Background: Aurora kinases show different localizations and play distinct roles, yet the mechanisms remain largely unknown.
Results: Different localization leads to functional divergence of the Auroras and their N termini also contribute to the localization.
Conclusion: Both N/C termini of Aurora A/B contribute to their spatial compartmentalization and function.
Significance: Functional divergence of Aurora kinases is largely determined by their localizations through binding with partners/substrates.
Keywords: cell cycle, checkpoint control, mitotic spindle, protein kinase, protein phosphorylation, TPX2, cell cycle, Aurora kinase, TPX2, INCENP, spindle assembly.
Abstract
Aurora kinase A and B share great similarity in sequences, structures, and phosphorylation motif, yet they show different localizations and play distinct crucial roles. The factors that determine such differences are largely unknown. Here we targeted Aurora A to the localization of Aurora B and found that Aurora A phosphorylates the substrate of Aurora B and substitutes its function in spindle checkpoint. In return, the centrosome targeting of Aurora B substitutes the function of Aurora A in the mitotic entry. Expressing the chimera proteins of the Auroras with exchanged N termini in cells indicates that the divergent N termini are also important for their spatiotemporal localizations and functions. Collectively, we demonstrate that functional divergence of Aurora kinases is determined by spatial compartmentalization, and their divergent N termini also contribute to their spatial and functional differentiation.
Introduction
The Aurora kinases belong to the serine/threonine kinase families and are essential for the cell cycle control in the eukaryotes (1, 2). Many low species have only one Aurora, whereas the mammals have at least three, Aurora A, B, and C, among which Aurora C resembles Aurora B but regulates meiosis and mitosis during early development (2–4). Aurora A and B display different localizations and functions during the cell cycle. Aurora A is located to the centrosomes throughout the cell cycle, spreads on the spindle microtubules in metaphase, and is relocated to the central spindle in anaphase and telophase (5–8). In contrast, Aurora B is mainly located on the centromere in early mitosis, where it serves as a component of the chromosome passenger complex, and on the midzone/midbody during the cytokinesis (3). In accordance with the spatiotemporal localization divergence, Aurora A and B perform distinct functions. Aurora A is required for centrosome maturation, mitotic entry, and centrosome separation, whereas Aurora B mainly regulates spindle assembly checkpoint, kinetochore attachment, and cytokinesis (3, 9–11).
Both molecules of Aurora A and B have a divergent N terminus and a conserved catalytic domain-containing C terminus in sequence (12–14). Aurora B combines with INCENP, Survivin, and Borealin to form the chromosome passenger complex, which is required for activation and specialized localization of Aurora B (3). Aurora A interacts with a microtubule-associated protein TPX2 that not only regulates the localization but also the activation of Aurora A on the spindle in prometaphase and metaphase (15). Aurora A also interacts with a variety of binding partners/substrates including centrosome-localized Ajuba, Bora, and PAK1 and functions on the centrosomes (10, 16, 17).
In the known cases of yeast (Saccharomyces cerevisiae and Schizosaccharomyces pombe), mycetozoa (Dictyostelium discoideum), primitive deuterostome (starfish, ascidian, and urchin), and so on, the only Aurora kinase shows the localization and function of both Aurora A and B of high species (18–20). Phylogenetic analysis shows that both Aurora A and B are likely evolved from a primitive bifunctional Aurora kinase, although how the differentiation of their Auroras is formed remains largely unknown (21). It is well accepted that duplication and divergence are the primary means by which new proteins and pathways are created. There are many factors including the substrate specificity, the specified binding of proteins, and the spatial mechanisms like the scaffolds, the regulation of reactions, and the formation of macromolecular complexes that contribute to the evolution and divergence of the proteins (22, 23). Generally, fully conserved positions of proteins may confer the proteins' common functional features, whereas less conserved specificity-determining positions are related to their divergent functions. It has been found that both Aurora A and B may have similar optimal phosphorylation motifs (24) and that they may share some common substrates such as CENP-E, MCAK, Kif2, and RASSF1A on the spindles in cells (25–29). However, the paradox is why Aurora A and B show very different localizations and play distinct roles during the cell cycle with such a similarity in sequence and phosphorylation motif affinity.
In this study we set out to determine how Aurora A and B are spatially compartmentalized for their functional divergence. We found that, in addition to the catalytic conservative C termini, the divergent N termini also regulate their localization, and the resultant spatial compartmentalization further leads to functional divergence.
Experimental Procedures
Molecular Cloning
For construction of fusion proteins, GFP-H2B-Aurora A, GFP-CENPB-Aurora A, GFP-Hec1-Aurora A, GFP-PLK4-Aurora B, wild-type Aurora A, or Aurora B was inserted into pEGFP-C1 plasmid. Full-length H2B, CENP-B aa4 1–158 centromere targeting sequence, and full-length Hec1 were cloned by RT-PCR from HeLa cell lysate and inserted to the N terminus of Aurora A sequence. PLK4 aa 570–820 centrosome-targeting sequence was inserted in the N-terminus of Aurora B sequence. Kinase dead mutation was generated by point mutation PCR. N-terminal and C-terminal truncation mutant of Aurora A and Aurora B was cloned to pEGFP-C1 plasmid. And fusion proteins GFP-Aurora A-B and GFP-Aurora B-A were generated as in Fig. 1 by overlapping PCR.
FIGURE 1.
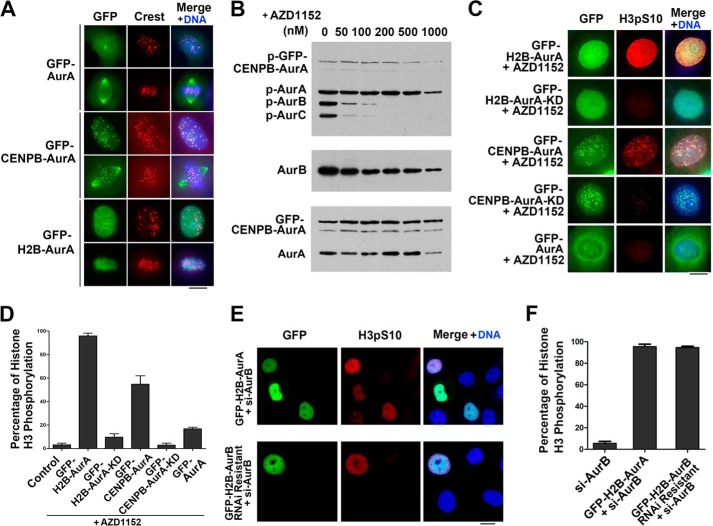
Chromatin-localized Aurora A phosphorylates the Aurora B substrate histone H3 in vivo. A, immunofluorescence labeling of HeLa cells transfected with GFP-Aurora A (AurA), GFP-CENPB-Aurora A, or GFP-H2B-Aurora A with a CREST antibody. The DNA was stained by DAPI. Note that the wild-type Aurora A was localized to the centrosome in interphase, whereas GFP-CENPB-Aurora A was situated to the centromere and GFP-H2B-Aurora A to the chromatin/chromosome both in interphase and mitosis. Scale bar, 10 μm. B, HeLa cells transfected with GFP-CENPB-Aurora A were synchronized to prometaphase and treated with different concentrations of Aurora B inhibitor AZD1152 for 1 h. Cell extracts were subjected to Western blot analysis with antibodies to phospho-Aurora A/-B/-C, Aurora B (AurB) and Aurora A. Note that AZD1152 at 200 nm and above totally inhibited the activation of Aurora B and Aurora C (AurC) indicated by autophosphorylation, whereas 200 nm AZD1152 did not inhibit the activation of GFP-CENPB-Aurora A. C, histone H3 Ser(P)-10 immunofluorescence labeling of HeLa cells transfected with GFP-H2B-Aurora A, GFP-H2B-Aurora A-KD, GFP-CENPB-Aurora A, GFP-CENPB-Aurora A-KD, or GFP-Aurora A followed by treatment with 200 nm AZD1152 for 1 h. The DNA was stained with DAPI. Note that GFP-H2B-Aurora A and GFP-CENPB-Aurora A expression resulted in phosphorylation of, and colocalized with histone H3 Ser-10 (H3pS10), whereas GFP-H2B-Aurora A-KD, GFP-CENPB-Aurora A-KD, and GFP-Aurora A did not induce histone H3 Ser-10 phosphorylation. Scale bar, 10 μm. D, quantitative characterization of HeLa cells treated as in C. Positive staining cells are defined as strongly staining with clear edge. Cells with no stain or weak vague staining are not counted as positive staining cells. Bars represent the percentage of the cells with histone H3 Ser-10 phosphorylation. Each data point represents 3 independent experiments with each measuring 100 cells, and error bars indicate S.D. E, histone H3 Ser(P)-10 immunofluorescence labeling of HeLa cells co-transfected with Aurora B siRNA to knock down endogenous Aurora B and GFP-H2B-Aurora A or RNAi resistant GFP-H2B-Aurora B. Note that both GFP-H2B-Aurora B and GFP-H2B-Aurora A phosphorylated histone H3 Ser-10 (H3pS10). F, quantitative characterization of HeLa cells treated as in E. Bars represented the percentage of the cells with histone H3 Ser-10 phosphorylation. Each data point represents three independent experiments with each measuring 100 cells, and error bars indicate S.D. Scale bar, 10 μm.
