Background: In Xenopus embryos, zygotically activated TGFβ signaling regulates the formation of three germ layers.
Results: Vegetally enriched SCP3 is required for TGFβ signaling and germ layer induction and functions by removing the inhibitory linker phosphorylation on R-Smads.
Conclusion: SCP3 ensures TGFβ-mediated germ layer formation.
Significance: This work presents a novel mechanism by which maternal factors control the response of embryonic blastomeres toward morphogen signals.
Keywords: embryo, phosphatase, SMAD transcription factor, transforming growth factor beta (TGF-beta), Xenopus, SCP3, germ layer induction
Abstract
Germ layer induction is one of the earliest events shortly after fertilization that initiates body formation of vertebrate embryos. In Xenopus, the maternally deposited transcriptional factor VegT promotes the expression of zygotic Nodal/Activin ligands that further form a morphogen gradient along the vegetal-animal axis and trigger the induction of the three germ layers. Here we found that SCP3 (small C-terminal domain phosphatase 3) is maternally expressed and vegetally enriched in Xenopus embryos and is essential for the timely induction of germ layers. SCP3 is required for the full activation of Nodal/Activin and bone morphogenetic protein signals and functions via dephosphorylation in the linker regions of receptor-regulated Smads. Consistently, the linker regions of receptor-regulated Smads are heavily phosphorylated in fertilized eggs, and this phosphorylation is gradually removed when embryos approach the midblastula transition. Knockdown of maternal SCP3 attenuates these dephosphorylation events and the activation of Nodal/Activin and bone morphogenetic protein signals after midblastula transition. This study thus suggested that the maternal SCP3 serves as a vegetally enriched, intrinsic factor to ensure a prepared status of Smads for their activation by the upcoming ligands during germ layer induction of Xenopus embryos.
Introduction
During vertebrate embryogenesis, the specification of three germ layers is one of the earliest and most important events shortly after fertilization. In Xenopus embryos, the maternally deposited VegT, a T-box family transcriptional factor, is localized asymmetrically along the animal-vegetal axis. After midblastula transition (MBT),5 when zygotic gene expression starts, VegT-localized vegetal blastomeres will initiate the expression of the transforming growth factor-β (TGFβ) superfamily genes, including Xnr1, -2, -4, -5, and -6 and derriere, under the direct control of VegT (1–4). As such, a vegetal to animal gradient of TGFβ signal is generated, which triggers the induction of three germ layers along the animal-vegetal axis (1, 5).
The members of the TGFβ superfamily that play key roles in Xenopus embryogenesis are the Nodal/Activin proteins and bone morphogenetic proteins (BMPs). During Xenopus early development, Nodal/Activin signaling specifies mesoderm and endoderm, whereas BMP signaling patterns ventral and lateral mesoderm and determines the formation of epidermal versus neural ectodermal cells (6, 7). TGFβ signaling is initiated when ligands bind and activate receptor serine/threonine kinases, and the receptor complexes propagate the signal through phosphorylation in the C-terminal SXS motif of receptor-regulated Smads (R-Smads). R-Smads are the key components of TGFβ signals, with Smad2 and -3 mediating Nodal/Activin signaling and Smad1, -5, and -8 mediating BMP signaling. The C-terminal phosphorylated R-Smads translocate from the cytoplasm into the nucleus and modulate target gene transcription (6–9).
The C-terminal phosphorylation of R-Smads is a key event in pathway activation, and it also serves as readout of TGFβ signal activity. Temporally, the C-terminal site-phosphorylated Smad2 and Smad1/5/8 are detected after, but not before, the MBT in Xenopus embryos. Spatially, they are asymmetrically distributed across the animal-vegetal and dorsal-ventral axes with a coincidence that early activated Smad1 and Smad2 localize in the vegetal part (10–12). These timely and regionally activated R-Smads turn on particular combinations of downstream genes and thus instruct different groups of cells to adopt distinct fates.
Besides the C terminus, multiple sites in the linker regions of R-Smads are also phosphorylated, and these phosphorylations control the activity, stability, and transportation of R-Smads (13). Studies in mammalian cells demonstrate that phosphorylation in the linker regions of R-Smads plays both positive and negative roles in TGFβ signaling (14). Mitogen-activated protein kinases (MAPKs) trigger the phosphorylation of multiple proline-directed Ser/Thr residues in the linker regions, including Ser-187, Ser-195, Ser-206, and Ser-214 in Smad1 and Thr-179, Ser-204, Ser-208, and Ser-213 in Smad3 (14–16). Following these priming phosphorylations by MAPKs, GSK3 can further phosphorylate Ser-210, Thr-202, Ser-198, and Ser-191 in Smad1, resulting in Smurf1-mediated Smad1 ubiquitination and cytoplasmic retention (15). Similarly, linker phosphorylation of Smad2/3 triggers its recognition and polyubiquitination by Nedd4L and some other unidentified E3 ligases (16). Moreover, ERK-mediated Smad2 phosphorylation at Ser-245/250/255 and Thr-220 as well as Smad3 phosphorylation at Ser-204/208 and Thr-179 inhibit the transcriptional activity of Smad2/3 (17). Thr-179 and Ser-213 of Smad3 could be phosphorylated by CDK2/4, leading to inhibition of the transcriptional activity (18). In contrast, p38, Rho kinase and c-Jun N-terminal kinase also phosphorylate Smad2/3 at multiple sites but enhance their transcriptional activity (19–21).
In Xenopus gastrula embryos, the linker region of Smad1 is sequentially phosphorylated by MAPK and GSK3 (22, 23). These events lead to Smad1 polyubiquitination/degradation and restriction of BMP signaling. Therefore, the linker phosphorylation was proposed as a mechanism of integrating BMP and FGF/insulin-like growth factor/Wnt signals during neural induction and patterning. Moreover, linker phosphorylation of Smad2/3 in the middle gastrula stage causes Smad2/3 cytosolic retention and termination of Nodal/Activin signaling. This was suggested as a mechanism to control the duration of cell competence to TGFβ signaling in Xenopus gastrula embryos (24). These studies all investigated the alterations of R-Smads in gastrula embryos, whereas whether the linker phosphorylation is regulated during cleavage embryos was unknown.
SCP3 (small C-terminal domain phosphatase 3), also called SCPL (small C-terminal domain phosphatase-like), CTDSP3, or CTDSPL, belongs to the FCP/SCP family of Ser/Thr phosphatases (25, 26). SCP1, SCP2, SCP3, and SCPL2 are related to the FCP1, which is the highly conserved, essential enzyme that dephosphorylates the C-terminal domain of RNA polymerase II (27). Although the SCPs can dephosphorylate the C-terminal domain of polymerase II in vitro, a role for them as general regulators of transcription has not been confirmed (28). Previous reports suggested that by dephosphorylating Smad2/3 at the linker (inhibitory) but not the C-terminal (activating) site, SCP1 and SCP2 can enhance TGFβ signaling, whereas in BMP signaling, they were proposed to reset Smad1 to the basal unphosphorylated state by dephosphorylation at both the linker and C-terminal sites (26, 28, 29). More recently, SCPL2 was shown to specifically dephosphorylate Smad1/5/8 in the C-terminal region and, as a result, attenuate BMP-induced transcriptional responses (30). The in vivo physiological function of the SCPs has not been extensively studied.
Using RNA-seq technology, we found that scp3 is preferentially distributed in vegetal blastomeres of Xenopus cleavage embryos. Further analysis indicated that the maternal deposited SCP3 is required for the full activation of zygotic Nodal/Activin and BMP signals and functions by dephosphorylating the linker regions of Smad2 and Smad1. Consistently, the level of R-Smad linker phosphorylation gradually declined after fertilization, and this event was attenuated by knockdown of SCP3. Thus, maternal SCP3 functions to ensure a prepared status of Smads for their activation by the upcoming ligands. Our results suggest that it is not only the expression of TGFβ ligands but also the release of inhibition on R-Smads at the predetermined time and region that ensures the activation of cellular signals and thus germ layer specification.
