Abstract
Viral metagenomics has recently revealed the ubiquitous and diverse nature of single-stranded DNA (ssDNA) viruses that encode a conserved replication initiator protein (Rep) in the marine environment. Although eukaryotic circular Rep-encoding ssDNA (CRESS-DNA) viruses were originally thought to only infect plants and vertebrates, recent studies have identified these viruses in a number of invertebrates. To further explore CRESS-DNA viruses in the marine environment, this study surveyed CRESS-DNA viruses in various marine invertebrate species. A total of 27 novel CRESS-DNA genomes, with Reps that share less than 60.1% identity with previously reported viruses, were recovered from 21 invertebrate species, mainly crustaceans. Phylogenetic analysis based on the Rep revealed a novel clade of CRESS-DNA viruses that included approximately one third of the marine invertebrate associated viruses identified here and whose members may represent a novel family. Investigation of putative capsid proteins (Cap) encoded within the eukaryotic CRESS-DNA viral genomes from this study and those in GenBank demonstrated conserved patterns of predicted intrinsically disordered regions (IDRs), which can be used to complement similarity-based searches to identify divergent structural proteins within novel genomes. Overall, this study expands our knowledge of CRESS-DNA viruses associated with invertebrates and explores a new tool to evaluate divergent structural proteins encoded by these viruses.
Keywords: single-stranded DNA virus, CRESS-DNA virus, circular DNA virus, intrinsically disordered proteins (IDPs), intrinsically disordered regions (IDRs), marine invertebrate, crustaceans
Introduction
Viral metagenomics, or shotgun sequencing of total nucleic acids from purified virus particles, enables examination of viral communities without prior knowledge of the viruses present, thus resulting in an unprecedented view of viral diversity (Breitbart et al., 2002; Edwards and Rohwer, 2005; Angly et al., 2006). This technique has uncovered many novel viral types and extended the environmental distribution of known viral groups (Delwart, 2007; Rosario and Breitbart, 2011). In particular, the incorporation of rolling circle amplification (RCA) into viral metagenomic studies has unearthed a high diversity and wide distribution of eukaryotic viruses with circular, single-stranded DNA (ssDNA) genomes that encode a conserved replication initiator protein (Rep; Delwart and Li, 2012; Rosario et al., 2012a). Before the metagenomics era, eukaryotic circular Rep-encoding ssDNA (CRESS-DNA) viruses were only known in agricultural and medical fields since they are known plant (Geminiviridae and Nanoviridae) and vertebrate (Circoviridae) pathogens. However, over the past decade metagenomic approaches have revealed the ubiquitous nature of eukaryotic CRESS-DNA viruses, with reports from various environments, including deep-sea vents (Yoshida et al., 2013), Antarctic lakes and ponds (López-Bueno et al., 2009; Zawar-Reza et al., 2014), wastewater (Rosario et al., 2009b; Roux et al., 2013; Kraberger et al., 2015; Phan et al., 2015), freshwater lakes (Roux et al., 2012, 2013), oceans (Rosario et al., 2009a; Labonte and Suttle, 2013; Roux et al., 2013), hot springs (Diemer and Stedman, 2012), the near-surface atmosphere (Whon et al., 2012; Roux et al., 2013), and soils (Kim et al., 2008; Reavy et al., 2015). Novel CRESS-DNA viruses have also been discovered from fecal samples of a variety of vertebrates (Blinkova et al., 2010; Li et al., 2010a,b; Phan et al., 2011; Ge et al., 2012; Ng et al., 2012; Sachsenroder et al., 2012; van den Brand et al., 2012; Cheung et al., 2013, 2014; Sikorski et al., 2013a; Garigliany et al., 2014; Lian et al., 2014; Smits et al., 2014; Zhang et al., 2014; Sasaki et al., 2015). Notably, CRESS-DNA viruses similar to circoviruses, which were previously thought to only infect vertebrates, have now been identified in a myriad of invertebrates, including insects (Ng et al., 2011; Rosario et al., 2011, 2012b; Dayaram et al., 2013; Padilla-Rodriguez et al., 2013; Pham et al., 2013a,b; Garigliany et al., 2015), crustaceans (Dunlap et al., 2013; Hewson et al., 2013a,b; Ng et al., 2013; Pham et al., 2014), cnidarians (Soffer et al., 2014), and gastropods (Dayaram et al., 2015a), suggesting that CRESS-DNA viruses may be prevalent amongst unexplored taxa.
Well-studied viruses from the Circoviridae, Nanoviridae, and Geminiviridae families demonstrate the rapid evolutionary potential of CRESS-DNA viruses due to high nucleotide substitution rates (Duffy et al., 2008; Duffy and Holmes, 2009) as well as mechanistic predispositions to recombination (Lefeuvre et al., 2009; Martin et al., 2011). These characteristics, combined with the high level of recently reported diversity, highlight the need to continually revisit taxonomic classification of this viral group to add new species, genera and/or families. However, this task is complicated by the fact that many of the CRESS-DNA virus genomes exhibit novel genome architectures, only share similarities to the highly conserved Rep of known viruses, and have similarities to viruses belonging to multiple different taxonomic groups (Rosario et al., 2012a; Roux et al., 2013). In addition, the definitive hosts for many of these CRESS-DNA viruses remain unknown, hindering their classification according to traditional standards.