Cell Culture and Drug Treatment
HeLa cells were cultured at 37 °C under 5% CO2 in DMEM supplemented with 10% fetal bovine serum. To synchronize HeLa cell to mitosis, the cell cycle was blocked by double thymidine treatment to G1-S phase and then released for 10 h. To generate mitosis cells with misaligned chromosomes, HeLa was released from single thymidine block and then treated with S-trityl-l-cysteine (STLC) for 10 h. Kinase activity of Aurora A is inhibited by 50 nm MLN8237 (Alisertib). Kinase activity of Aurora B is inhibited by 200 nm AZD1152.
Immunofluorescence Microscopy
Cells grown on coverslips were fixed with precooled methanol for 5 min. Then the coverslips were incubated with primary antibody diluted in 3% BSA overnight at 4°C. After washing 3 times in PBS, coverslips were incubated with secondary antibody diluted in 3% BSA for 1 h at room temperature. The coverslips were further washed in PBS and mounted with Mowiol added with 1 mg/ml DAPI for DNA staining. The antibodies used in immunofluorescence microscopy include anti-centromere antibody from patients with CREST syndrome (MBL), anti-H3pS10 (Cell Signaling), anti-BubR1 (Abcam), anti-γ-tubulin (Sigma), anti-cyclin B1 (Santa Cruz).
Western Blotting
Cell lysates in sampling buffer were loaded onto 10% SDS-PAGE gel for electrophoresis and transferred onto nitrocellulose membranes. The membranes were blocked in TTBS (TBS plus 0.1% Tween 20) containing 1.5% milk for 1 h at room temperature and incubated with primary antibody diluted in TTBS/milk overnight at 4°C. The membranes were washed 3 times in TTBS and blocked in TTBS/milk for 1 h, incubated with HRP-conjugated secondary antibody diluted in TTBS/milk for 1 h at room temperature, and then washed 3 times. Finally, the blots were developed by chemiluminescence. The antibodies used in Western blotting include anti-phospho-Aurora A/Aurora B/Aurora-C (Cell Signaling), anti-Aurora B (BD Biosciences), anti-Aurora A (produced in our laboratory), and anti-GAPDH (Proteintech).
RNA Interference
For siRNA treatment, duplexed siRNA were introduced using Lipofectamine 2000. Cells were processed for immunofluorescence microscopy 56 h after transfection. siRNAs used include si-Aurora A (5′-GCAGAGAACTGCTACTTAT-3′) and si-Aurora B (5′-GGATCTACTTGATTCTAGA-3′).
Live Cell Imaging
Cells were plated in glass-bottom dishes. RFP-H2B was transfected to indicate the chromosomes. Data were acquired by a DeltaVision live cell imaging system (Applied Precision) equipped with an Olympus IX-71 inverted micro-scope and a 60×/1.42 oil objective.
Statistical Analysis
Each data point represents three independent experiments, and error bars indicate S.D. The statistical significance of differences was calculated with a two-tailed Student's t test. Differences were considered significant at p < 0.05. *, **, and *** indicate p < 0.05, p < 0.01, and p < 0.001, respectively.
Results
Chromatin-localized Aurora A Phosphorylates Histone H3 in Vivo
Because Aurora A and B have common substrates and functions on the spindle (30) and some of Aurora B substrates including histone H3, INCENP, and Survivin can be phosphorylated by Aurora A in vitro (31), we set out to test whether special functions of Aurora A and B are determined by their distinct localizations. We speculated that, if the functional divergence of Aurora A and B is achieved by their spatial compartmentalization through specifically binding their substrates and binding partners, the forced localization exchange of the both would substitute the functions of each other. By fusing Aurora A with either histone H2B or the centromere protein truncate CENP-B1–158 tagged with GFP (GFP-H2B-Aurora A and GFP-CENPB-Aurora A) and transiently expressing these fusion proteins in cells, we found that the fusion protein GFP-CENPB-Aurora A was localized to the nucleus and primarily the centromere during the cell cycle, and a fraction of it was relocated to the spindle/poles as did the wild-type Aurora A from prophase to metaphase and that the fusion protein GFP-H2B-Aurora A was located primarily on the chromatin/chromosomes during the cell cycle (Fig. 1A and supplemental Movies S1 and S2). As it is known that Aurora B is localized in interphase nucleus and relocated to the centromere in early mitosis, we concluded that GFP-H2B-Aurora A and GFP-CENPB-Aurora A proteins had been localized to the areas to which Aurora B is preferentially localized. By probing the active phosphorylation status of Aurora A at Ser-232 using a phospho-specific antibody, we also found that these two fusion proteins were also activated on their T-loops like the wild-type Aurora A (data not shown). To evaluate whether these fusion proteins may substitute the functions of Aurora B, we eliminated the kinase activity of endogenous Aurora B by treating HeLa cells with a serial concentration of an Aurora B-specific inhibitor AZD1152. The inhibition efficiency was tested by detecting the active phosphorylation status of Aurora proteins using the phospho-specific antibody. The result showed that, in the presence of AZD1152 at the concentration of 200 nm and above, the kinase activity of Aurora B was totally inhibited, whereas the kinase activity of GFP-CENPB-Aurora A was less affected by AZD1152 (Fig. 1B). Then the cells expressing GFP-H2B-Aurora A and GFP-CENPB-Aurora A were treated with 200 nm AZD1152 for 1 h and processed for immunofluorescence microscopy. The result showed that the Aurora B substrate histone H3 Ser-10 was extensively phosphorylated in almost all transfected cells by GFP-H2B-Aurora A regardless the period of the cell cycle (96.52%). In contrast, only a very small amount of cells transfected with GFP-Aurora A showed slight phosphorylation of histone H3 Ser-10. Moreover, in cells expressing GFP-CENPB-Aurora A that was located at the centromere, the phosphorylated histone H3 Ser-10 was only restricted at the centromere (Fig. 1, C and D). On the contrary, the kinase dead mutants of GFP-H2B-Aurora A and GFP-CENPB-Aurora A (GFP-H2B-Aurora A-KD and GFP-CENPB-Aurora A-KD) barely phosphorylated histone H3 Ser-10 (Fig. 1, C and D). Together, these results strongly indicated that histone H3 Ser-10 was specifically phosphorylated by GFP-H2B-Aurora A or GFP-CENPB-Aurora A. To further confirm whether GFP-H2B-Aurora A performed the same function as Aurora B, endogenous Aurora B was knocked down in HeLa cells by siRNA, and GFP-H2B-Aurora A or GFP-H2B-Aurora B was simultaneously expressed. By immunofluorescence microscopy, we found that histone H3 at Ser-10 was phosphorylated in almost all of the cells expressing GFP-H2B-Aurora A or GFP-H2B-Aurora B (Fig. 1, E and F). Collectively, these results demonstrated that, when forced to locate to the same place of Aurora B, Aurora A may phosphorylate the Aurora B substrate histone H3. Thus, we conclude that the specific phosphorylation of histone H3 Ser-10 by Aurora B rather than Aurora A is indeed caused by the spatial compartmentalization of the Auroras and that other substrates of Aurora B could also be phosphorylated by Aurora A provided that the spatial compartmentalization of these two Aurora kinases is eliminated.