Experimental Procedures
Identification of Asymmetrically Localized Maternal mRNAs in Xenopus
For the dissociation of blastomeres, embryos were transferred to 80% phosphate-buffered saline lacking calcium and magnesium and manually demembranated. Animal and vegetal blastomeres were collected at the 8-cell stage (31), and RNA of animal and vegetal blastomeres was isolated using TRIzol reagent (Invitrogen). NEBNext mRNA library Prep Master mix set for Illumina (New England Biolabs) was used for the construction of sequencing libraries. The library was subjected to paired-end sequencing on a Genome Analyzer IIx (Illumina) with 100-bp read length. We used Bowtie (32) to map reads to the RefSeq genes and then detected differentially expressed genes between vegetal and animal blastomeres by DESeq (33).
Plasmid Constructs
For in vitro synthesis of mRNA, full-length Xenopus laevis scp3, smad1, smad2, Xnr1, and bmp4 were amplified from cDNA of X. laevis embryos by PCR and subcloned into the pCS2+ vectors, and for detection, a FLAG tag was added to the C terminus of Xenopus scp3. For in situ hybridization, the partial cDNAs of X. laevis scp3, edd, and lim5 were subcloned into pEASY-T3 cloning vector (TransGen Biotech). The activin-responsive element (ARE)-luciferase and BMP-responsive element (BRE)-luciferase constructs were originally described (34, 35). PCR-based site-directed mutagenesis (TransGen Biotech) was employed to generate mutants.
Oocytes, Embryos, Microinjection, and Explants
Full-grown oocytes (stage VI) were manually defolliculated and cultured in oocyte culture medium, as described (36). Oocytes were uninjected or injected with morpholino (MO) and cultured for a total of 48 h at 18 °C before fertilization. In preparation for fertilization, they were stimulated to mature by the addition of 2 μm progesterone to the culture medium and cultured for 10–12 h. Oocytes were then labeled with vital dyes and introduced into a stimulated female host using the host transfer technique described previously (37, 38). In vitro fertilization, embryo culture, staging, microinjection, and culture of Xenopus embryo explants were carried out as described (39).
RNA Synthesis and in Situ Hybridization
To in vitro synthesis capped mRNAs, the template plasmids were linearized and transcribed using a mMESSAGE mMACHINE SP6 kit (Ambion, Austin, TX) according to the manufacturer's instructions. For digoxigenin-RNA probes, the linearized plasmids were transcribed with T7, T3, or SP6 RNA polymerase (Promega). The in situ hybridizations were performed following the standard protocol (40). For hemisections, rehydrated embryos were bisected through the animal-vegetal axis with a razor blade. Bisected embryos were refixed with MEMFA (0.1 m MOPS, 2 mm EGTA, 1 mm MgSO4, 3.7% formaldehyde) for 30 min before hybridization (41).
Cell Culture, Transfections, and Stimulations
Hep3B cells were cultured in Minimal Essential Medium supplied with 10% fetal bovine serum (FBS) at 37 °C with 5% CO2. The transfection of plasmids and siRNA oligonucleotides was performed using Lipofectamine 2000 reagent (Invitrogen). Cells were treated with BMP4 (20 ng/ml; R&D Systems) or Activin A (20 ng/ml; R&D Systems) dissolved in medium containing 0.2% FBS.
MOs and siRNAs
Antisense and standard control MO (std MO) oligonucleotides were purchased from Gene Tools, LLC (Philomath, OR). MO sequences of X. laevis scp3 were as follows: MO-1, CATGGAAACGAATTGGCTGTTGTTC; MO-2, TTGTCCTCTTTATCTAATCCGCAGT. The siRNA oligonucleotides were designed and synthesized by RiboBio Co., Ltd. (Guangzhou, China). Human SCP3 siRNA target sequences were the following: siRNA-1, GCCTATTAGTAATGCTGAT; siRNA-2, GCTGCACAGACTCTGCAAT.
Total RNA Isolation, Reverse Transcription PCR, and Quantitative Real-time PCR (qPCR)
Total RNA was isolated using TRIzol reagent (Invitrogen), and reverse transcription was performed using the Reverse Transcription System (Promega, Fitchburg, WI). For qPCR, the CFX96 TouchTM real-time PCR system (Bio-Rad), Fast-Plus EvaGreen Master Mix (Biotium), and 40 cycles of amplification were used. X. laevis ornithine decarboxylase (odc) and human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as the loading controls, respectively. The statistical significance was analyzed by Student's t test, and all qPCR data are presented as mean ± S.D. The qPCR primers are listed in supplemental Table S1.
Reporter Assays
For the luciferase reporter assays in Xenopus embryos, embryos were injected with reporter constructs at the 2-cell stage and collected at stage 10.5. For the luciferase reporter assay in Hep3B cells, the experiment was performed as described previously (42). Luciferase reporter assays were performed using a Dual-Luciferase assay kit (Vigorous). Data are represented as mean ± S.D. after being normalized with Renilla activity (n = 3).
Antibodies and Immunoblotting
Immunoblot analysis was performed as described with minor modifications for frog embryo lysates; 50 μg of protein was loaded per lane, and for Xenopus Smad2-CP Western blot analysis, the membrane was blocked for at least 10 h at room temperature in 5% polyvinylpyrrolidone in TBST (Tris-buffered saline plus Tween 20) (26, 39). Antibodies were obtained commercially: Smad1 (6944; Cell Signaling, Boston, MA), Smad1-LP (9553; Cell Signaling), Smad1-CP (9511; Cell Signaling), Smad2 (610842; BD Biosciences), Smad2-LP (3104; Cell Signaling), Smad2-CP (3101; Cell Signaling), Actin (M20010; Abmart, Shanghai, China), and FLAG (F3165; Sigma).
Results
Asymmetrically Localized Maternal mRNAs in 8-cell Stage Xenopus Embryos
To identify asymmetrically localized maternal mRNAs along the animal-vegetal axis in cleavage Xenopus embryos, we isolated animal and vegetal blastomeres at the 8-cell stage, extracted the maternal mRNA from each, and analyzed them by RNA-seq technology. RNA-seq information is provided in Table 1, and the data have been deposited in the Gene Expression Omnibus (GEO) database (accession number GSE68972). In two independent experiments, 43 maternal transcripts were identified to be preferentially distributed in the vegetal region, but transcripts enriched in the animal region were not found. Among these genes, 30 transcripts have been previously demonstrated as vegetally enriched mRNAs in the oocytes, eggs, or embryos before MBT (supplemental Table S2), confirming the credibility of our RNA sequencing analysis. The functions of the asymmetrically localized candidates include germ line development, germ layer differentiation, pattern formation, signal transduction, etc. (supplemental Table S2).
TABLE 1.
RNA-seq information
Samples from two independent experiments are shown. V1 and A1, vegetal and animal samples, respectively, from one experiment; V2 and A2, vegetal and animal samples from another experiment.