CRESS-DNA viruses are characterized by small genomes (∼1.7–3 kb) that contain 2–6 protein-encoding genes. The smallest monopartite CRESS-DNA viruses, members of the Circoviridae family, exhibit only two major open reading frames (ORFs), which encode a Rep and a capsid protein (Cap). Many of the novel eukaryotic CRESS-DNA viral genomes obtained from environmental samples or individual organisms through either metagenomic sequencing or degenerate PCR (herein referred to as “metagenomic CRESS-DNA viruses”) exhibit similarities to circoviruses and have been referred to as ‘circo-like’ viruses. Although many of the metagenomic circo-like virus genomes are highly divergent, these surveys have uncovered a novel CRESS-DNA viral group, the proposed Cyclovirus genus (Li et al., 2010a). Cycloviruses, which form a sister group to the Circovirus genus within the family Circoviridae, have been identified from both vertebrates (Li et al., 2010a; Smits et al., 2013; Tan Le et al., 2013; Garigliany et al., 2014; Zhang et al., 2014) and invertebrates (Rosario et al., 2011, 2012b; Dayaram et al., 2013, 2014, 2015b; Padilla-Rodriguez et al., 2013).
Similarities to circoviruses are mainly based on the Rep whereas the second major ORF in novel circo-like metagenomic CRESS-DNA viruses generally does not have any significant matches in the database but is assumed to encode for a structural protein based on the genomic architecture of known circoviruses. In lieu of significant matches to known structural proteins in the GenBank database, it is important to investigate putative novel Caps in CRESS-DNA viruses to provide evidence regarding their structural function. A potential avenue to identify conserved patterns in highly divergent structural proteins, such as those observed in novel metagenomic CRESS-DNA viruses, is to investigate the presence of predicted intrinsically disordered regions (IDRs). IDRs are regions within a protein that lack a rigid or fixed (i.e., ordered) structure, allowing a protein to exist in different states depending on the substrate with which it is interacting (Dunker et al., 2001; Brown et al., 2011). Research examining IDRs within viral proteomes has revealed that smaller viral genomes, such as those of CRESS-DNA viruses, contain a higher proportion of predicted disordered residues than larger viruses (Xue et al., 2012, 2014; Pushker et al., 2013). Therefore it has been suggested that small viruses may exploit IDRs to encode multifunctional proteins (Xue et al., 2012, 2014; Pushker et al., 2013). Since structural proteins in several viral families commonly contain IDRs (Chen et al., 2006; Goh et al., 2008a,b; Chang et al., 2009; Jensen et al., 2011), the presence of similar patterns of predicted disorder amongst unidentified CRESS-DNA proteins may provide one line of evidence for these proteins representing putative Caps.
To contribute to efforts exploring the diversity of CRESS-DNA viruses in invertebrates, this study investigated various marine invertebrate species for the presence of these viruses. A total of 27 novel CRESS-DNA genomes were recovered from 21 invertebrate species, expanding the known diversity of CRESS-DNA viruses associated with marine organisms and providing the first evidence of viruses associated with some under-sampled taxa. The well-conserved Rep of CRESS-DNA viruses was used to explore the relationships between these novel viruses and previously reported eukaryotic CRESS-DNA viruses in GenBank, including metagenomic CRESS-DNA viruses. In addition, the non-Rep-encoding ORFs (i.e., putative Caps) within these genomes were investigated for IDRs. Disorder prediction methods suggest that CRESS-DNA viral Caps exhibit conserved patterns of predicted disorder, which can be used to complement similarity-based searches to identify structural proteins within novel CRESS-DNA viral genomes.
Materials and Methods
Sample Processing and Genome Discovery
CRESS-DNA viruses were investigated in a variety of marine invertebrate species that were collected as samples of opportunity (Table 1 and Supplementary Table S1). Specimens were identified with the highest degree of taxonomic resolution possible based on morphology. Whole organisms or tissue sections were serially rinsed three times using sterile SM Buffer [0.1 M NaCl, 50 mM Tris-HCl (pH 7.5), 10 mM MgSO4]. Viral particles were partially purified from each specimen prior to DNA extraction. For this purpose, samples were homogenized in one of two ways depending on the size of the specimen. Smaller organisms or dissected tissues that could be placed in a 1.5 ml microcentrifuge tube were homogenized in 1 ml of sterile SM Buffer through bead-beating using 1.0 mm sterile glass beads in a bead beater (Biospec Products). Homogenates were then centrifuged at 6000 × g for 6 min. Larger organisms or tissues of dissected organisms, such as muscle or gonads, were placed in a gentleMACSTM M tube (Miltenyl Biotec) containing 3 ml of sterile SM buffer. Samples were then homogenized using a gentleMACS dissociator (Miltenyl Biotec) followed by centrifugation at 6000 × g for 9 min. The supernatant from both homogenization methods was filtered through a 0.45 μm Sterivex filter (Millipore) and nucleic acids were extracted from 200 μl of filtrate using the QIAmp MinElute Virus Spin Kit (Qiagen).
Table 1.