Kinetochore-targeted Aurora A Performs the Function of Aurora B on the Spindle Checkpoint Regulation
One of the most important functions of Aurora B in mitosis is to regulate the kinetochore-microtubule attachment and the spindle checkpoint. The activation of the spindle checkpoint is stimulated by Aurora B in two pathways, one of which is that Aurora B destabilizes erroneous kinetochore-microtubule attachment and indirectly induce spindle checkpoint. The centromere-located Aurora B gives rise to a phosphorylation gradient around the centromere, and the phosphorylation of the Aurora B substrates at the kinetochores depends on their distance from Aurora B at the centromere (32, 33). If microtubules from two opposite poles fail to bind the kinetochores correctly, the tension between the paired kinetochores is low, and Aurora B phosphorylates its multiple substrates of the KMN network (including Hec1, DSN1, Knl1) on the kinetochores, as the distance from the substrates to Aurora B is short and thus activates the spindle assembly checkpoint by releasing the microtubules from the kinetochores (34, 35). Another pathway is that Aurora B functions synergically with MPS1 to induce kinetochore localization of spindle checkpoint proteins BubR1 and mad2 (36, 37). To further explore whether the forced localization of Aurora A on the centromere can substitute Aurora B for the function of the spindle checkpoint regulation, we generated and expressed fusion proteins of Aurora A or its kinase dead mutant with Hec1 tagged with GFP (GFP-Hec1-AurA and GFP-Hec1-AurA-KD) to target Aurora A to the kinetochores (Fig. 2A and supplemental Movie S3). Furthermore, we treated HeLa cells by STLC, a specific Eg5 inhibitor that weakens the interaction of Eg5 with microtubules resulting in the failure of bipolar spindle formation and mitotic arrest (38, 39), to synchronize the cells in prometaphase and then with 200 nm AZD1152 for 1 h to inhibit their endogenous Aurora B, and immunostained the cells for the spindle checkpoint protein BubR1. We observed that AZD1152 treatment abolished the kinetochore localization of this spindle checkpoint protein, and the chromosomes were misaligned, whereas the DMSO-treated control cells clearly had BubR1 on their kinetochores (Fig. 2, B and D). Then the cells expressing GFP-CENPB-Aurora A, GFP-Hec1-Aurora A, GFP-Hec1-Aurora A-KD, and GFP-Aurora A were synchronized to prometaphase by STLC and treated with 200 nm AZD1152 for 1 h. The results showed that when GFP-Hec1-Aurora A was expressed, the spindle checkpoint was rescued, as indicated by the localization of BubR1 on the kinetochores (Fig. 2, C and D). When GFP-Hec1-Aurora A-KD was expressed, BubR1 localization on the kinetochores was abolished in the presence of AZD1152, indicating no functions of GFP-Hec1-Aurora A-KD for the localization of BubR1. Moreover, expressing GFP-CENPB-Aurora A and GFP-Aurora A that could not localize to the kinetochores also could not rescue the localization of BubR1 on the kinetochore in the presence of AZD1152 (Fig. 2, C and D). Thus, the forced localization of functional Aurora A on the kinetochores rather than the other places, including the centromere, is sufficient to substitute Aurora B for the function of activating the spindle checkpoint.
FIGURE 2.
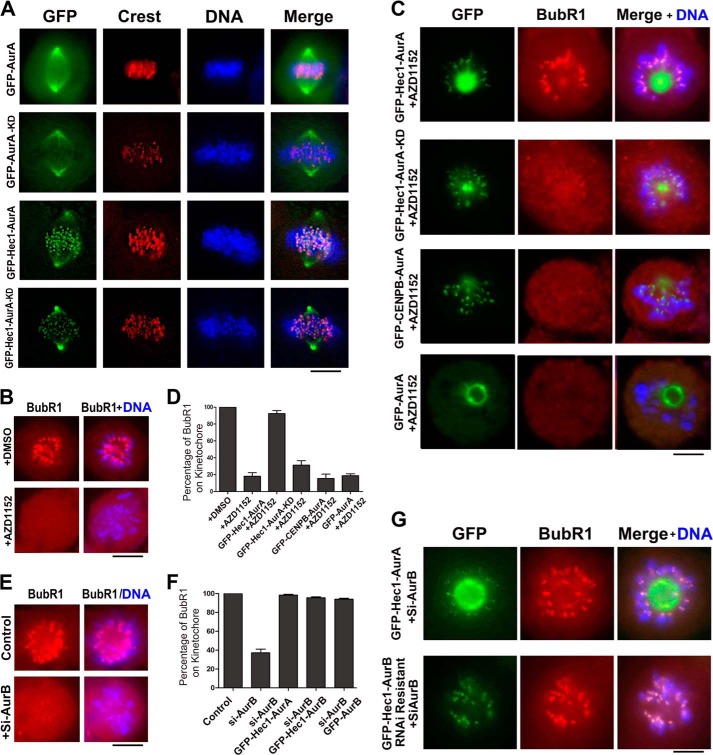
Forced kinetochore localization of Aurora A performs the function of Aurora B on the spindle checkpoint regulation. A, immunofluorescence photomicrographs of HeLa cells transfected with GFP-Aurora A or GFP-Hec1-Aurora A and stained with a CREST antibody. Note that Aurora A (Aura) and its kinase-dead mutant (KD) were localized to the spindle and centrosomes in mitosis, whereas part of Hec1-Aurora A and its relevant kinase-dead mutant were localized to the kinetochores. Scale bar, 10 μm. B, HeLa cells were synchronized to prometaphase by STLC and treated by 200 nm AZD1152 for 1 h followed by immunofluorescence labeling with an anti-BubR1 antibody. Note that inhibiting Aurora B kinase activity resulted in the loss of BubR1 on the kinetochores. Scale bar, 10 μm. C, HeLa cells were transfected with GFP-Hec1-Aurora A, GFP-Hec1-Aurora A-KD, GFP-CENPB-Aurora A, or GFP-Aurora A, synchronized to prometaphase by STLC and treated with 200 nm AZD1152 for 1 h followed by immunostaining with an anti-BubR1 antibody. Note that expressing GFP-Hec1-Aurora A could rescue the loss of BubR1 on the kinetochores induced by AZD1152 treatment, whereas GFP-Hec1-Aurora A-KD, GFP-CENPB-Aurora A, and GFP-Aurora A could not. Scale bar, 10 μm. D, quantitative characterization of HeLa cells treated as in B and C. Bars represented the percentage of the cells with BubR1 on kinetochores. Each data point represents three independent experiments with each measuring 50 cells, and error bars indicate S.D. E, Aurora B knockdown by siRNA resulted in the loss of BubR1 on kinetochores. HeLa cells were transfected with Aurora B siRNA and processed for immunostaining with an anti-BubR1 antibody. Scale bar, 10 μm. F, quantitative characterization of HeLa cells treated as in E and G. Bars represent the percentage of the cells with BubR1 on kinetochores. Each data point represents 3 independent experiments with each measuring 50 cells, and error bars indicate S.D. G, BubR1 immunofluorescence labeling of HeLa cells co-transfected with Aurora B siRNA and GFP-Hec1-Aurora A or RNAi-resistant GFP-Hec1-Aurora B. Note that expression of either GFP-Hec1-Aurora B or GFP-Hec1-Aurora B rescued the loss of BubR1 on kinetochores caused by endogenous Aurora B knockdown. Scale bar, 10 μm.
We further knocked down Aurora B by siRNA to make the functional loss defect of Aurora B. The result showed that the Aurora B knockdown significantly reduced the localization of BubR1 on the kinetochores by ∼70% in comparison with the irrelevant control knockdown (Fig. 2, E and F). By expressing exogenous Aurora B, we performed the rescue experiment and found that exogenous GFP-Aurora B expression could rescue the defect by up to 95%. When GFP-Hec1-Aurora B was expressed, it could rescue the defect by up to >95% (Fig. 2, E–G). We also expressed GFP-Hec1-Aurora A in Aurora B knockdown cells. Interestingly, we observed that GFP-Hec1-Aurora A could almost fully rescue the defect caused by Aurora B knockdown (Fig. 2, F and G). In contrast, GFP-CENPB-Aurora A expression was not able to rescue the defect (Fig. 2, C and D), indicating that the centromere-targeted GFP-CENPB-Aurora A could not be able to reach the substrates of Aurora B at the kinetochores. Together, these results demonstrate that the spindle checkpoint defect caused by Aurora B knockdown can be fully rescued by targeting Aurora A to the kinetochores to substitute endogenous Aurora B for regulating the kinetochore localization of BubR1.