| V1 | V2 | A1 | A2 | |
|---|---|---|---|---|
| Blastomere | Vegetal | Vegetal | Animal | Animal |
| No. of read pairs | 44,735,165 | 45,132, 556 | 44,083,587 | 45,784,433 |
| No. of read pairs mapped to RefSeq genes | 18,087,489 (40.43%) | 17,126,196 (37.95%) | 18,595,352 (42.18%) | 19,094,952 (41.71%) |
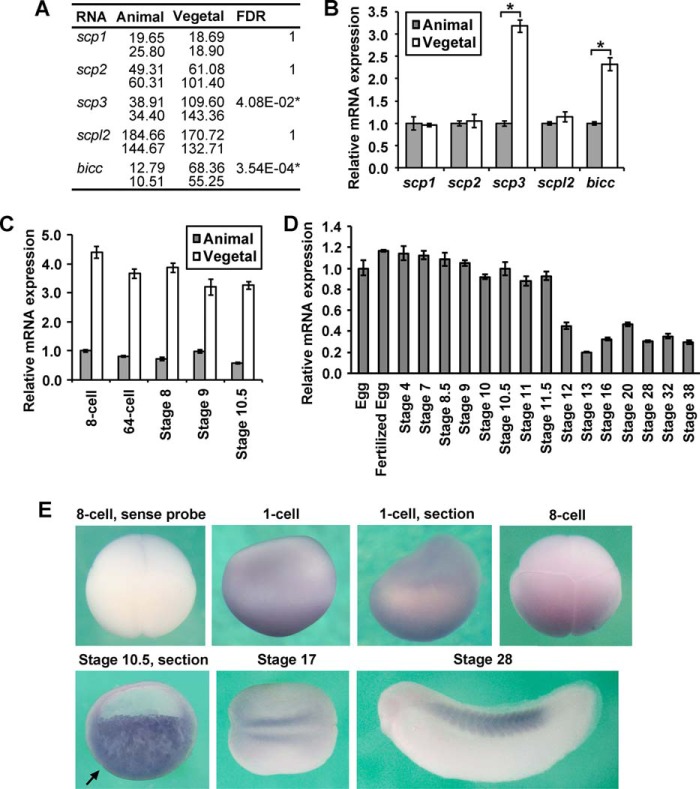
Vegetally Enriched scp3
The RNA-seq results indicated that among the SCPs, only scp3 is expressed asymmetrically along the animal-vegetal axis, being over 3-fold higher in vegetal blastomeres (Fig. 1A). This asymmetrical expression pattern was further confirmed by qPCR using independent embryonic samples (Fig. 1B), and it is maintained throughout the cleavage and blastula stages (Fig. 1C). Throughout embryonic development, scp3 mRNA level remained consistent until stage 11.5, and its zygotic expression after that point was relatively low until stage 38 (Fig. 1D). By in situ hybridization (Fig. 1E), scp3 mRNA was more significantly detected in the vegetal region of the embryos in 1-cell stage, 8-cell stage, and stage 10.5, consistent with the RNA-seq and qPCR results (Fig. 1, A–C). In the section, the staining was detected deep in the cytosol (Fig. 1E, section). At later stages, weak expression was detected in the regions of presomitic mesoderm and somites (Fig. 1E). The expression pattern in cleavage Xenopus embryos was consistent with the previous report showing that scp3 mRNA was localized in the vegetal cortex of the oocytes (43). The high level of maternal expression and vegetally enriched localization suggest an important role of SCP3 in blastula and gastrula embryos.
FIGURE 1.
Expression pattern of scp3 in Xenopus. A and B, at the 8-cell stage, total mRNA from animal and vegetal blastomeres was extracted and analyzed by RNA-seq (A) or qPCR (B). The numbers in A indicate the reads/kb/million mapped reads of sequencing in two independent experiments (54). Vegetally enriched transcript bicc was used as a control. *, significantly different (A, false discovery rate <0.05; B, p < 0.05) between animal and vegetal samples. C, at the indicated stages, RNA from animal and vegetal cells was extracted, and the relative mRNA abundance of scp3 was analyzed by qPCR. D, qPCR shows the temporal expression pattern of scp3. E, expression profile of scp3 verified by in situ hybridization. The embryos from the 1-cell stage to stage 10.5 are shown in a lateral view. A stage 17 embryo is shown in a dorsal view with the anterior end toward the left. A stage 28 embryo is shown in a lateral view with its head toward the left. The arrow indicates the dorsal lip. Error bars, S.D.
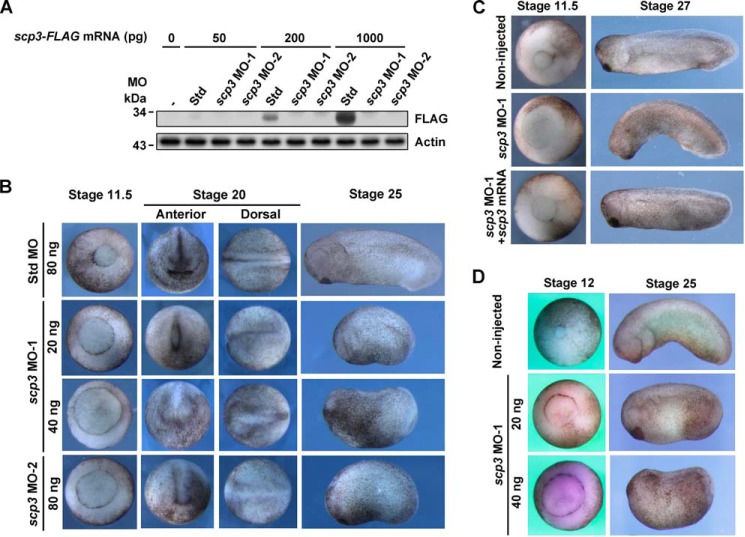
SCP3 Is Essential for Xenopus Embryogenesis
To study the function of SCP3, we tested two independent scp3 MOs by coinjecting MOs with FLAG-tagged scp3 mRNA, and the results indicated that the MOs were effective (Fig. 2A). We then injected the MOs into 1-cell stage Xenopus embryos. Compared with the std MO-injected embryos, the scp3 morphants exhibited delayed gastrulation, abnormal formation of neural plate, short anterior-posterior axis, and general development retardation (n = 91, 100%) (Fig. 2B). The phenotypes were specific, because they could be fully rescued by Xenopus scp3 mRNA at stage 11.5 (750 pg/embryo, n = 48, 87.5%) and stage 27 (200 pg/embryo, n = 55, 85.5%) (Fig. 2C). To further confirm these results, MOs were injected into in vitro cultured oocytes, and the embryos were obtained via the host transfer technology (36). A very similar phenotype was observed (Fig. 2D, n = 32, 100%).
FIGURE 2.
SCP3 is essential for Xenopus embryogenesis. A, MO test in Xenopus embryos. Two-cell stage embryos were coinjected with 40 ng of std MO or scp3 MOs and mRNA coding for X. laevis SCP3-FLAG with the indicated doses. At stage 10.5, the embryos were harvested for Western blotting analysis. B, std MO or scp3 MOs was injected into 1-cell stage embryos with the indicated doses. C, 2-cell stage embryos were injected with 40 ng of std MO or scp3 MO-1 with or without 750 pg (left) or 200 pg (right) of X. laevis scp3 mRNA as indicated. D, uninjected oocytes and scp3 MO-1-injected oocytes were processed by the host transfer technique and allowed to develop until the indicated stages.
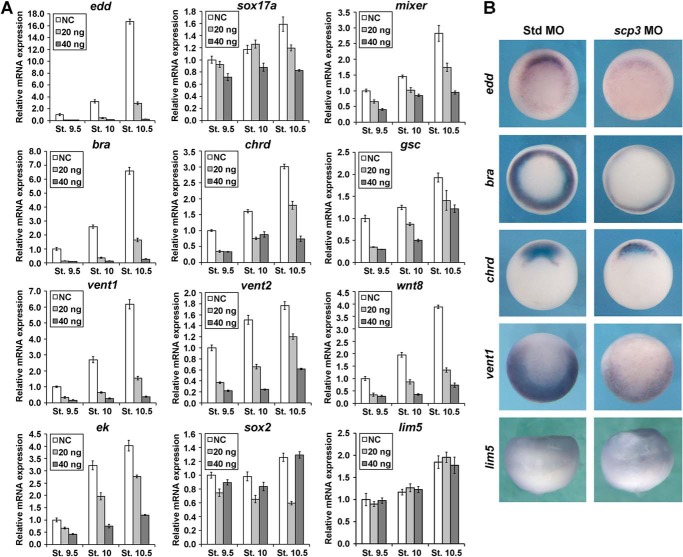
Considering that the asymmetrical distribution of scp3 mRNA may impact the germ layer differentiation and pattern formation, we analyzed the expression of related marker genes. Embryos derived from uninjected oocytes and oocytes injected with 20 or 40 ng of scp3 MO-1 were collected at stages 9.5, 10, and 10.5 for qPCR (Fig. 3A). In scp3 morphants, the expression of endodermal markers endodermin (edd), sox17a, and mixer; pan-mesodermal marker brachyury (bra); dorsal-mesodermal markers chordin (chrd) and goosecoid (gsc); and ventral-mesodermal markers vent1, vent2, and wnt8 was down-regulated in a dose-dependent manner. In terms of ectoderm, the expression level of the epidermal marker epidermal keratin (ek) decreased. The expression of dorsal ectoderm marker sox2 was slightly down-regulated by a low dose of scp3 MO-1 but appeared to be the same as the control sample after the injection of a higher dose. lim5, a gene that specifically regulates differential cell adhesion behavior of the ectoderm but not the cell fate, was not affected (Fig. 3A). Moreover, 2-cell stage embryos were injected with scp3 MO-1 and analyzed at stage 10.5 using whole mount in situ hybridization. In scp3 morphants, the expression of edd, bra, chrd, and vent1 was drastically inhibited, but lim5 was unaffected (Fig. 3B). These results reveal an important role for SCP3 in the formation of mesendodermal and epidermal structures during Xenopus embryogenesis.