CRESS-DNA genomes identified in this study, the organism they were obtained from, and genome details (acronym, genome length, nonanucleotide motif, genome type, and ORFs identified).
| Genome1 | Organism | Tissue type | Genome (bp) | Genomic architecture | Nonanucleotide2 | Cap3 | Rep |
|---|---|---|---|---|---|---|---|
| P. diogenes Giant Hermit Crab aCV(I0004A) | Petrochirus diogenes | Abdomen | 1815 | Type V | TAGTATTAC | X∗ | X |
| Palaemonete sp. Common Grass Shrimp aCV (I0006H) | Palaemonete sp. | Hepatopancreas | 2257 | Type II | TAGTATTAC | X∗ | X |
| Aiptasia sp. Sea Anemone aCV (I0007C2) | Aiptasia sp. | Whole organism | 1901 | Type I | CATTATTAC | X | X |
| Aiptasia sp. Sea Anemone aCV (I0007C3) | Aiptasia sp. | Whole organism | 1942 | Type I | CATTATTAC | X | X |
| L. variegatus Variable Sea Urchin aCV (I0021) | Lytechinus variegatus | Gonads | 2167 | Type III | GACTATTAC∗ | X∗ | X |
| Didemnum sp. Sea Squirt aCV (I0026A4) | Didemnum sp. | Whole organism | 2061 | Type IV | CAGTATTAC | X | X |
| Didemnum sp. Sea Squirt aCV (I0026A7) | Didemnum sp. | Whole organism | 2143 | Type I | CAGTATTAC | X∗ | X |
| Littorina sp. Snail aCV (I0041) | Littorina sp. | Whole organism | 2237 | Type II | CAGTATTAC | X | X |
| C. ornatus Ornate Blue Crab aCV (I0054) | Callinectes ornatus | Gonads | 1241 | Type I | CAGTATTAC | X | X |
| C. sapidus Atlantic Blue Crab aCV (I0056) | Callinectes sapidus | Gonads | 1876 | Type I | CAGTATTAC | X | X |
| P. intermedius Brackish Grass Shrimp aCV (I0059) | Palaemonetes intermedius | Whole organism | 2293 | Type I | CAGTATTAC | X∗ | X |
| F. duorarum Pink Shrimp aCV (I0066) | Farfantepenaeus duorarum | Whole organism | 1799 | Type I | CAGTATTAC | X | X |
| F. duorarum Pink Shrimp aCV (I0069) | Farfantepenaeus duorarum | Whole organism | 1966 | Type I | CAGTATTAC | X∗ | X |
| Marine Snail aCV (I0084) | Marine Snail | Whole organism | 2305 | Type I | TAGTATTAC | X∗ | X |
| Hermit Crab aCV (I0085A4) | Hermit Crab | Abdomen | 2291 | Type I | TAGTATTAC | X∗ | X |
| Hermit Crab aCV (I0085A5) | Hermit Crab | Abdomen | 2291 | Type I | TAGTATTAC | X∗ | X |
| Hermit Crab aCG (I0085b) | Hermit Crab | Abdomen | 1063 | Type VII | CAGTATTAC | X | |
| Fiddler Crab aCV (I0086a) | Fiddler Crab | Gonads and claw muscle | 1635 | Type II | GATTATTAC | X | X |
| Fiddler Crab aCV (I0086b) | Fiddler Crab | Gonads and claw muscle | 1511 | Type V | AAGTATTAC | X | X |
| P. kadiakensis Mississippi Grass Shrimp aCV (I0099) | Palaemonetes kadiakensis | Whole organism | 1895 | N/A | None | X∗ | X |
| Gammarus sp. Amphipod aCV (I0153) | Gammarus sp. | Whole organism | 1999 | Type I | TAGTATTAC | X∗ | X |
| Mytilus sp. Clam aCV (I0169) | Mytilus sp. | Whole organism | 1894 | Type I | TAGTATTAC | X | X |
| Calanoida sp. Copepod aCV (I0298) | Calanoida sp. | Whole organism | 2469 | Type II | TAGTATTAC | X | X |
| A. melana Sponge aCG (I0307) | Artemia melana | Tissue segment | 1826 | Type VII | TAGTATTAC | X | |
| P. pacifica Coral aCV (I0345) | Primnoa pacifica | Polyps | 1240 | N/A | None | X∗ | X |
| P. placomus Coral aCV (I0351) | Paramuricea placomus | Polyps | 2292 | Type II | TAGTATTAC | X∗ | X |
| S. brevirostris Brown Rock Shrimp aCV (I0722) | Sicyonia brevirostris | Gonads | 1600 | Type V | TAATATTAC∗ | X | X |
1Genome names contain abbreviation aCV for associated circular virus or aCG for associated circular genome. ID within parentheses corresponds to ID used throughout the paper.
2Nonanucleotide motif sequences that were not identified within a stem-loop structure are denoted with an asterisk (∗).
3Non-Rep encoding ORFs were identified as putative capsid proteins based on BLAST results. However, many non-Rep-encoding ORFs did not exhibit any significant matches (marked with an asterisk∗).
DNA extracts were amplified through RCA using the illustra TempliPhi Amplification kit (GE Healthcare) to enrich for small circular templates (Kim et al., 2008; Kim and Bae, 2011). RCA-amplified DNA was digested with a suite of FastDigest restriction enzymes (Life Technologies; BamHI, EcoRV, PdmI, HindIII, KpnI, PstI, XhoI, SmaI, BgiII, EcoRI, XbaI, and NcoI) following manufacturer’s instructions in separate reactions to obtain complete, unit-length genomes for downstream cloning and sequencing. Restriction enzyme digested products were resolved on an agarose gel and bands ranging in size from 1000 to 4000 bp were excised and cleaned using the Zymoclean Gel DNA Recovery Kit (Zymo Research). Products resulting from blunt-cutting enzyme digestions were cloned using the CloneJET PCR Cloning kit (Life Technologies), whereas products containing sticky ends were cloned using pGEM-3Zf(+) vectors (Promega) pre-digested with the appropriate enzyme. All clones were commercially Sanger sequenced using vector primers and genomes exhibiting significant similarities to eukaryotic CRESS-DNA viruses were completed through primer walking.