Centrosome-localized Aurora B Can Substitute Aurora A in Promoting the Mitotic Entry
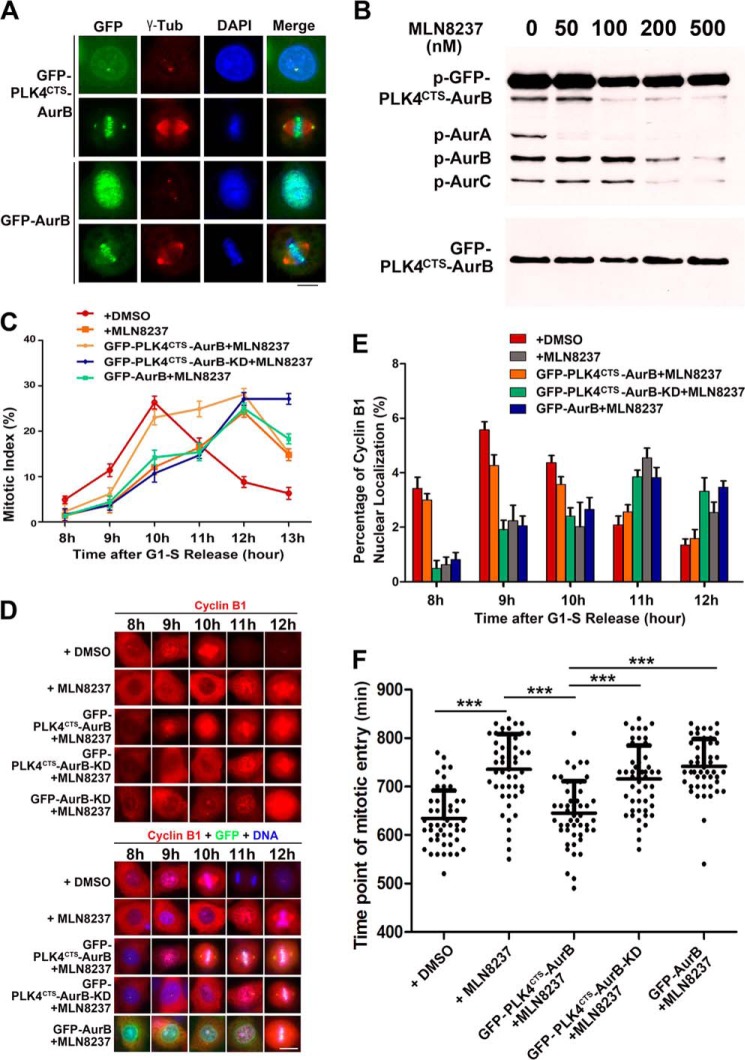
The active Aurora A at the centrosome is required for the timely mitotic entry. In G2 phase, Aurora A directly phosphorylates Cdc25B at Ser-353 and regulates its localization at the centrosome to activate CDK1 kinase (40). Aurora A, with its co-activator Bora at the centrosome, also activates Plk1 by phosphorylating it at Thr-210. Then, Plk1, Aurora A, Cdc25B, and Cdk1 form a feedback loop and positively regulate each other's activity for the mitotic entry (41). To investigate whether Aurora B can substitute Aurora A on the centrosome to regulate the mitotic entry, we constructed a fusion protein GFP-PLK4CTS-Aurora B, which contains a PLK4 centrosome targeting sequence (CTS, aa 570–820) that may lead the protein to the centrosomes (42), and expressed it in HeLa cells. We observed that GFP-PLK4CTS-Aurora B was specifically localized to the centrosomes both in mitotic and interphase cells in addition to a fraction of it in the nucleus and the centromeres (Fig. 3A and supplemental Movie S4). The phosphorylation status identification of Aurora B at the T-loop indicated that GFP-PLK4CTS-Aurora B was autophosphorylated and activated (Fig. 3B). The cells expressing the GFP-PLK4CTS-Aurora B fusion proteins were treated with the Aurora A-specific inhibitor MLN8237 to eliminate the kinase activity of endogenous Aurora A. The result showed that, in the presence of MLN8237 at the final concentration of 50 nm and above, the endogenous Aurora A kinase activity was totally inhibited, indicated by the phosphorylation of its T-loop, but the kinase activity of GFP-PLK4CTS-AurB was not affected (Fig. 3B). Then the cells expressing the fusion proteins were synchronized to G1-S phase and released for 8–13 h followed by the drug treatment. It was found that, in DMSO-treated control cells, the mitotic index peaked at 10 h after the release, whereas the mitotic index of MLN8237-treated cells reached the peak level at 12 h (Fig. 3C). These indicate that inhibiting Aurora A kinase activity delayed the mitotic entry. Interestingly, when the cells expressing GFP-PLK4CTS-Aurora B were treated with MLN8237, the mitotic delay by MLN8237 was largely rescued. In contrast, the kinase dead mutant GFP-PLK4CTS-Aurora B-KD, that had the similar centrosome localization and GFP-Aurora B, failed to alleviate the mitotic delay by MLN8237 treatment. In the cells expressing the fusion proteins and treated with MLN8237 we also checked the nuclear translocation of cyclin B1 that precedes the nuclear envelope breakdown and is required for the G2-M transition (43). Consistent with the mitotic index data (Fig. 3C), the timing of cyclin B1 nuclear translocation in MLN8237-treated cells was largely delayed, and the delay could be rescued by expressing GFP-PLK4CTS-Aurora B but not GFP-PLK4CTS-Aurora B-KD or GFP-Aurora B (Fig. 3, D and E). Furthermore, we directly determined the time of mitotic entry by examining the duration from the time point of G1-S phase release to nuclear envelope breakdown in cells expressing the fusion proteins and treated with MLN8237. Consistent with the results above, we observed that GFP-PLK4CTS-Aurora B indeed could rescue the mitotic delay (Fig. 3F). Together, these results demonstrate that when Aurora B is artificially forced to locate to the centrosomes, it can substitute Aurora A to regulate the mitotic entry.
FIGURE 3.
Forced localization of Aurora B to centrosome could perform the function of Aurora A in promoting the mitotic entry. A, immunofluorescence photomicrographs of HeLa cells transfected with GFP-Aurora B or GFP-PLK4CTS-Aurora B (GFP-PLK4CTSAurB) and stained with a γ-tubulin (γ-tub) antibody. Note that GFP-PLK4CTS-Aurora B was localized to the centrosomes, whereas GFP-Aurora B was localized to the nucleus and the centromere in interphase and mitosis. Scale bar, 10 μm. B, HeLa cells transfected with GFP-PLK4CTS-Aurora B were synchronized to mitosis and treated with different concentrations of Aurora A inhibitor MLN8237 for 1 h. Cell extracts were subjected to immunoblot analysis with antibodies to phospho-Aurora A/Aurora B/Aurora-C and Aurora A. Note that the activity of Aurora A was inhibited by MLN8237 at 50 nm, whereas the activity of GFP-PLK4CTS-Aurora B was not affected. C, quantitative characterization of mitotic index of the cells expressed GFP-Aurora B, GFP-PLK4CTS-Aurora B, or GFP-PLK4CTS-Aurora B-KD. HeLa cells were synchronized to G1-S phase by thymidine block and released for 8–13 h. Before immunofluorescence analysis, 50 nm MLN8237 was added for 1 h. DMSO was used as solvent control. Each data point represents three independent experiments with each measuring 50 cells, and error bars indicate S.D. D and E, immunofluorescence staining and quantitative characterization of cyclin B1 nuclear localization in HeLa cells expressed GFP-Aurora B, GFP-PLK4CTS-Aurora B, or GFP-PLK4CTS-Aurora B-KD. HeLa cells were synchronized and treated with MLN8237 as in C and stained with cyclin B1 antibody. Cyclin B1 nuclear localization was defined by its staining and is stronger in the nucleus than in the cytoplasm. Each data point represents three independent experiments with each measuring 50 cells, and error bars indicate S.D. Scale bar, 10 μm. F, quantitative characterizations of time of mitotic entry. Cells expressed GFP-Aurora B, GFP-PLK4CTS-Aurora B, or GFP-PLK4CTS-Aurora B-KD were synchronized to G1-S phase by thymidine block and released followed by live cell imaging to measure the duration from the time point of G1-S phase release to nuclear envelope breakdown. n = 50 cells per group. ***, p < 0.001. Error bars indicate S.D.