FIGURE 3.
SCP3 is required for the expression of Nodal/Activin and BMP target genes during early Xenopus embryogenesis. A, oocytes were injected with 20 or 40 ng of scp3 MO-1. After the maturation, host transfer, and fertilization, the embryos were collected at stages 9.5, 10, and 10.5 for qPCR. NC, negative control, uninjected embryos treated in the same manner as injected ones. B, 2-cell stage embryos were injected with 60 ng of std MO or scp3 MO-1 and analyzed at stage 10.5 using in situ hybridization for the indicated genes. Error bars, S.D.
SCP3 Is Required for the Expression of Nodal/Activin and BMP Target Genes in Gastrula Embryos
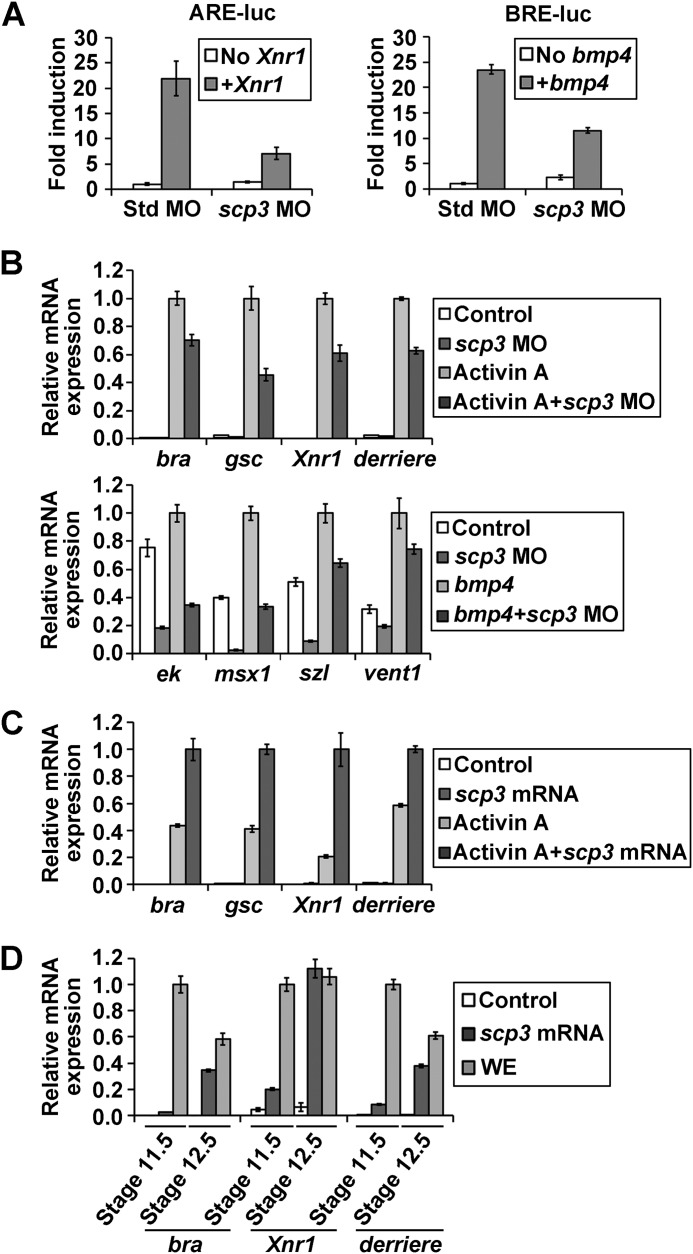
Three lines of evidence indicate that SCP3 functions via Nodal/Activin and/or BMP signals. First, the abundant vegetal distribution overlapped with the localization of early activated Smad2 and Smad1 (10). Second, the phenotype of scp3 morphants is highly similar to that of smad2, smad3, or smad4 morphants (44). Third, target genes of both Nodal/Activin (endodermal and dorsal-mesodermal markers) and BMP (ventral-mesodermal and epidermal markers) were down-regulated by scp3 MO (Fig. 3, A and B). Therefore, we investigated whether SCP3 could regulate Nodal/Activin or BMP signal in Xenopus embryos using ARE and BRE reporters. The ARE and BRE activity was stimulated by injection of mRNA encoding Xnr1 and BMP4, respectively. When scp3 MO-1 was coinjected, the expression of both Nodal and BMP reporters was severely inhibited (Fig. 4A).
FIGURE 4.
SCP3 is required for the Nodal/Activin and BMP signals. A, reporter assays with ARE and BRE. Two-cell stage embryos were injected in the equatorial region with the ARE or BRE luciferase reporter constructs (20 pg), the Renilla luciferase reporter construct (2 pg), std MO or scp3 MO-1 (60 ng), and the indicated mRNAs: Xnr1 (100 pg) and bmp4 (1 ng). Embryos were harvested at stage 10.5 and assayed for luciferase activity. B, 2-cell stage embryos were injected in the animal pole with std MO or scp3 MO-1 (60 ng). Animal caps were excised at stage 8.5 and assayed by qPCR for the expression of markers at stage 10.5. For the Activin target assay, animal cap explants were treated with 5 ng/ml Activin A, and for the BMP target assay, 2 ng of bmp4 mRNA was coinjected. C, 2-cell stage embryos were injected in the animal pole with water or 300 pg of scp3 mRNA. Animal caps were excised at stage 8.5, treated with 5 ng/ml of Activin A, and assayed by qPCR for the expression of markers at stage 10.5. D, 2-cell stage embryos were injected in the animal pole with water or 300 pg of scp3 mRNA. Animal caps were excised at stage 8.5 and assayed by qPCR for the expression of markers at the indicated stages. WE, whole embryo. Error bars, S.D.
We further carried out animal cap assays to dissect the function of SCP3 in Nodal/Activin and BMP signals. Animal caps from embryos injected with either std MO or scp3 MO-1 were excised at stage 8.5 and treated with Activin A. At stage 10.5, the caps were collected and analyzed by qPCR. The induction of mesodermal marker genes bra, gsc, Xnr1, and derriere by Activin A was significantly reduced in scp3 MO-injected animal caps (Fig. 4B, top). Similarly, scp3 MO-1 caused the down-regulation of BMP target genes ek, msx1, sizzled (szl), and vent1 with and without the stimulation of BMP4 (Fig. 4B, bottom).
Consistent with these loss-of-function results, overexpression of SCP3 by injecting its mRNA could enhance the expression of mesodermal markers induced by Activin A in stage 10.5 animal caps (Fig. 4C). Furthermore, SCP3 alone was able to trigger the expression of bra, Xnr1, and derriere in later stage animal cap cells (Fig. 4D). Taken together, these data strongly suggest that SCP3 is required for the full activation of Nodal/Activin and BMP signals in Xenopus embryos.