Genome Annotation
Genomes were assembled using Sequencher 4.1.4 (Gene Codes Corporation). Putative ORFs >100 amino acids were identified and annotated using SeqBuilder version 11.2.1 (Lasergene). Partial genes or genes that seemed interrupted were analyzed for potential introns using GENSCAN (Burge and Karlin, 1997). The potential origin of replication (ori) for each genome was identified by locating a canonical nonanucleotide motif (NANTATTAC; Rosario et al., 2012a) and confirming predicted stem-loop structures using Mfold with constraints applied to prevent hairpin formation within the nonanucleotide motif and a folding temperature set at 17°C (Zuker, 2003). Final annotated genomes have been deposited to GenBank with accession numbers KR528543–KR528569.
Database Sequences and Sequence Analysis
To conduct sequence comparisons, members of the Circovirus genus, as well as complete eukaryotic CRESS-DNA viral genomes obtained from environmental samples or individual organisms through either metagenomic sequencing or degenerate PCR (herein referred to as “metagenomic CRESS-DNA viruses”) were retrieved from GenBank. Since the Rep is the only conserved protein among CRESS-DNA viruses (Ilyina and Koonin, 1992; Rosario et al., 2012a) this protein was used to compare the different genomes. Rep pairwise identities were calculated using SDT v1.2 (Muhire et al., 2014) and summarized using heat maps generated in R (R Core Team, 2014). A maximum likelihood (ML) phylogenetic tree based on Rep amino acid sequences was also constructed. For this purpose, alignments were performed in MEGA 6.06 (Tamura et al., 2013) using the MUSCLE algorithm (Edgar, 2004) and manually edited. Sequences were inspected for the presence of conserved amino acid motifs that have been shown to play a role in rolling circle replication (RCR) of eukaryotic CRESS-DNA viruses, including three RCR and three superfamily 3 (SF3) helicase motifs (Gorbalenya et al., 1990; Ilyina and Koonin, 1992; Gorbalenya and Koonin, 1993; Rosario et al., 2012a). Although all the recently reported CRESS-DNA viruses are included in the heatmap, only sequences exhibiting all six motifs are included in the phylogenetic analysis. In addition, divergent regions that were poorly aligned, as shown by a high percentage of gaps, were removed from the alignment (Supplementary Data Sheet 1). Since the Nanoviridae and Geminiviridae are also CRESS-DNA viral families that are evolutionarily related to the Circoviridae (Ilyina and Koonin, 1992; Rosario et al., 2012a), select representatives of these families were included in the phylogenetic analysis. The ML phylogenetic tree was inferred using PHYML (Guindon et al., 2010) implementing the best substitution model (rtRev+I+G+F; Dimmic et al., 2002) according to ProtTest (Abascal et al., 2005). Branch support was assessed using the approximate likelihood ratio test (aLRT) SH-like method (Anisimova and Gascuel, 2006).
Intrinsically Disordered Region (IDR) Analysis of Putative Capsid Proteins
To determine if the non-Rep-encoding ORFs from the CRESS-DNA viral genomes presented here (n = 25), circoviruses (n = 15), and metagenomic CRESS-DNA viruses (n = 259; including 37 cycloviruses) represent putative Caps, these proteins were evaluated for IDRs. Disordered protein regions were predicted using the DisProt VL3 disorder predictor (Obradovic et al., 2003; Sickmeier et al., 2007). This artificial neural network utilizes an ensemble of feed forward neural networks with 20 attributes (18 amino acid frequencies, average flexibility, and sequence complexity; Obradovic et al., 2003). Disorder disposition scores above a 0.5 threshold indicate intrinsic disorder. Counts and statistical analysis for the fraction of disorder- and order-promoting amino acid residues was conducted using R with the “seqinr” package (Charif and Lobry, 2007).
Results
A total of 27 CRESS-DNA genomes were recovered from 21 marine invertebrates (Table 1). Most of the recovered genomes (66.7%) were identified from Crustacea, mainly from the order Decapoda. Recovered genomes ranged in size from 1063 to 2469 nt and exhibited a variety of genome architectures. Of the 27 genomes identified, 23 exhibited a common putative ori marked by a conserved nonanucleotide motif (NANTATTAC) at the apex of a predicted stem-loop structure (Table 1). The remaining four genomes lacked a stem-loop structure (n = 2) or a stem-loop structure and a nonanucleotide motif (n = 2). Genomes lacking the canonical nonanucleotide motif could not be assigned to any genome type; therefore only 25 genomes were assigned to genomic architecture types previously described by Rosario et al. (2012a) (Figure 1). The predominant genomic architecture observed was Type I (n = 13), which is typical of members of the Circovirus genus. However, other genomic architectures were observed including Types II (n = 5), III (n = 1), IV (n = 1), V (n = 3), and VII (n = 2) (Figure 1). It is important to note that genomes exhibiting a Type VII genome architecture only exhibit a single major ORF encoding a Rep. This type of architecture is observed in genomic components of multipartite viruses from the Nanoviridae family and satellite DNA molecules that require helper viruses for encapsidation (Gronenborn, 2004; Briddon and Stanley, 2006). Therefore genomes exhibiting only a single major ORF may represent partial genomes of multipartite viruses or non-viral mobile genetic elements such as plasmids (Rosario et al., 2012a).
FIGURE 1.
Genome types of novel CRESS-DNA genomes identified in this study (Rosario et al., 2012a). Genome schematics illustrate a major ORF encoding the replication initiator protein (Rep), putative origin of replication (ori) marked by stem-loop structure, and a second major ORF.