Both N Termini of Aurora A and B Contribute to Their Spatial Localizations
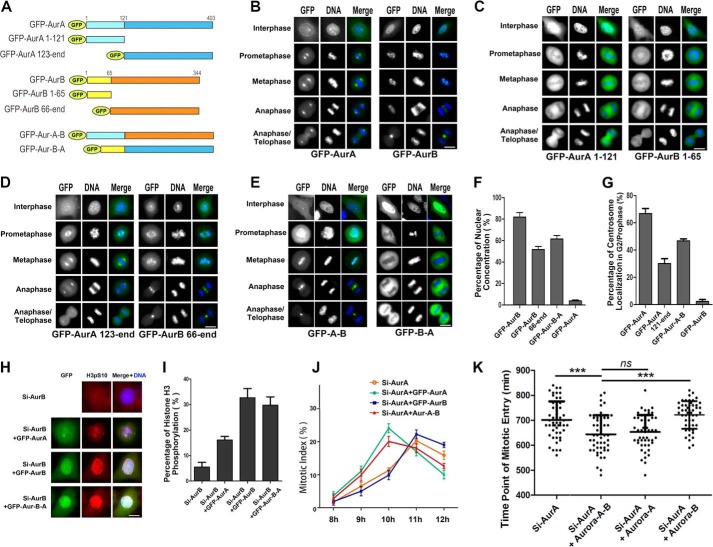
It has been reported that the critical domains for localization and function of Aurora A and B reside within their C termini (2, 4), and that the C termini alone could localize to the equivalent sites of and function as the full-length Aurora A and B(44); however, the functions of the N termini remain unknown. To determine whether the N termini contribute to their localizations and functions, we constructed a series of GFP-tagged truncation mutants of the human Auroras and examined their localizations (Fig. 4A). The result showed that GFP-tagged truncates Aurora A1–121 (GFP-Aurora A aa 1–121) showed a diffuse pattern with a significant fraction on the spindle and spindle poles in HeLa cells, in comparison with the centrosomal localization of Xenopus Aurora A N-terminal truncate in Xenopus cells XL2 and egg extracts (45) and that Aurora B1–65 (GFP-Aurora B aa 1–65) was largely in the nucleus in G2/prophase and smeared in the cytoplasm but not in the centromere in mitosis (Fig. 4, B and C, and supplemental Movies S5 and S6). We also observed that the catalytic C-terminal truncates GFP-Aurora A aa 123-end and GFP-Aurora B 66-end showed the similar localizations with the wild-type Aurora A and B in mitosis, respectively (Fig. 4D), as reported (46). However, when the N terminus of Aurora A was substituted by the N terminus of Aurora B to construct a chimera protein GFP-Aurora B-A and expressed in cells, this chimera protein was found to be largely localized to the nucleus like Aurora B in G2/prophase and relocated to the area of the wild-type Aurora A in mitosis (Fig. 4, E and F, and supplemental Movie S7), indicating that the N terminus of Aurora B may contribute to its nuclear localization before the mitotic entry. In comparison, we fused the N terminus of Aurora A with the C terminus of Aurora B (GFP-Aurora A-B) and expressed it in cells. Surprisingly, we observed that this chimera protein showed the centrosomal localization in G2/prophase like Aurora A, and this localization was enhanced before the mitotic entry followed by a typical localization of Aurora B on the centromere and midzone/midbody in mitosis (Fig. 4, D and G, and supplemental Movie S8). Consistently, we also noticed that Aurora A depleted of its N terminus showed the reduced centrosomal localization and Aurora B depleted of its N terminus showed the reduced nuclear concentration (Fig. 4, D, F, and G). These indicate that the N terminus of Aurora A and B may contribute to their respective localization before the mitotic entry. Together, these results demonstrate that the divergent N termini of the Auroras may contribute to their localizations in interphase and that once the cell enters mitosis, the C termini may mainly determine the canonical localizations of the Aurora A and B proteins.
FIGURE 4.
N termini of both Aurora A and B contribute to their spatial localizations and functions. A, schematic depiction of the truncate and chimera protein constructs of N and C termini of Aurora A (AurA) and B (AurB) tagged with GFP. GFP-Aurora A-B is composed of the N terminus of Aurora A and C terminus of Aurora B. Conversely, GFP-Aurora B-A is composed of the N terminus of Aurora B and C terminus of Aurora A. B, localizations of the wild-type full-length GFP-Aurora A and GFP-Aurora B in the cell cycle. Scale bar, 10 μm. C, localizations of the N-terminal truncates of Aurora A and B in the cell cycle. Scale bar, 10 μm. D, localizations of the C-terminal truncates of Aurora A and B in the cell cycle. Note the reduced localization of the C terminus of Aurora A on the centrosome in interphase cells compared with the full-length wild-type Aurora A in B. Scale bar, 10 μm. E, localization of the chimera proteins GFP-Aurora A-B and GFP-Aurora B-A in the cell cycle. Note that GFP-Aurora A-B was localized on the centrosomes in interphase cells like wild-type Aurora A and in the centromeres of mitotic cells like wild-type Aurora B, whereas GFP-Aurora B-A was concentrated in late interphase nucleus like wild-type of Aurora B and on the spindle/poles in mitosis like wild-type Aurora A. Scale bar, 10 μm. F, quantitative characterizations of the nuclear accumulation of GFP-Aurora B, GFP-Aurora B 66-end, GFP-Aurora B-A, and GFP-Aurora A. Nuclear accumulation was defined by its stronger staining in the nucleus than in the cytoplasm. G, quantitative characterizations of the centrosome localization of GFP-Aurora A, GFP-Aurora A 123-end, GFP-Aurora A-B, and GFP-Aurora B. Each data point represents 3 independent experiments with each measuring 100 cells, and error bars indicate S.D. H, immunofluorescence photomicrographs of HeLa cells co-transfected with Aurora B siRNA and GFP-Aurora A, RNAi-resistant GFP-Aurora B, or GFP-Aurora B-A and stained with anti-H3pS10 antibody. Scale bar, 10 μm. I, quantitative characterization of HeLa cells treated as in H. Bars represent the percentage of the cells with histone H3 Ser-10 phosphorylation. Each data point represents three independent experiments with each measuring 200 cells, and error bars indicate S.D. J, quantitative characterization of mitotic index of HeLa cells co-transfected with Aurora A siRNA and RNAi resistant GFP-Aurora A, GFP-Aurora B, or GFP-Aurora A-B. The cells were synchronized to G1-S phase by thymidine block and released for 8–13 h. Each data point represents three independent experiments with each measuring 100 cells, and error bars indicate S.D. K, quantitative characterizations of the time of the mitotic entry. Cells treated as in J were released from G1-S followed by live cell imaging to measure the duration from the time point of G1-S phase release to nuclear envelope breakdown. n = 50 cells per group; ***, p < 0.001. Error bars indicate S.D. ns, not significant.
Next, we set to test the functions of the chimera proteins. By knocking down endogenous Aurora B and expressing GFP-tagged exogenous Aurora B, Aurora A, or Aurora B-A, we found that both exogenous Aurora B and Aurora B-A, but not Aurora A, could efficiently rescue the phosphorylation defect of histone H3 at Ser-10 in the nucleus caused by endogenous Aurora B knockdown (Fig. 4, H and I). This indicates that the N terminus of Aurora B may contribute its localization and function. By knocking down endogenous Aurora A and expressing GFP-tagged exogenous Aurora A, Aurora B, or Aurora A-B, we tested the function of the Aurora A N terminus through examining the mitotic index. We observed that the mitotic entry was delayed when Aurora A was knocked down and the delay could be rescued by GFP-Aurora A and GFP-Aurora A-B but not GFP-Aurora B (Fig. 4J). Furthermore, we measured the duration from the time point of G1-S phase release to nuclear envelope breakdown in these cells. Consistent with previous result, we found that, like GFP-Aurora A, GFP-Aurora A-B also rescues mitotic delay, indicating that the N terminus of Aurora A targeted the chimera protein GFP-Aurora A-B to the typical localization site of Aurora A, where kinase activity rescued the defect of Aurora A knockdown (Fig. 4K). Collectively, we conclude that the N termini of both Aurora A and B contribute to their localizations before mitosis and hence functions of the both kinases.
N-terminal Deletion of Aurora A Resulted in Prolonged Mitosis and Defect in Spindle Bipolarity
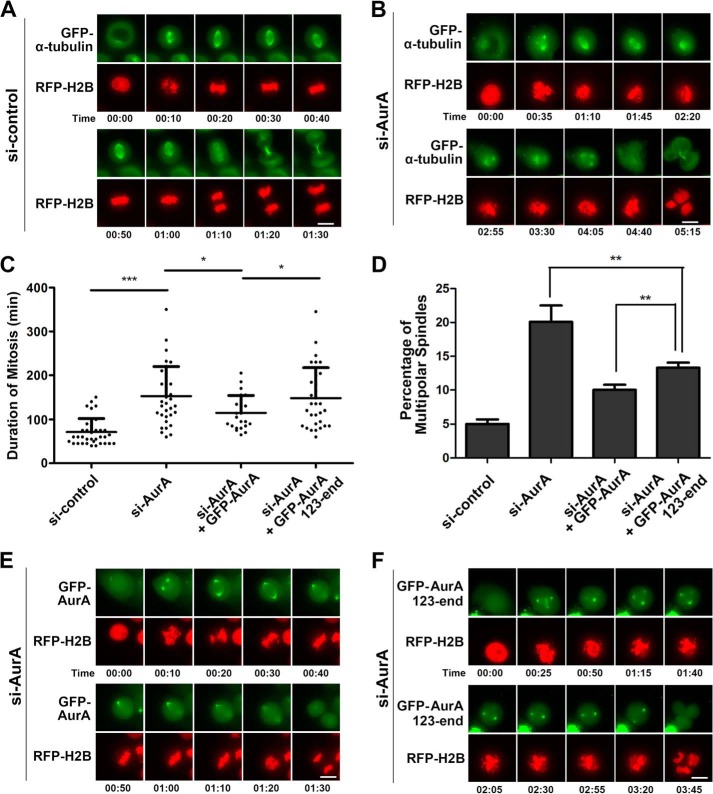
Next, we set to further study the roles of the N terminus of Aurora A by depleting endogenous Aurora A and rescuing it with different constructs. First, we knocked down Aurora A in HeLa cells that stably expressed GFP-α-tubulin and observed by time-lapse microscopy that, in comparison with the control cells, the spindle assembly in Aurora A knockdown cells was abnormal with extra poles, and the mitosis was prolonged (Fig. 5, A–D), consisting with previously reports (47, 48). Then, we depleted Aurora A from HeLa cells and transiently expressed full-length GFP-Aurora A. We observed that the cells were able to form normal bipolar spindle and divided normally. In contrast, the C terminus of Aurora A (GFP-Aurora A aa 123-end) could not efficiently rescue the defects caused by endogenous Aurora A knockdown (Fig. 5, C, E, and F). The cells assembled extra poles, the chromosomal congression was defective, the mitosis lasted significantly longer, and the cell division was abnormal (Fig. 5F). Taken together, we conclude that the N terminus contributes to the function of Aurora A in regulating the spindle bipolarity.