SCP3 Potentiates Nodal/Activin and BMP Signals by Dephosphorylating Linker Sites of R-Smads in Blastula and Gastrula Embryos
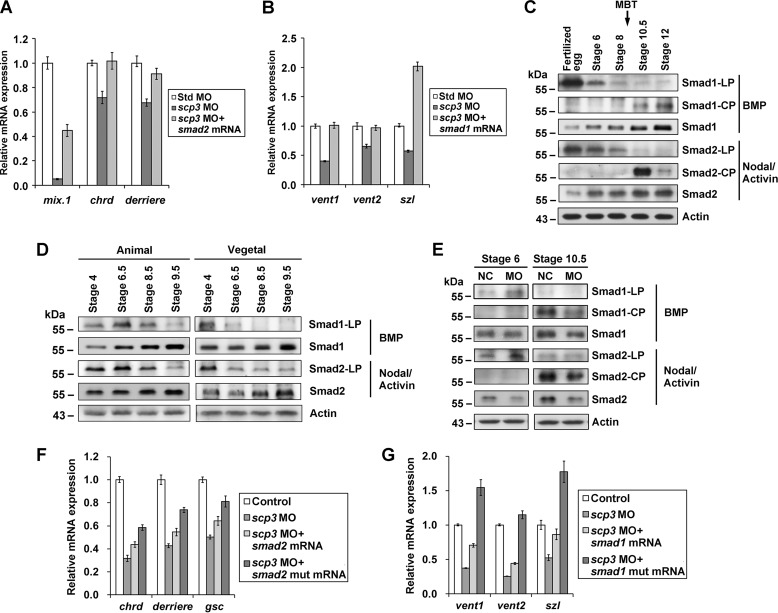
It was reported that SCPs could interact with and dephosphorylate Smad2 and Smad1 (26, 28–30); we therefore hypothesized that SCP3 regulated Nodal/Activin and BMP signals in Xenopus via R-Smads. The injection of scp3 MO-1 caused down-regulation of Nodal/Activin target genes (mix.1, chrd, and derriere) as well as BMP target genes (vent1, vent2, and szl), and this was significantly rescued by coinjection of X. laevis smad2 or smad1 mRNA, respectively (Fig. 5, A and B). These results suggest that the R-Smads are the components downstream from SCP3 along the Nodal/Activin and BMP pathways.
FIGURE 5.
SCP3 potentiates Nodal/Activin and BMP signals by dephosphorylating linker sites of R-Smads in Xenopus early embryos. A, 2-cell stage embryos were coinjected with 40 ng of std MO or scp3 MO-1 and mRNA coding for X. laevis Smad2 (2 ng). At stage 10.5, the embryos were harvested to measure the expression of indicated marker genes by qPCR. B, 2-cell stage embryos were coinjected with 40 ng of std MO or scp3 MO-1 and mRNA coding for X. laevis Smad1 (1 ng). At stage 10.5, the embryos were harvested to measure the expression of the indicated marker genes by qPCR. C, dynamics of Smad1 and Smad2 phosphorylation during the Xenopus embryogenesis. Embryos were harvested at the indicated developmental stages, and their lysates were analyzed by Western blotting using specific antibodies. LP, linker phosphorylation; CP, C-terminal phosphorylation. Nodal/Activin- or BMP-responsive Smads are indicted on the right. D, animal and vegetal cells were separated and collected at the indicated stages, and their lysates were analyzed by Western blotting using specific antibodies. E, oocytes were uninjected or injected with 80 ng of scp3 MO-1. After the maturation, host transfer, and fertilization, the embryos were collected at indicated stages and analyzed using specific antibodies. NC, negative control, uninjected embryos treated the same as injected ones. F, 2-cell stage embryos were coinjected with 60 ng of std MO or scp3 MO-1 and 2 ng of mRNA coding for wild-type X. laevis Smad2 or mutant Smad2 (S245A/S250A/S255A). At stage 10, the embryos were harvested to measure the expression of indicated marker genes by qPCR. G, 2-cell stage embryos were coinjected with 60 ng of std MO or scp3 MO-1 and 1.5 ng of mRNA coding for wild-type X. laevis Smad1 or mutant Smad1 (S205A). At stage 10, the embryos were harvested to measure the expression of the indicated marker genes by qPCR. Error bars, S.D.
During Xenopus embryogenesis, the Nodal/Activin and BMP pathways are activated by zygotically expressed ligands, and this happens shortly after the MBT. Consistently, the phosphorylation of R-Smads at C-terminal sites, as an indicator of signal activation, was detected after MBT (Fig. 5C) (10). In contrast, in fertilized eggs, the Smad1 was heavily phosphorylated at Ser-205 in the linker region (corresponding to Ser-206 of human Smad1), and phosphorylation on this site is believed to be inhibitory (15, 45). Extensive phosphorylation of Smad2 at Ser-245/250/255 was also detected (Fig. 5C), and these sites are also considered inhibitory upon phosphorylation (16, 17). More importantly, this linker phosphorylation was gradually reduced when the embryos were approaching the MBT, and this was accompanied by an increase of R-Smads protein level (Fig. 5C). These results suggested that in fertilized eggs, R-Smads are contained by inhibitory linker phosphorylation. Along with cleavage, this phosphorylation is gradually removed, and R-Smad proteins accumulate.
Next, we addressed whether the reduction of these inhibitory phosphorylations is spatially different. Animal and vegetal regions at stages 4, 6.5, 8.5, and 9.5 were collected, and the phosphorylation level of Smad2 and Smad1 in the linker regions was verified. The results indicated that, from stage 4 to stage 6.5, the phosphorylation level of both Smad2 and Smad1 decreased significantly in vegetal blastomeres but not in animal blastomeres (Fig. 5D). From stage 6.5 to stage 9.5, the phosphorylation level in animal blastomeres also decreased gradually, resulting in the overall low level of phosphorylation in R-Smad linker regions in gastrula embryos (Fig. 5C). This result suggested that some asymmetrically localized phosphatase(s) may lead to asymmetrical dephosphorylation.
To test whether SCP3 plays a role in dephosphorylating these linker sites of R-Smads, we injected scp3 MO-1 into oocytes to knock down the maternal SCP3 protein and harvested embryos at stages 6 and 10.5 for immunoblotting using antibodies that specifically recognize the phosphorylated linker sites or C-terminal tails of Smad1 or Smad2. Consistent with the reduced Nodal/Activin and BMP signals and the down-regulation of their targets, in stage 10.5 scp3 morphants, the phosphorylation of the C-terminal tails of R-Smads was clearly weakened, and this was co-related with reduced protein level of both Smad2 and Smad1 (Fig. 5E). At stage 6, however, the phosphorylation at linker sites of both Smads was significantly elevated, although the protein level of Smads was reduced in scp3 morphants. These results indicate that knockdown of maternal SCP3 attenuated the dephosphorylation of linker regions in Smad1/2 and reduced the accumulation of total Smad1/2 during cleavage stages. This is consistent with the above finding that removal of maternal SCP3 reduced embryos' response to the upcoming zygotic Nodal/Activin and BMP signals after MBT.
Furthermore, to confirm that SCP3 can potentiate Nodal/Activin and BMP signals by dephosphorylating linker sites of R-Smads in embryos, we constructed R-Smad phosphorylation-resistant mutants. The mutant Smad2 differs by the three phosphorylation sites in the linker region (S245A/S250A/S255A) from Smad2, and the mutant Smad1 differs by a single amino acid (S205A) from Smad1. mRNAs encoding mutant R-Smads were coinjected with scp3 MO-1, and their activity was compared with that of wild-type R-Smads. The results indicated that the phosphorylation-deficient Smad2 caused more significant rescue of target genes in comparison with the wild-type Smad2 (Fig. 5F). Similarly, phosphorylation-insensitive Smad1 showed a better rescue effect (Fig. 5G). Taken together, these results strongly suggest that the vegetally enriched maternal SCP3 gradually removes the linker phosphorylation of both R-Smads before MBT and thus leads to preparation for the activation by the upcoming ligands shortly after MBT.