The majority of the CRESS-DNA viruses detected in marine invertebrates were most similar to viral sequences identified through metagenomic surveys of marine samples (Supplementary Table S1). However, one of genomes, Lytechinus variegatus variable sea urchin associated circular virus_I0021, was most similar to plant viruses from the Geminiviridae family. Most of the viral genomes had database similarities for the Rep; except for Sicyonia brevirostris brown rock shrimp associated circular virus_I0722, which only had similarities for the putative Cap (Supplementary Table S1). Similar to several previously described CRESS-DNA viruses (Li et al., 2010a; Rosario et al., 2012b; van den Brand et al., 2012; Sikorski et al., 2013b; Du et al., 2014; Ng et al., 2014; Dayaram et al., 2015a,b; Kraberger et al., 2015), three viral genomes (Artemia melana sponge associated circular virus_I0307, Didemnum sp. sea squirt associated circular virus_I0026_A7, and Palaemonetes kadiakensis Mississippi grass shrimp associated circular virus_I0099) exhibited Reps interrupted by introns (Supplementary Table S1).
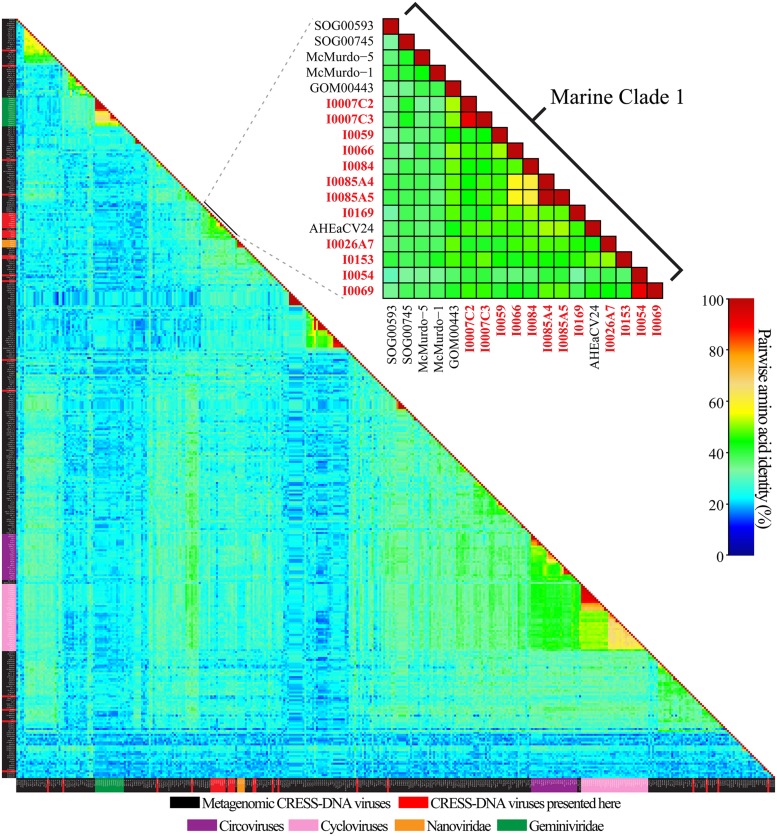
Pairwise identities indicate that the CRESS-DNA viruses detected in marine invertebrates share less than 60.1% sequence identity (average sequence identity = 26.04%) with previously identified Reps from CRESS-DNA viruses in GenBank, indicating that these viruses represent novel species (Figure 2). Twenty-one of the 27 recovered Reps contained all six conserved RCR and helicase motifs (see Materials and Methods) and were used for phylogenetic analysis. Analysis of these Reps with representative CRESS-DNA viral Reps from GenBank, including available metagenomic CRESS-DNA viral Reps, show that most of the sequences from marine invertebrate associated viruses detected here are more closely related to circo-like viruses recovered through metagenomic surveys of the marine environment than to previously defined CRESS-DNA viral groups (Figure 3). Eleven of the 21 Reps from marine invertebrate associated viruses do not form distinct clusters with each other or any known sequences (Figure 3). However, ten of the Reps form a well-supported clade that also includes sequences detected in the Gulf of Mexico (GOM00443; JX904231.1), Straight of Georgia (JX904106.1), McMurdo Ice Shelf (YP_009047125.1; YP_009047137.1), and a semi-enclosed shallow estuary (Avon-Heathcote Estuary associated circular virus 24; AJP36460.1). Pairwise identity scores indicate that all members of this clade, named Marine Clade 1 for the purposes of this study, share more than 32.7% identity, with an average pairwise identity score of 47.2% (Figure 2). Members of the Marine Clade 1 seem to be more closely related to members of the Nanoviridae (31.95% average pairwise identity) than any other known CRESS-DNA viral group; however, members of this clade exhibit different genomic architectures compared to these plant viruses. CRESS-DNA viral genomes from the Marine Clade 1 encode two major ORFs in an ambisense organization (i.e., Type I architecture), which is similar to members of the Circoviridae, rather than the single ORF, Type VII genome organization observed in genomic components from the Nanoviridae.
FIGURE 2.
Graphical representation of pairwise amino acid identities of the replication initiator proteins (Rep) from CRESS-DNA genomes from this study, metagenomic CRESS-DNA viruses, cycloviruses, circoviruses, and select members of the Nanoviridae and Geminiviridae families. Reps identified from this study within the Marine Clade 1 are in red font. Description of acronyms and the matrix used to generate the heatmap can be found in Supplementary Tables S2 and S3, respectively.
FIGURE 3.
