approaches can yield important insights into the mechanism that governs the binding reaction, and they can contribute to the design of even more effective stimulators of type I NKT cells. For example, it is interesting to note that although the linker of NU-αGC has been altered in the NC-αGC lipid, allowing for more flexibility of the naphthyl group, the two crystal structures show no obvious differences, and the overlay indicates the conserved binding orientation of the type I NKT cell TCR trumps flexibility in these models. NC-αGC is a very strong antigen in vivo in mice and in vitro for humans. Although it lacks activity in the in vitro cell presentation assay, we hypothesize that this may reflect reduced in vitro CD1d loading efficiency, due to the aromatic naphthyl moiety, which reduces water solubility, or possibly the absence of a crucial lipid binding or transport protein derived from the antigen-presenting cells, subsequently leading to less stimulatory CD1d molecules and thus less T cell activation. Conversely, EF77, the plakoside A GSL, demonstrates potent activity in all assays. 7DW8-5 activates mouse and human type I NKT cells in vitro in the presence of antigen-presenting cells, and the ability of 7DW8-5 to activate human type I NKT cells correlates with previously reported data that this GSL is able to serve as a vaccine adjuvant in primate models. Although this GSL does not cause robust IFN-γ cytokine secretion in our mouse in vivo system, this could be due to the fact that this lipid may biodistribute differently than other GSLs after i.v. injection, and perhaps a different route is used in primates. It has been noted that mouse intramuscular injection of this lipid causes it to localize to local lymph nodes, whereas αGalCer seems to go systemic.8 The EF77 crystal structure showed similar features to its sister ligand SMC124. Both are related to plakoside A, but the EF77 ceramide has an αGalCer-like sphingosine and the plakoside A alkyl chain. The plakoside A acyl chain, with its cyclopropyl group, binds in the hydrophobic A′ pocket of CD1d. SMC124 has the opposite configuration, with an αGalCer acyl chain and the plakoside A sphingoid base, which also has a cyclopropyl group, buried in the F′ pocket (16). Similar to EF77, the acyl chain-modified 7DW8-5 ligand shows the phenyl ring localizing to the A′ groove; however, this GSL proved more difficult to model. Our crystal structure shows a duel binding motif of the acyl chain in the A′ pocket indicating that the fatty acid chain may orient both clockwise and counterclockwise around C12, possibly altering its interaction with CD1d, rather than the TCR.
The GCK127 and GCK152 have the weakest affinity in the mouse SPR studies, with rapid off-rates, which may in part be due to the inability of the CD1d F′ roof to pre-form prior to TCR engagement (48). This F′ roof formation likely occurs upon binding of the other GSLs used in this study, accounting for the 10-fold difference in dissociation rates in the mouse SPR studies. Because this pre-formation does not occur in human CD1d, we do not see a disparity in the dissociation rates of the human type I NKT cell TCR. The GCK152 GSL also seems to fail to form some of the hydrogen bonds seen between αGalCer and CD1d and TCR, also accounting for a faster off-rate of the TCR. Although the human TCR affinities would predict a human type I NKT cell response, GCK127 and GCK152 do not strongly activate the human cell lines in our hands similar to what was observed for the related α-C-GalCer. It is perhaps this alteration from an O-glycoside bond to a carbon linkage that slightly alters the binding of the glycolipid within CD1d. A rotation of the 3′-OH of the sphingoid base followed by concomitant loss of a hydrogen bond with Asp-80 of CD1d has been observed in two separate crystal structures (36, 49). Nonetheless, lipids of this nature do not appear to exhibit the same therapeutic potential than the other lipids in our assays. The buried surface area contacts between the TCRα loop and CD1d may shed some light on the variation in the biological and biochemical parameters. In conclusion, these different GSLs have the capacity to form interactions between CD1d and type I NKT cells, but they have various responses in the mouse and human studies, indicating a lack of one-to-one correlation.
Recent studies suggested that kinetic data obtained from TCR and peptide-MHC molecules in solution (e.g. by SPR) do not always correlate with T cell signaling outcome (50). Instead, measuring the interaction in two dimensions, where both MHC and TCR are each presented on a cell, is a better predictor of the potency of an antigen. In the two-dimensional system, a pulling force can be applied to mimic naturally occurring forces that occur when cells are in contact. Under those conditions, the TCR dissociation is increased 3–4 orders of magnitude compared with measurements carried out in solution (38, 50). Applying these forces to the CD1d system would allow us to assess the role of the CD1d-lipid interaction in modulating the overall stability of the ternary complex, as interactions among all three molecules (antigen-CD1d, antigen-TCR, and CD1d-TCR) are likely important in the two-dimensional system. When performing SPR studies, the stability of the CD1d-lipid complex does not affect TCR affinity, likely because no pulling forces are applied. Future studies could include the study of the two-dimensional interaction using pulling force, while systematically changing the terminal lipid backbone, to alter its interaction with CD1d (50). Those assays would shed light on the contribution of the CD1d-lipid interaction in T cell triggering through modulating the stability and duration of their interaction. In addition, future studies that can accurately measure the pharmacokinetic properties of the GSLs, binding affinities for the GSL (see above) and CD1d molecules and biodistribution of different GSLs would be particularly interesting to address this outstanding issue.