FIGURE 5.
N-terminal deletion of Aurora A results in prolonged mitosis and multipolar spindle assembly. A and B, HeLa cells stably expressing GFP-α-tubulin were transfected with irrelevant (A) or Aurora A (AurA) siRNA (B) and subjected to time-lapse microscopy. RFP-H2B was transiently expressed as a chromatin/chromosome indicator. Note that Aurora A depletion resulted in multipolar cell division. Scale bars, 10 μm. C and D, quantitative characterizations of the duration of mitosis with live cells (C) and percentage of the cells with abnormal spindle assembly with fixed cells (D) shown in (A, B, E, and F) (see below). *, p < 0.05; **, p < 0.01; ***, p < 0.0001. The duration of mitosis is determined from the nuclear envelope breakdown to two daughter cells formation. Each data point represents 3 independent experiments with each measuring 50 cells, and error bars indicate S.D. E and F, HeLa cells co-transfected with Aurora A siRNA-, RFP-H2B-, and RNAi-resistant GFP-Aurora A (E) or GFP-Aurora A aa123-end (F) were viewed by time-lapse microscopy. Note that Aurora A aa 123-end could not rescue Aurora A depletion. Scale bars, 10 μm.
N-terminal Deletion of Aurora B Results in Reduced Phosphorylation of Hec1 and Abnormal Mitosis
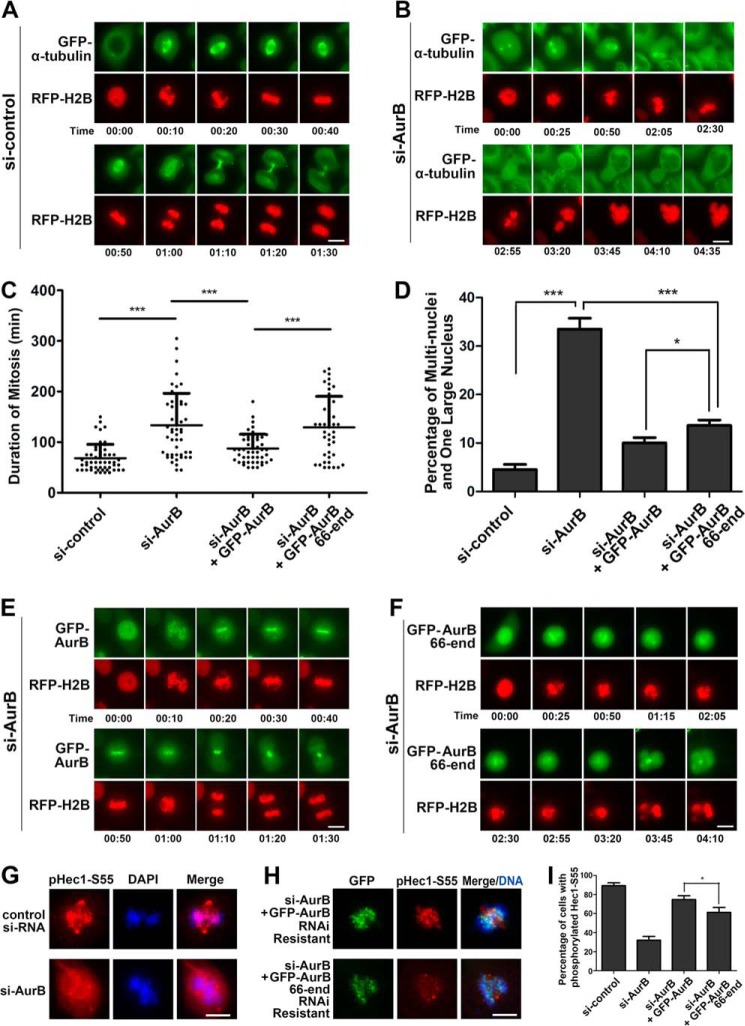
We also further studied the function of the N terminus of Aurora B with similar knockdown and rescue experiments. When Aurora B was depleted from HeLa cells that stably expressed GFP-α-tubulin, bipolar spindles could form, but the chromosome congression was slow; the cells were finally committed to exit mitosis after a long arrest and without successful division (Fig. 6, A–D). These cells contained either a single large nucleus or several small nuclei. Then we performed the rescue experiments in HeLa cells using either the full-length GFP-Aurora B or the C terminus truncate aa 66-end of GFP-Aurora B. The results showed that the full-length GFP-Aurora B successfully rescued the defect caused by endogenous Aurora B knockdown; in contrast, the N terminus truncate was less efficient in rescuing these defects, and the chromosome congression still failed leading to one large nucleus or multinuclei formation (Fig. 6, C–F). We further examined the phosphorylation of Hec1 at Ser-55 on the kinetochores, which is thought to be regulated by Aurora B (42). We observed that when Aurora B was knocked down by siRNA, the phosphorylation of Hec1 Ser-55 on the kinetochores was significantly reduced and that this defect could be rescued by full-length wild-type Aurora B but not the C terminus truncate aa 66-end of GFP-Aurora B (Fig. 6, G–I). Together, we conclude that the N terminus is required for the full function of Aurora B in phosphorylating its substrates on the kinetochores, which is required for normal cell division. This also indicates that the N terminus of Aurora B may play an important role in bringing centromere located Aurora B to the kinetochores to phosphorylate its substrates such as Hec1, DSN1, and KNL1, suggesting that N terminus-mediated relocation of Aurora B from the centromere to kinetochore is needed for the regulation of kinetochore-microtubule connections.
FIGURE 6.
N-terminal deletion of Aurora B results in reduced phosphorylation of Hec1 on kinetochores and abnormal mitotic exit with formation of multinuclei or single large nucleus. A and B, HeLa cells stably expressing GFP-α-tubulin were transfected with irrelevant (A) or Aurora B (AurB) siRNA (B) and subjected to time-lapse microscopy. RFP-H2B was transiently expressed as a chromatin/chromosome indicator. Note that Aurora B depletion resulted in abnormal mitotic exit with formation of multinuclei or single large nucleus. Scale bars, 10 μm. C and D, quantitative characterizations of the duration of mitosis with live cells (C) and percentage of the cells with multinuclei or a large nucleus (D). *, p < 0.05; ***, p < 0.0001. Duration of mitosis is determined from the nuclear envelope breakdown to two daughter cells formation. Each data point represents 3 independent experiments with each measuring 50 cells, and error bars indicate S.D. (E and F) HeLa cells were co-transfected with Aurora B siRNA, RFP-H2B and RNAi resistant GFP-Aurora B (E) or GFP-Aurora B aa 66-end (F) and viewed by time-lapse microscopy. Note that Aurora B aa 66-end could not rescue Aurora B depletion. Scale bars, 10 μm. G, Aurora B knockdown by siRNA resulted in decreased phosphorylation of Hec1 on kinetochores. HeLa cells were transfected with Aurora B siRNA and processed for immunostaining with an antibody to phospho-Hec1-Ser-55. Scale bars, 10 μm. H, HeLa cells co-transfected with Aurora B siRNA and RNAi-resistant GFP-Aurora B or RNAi-resistant GFP-Aurora B aa 66-end and processed for immunostaining with an antibody to phospho-Hec1-Ser-55. Note that expression of GFP-Aurora B aa 66-end could not fully rescue the phosphorylation status of Hec1. Scale bars, 10 μm. I, quantitative characterization of HeLa cells treated as in G and H. Bars represent the percentage of cells with phosphorylated Hec1 on kinetochores. Each data point represents three independent experiments with each measuring 50 cells. Error bars indicate S.D. *, p < 0.05.