Multifurcation maximum likelihood phylogenetic reconstruction based on the Reps of CRESS-DNA genomes recovered here, metagenomic CRESS-DNA viruses, cycloviruses, circoviruses, and representative members of the Nanoviridae and Geminiviridae families. Reps obtained from CRESS-DNA genomes obtained in this study are highlighted in blue font. Branches are colored for the different CRESS-DNA viral groups including the Marine Clade 1 (red), circoviruses (purple), cycloviruses (pink), nanoviruses (orange), and geminiviruses (green). Representative nanoviruses (n = 4) and geminiviruses (n = 15) have been condensed into their family names. Reps from genomes exhibiting a single ORF are highlighted using an asterisk (∗). Branches with less than 60% aLRT branch support have been collapsed. Description of acronyms used can be found in Supplementary Table S4.
Capsid Analysis
Only half of the CRESS-DNA viral genomes described here contained an ORF that had significant BLASTX matches (e-value < 0.001; amino acid identities ranging from 26–54%) to proteins annotated as putative Caps in GenBank (Table 1). Furthermore, most of the matches in the database were to putative CRESS-DNA viral Caps detected through metagenomic surveys, which are not supported by biochemical data and have not necessarily been well curated. Therefore, alternative methods were explored to investigate non-Rep-encoding ORFs (i.e., putative Caps) found in CRESS-DNA viral genomes.
The majority of metagenomic CRESS-DNA viruses reported from marine invertebrates in this study and in GenBank are most similar to previously described circoviruses. Therefore, the predicted IDP profiles of well-characterized members of the Circovirus genus were examined in an effort to identify conserved patterns in structural proteins encoded by these viruses. These circovirus IDP profiles were then compared against profiles observed in cycloviruses (the proposed sister group to the circoviruses, which exhibit conserved features and share high identities with circoviruses) and other metagenomic CRESS-DNA viruses.
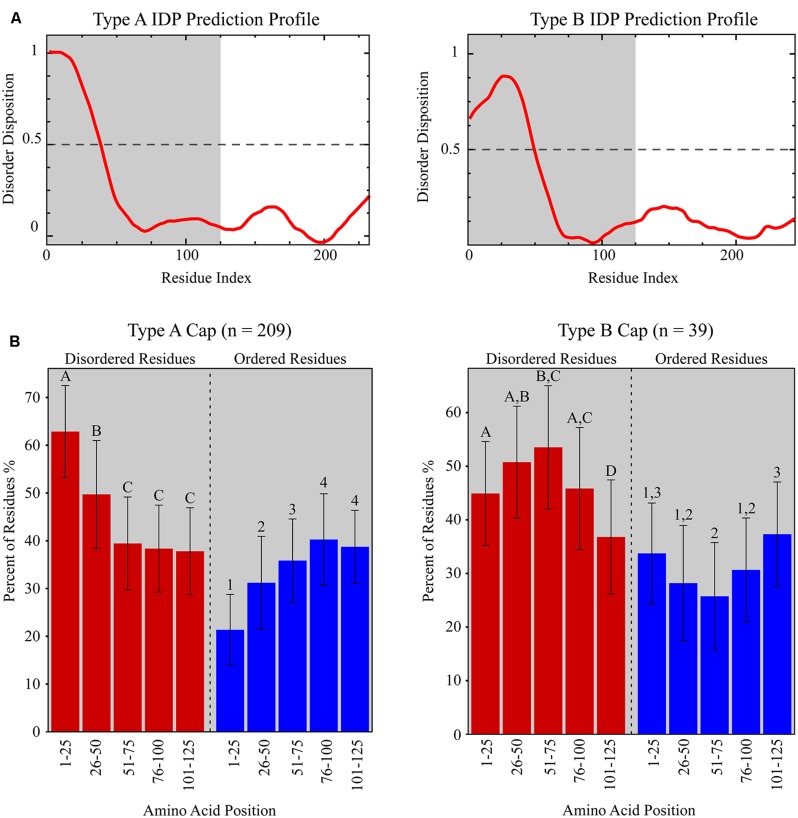
The DisProt VL3 disorder prediction analysis revealed that Caps encoded by members of the Circovirus genus (n = 15) exhibit one of two protein disorder profiles, distinguished here as Type A or Type B, based on the first 125 amino acids of these proteins (Figure 4A). Type A Caps exhibit IDP profiles that are predicted to have the highest degree of disorder closest to the N-terminus (i.e., amino acid residues 1–50) before the profile tapers to a structured region with variable predicted disorder. Type A Caps exhibit significant enrichment for amino acid residues that promote disorder (R, K, E, P, S, Q, and A) within the first 50 residues relative to amino acid residues 51–125 (ANOVA with post hoc Tukey’s HSD; p < 0.05) and a depletion of order promoting amino acid residues (W, C, F, I, Y, V, L, and N) within the first 25 residues relative to amino acid residues 26–125 (ANOVA with post hoc Tukey’s HSD; p < 0.05; Figure 4B). On the other hand, Type B Caps exhibit IDP profiles that peak in predicted disorder between amino acid residues 26–75. Type B Caps show an enrichment of disorder promoting residues between residue positions 26 through 75, whereas there is a depletion of predicted order promoting residues in this region compared to residues 1–25 and 76–125 (Figure 4B). Beyond 125 amino acids, IDP profiles exhibited more structured regions for both Types A and B Caps, with no distinguishable predicted disorder pattern (Figure 4A).
FIGURE 4.