Acknowledgments
We thank the Stanford Synchrotron Radiation Lightsource and the Advanced Light Source for access to remote data collection. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research, and by National Institutes of Health Grant P41GM103393 from NIGMS. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, United States Department of Energy under Contract No. DE-AC02-05CH11231.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 AI074952 and R21 AI107318 (to D. M. Z.), RO1 AI45053 and AI71922 (to M. K.), RO1 GM087136 (to A. H.), and RO1 AI070258 (to M. T.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 4Y4F, 4Y4H, 4Y16, 4Y4K, and 4Y2D) have been deposited in the Protein Data Bank (http://wwpdb.org/).
M. Tsuji, personal communication.
- NKT
- natural killer T cell
- αGalCer
- α-galactosylceramide
- GSL
- glycosphingolipid
- SPR
- surface plasmon resonance
- TCR
- T cell receptor
- Th1
- T helper type 1
- Th2
- T helper type 2
- NU
- naphthylurea
- PBMC
- peripheral blood mononuclear cell
- NC-αGC
- naphthylcarba-αGalCer.
References
- 1. Robertson F. C., Berzofsky J. A., Terabe M. (2014) NKT cell networks in the regulation of tumor immunity. Front. Immunol. 5, 543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roozbeh M., Mohammadpour H., Azizi G., Ghobadzadeh S., Mirshafiey A. (2014) The potential role of iNKT cells in experimental allergic encephalitis and multiple sclerosis. Immunopharmacol. Immunotoxicol. 36, 105–113 [DOI] [PubMed] [Google Scholar]
- 3. Bendelac A., Savage P. B., Teyton L. (2007) The biology of NKT cells. Annu. Rev. Immunol. 25, 297–336 [DOI] [PubMed] [Google Scholar]
- 4. Brennan P. J., Brigl M., Brenner M. B. (2013) Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat. Rev. Immunol. 13, 101–117 [DOI] [PubMed] [Google Scholar]
- 5. Yu K. O., Porcelli S. A. (2005) The diverse functions of CD1d-restricted NKT cells and their potential for immunotherapy. Immunol. Lett. 100, 42–55 [DOI] [PubMed] [Google Scholar]
- 6. Calabi F., Milstein C. (1986) A novel family of human major histocompatibility complex-related genes not mapping to chromosome 6. Nature 323, 540–543 [DOI] [PubMed] [Google Scholar]
- 7. Zajonc D. M., Wilson I. A. (2007) Architecture of CD1 proteins. T cell activation by CD1 and lipid antigens. Curr. Top. Microbiol. Immunol. 314, 27–50 [DOI] [PubMed] [Google Scholar]
- 8. Borg N. A., Wun K. S., Kjer-Nielsen L., Wilce M. C., Pellicci D. G., Koh R., Besra G. S., Bharadwaj M., Godfrey D. I., McCluskey J., Rossjohn J. (2007) CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature 448, 44–49 [DOI] [PubMed] [Google Scholar]
- 9. Pellicci D. G., Patel O., Kjer-Nielsen L., Pang S. S., Sullivan L. C., Kyparissoudis K., Brooks A. G., Reid H. H., Gras S., Lucet I. S., Koh R., Smyth M. J., Mallevaey T., Matsuda J. L., Gapin L., et al. (2009) Differential recognition of Cd1d-α-Galactosyl ceramide by the vb8.2 and Vb7 semi-invariant Nkt T cell receptors. Immunity 31, 47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morita M., Motoki K., Akimoto K., Natori T., Sakai T., Sawa E., Yamaji K., Koezuka Y., Kobayashi E., Fukushima H. (1995) Structure-activity relationship of α-galactosylceramides against B16-bearing mice. J. Med. Chem. 38, 2176–2187 [DOI] [PubMed] [Google Scholar]
- 11. Banchet-Cadeddu A., Hénon E., Dauchez M., Renault J.-H., Monneaux F., Haudrechy A. (2011) The stimulating adventure of KRN 7000. Org. Biomol. Chem. 9, 3080–3104 [DOI] [PubMed] [Google Scholar]
- 12. Anderson B. L., Teyton L., Bendelac A., Savage P. B. (2013) Stimulation of natural killer T cells by glycolipids. Molecules 18, 15662–15688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmieg J., Yang G., Franck R. W., Tsuji M. (2003) Superior protection against malaria and melanoma metastases by a C-glycoside analogue of the natural killer T cell ligand-galactosylceramide. J. Exp. Med. 198, 1631–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kakuta S., Tagawa Y., Shibata S., Nanno M., Iwakura Y. (2002) Inhibition of B16 melanoma experimental metastasis by interferon-γ through direct inhibition of cell proliferation and activation of antitumour host mechanisms. Immunology 105, 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petrovsky N., Aguilar J. C. (2004) Vaccine adjuvants: current state and future trends. Immunol. Cell Biol. 82, 488–496 [DOI] [PubMed] [Google Scholar]