SCP3 Potentiates Nodal/Activin and BMP Signals in Human Cells
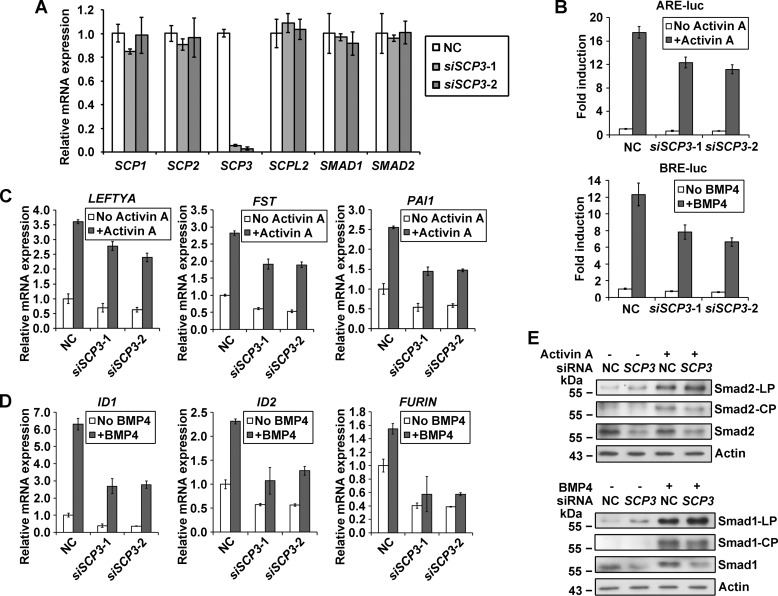
To further test the role of SCP3 on TGFβ signaling in human cells, we performed a series of loss-of-function analyses by introducing siRNA oligonucleotides targeting SCP3 in Hep3B cells (Fig. 6A). In reporter assays, transfection with SCP3 siRNA decreased the ability of Activin A and BMP4 to induce ARE reporter and BRE reporter expression, respectively (Fig. 6B). In target gene tests, no matter whether the cells were treated or untreated with ligands, SCP3 siRNA caused a marked decrease in the expression of Nodal/Activin target genes (LEFTYA, FST, and PAI1) and BMP target genes (ID1, ID2, and FURIN) (Fig. 6, C and D). At the phosphorylation level, transfection of SCP3 siRNA resulted in accumulation of linker-phosphorylated Smad2 and Smad1 both with and without the treatment of ligands (Fig. 6E). SCP3 siRNA also caused a significant decrease in the amount of total Smad2 and Smad1. The ligand-induced C-terminal phosphorylation, however, was weaker in SCP3 siRNA-transfected cells in comparison with that in control siRNA-transfected cells (Fig. 6E). qPCR results confirmed that the mRNA level of both SMAD1 and SMAD2 was not affected by siRNAs against SCP3 (Fig. 6A), and the down-regulation of R-Smad proteins is probably due to linker phosphorylation-mediated degradation (15, 16). Thus, in agreement with the results in Xenopus, the data from human cells revealed that SCP3 plays a positive role in both Nodal/Activin and BMP signals by dephosphorylating linker sites of R-Smads.
FIGURE 6.
SCP3 positively regulates Nodal/Activin and BMP signals in Hep3B cells. A, Hep3B cells were transfected with 50 nm NC or SCP3 siRNAs, and 40 h later, cells were collected for qPCR. B, reporter assays in Hep3B cells. The negative control (NC) siRNA or human SCP3 siRNAs (50 nm) were co-transfected with ARE or BRE luciferase reporter constructs, and 12 h later, cells were left untreated or treated with Activin A or BMP4 for 24 h and then collected for reporter activity measurement. C, cells were transfected with 50 nm NC or SCP3 siRNAs, and 40 h later, cells were treated with Activin A for 4 h. mRNA levels of Activin-induced genes were determined by qPCR. D, cells were transfected with 50 nm NC or SCP3 siRNAs, and 40 h later, cells were treated with BMP4 for 4 h. mRNA levels of BMP-induced genes were determined by qPCR. E, cells were transfected with 100 nm NC or SCP3 siRNA-1 and cultured for 48 h. After treatment with Activin A or BMP4 for 1 h, the cells were collected for Western blotting analysis. Error bars, S.D.
Discussion
SCP3 Positively Regulates TGFβ Signaling by Dephosphorylating Linker Regions of R-Smads
As increasing evidence indicates the importance of linker phosphorylation in R-Smads, protein phosphatases have been identified to oppositely control this dynamic process (14, 46). In the context of the cleavage Xenopus embryo, we propose that SCP3 plays positive roles in Nodal/Activin and BMP signals. To draw this conclusion, we rely on the following results. First, the phenotype of scp3 morphants is highly similar to that of smad4 morphants. Second, MO-mediated knockdown of SCP3 leads to down-regulation of Nodal/Activin and BMP target genes, which can be rescued by smad2 and smad1 mRNA, respectively. For some dorsal marker genes that are oppositely regulated by Nodal/Activin and BMP signals, the change of their expression is probably due to a combined deficiency of both pathways. Third, in reporter assays, scp3 MO decreases the expression of ARE and BRE reporters. Fourth, in animal cap assays, the induction of mesodermal marker genes and expression of BMP target genes are inhibited by scp3 MO. Fifth, injection of scp3 MO reduces the levels of C-terminal phosphorylation of both Smad2 and Smad1 in gastrula embryos. In our study, SCP3 enhances the Nodal/Activin and BMP signals at least partially by releasing the inhibitory phosphorylation at Ser-245/250/255 in Smad2 and Ser-205 in Smad1, respectively. Furthermore, we found that knocking down SCP3 reduced the total amount of Smad2 and Smad1. This is consistent with the previous reports showing that the phosphorylation of R-Smads at some linker sites triggers the recognition and polyubiquitination by E3 ligase and results in the degradation of R-Smads (15, 16).
So far, only SCPs have been identified as phosphatases of R-Smad linker sites (14, 46). In Nodal/Activin signaling, SCP3 enhances TGFβ signaling by dephosphorylating Smad2 in the linker region, just as the SCP1 and SCP2 do (28, 29). In BMP signaling, SCP1 and SCP2 were suggested to dephosphorylate Smad1 at both linker and C-terminal sites, resulting in the attenuation of BMP pathway (26, 28, 29). Our results, from both Xenopus embryos and human cells, confirmed that SCP3 is involved in dephosphorylating the linker sites of Smad1. However, whether SCP3 functions to remove the C-terminal phosphorylation of Smad1 is uncertain because the total Smad1 protein level was reduced when SCP3 was down-regulated. Nevertheless, the net outcome of SCP3 knockdown is a lower BMP signaling, suggesting a rather positive role of SCP3. Interestingly, a recent study suggested that SCPL2 specifically dephosphorylates the C terminus of Smad1/5/8 and therefore attenuates the BMP signaling (30).
Maternal SCP3 Ensures Zygotic TGFβ Signaling for Germ Layer Formation in Xenopus Embryos
In Xenopus embryos, the Nodal/Activin signal is strongly activated shortly after MBT, when zygotic transcription is initiated (10, 12). The zygotically expressed activin-like ligands, including Xnr1, -2, -4, -5, -6, and derriere, stimulate the signal and specify meso-endodermal cell fates (1, 2, 4). The maternally expressed transcriptional factor VegT activates the zygotic transcription of these Activin-like ligands, and ectopic expression of these ligands can rescue mesoderm induction following depletion of maternal VegT, illustrating a relatively linear cascade (4, 47, 48). Despite the timely and regional expression of ligands, however, the competence of the responding cells may also be a determinant of early patterning.