(A) Representative IDP prediction profiles for Type A and Type B capsid proteins (Caps) from the Disprot VL3 predictor. Type A and Type B IDP prediction profiles are based on the Porcine circovirus 2 Cap (NP_937957.1) and the Beak and feather disease virus Cap (NP_047277.1), respectively. The grey shaded area represents the amino acid residue interval used in (B). (B) Graphs showing the fraction of predicted disordered (red bars) and ordered (blue bars) residues within discrete amino acid intervals for Type A and Type B Caps identified from all CRESS-DNA viral genomes analyzed in this study. Significantly different amino acid intervals for each Cap type are distinguished using letters (“A”, “B”, “C”, “D” for statistics based on percentage of predicted disordered residues) or numbers (“1”, “2”, “3”, “4” for statistics based on percentage of predicted ordered residues; ANOVA with post hoc Tukey’s HSD; p < 0.05). Note that the percentage of predicted disordered and ordered residues does not add to 100% due to the presence of residues that are not considered either disordered or ordered (i.e., H, M, T, and D).
The overwhelming majority of Caps from the Circovirus genus (86.7%) exhibited Type A IDP profiles; however, two avian circoviruses, Finch circovirus (YP_803551.1) and Beak and feather disease virus (NP_047277.1), had Type B IDP profiles (Table 2 and Supplementary Table S5). Similarly, 97.3% of cyclovirus putative Caps (n = 37) exhibited Type A IDP profiles. Comparison of IDP profiles showed that a majority of metagenomic CRESS-DNA viruses also contained patterns of increased predicted disorder at the N-terminus of the putative Cap, consistent with the Circoviridae. Interestingly, Type B IDP profiles were more prevalent among putative Caps from metagenomic CRESS-DNA viral genomes in GenBank (10.8%; n = 222) and the novel genomes reported in this study (56%; n = 25). Notably, 7 of the 10 viruses found in the Marine Clade 1 described here exhibit Type B Caps. Among the total 299 CRESS-DNA genome sequences analyzed, most putative Caps exhibit Type A IDP profiles (69.9%), followed by Type B (13%). Notably, most of the putative Caps lacking a significant match in the database exhibited one these profiles.
Table 2.
Intrinsically disordered protein (IDP) profile types identified in non-Rep encoding ORFs of CRESS-DNA viruses.
| Group | Total sequences | IDP Cap type |
||
|---|---|---|---|---|
| Type A | Type B | No type | ||
| Circoviruses | 15 | 86.7% | 13.3% | 0.0% |
| Cycloviruses | 37 | 97.3% | 0.0% | 2.7% |
| Metagenomic CRESS-DNA viruses | 222 | 67.6% | 10.8% | 21.6% |
| This study | 25 | 40.0% | 56.0% | 4.0% |
| Total | 299 | 69.9% | 13.0% | 17.1% |
Discussion
Metagenomic studies have revealed a prodigious amount of diversity in eukaryotic CRESS-DNA viruses in the marine environment (Rosario et al., 2009a; Rosario and Breitbart, 2011; Labonte and Suttle, 2013; McDaniel et al., 2014). However, few studies have isolated these viruses directly from organisms. Building upon recent studies suggesting that CRESS-DNA viruses are associated with marine invertebrates (Dunlap et al., 2013; Hewson et al., 2013a,b; Ng et al., 2013; Pham et al., 2014; Soffer et al., 2014; Dayaram et al., 2015a), this study investigated a variety of marine invertebrates, including under sampled taxa, for the presence of these viruses. Viral genomes presented here were primarily recovered from Crustacea, suggesting that this subphylum harbors a rich diversity of CRESS-DNA viruses. This is consistent with previous research that identified CRESS-DNA viruses in copepods (Dunlap et al., 2013), which are the most abundant members of mesozooplankton (Kleppel et al., 1996), as well as different species of shrimp (Ng et al., 2013; Pham et al., 2014), which comprise some of the world’s most important food sources (Goss et al., 2000; Paezosuna, 2003). In addition, this is the first study to report viruses associated with marine snails, anemones, sea squirts, and several crab species. Although a definitive host for these viruses cannot be assigned with the present data, this study reveals the need for further examination of viruses associated with common marine invertebrates and experiments to determine their potential impact, if any, on the ecology of these organisms. The grouping of the invertebrate-associated CRESS-DNA viruses reported here with metagenomic CRESS-DNA viruses implies that marine invertebrates may serve as hosts for many of the sequences obtained from marine environments.
The marine invertebrate associated CRESS-DNA viruses identified here are only distantly related to known members of the Circoviridae and may represent novel groups. Approximately one third of the novel sequences reported here belong to the Marine Clade 1, whose members share an average pairwise identity of 47.2%. Members of this viral clade share an average pairwise identity score of 27.5% with members of the Circoviridae, whose members (genus Circovirus and proposed genus Cyclovirus) share 48.9% average pairwise identity. Although members of the Marine Clade 1 share slightly higher average pairwise identity with the Nanoviridae (31.2%), their genome architecture is clearly distinct from these plant-infecting viruses. Therefore, genomic architectures and comparative Rep analyses suggest that members of the Marine Clade 1 may represent a novel CRESS-DNA viral family.
The highly conserved Rep enables its straightforward identification through similarity-based searches; however, there is currently no reliable method for characterizing highly divergent putative Caps for metagenomic CRESS-DNA viruses. Since many of the novel metagenomic CRESS-DNA viruses are most similar to members of the Circoviridae, which only contain two major ORFs encoding a Rep and Cap, the putative Cap is often assigned simply based on the conserved genome architectures exhibited by this group.