- 16. Tyznik A. J., Farber E., Girardi E., Birkholz A., Li Y., Chitale S., So R., Arora P., Khurana A., Wang J., Porcelli S. A., Zajonc D. M., Kronenberg M., Howell A. R. (2011) Glycolipids that elicit IFN-γ-biased responses from natural killer T cells. Chem. Biol. 18, 1620–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li X., Shiratsuchi T., Chen G., Dellabona P., Casorati G., Franck R. W., Tsuji M. (2009) Invariant TCR rather than CD1d shapes the preferential activities of C-glycoside analogues against human versus murine invariant NKT cells. J. Immunol. 183, 4415–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aspeslagh S., Nemcovic M., Pauwels N., Venken K., Wang J., Van Calenbergh S., Zajonc D. M., Elewaut D. (2013) Enhanced TCR footprint by a novel glycolipid increases NKT-dependent tumor protection. J. Immunol. 191, 2916–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li X., Fujio M., Imamura M., Wu D., Vasan S., Wong C.-H., Ho D. D., Tsuji M. (2010) Design of a potent CD1d-binding NKT cell ligand as a vaccine adjuvant. Proc. Natl. Acad. Sci. U.S.A. 107, 13010–13015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Padte N. N., Li X., Tsuji M., Vasan S. (2011) Clinical development of a novel CD1d-binding NKT cell ligand as a vaccine adjuvant. Clin. Immunol. 140, 142–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Padte N. N., Boente-Carrera M., Andrews C. D., McManus J., Grasperge B. F., Gettie A., Coelho-dos-Reis J. G., Li X., Wu D., Bruder J. T., Sedegah M., Patterson N., Richie T. L., Wong C.-H., Ho D. D., et al. (2013) A glycolipid adjuvant, 7DW8-5, enhances CD8+ T cell responses induced by an adenovirus-vectored malaria vaccine in non-human primates. PLoS ONE 8, e78407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Venkataswamy M. M., Ng T. W., Kharkwal S. S., Carreño L. J., Johnson A. J., Kunnath-Velayudhan S., Liu Z., Bittman R., Jervis P. J., Cox L. R., Besra G. S., Wen X., Yuan W., Tsuji M., Li X., et al. (2014) Improving mycobacterium bovis bacillus Calmette-Guèrin as a vaccine delivery vector for viral antigens by incorporation of glycolipid activators of NKT cells. PLoS ONE 9, e108383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu X., Hegazy W. A., Guo L., Gao X., Courtney A. N., Kurbanov S., Liu D., Tian G., Manuel E. R., Diamond D. J., Hensel M., Metelitsa L. S. (2014) Effective cancer vaccine platform based on attenuated salmonella and a type III secretion system. Cancer Res. 74, 6260–6270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Naidenko O. V., Maher J. K., Ernst W. A., Sakai T., Modlin R. L., Kronenberg M. (1999) Binding and antigen presentation of ceramide-containing glycolipids by soluble mouse and human CD1d molecules. J. Exp. Med. 190, 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lawton A. P., Prigozy T. I., Brossay L., Pei B., Khurana A., Martin D., Zhu T., Späte K., Ozga M., Höning S., Bakke O., Kronenberg M. (2005) The mouse CD1d cytoplasmic tail mediates CD1d trafficking and antigen presentation by adaptor protein 3-dependent and -independent mechanisms. J. Immunol. 174, 3179–3186 [DOI] [PubMed] [Google Scholar]
- 26. Brossay L., Tangri S., Bix M., Cardell S., Locksley R., Kronenberg M. (1998) Mouse CD1-autoreactive T cells have diverse patterns of reactivity to CD1+ targets. J. Immunol. 160, 3681–3688 [PubMed] [Google Scholar]
- 27. Rogers P. R., Matsumoto A., Naidenko O., Kronenberg M., Mikayama T., Kato S. (2004) Expansion of human Vα24+ NKT cells by repeated stimulation with KRN7000. J. Immunol. Methods 285, 197–214 [DOI] [PubMed] [Google Scholar]