Competence represents the degree to which a cell responds to an external signal (49). Numerous studies indicate that cleavage Xenopus embryos lack competence to the Nodal/Activin and BMP signals. Using heterochronic Nieuwkoop recombinants of animal-vegetal explants, Jones and Woodland (50) found that only from stage 5 onward do vegetal cells become competent to the Nodal/Activin signal that induces mesoderm. An elegant experiment using BiFC (bimolecular fluorescence complementation) technology in Xenopus embryos demonstrated that treatment of Activin at any time before MBT does not result in nuclear accumulation of Smad2-Smad4 complexes (51). Consistently, endogenous Smad2 phosphorylation in the C-terminal region can be detected only after MBT (10). In terms of the BMP signal, although some BMPs, such as BMP2 and BMP7, are maternally expressed, the C-terminally phosphorylated Smad1 is not detected in embryos until stage 8 (1, 11, 12, 52). Ectopic expression of BMP ligands cannot stimulate Smad1 phosphorylation at stage 7 (10). Furthermore, this timely Smad1 activation is independent of zygotic transcription because polymerase II inhibitor α-amanitin does not prevent the appearance of Smad1 C-terminal phosphorylation in response to BMP at MBT (10, 53).
In this work, we found that the linker regions of R-Smads are heavily phosphorylated, as a signaling-locked status, in fertilized eggs. Throughout the cleavage of blastomeres, the maternally deposited SCP3 functions to remove this inhibitory phosphorylation gradually. These observations suggest that the inhibitory linker phosphorylation in R-Smads may serve as a maternal blocker of signaling activation. The gradual dephosphorylation of these inhibitory linker regions by SCP3 is probably a mechanism to ensure the response to upcoming ligands. As such, shortly after the MBT, a good supply of responsive R-Smad allows for the rapid activation of TGFβ signaling.
Spatially, SCP3 is enriched vegetally, consistent with the vegetally enriched active R-Smads at stage 9.5–10, during which Nodal/Activin and BMP signals start to be highly activated (1, 7, 10, 12). Compared with SCP3 in the vegetal part, the lower level of SCP3 in the animal part causes weaker but necessary dephosphorylation effects, possibly contributing to activation of R-Smads not restricted in the vegetal region, such as activated Smad1 in the ventral side across the animal-vegetal axis during gastrulation (10–12). Interestingly, we do not observe any difference in SCP3 expression level across the dorsal-ventral axis, indicating a general function of SCP3 for dorsally enriched Nodal/Activin signal and ventrally enriched BMP signal in gastrula embryos.
Taken together, we have illustrated a phosphatase-mediated mechanism in the regulation of cell competence, occurring at the predetermined time and region of Xenopus embryogenesis. Our findings suggest that vegetally enriched maternal SCP3 may cause a spatially restricted linker-dephosphorylation of R-Smads, ensuring a prepared status for their activation by the asymmetrically produced ligands. Challenges for the future will be to understand what component or pathway is the trigger to activate SCP3. It is possible to speculate that this activation starts from fertilization, indicating the elaborate and forward-looking modulation of life.
Author Contributions
W. W., G. S., and Q. T. designed the study; G. S., Z. G., H. S., and Y. F. carried out most experiments and analyzed the data; X. Y. and Y. G. C. carried out experiments in human cell line; M. Q. Z., Z. H., and X. M. performed the RNA-seq and related bioinformatics analysis; Q. T. and Z. M. performed the host transfer experiments; W. W. and G. S wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Drs. Fuchou Tang and Pengliang Xu from Peking University and Liyang Liu from Tsinghua University for technical assistance with RNA-seq.
This work was supported by MOST Grant 2011CB943803 and National Natural Science Foundation of China (NSFC) Grant 31221064 (to W. W.), MOST Grant 2011CB943800 (to Q. T.), and NSFC Grant 91019016 and National Basic Research Foundation of China (NBRPC) Grant 2012CB316503 (to M. Q. Z.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables S1 and S2.
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE68972).
- MBT
- midblastula transition
- BMP
- bone morphogenetic protein
- R-Smad
- receptor-regulated Smad
- RNA-seq
- RNA sequencing
- ARE
- activin-responsive element
- BRE
- BMP-responsive element
- MO
- morpholino
- std MO
- standard control MO
- qPCR
- quantitative PCR.
References
- 1. Heasman J. (2006) Maternal determinants of embryonic cell fate. Semin. Cell Dev. Biol. 17, 93–98 [DOI] [PubMed] [Google Scholar]
- 2. Loose M., Patient R. (2004) A genetic regulatory network for Xenopus mesendoderm formation. Dev. Biol. 271, 467–478 [DOI] [PubMed] [Google Scholar]
- 3. Taverner N. V., Kofron M., Shin Y., Kabitschke C., Gilchrist M. J., Wylie C., Cho K. W., Heasman J., Smith J. C. (2005) Microarray-based identification of VegT targets in Xenopus. Mech. Dev. 122, 333–354 [DOI] [PubMed] [Google Scholar]
- 4. Zhang J., Houston D. W., King M. L., Payne C., Wylie C., Heasman J. (1998) The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell 94, 515–524 [DOI] [PubMed] [Google Scholar]
- 5. Wardle F. C., Smith J. C. (2006) Transcriptional regulation of mesendoderm formation in Xenopus. Semin. Cell Dev. Biol. 17, 99–109 [DOI] [PubMed] [Google Scholar]
- 6. Dale L., Jones C. M. (1999) BMP signalling in early Xenopus development. BioEssays 21, 751–760 [DOI] [PubMed] [Google Scholar]
- 7. Hill C. S. (2001) TGF-β signalling pathways in early Xenopus development. Curr. Opin. Genet. Dev. 11, 533–540 [DOI] [PubMed] [Google Scholar]
- 8. Bandyopadhyay A., Yadav P. S., Prashar P. (2013) BMP signaling in development and diseases: a pharmacological perspective. Biochem. Pharmacol. 85, 857–864 [DOI] [PubMed] [Google Scholar]
- 9. Massagué J. (2012) TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Faure S., Lee M. A., Keller T., ten Dijke P., Whitman M. (2000) Endogenous patterns of TGFβ superfamily signaling during early Xenopus development. Development 127, 2917–2931 [DOI] [PubMed] [Google Scholar]
- 11. Kurata T., Nakabayashi J., Yamamoto T. S., Mochii M., Ueno N. (2001) Visualization of endogenous BMP signaling during Xenopus development. Differentiation 67, 33–40 [DOI] [PubMed] [Google Scholar]
- 12. Schohl A., Fagotto F. (2002) β-Catenin, MAPK and Smad signaling during early Xenopus development. Development 129, 37–52 [DOI] [PubMed] [Google Scholar]
- 13. Kamato D., Burch M. L., Piva T. J., Rezaei H. B., Rostam M. A., Xu S., Zheng W., Little P. J., Osman N. (2013) Transforming growth factor-β signalling: role and consequences of Smad linker region phosphorylation. Cell. Signal. 25, 2017–2024 [DOI] [PubMed] [Google Scholar]
- 14. Liu T., Feng X. H. (2010) Regulation of TGF-β signalling by protein phosphatases. Biochem. J. 430, 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sapkota G., Alarcón C., Spagnoli F. M., Brivanlou A. H., Massagué J. (2007) Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol. Cell 25, 441–454 [DOI] [PubMed] [Google Scholar]
- 16. Gao S., Alarcón C., Sapkota G., Rahman S., Chen P. Y., Goerner N., Macias M. J., Erdjument-Bromage H., Tempst P., Massagué J. (2009) Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-β signaling. Mol. Cell 36, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kretzschmar M., Doody J., Timokhina I., Massagué J. (1999) A mechanism of repression of TGFβ/Smad signaling by oncogenic Ras. Genes Dev. 13, 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matsuura I., Denissova N. G., Wang G., He D., Long J., Liu F. (2004) Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature 430, 226–231 [DOI] [PubMed] [Google Scholar]