Discussion
In this study we reveal the relations between the spatiotemporal compartmentalizations and functions of Aurora A and B and show that one may carry out the functions of the other when one is artificially localized to the places of the other. We also reveal that the divergent N termini of the Auroras have roles for their spatiotemporal localizations. When the N termini of Aurora A was fused to the C termini of Aurora B, the chimera proteins could localize and function like Aurora A in addition to like Aurora B and vice versa. Consistent with the previous reports, our results have shown that Aurora A and B have similar potential substrates and kinase activity, and their specific functions are likely determined by their different localizations. Therefore, the functional divergence of Aurora A and B is likely determined by the spatial compartmentalization of their binding partners that binds both the conservative C and the divergent N termini of the Aurora kinases.
Previously we and others have uncovered that the localizations of both Aurora A and B are mainly determined by their substrates and binding partners, and their functions rely on their catalytic C termini (4, 44). We revealed that a single amino acid change on residue Gly-198 can convert Aurora A into Aurora B-like kinase (4). The mutant Aurora AG198N exhibits the Aurora B-like localization on the centromere and midzone, where it interacts with components of the chromosome passenger complex and rescues the chromosome misalignment caused by Aurora B knockdown. A longer side chain on residue 198 of mutant Aurora AG198N prevents its attachment to TPX2, and increased hydrophilicity of residue 198 also promotes Aurora AG198N to interact with INCENP. This indicates that the slight difference in structure of Aurora A and B leads to differential interaction with the binding partners of each other and thus results in differentiation of localization and function. In this work we further find that the divergent N termini of both Aurora A and B may have their own binding partners and contribute to the distinct localizations. Without the N terminus, the ability of Aurora A in regulating the mitotic spindle bipolarity is impaired, whereas Aurora B cannot fully phosphorylate its substrates on kinetochores and regulate the kinetochore-microtubule connection required for faithful chromosome separation and cell division.
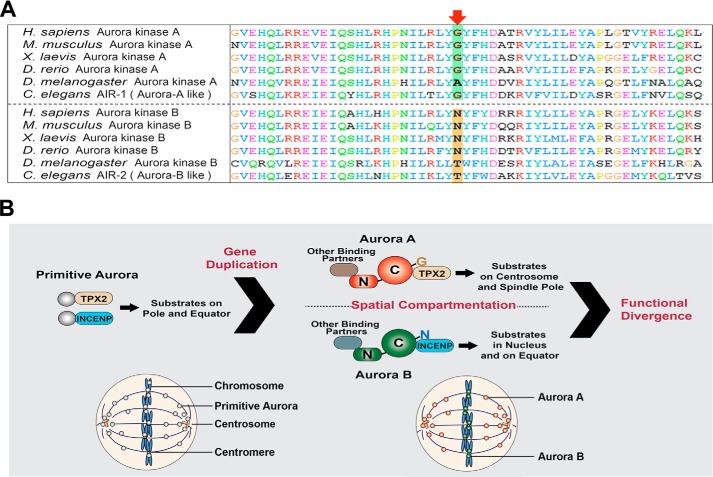
In the evolution process both Aurora A and B may originate from a single Aurora protein. Phylogenetic analysis suggests that the duplication of Aurora genes has independently occurred in vertebrates, invertebrates, and plants (21, 30). The polar Aurora kinases (Aurora A-like) and the equatorial Aurora kinases (Aurora B-like) in invertebrates (such as Caenorhabditis elegans and Drosophila melanogaster) have the similar localizations and functions with their vertebrate counterpart, although they are not orthologous with the respective vertebrate Aurora A and B. Interestingly, despite confusion in the phylogenetic trees, all polar Aurora kinases in model organisms share a common feature in that the amino acid sequence corresponding to human Aurora A residue 198 is glycine or alanine, which has a short and hydrophobic side chain (Fig. 7A). On the contrary, all equatorial Aurora kinases share an amino acid with a long and hydrophilic side chain such as asparagine or threonine at the residue corresponding to human Aurora A residue 198. Our previous and present studies together with others' have presented that Aurora A and B share great similarity in substrates and kinase activity, and the functional divergence of Aurora kinases is largely determined by their different localization defined by their binding affinity with specific binding partners. So, it seems that, after the gene duplication events in the evolutional process, the mutation on a single amino acid of the Auroras confers the differential interaction affinity with their binding partners such as TPX2 and INCENP; and in turn, the Auroras are brought to the polar or equatorial localization by the differential binding partners, respectively, and thus led to the differentiation of their functions (Fig. 7B). Furthermore, their elongated and low conservative N termini suggest that they may have differential binding sites for their additional binding partners that may bring them to their special locations to perform their distinct functions. To fully understand the evolution and function of the Auroras, it is worthy identifying these additional binding partners in the future.
FIGURE 7.
Model depicting the evolution process of Aurora kinases. A, the sequence alignment of a segment of the Auroras from various species showing the residue corresponding to Gly-198 of human Aurora A (Arrow) is divergent in polar Auroras (Aurora A like) and equatorial Auroras (Aurora B like). B, model depicting the evolution process of Aurora kinases. Primitive Aurora kinase is bifunctional and localized on both polar and equatorial regions of the cells. Gene duplication and divergence occurred independently in protostome and deuterostome. The divergence on the residue corresponding to Gly-198 of human Aurora A results in differential affinity to TPX2 and INCENP. The divergent N terminus along with the residue corresponding to Gly-198 of human Aurora A led to spatial differentiation of Aurora A and B and thus conferred further the functional divergence of the modern Auroras.
Author Contributions
C. M. Z., Q. J., F. D. Z. and S. L. conceived and coordinated the study and wrote the paper. S. L., Z. X. D., J. Y. F., C. Y. X., G. W. X., Z. G. W., J. L., G. W., S. L. Z., and B. Y. Z. designed, performed, and analyzed the experiments.
Supplementary Material
Acknowledgments
We thank all the other members in our laboratories for useful comments.
This work was supported by State Key Basic Research and Development Plan of the Ministry of Science and Technology of China Grants 2010CB833705 and 2014CB138402 and the National Natural Science Foundation of China Grants 31430051, 31371365, 91313302, and 31030044. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Movies S1–S8.
- aa
- amino acid(s)
- STLC
- S-trityl-l-cysteine
- CTS
- centrosome targeting sequence
- RFP
- red fluorescent protein.
References
- 1. Carmena M., Earnshaw W. C. (2003) The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 4, 842–854 [DOI] [PubMed] [Google Scholar]
- 2. Fu J., Bian M., Jiang Q., Zhang C. (2007) Roles of aurora kinases in mitosis and tumorigenesis. Mol Cancer Res. 5, 1–10 [DOI] [PubMed] [Google Scholar]
- 3. Carmena M., Wheelock M., Funabiki H., Earnshaw W. C. (2012) The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 13, 789–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fu J., Bian M., Liu J., Jiang Q., Zhang C. (2009) A single amino acid change converts Aurora A into Aurora B-like kinase in terms of partner specificity and cellular function. Proc. Natl. Acad. Sci. U.S.A. 106, 6939–6944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reboutier D., Troadec M. B., Cremet J. Y., Chauvin L., Guen V., Salaun P., Prigent C. (2013) Aurora A is involved in central spindle assembly through phosphorylation of Ser-19 in P150Glued. J. Cell Biol. 201, 65–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stenoien D. L., Sen S., Mancini M. A., Brinkley B. R. (2003) Dynamic association of a tumor amplified kinase, Aurora A, with the centrosome and mitotic spindle. Cell Motil. Cytoskeleton 55, 134–146 [DOI] [PubMed] [Google Scholar]
- 7. Fu W., Tao W., Zheng P., Fu J., Bian M., Jiang Q., Clarke P. R., Zhang C. (2010) Clathrin recruits phosphorylated TACC3 to spindle poles for bipolar spindle assembly and chromosome alignment. J. Cell Sci. 123, 3645–3651 [DOI] [PubMed] [Google Scholar]
- 8. Fu W., Chen H., Wang G., Luo J., Deng Z., Xin G., Xu N., Guo X., Lei J., Jiang Q., Zhang C. (2013) Self-assembly and sorting of acentrosomal microtubules by TACC3 facilitate kinetochore capture during the mitotic spindle assembly. Proc. Natl. Acad. Sci. U.S.A. 110, 15295–15300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giet R., Uzbekov R., Cubizolles F., Le Guellec K., Prigent C. (1999) The Xenopus laevis aurora-related protein kinase pEg2 associates with and phosphorylates the kinesin-related protein XlEg5. J. Biol. Chem. 274, 15005–15013 [DOI] [PubMed] [Google Scholar]
- 10. Hirota T., Kunitoku N., Sasayama T., Marumoto T., Zhang D., Nitta M., Hatakeyama K., Saya H. (2003) Aurora A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell 114, 585–598 [DOI] [PubMed] [Google Scholar]