- 28. Zajonc D. M., Maricic I., Wu D., Halder R., Roy K., Wong C.-H., Kumar V., Wilson I. A. (2005) Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J. Exp. Med. 202, 1517–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang J., Li Y., Kinjo Y., Mac T. T., Gibson D., Painter G. F., Kronenberg M., Zajonc D. M. (2010) Lipid binding orientation within CD1d affects recognition of Borrelia burgdorferi antigens by NKT cells. Proc. Natl. Acad. Sci. U.S.A. 107, 1535–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leslie A. G. (2006) The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 31. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 32. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vagin A. A., Steiner R. A., Lebedev A. A., Potterton L., McNicholas S., Long F., Murshudov G. N. (2004) REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 35. Schüttelkopf A. W., van Aalten D. M. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 36. Aspeslagh S., Li Y., Yu E. D., Pauwels N., Trappeniers M., Girardi E., Decruy T., Van Beneden K., Venken K., Drennan M., Leybaert L., Wang J., Franck R. W., Van Calenbergh S., Zajonc D. M., Elewaut D. (2011) Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis. EMBO J. 30, 2294–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Costantino V., Fattorusso E., Mangoni A., Di Rosa M., Ianaro A. (1997) Glycolipids from sponges. 6.1 Plakoside A and B, two unique prenylated glycosphingolipids with immunosuppressive activity from the marine sponge plakortis simplex. J. Am. Chem. Soc. 119, 12465–12470 [Google Scholar]
- 38. Liu B., Chen W., Evavold B. D., Zhu C. (2014) Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell 157, 357–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rudolph M. G., Stanfield R. L., Wilson I. A. (2006) How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24, 419–466 [DOI] [PubMed] [Google Scholar]
- 40. Rossjohn J., Gras S., Miles J. J., Turner S. J., Godfrey D. I., McCluskey J. (2015) T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 [DOI] [PubMed] [Google Scholar]
- 41. Li Y., Girardi E., Wang J., Yu E. D., Painter G. F., Kronenberg M., Zajonc D. M. (2010) The Vα14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. J. Exp. Med. 207, 2383–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Koch M., Stronge V. S., Shepherd D., Gadola S. D., Mathew B., Ritter G., Fersht A. R., Besra G. S., Schmidt R. R., Jones E. Y., Cerundolo V. (2005) The crystal structure of human CD1d with and without α-galactosylceramide. Nat. Immunol. 6, 819–826 [DOI] [PubMed] [Google Scholar]
- 43. Mallevaey T., Clarke A. J., Scott-Browne J. P., Young M. H., Roisman L. C., Pellicci D. G., Patel O., Vivian J. P., Matsuda J. L., McCluskey J., Godfrey D. I., Marrack P., Rossjohn J., Gapin L. (2011) A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity 34, 315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wun K. S., Cameron G., Patel O., Pang S. S., Pellicci D. G., Sullivan L. C., Keshipeddy S., Young M. H., Uldrich A. P., Thakur M. S., Richardson S. K., Howell A. R., Illarionov P. A., Brooks A. G., Besra G. S., et al. (2011) A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells. Immunity 34, 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Girardi E., Zajonc D. M. (2012) Molecular basis of lipid antigen presentation by CD1d and recognition by natural killer T cells. Immunol. Rev. 250, 167–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rossjohn J., Pellicci D. G., Patel O., Gapin L., Godfrey D. I. (2012) Recognition of CD1d-restricted antigens by natural killer T cells. Nat. Rev. Immunol. 12, 845–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wun K. S., Ross F., Patel O., Besra G. S., Porcelli S. A., Richardson S. K., Keshipeddy S., Howell A. R., Godfrey D. I., Rossjohn J. (2012) Human and mouse type I natural killer T cell antigen receptors exhibit different fine specificities for CD1d-antigen complex. J. Biol. Chem. 287, 39139–39148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Joyce S., Girardi E., Zajonc D. M. (2011) NKT cell ligand recognition logic: molecular basis for a synaptic duet and transmission of inflammatory effectors. J. Immunol. 187, 1081–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Patel O., Cameron G., Pellicci D. G., Liu Z., Byun H. S., Beddoe T., McCluskey J., Franck R. W., Castaño A. R., Harrak Y., Llebaria A., Bittman R., Porcelli S. A., Godfrey D. I., Rossjohn J. (2011) NKT TCR recognition of CD1d-α-C-galactosylceramide. J. Immunol. 187, 4705–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang J., Zarnitsyna V. I., Liu B., Edwards L. J., Jiang N., Evavold B. D., Zhu C. (2010) The kinetics of two-dimensional TCR and pMHC interactions determine T cell responsiveness. Nature 464, 932–936 [DOI] [PMC free article] [PubMed] [Google Scholar]