- 19. Kamaraju A. K., Roberts A. B. (2005) Role of Rho/ROCK and p38 MAP kinase pathways in transforming growth factor-β-mediated Smad-dependent growth inhibition of human breast carcinoma cells in vivo. J. Biol. Chem. 280, 1024–1036 [DOI] [PubMed] [Google Scholar]
- 20. Mori S., Matsuzaki K., Yoshida K., Furukawa F., Tahashi Y., Yamagata H., Sekimoto G., Seki T., Matsui H., Nishizawa M., Fujisawa J., Okazaki K. (2004) TGF-β and HGF transmit the signals through JNK-dependent Smad2/3 phosphorylation at the linker regions. Oncogene 23, 7416–7429 [DOI] [PubMed] [Google Scholar]
- 21. Engel M. E., McDonnell M. A., Law B. K., Moses H. L. (1999) Interdependent SMAD and JNK signaling in transforming growth factor-β-mediated transcription. J. Biol. Chem. 274, 37413–37420 [DOI] [PubMed] [Google Scholar]
- 22. Pera E. M., Ikeda A., Eivers E., De Robertis E. M. (2003) Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev. 17, 3023–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fuentealba L. C., Eivers E., Ikeda A., Hurtado C., Kuroda H., Pera E. M., De Robertis E. M. (2007) Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell 131, 980–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grimm O. H., Gurdon J. B. (2002) Nuclear exclusion of Smad2 is a mechanism leading to loss of competence. Nat. Cell Biol. 4, 519–522 [DOI] [PubMed] [Google Scholar]
- 25. Yeo M., Lin P. S., Dahmus M. E., Gill G. N. (2003) A novel RNA polymerase II C-terminal domain phosphatase that preferentially dephosphorylates serine 5. J. Biol. Chem. 278, 26078–26085 [DOI] [PubMed] [Google Scholar]
- 26. Knockaert M., Sapkota G., Alarcón C., Massagué J., Brivanlou A. H. (2006) Unique players in the BMP pathway: small C-terminal domain phosphatases dephosphorylate Smad1 to attenuate BMP signaling. Proc. Natl. Acad. Sci. U.S.A. 103, 11940–11945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Archambault J., Pan G., Dahmus G. K., Cartier M., Marshall N., Zhang S., Dahmus M. E., Greenblatt J. (1998) FCP1, the RAP74-interacting subunit of a human protein phosphatase that dephosphorylates the carboxyl-terminal domain of RNA polymerase IIO. J. Biol. Chem. 273, 27593–27601 [DOI] [PubMed] [Google Scholar]
- 28. Sapkota G., Knockaert M., Alarcón C., Montalvo E., Brivanlou A. H., Massagué J. (2006) Dephosphorylation of the linker regions of Smad1 and Smad2/3 by small C-terminal domain phosphatases has distinct outcomes for bone morphogenetic protein and transforming growth factor-β pathways. J. Biol. Chem. 281, 40412–40419 [DOI] [PubMed] [Google Scholar]
- 29. Wrighton K. H., Willis D., Long J., Liu F., Lin X., Feng X. H. (2006) Small C-terminal domain phosphatases dephosphorylate the regulatory linker regions of Smad2 and Smad3 to enhance transforming growth factor-β signaling. J. Biol. Chem. 281, 38365–38375 [DOI] [PubMed] [Google Scholar]
- 30. Zhao Y., Xiao M., Sun B., Zhang Z., Shen T., Duan X., Yu P. B., Feng X. H., Lin X. (2014) C-terminal domain (CTD) small phosphatase-like 2 modulates the canonical bone morphogenetic protein (BMP) signaling and mesenchymal differentiation via Smad dephosphorylation. J. Biol. Chem. 289, 26441–26450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kataoka K., Tazaki A., Kitayama A., Ueno N., Watanabe K., Mochii M. (2005) Identification of asymmetrically localized transcripts along the animal-vegetal axis of the Xenopus egg. Dev. Growth Differ. 47, 511–521 [DOI] [PubMed] [Google Scholar]
- 32. Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Anders S., Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hata A., Seoane J., Lagna G., Montalvo E., Hemmati-Brivanlou A., Massagué J. (2000) OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell 100, 229–240 [DOI] [PubMed] [Google Scholar]
- 35. Huang H. C., Murtaugh L. C., Vize P. D., Whitman M. (1995) Identification of a potential regulator of early transcriptional responses to mesoderm inducers in the frog embryo. EMBO J. 14, 5965–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wylie C., Kofron M., Payne C., Anderson R., Hosobuchi M., Joseph E., Heasman J. (1996) Maternal β-catenin establishes a “dorsal signal” in early Xenopus embryos. Development 122, 2987–2996 [DOI] [PubMed] [Google Scholar]
- 37. Heasman J., Holwill S., Wylie C. C. (1991) Fertilization of cultured Xenopus oocytes and use in studies of maternally inherited molecules. Methods Cell Biol. 36, 213–230 [DOI] [PubMed] [Google Scholar]
- 38. Olson D. J., Hulstrand A. M., Houston D. W. (2012) Maternal mRNA knock-down studies: antisense experiments using the host-transfer technique in Xenopus laevis and Xenopus tropicalis. Methods Mol. Biol. 917, 167–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y., Fu Y., Gao L., Zhu G., Liang J., Gao C., Huang B., Fenger U., Niehrs C., Chen Y. G., Wu W. (2010) Xenopus skip modulates Wnt/β-catenin signaling and functions in neural crest induction. J. Biol. Chem. 285, 10890–10901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Harland R. M. (1991) In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 36, 685–695 [DOI] [PubMed] [Google Scholar]
- 41. Sudou N., Yamamoto S., Ogino H., Taira M. (2012) Dynamic in vivo binding of transcription factors to cis-regulatory modules of cer and gsc in the stepwise formation of the Spemann-Mangold organizer. Development 139, 1651–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yan X., Lin Z., Chen F., Zhao X., Chen H., Ning Y., Chen Y. G. (2009) Human BAMBI cooperates with Smad7 to inhibit transforming growth factor-β signaling. J. Biol. Chem. 284, 30097–30104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Claussen M., Horvay K., Pieler T. (2004) Evidence for overlapping, but not identical, protein machineries operating in vegetal RNA localization along early and late pathways in Xenopus oocytes. Development 131, 4263–4273 [DOI] [PubMed] [Google Scholar]
- 44. Rana A. A., Collart C., Gilchrist M. J., Smith J. C. (2006) Defining synphenotype groups in Xenopus tropicalis by use of antisense morpholino oligonucleotides. PLoS Genet. 2, e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kretzschmar M., Doody J., Massagué J. (1997) Opposing BMP and EGF signalling pathways converge on the TGF-β family mediator Smad1. Nature 389, 618–622 [DOI] [PubMed] [Google Scholar]
- 46. Bruce D. L., Sapkota G. P. (2012) Phosphatases in SMAD regulation. FEBS Lett. 586, 1897–1905 [DOI] [PubMed] [Google Scholar]
- 47. Clements D., Friday R. V., Woodland H. R. (1999) Mode of action of VegT in mesoderm and endoderm formation. Development 126, 4903–4911 [DOI] [PubMed] [Google Scholar]
- 48. Kofron M., Demel T., Xanthos J., Lohr J., Sun B., Sive H., Osada S., Wright C., Wylie C., Heasman J. (1999) Mesoderm induction in Xenopus is a zygotic event regulated by maternal VegT via TGFβ growth factors. Development 126, 5759–5770 [DOI] [PubMed] [Google Scholar]
- 49. Kaneko K., Sato K., Michiue T., Okabayashi K., Ohnuma K., Danno H., Asashima M. (2008) Developmental potential for morphogenesis in vivo and in vitro. J. Exp. Zool. B Mol. Dev. Evol. 310, 492–503 [DOI] [PubMed] [Google Scholar]
- 50. Jones E. A., Woodland H. R. (1987) The development of animal cap cells in Xenopus: a measure of the start of animal cap competence to form mesoderm. Development 101, 557–563 [Google Scholar]
- 51. Saka Y., Hagemann A. I., Piepenburg O., Smith J. C. (2007) Nuclear accumulation of Smad complexes occurs only after the midblastula transition in Xenopus. Development 134, 4209–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fritz B. R., Sheets M. D. (2001) Regulation of the mRNAs encoding proteins of the BMP signaling pathway during the maternal stages of Xenopus development. Dev. Biol. 236, 230–243 [DOI] [PubMed] [Google Scholar]
- 53. Newport J., Kirschner M. (1982) A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell 30, 675–686 [DOI] [PubMed] [Google Scholar]
- 54. Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.