- 11. Wang G., Jiang Q., Zhang C. (2014) The role of mitotic kinases in coupling the centrosome cycle with the assembly of the mitotic spindle. J. Cell Sci. 127, 4111–4122 [DOI] [PubMed] [Google Scholar]
- 12. Bayliss R., Sardon T., Vernos I., Conti E. (2003) Structural basis of Aurora A activation by TPX2 at the mitotic spindle. Mol Cell 12, 851–862 [DOI] [PubMed] [Google Scholar]
- 13. Sessa F., Mapelli M., Ciferri C., Tarricone C., Areces L. B., Schneider T. R., Stukenberg P. T., Musacchio A. (2005) Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell 18, 379–391 [DOI] [PubMed] [Google Scholar]
- 14. Glover D. (2003) Aurora A on the mitotic spindle is activated by the way it holds its partner. Mol. Cell 12, 797–799. [DOI] [PubMed] [Google Scholar]
- 15. Eyers P. A., Erikson E., Chen L. G., Maller J. L. (2003) A novel mechanism for activation of the protein kinase Aurora A. Curr. Biol. 13, 691–697 [DOI] [PubMed] [Google Scholar]
- 16. Hutterer A., Berdnik D., Wirtz-Peitz F., Zigman M., Schleiffer A., Knoblich J. A. (2006) Mitotic activation of the kinase Aurora A requires its binding partner Bora. Dev. Cell 11, 147–157 [DOI] [PubMed] [Google Scholar]
- 17. Zhao Z. S., Lim J. P., Ng Y. W., Lim L., Manser E. (2005) The GIT-associated kinase PAK targets to the centrosome and regulates Aurora A. Mol. Cell 20, 237–249 [DOI] [PubMed] [Google Scholar]
- 18. Abe Y., Okumura E., Hosoya T., Hirota T., Kishimoto T. (2010) A single starfish Aurora kinase performs the combined functions of Aurora A and Aurora B in human cells. J. Cell Sci. 123, 3978–3988 [DOI] [PubMed] [Google Scholar]
- 19. Hebras C., McDougall A. (2012) Urochordate ascidians possess a single isoform of Aurora kinase that localizes to the midbody via TPX2 in eggs and cleavage stage embryos. PLoS ONE 7, e45431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li H., Chen Q., Kaller M., Nellen W., Gräf R., De Lozanne A. (2008) Dictyostelium Aurora kinase has properties of both Aurora A and Aurora B kinases. Eukaryot. Cell 7, 894–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brown J. R., Koretke K. K., Birkeland M. L., Sanseau P., Patrick D. R. (2004) Evolutionary relationships of Aurora kinases: implications for model organism studies and the development of anti-cancer drugs. BMC Evol. Biol. 4, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Capra E. J., Perchuk B. S., Skerker J. M., Laub M. T. (2012) Adaptive mutations that prevent crosstalk enable the expansion of paralogous signaling protein families. Cell 150, 222–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rausell A., Juan D., Pazos F., Valencia A. (2010) Protein interactions and ligand binding: from protein subfamilies to functional specificity. Proc. Natl. Acad. Sci. U.S.A. 107, 1995–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alexander J., Lim D., Joughin B. A., Hegemann B., Hutchins J. R., Ehrenberger T., Ivins F., Sessa F., Hudecz O., Nigg E. A., Fry A. M., Musacchio A., Stukenberg P. T., Mechtler K., Peters J. M., Smerdon S. J., Yaffe M. B. (2011) Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci. Signal. 4, ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim Y., Holland A. J., Lan W., Cleveland D. W. (2010) Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell 142, 444–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tanenbaum M. E., Macurek L., van der Vaart B., Galli M., Akhmanova A., Medema R. H. (2011) A complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by Aurora kinases. Curr. Biol. 21, 1356–1365 [DOI] [PubMed] [Google Scholar]
- 27. Knowlton A. L., Vorozhko V. V., Lan W., Gorbsky G. J., Stukenberg P. T. (2009) ICIS and Aurora B coregulate the microtubule depolymerase Kif2a. Curr. Biol. 19, 758–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jang C. Y., Coppinger J. A., Seki A., Yates J. R., 3rd, Fang G. (2009) Plk1 and Aurora A regulate the depolymerase activity and the cellular localization of Kif2a. J. Cell Sci. 122, 1334–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Song S. J., Song M. S., Kim S. J., Kim S. Y., Kwon S. H., Kim J. G., Calvisi D. F., Kang D., Lim D. S. (2009) Aurora A regulates prometaphase progression by inhibiting the ability of RASSF1A to suppress APC-Cdc20 activity. Cancer Res. 69, 2314–2323 [DOI] [PubMed] [Google Scholar]
- 30. Hochegger H., Hégarat N., Pereira-Leal J. B. (2013) Aurora at the pole and equator: overlapping functions of Aurora kinases in the mitotic spindle. Open Biol. 3, 120185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carmena M., Ruchaud S., Earnshaw W. C. (2009) Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell Biol. 21, 796–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu D., Vader G., Vromans M. J., Lampson M. A., Lens S. M. (2009) Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science 323, 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang E., Ballister E. R., Lampson M. A. (2011) Aurora B dynamics at centromeres create a diffusion-based phosphorylation gradient. J. Cell Biol. 194, 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lampson M. A., Cheeseman I. M. (2011) Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 21, 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Przewloka M. R., Glover D. M. (2009) The Kinetochore and the centromere: a working long distance relationship. Ann. Rev. Genet. 43, 439–465 [DOI] [PubMed] [Google Scholar]
- 36. Santaguida S., Vernieri C., Villa F., Ciliberto A., Musacchio A. (2011) Evidence that Aurora B is implicated in spindle checkpoint signalling independently of error correction. EMBO J. 30, 1508–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saurin A. T., van der Waal M. S., Medema R. H., Lens S. M., Kops G. J. (2011) Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat. Commun. 2, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. DeBonis S., Skoufias D. A., Lebeau L., Lopez R., Robin G., Margolis R. L., Wade R. H., Kozielski F. (2004) In vitro screening for inhibitors of the human mitotic kinesin Eg5 with antimitotic and antitumor activities. Mol Cancer Ther. 3, 1079–1090 [PubMed] [Google Scholar]
- 39. Skoufias D. A., DeBonis S., Saoudi Y., Lebeau L., Crevel I., Cross R., Wade R. H., Hackney D., Kozielski F. (2006) S-trityl-l-cysteine is a reversible, tight binding inhibitor of the human kinesin Eg5 that specifically blocks mitotic progression. J. Biol. Chem. 281, 17559–17569 [DOI] [PubMed] [Google Scholar]
- 40. Dutertre S., Cazales M., Quaranta M., Froment C., Trabut V., Dozier C., Mirey G., Bouché J. P., Theis-Febvre N., Schmitt E., Monsarrat B., Prigent C., Ducommun B. (2004) Phosphorylation of CDC25B by Aurora A at the centrosome contributes to the G2-M transition. J. Cell Sci. 117, 2523–2531 [DOI] [PubMed] [Google Scholar]
- 41. Seki A., Coppinger J. A., Jang C. Y., Yates J. R., Fang G. (2008) Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science 320, 1655–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sonnen K. F., Gabryjonczyk A.-M., Anselm E., Stierhof Y.-D., Nigg E. A. (2013) Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J. Cell Sci. 126, 3223–3233 [DOI] [PubMed] [Google Scholar]
- 43. Li J., Meyer A. N., Donoghue D. J. (1997) Nuclear localization of cyclin B1 mediates its biological activity and is regulated by phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 94, 502–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hans F., Skoufias D. A., Dimitrov S., Margolis R. L. (2009) Molecular distinctions between Aurora A and B: a single residue change transforms aurora A into correctly localized and functional Aurora B. Mol. Biol. Cell 20, 3491–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Giet R., Prigent C. (2001) The non-catalytic domain of the Xenopus laevis Aurora A kinase localises the protein to the centrosome. J. Cell Sci. 114, 2095–2104 [DOI] [PubMed] [Google Scholar]
- 46. Scrittori L., Skoufias D. A., Hans F., Gerson V., Sassone-Corsi P., Dimitrov S., Margolis R. L. (2005) A small C-terminal sequence of Aurora B is responsible for localization and function. Mol. Biol. Cell 16, 292–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marumoto T., Honda S., Hara T., Nitta M., Hirota T., Kohmura E., Saya H. (2003) Aurora A kinase maintains the fidelity of early and late mitotic events in HeLa cells. J. Biol. Chem. 278, 51786–51795 [DOI] [PubMed] [Google Scholar]
- 48. Bian M., Fu J., Yan Y., Chen Q., Yang C., Shi Q., Jiang Q., Zhang C. (2010) Short exposure to paclitaxel induces multipolar spindle formation and aneuploidy through promotion of acentrosomal pole assembly. Sci. China Life Sci. 53, 1322–1329 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.