This study investigated the IDP profiles of all available circo-like CRESS-DNA viruses to evaluate if putative Caps exhibit conserved patterns that could be used to identify this structural protein even in the absence of significant similarities in the database. The Cap of Porcine circovirus 2 represents a Type A IDP profile and that of Beak and feather disease virus represents a Type B IDP profile. Since the non-Rep-encoding ORF for both of these circoviruses have been shown to be structural (Nawagitgul et al., 2000; Patterson et al., 2013), this provides evidence that both the Type A and Type B IDP profiles represent a Cap. These Cap IDP profiles may be driven by the arginine and/or lysine rich region at the N-terminus of the Cap (Niagro et al., 1998), as both of these amino acids are considered disorder-promoting residues by the DisProt VL3 neural network. In addition to characterizing IDP profiles of circo-like CRESS-DNA viruses, analysis of select Geminiviridae and Nanoviridae Caps demonstrated that these viruses also exhibit Type A and Type B IDP profiles (Supplementary Table S5). Although further research into these plant virus families is needed, these findings suggest that the IDP patterns identified here may be conserved across Caps from the different families of eukaryotic CRESS-DNA viruses.
Thirteen of the eukaryotic CRESS-DNA viruses presented here had a non-Rep-encoding ORF without any database similarities, which were characterized as a putative Cap based on IDP profiles. Likewise, hypothetical proteins from 32 metagenomic CRESS-DNA viruses were identified as putative Caps using this method (Supplementary Table S5). While the Caps in the database were dominated by Type A IDP profiles, the majority of the new marine invertebrate associated genomes presented here exhibited Type B IDP profiles. In addition, 50 of the CRESS-DNA genomes analyzed here (17.1%; n = 299), including the Primnoa pacifica coral associated circular virus I0345 identified here, contained a non-Rep-encoding ORF that did not exhibit either the Type A or Type B profile. While it is possible that other IDP profiles representative of novel Caps exist, caution should be used in annotating these ORFs as putative Caps without supporting evidence. Finally, while examining metagenomic sequences annotated as CRESS-DNA viruses in GenBank, numerous genomes were identified that only contained a single ORF, which encoded a Rep. These sequences (Supplementary Table S5), along with the two Type VII genomes found in this study, most likely represent partial viral genomes [i.e., a single component of a multipartite virus (Gutierrez, 1999; Gronenborn, 2004)], satellite DNA molecules (Briddon and Stanley, 2006), or non-viral mobile genetic elements (Rosario et al., 2012a). Genomes exhibiting a single ORF cannot be distinguished phylogenetically from complete viral genomes based on the Rep (Figure 3). Therefore, it is important to investigate complete genomes of CRESS-DNA viruses rather than partial sequences.
The IDP analysis has interesting implications for understanding the evolutionary pressures acting upon the Rep and Cap of CRESS-DNA viruses, which include the smallest known eukaryotic viral pathogens. Small viruses exhibit a higher proportion of predicted disordered residues than larger viruses and may exploit IDRs to encode multifunctional proteins (Xue et al., 2012, 2014; Pushker et al., 2013). Rep proteins encoded by CRESS-DNA viruses exhibited low disposition for predicted disorder promoting amino acid residues or an inconsistency in predicted disorder patterns (data not shown), while the Caps consistently exhibited profiles with increased predicted disorder at the N-terminus, suggesting that the high proportion of predicted disordered regions in these small viruses may be driven by the Cap. IDRs have a tendency to evolve more rapidly than structured regions (Brown et al., 2002, 2011; Chen et al., 2006; Bellay et al., 2011; Nilsson et al., 2011; van der Lee et al., 2014); consequently, IDRs may hinder our ability to perform phylogenetic reconstructions based on the Cap. Although we are unable to perform reliable Cap alignments, the ability to classify these proteins within CRESS-DNA virus genomes due to conserved predicted disorder profiles reveals that these viruses exhibit regions in which disorder is conserved despite rapidly evolving amino acids (i.e., flexible disorder; van der Lee et al., 2014).
Although the functional significance of predicted IDP profiles detected in this study has yet to be determined, the identification of conserved IDP profiles may prove useful to identify divergent structural proteins encoded by CRESS-DNA viruses. The identification of a given IDP profile (Type A or B) for a putative ORF in a genomic context may allow the recognition of novel CRESS-DNA viral structural proteins that cannot be identified by standard BLAST searches. The IDP profile analysis needs to be complemented by other genomic features that are characteristic of CRESS-DNA viruses, including the presence of a Rep exhibiting RCR and helicase motifs and a putative ori marked by a conserved nonanucleotide motif (NANTATTAC) at the apex of a stem-loop structure. Future work needs to evaluate if the high proportion of IDRs observed in CRESS-DNA viruses and other small viruses is indeed mainly driven by structural proteins. If this observation is validated, IDP profile analysis of hypothetical proteins may provide a reliable tool to identify structural proteins encoded by small viruses.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Ian Hewson, Renee Bishop-Pierce, Christina Kellogg, Robert W. Thacker, Stan Rice, Sandra Gilchrist, Brandan Cole, Brittany Hall, Ernst Peebles, Ralph Kitzmiller, Scott Burghart, and Elise Pickett for sample donations. We thank Bin Xue for his guidance in the intrinsically disordered protein analysis. This work was funded through grant DEB-1239976 from the National Science Foundation’s Assembling the Tree of Life Program to KR and MB.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00696
References
- Abascal F., Zardoya R., Posada D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21 2104–2105. 10.1093/bioinformatics/bti263 [DOI] [PubMed] [Google Scholar]
- Angly F. E., Felts B., Breitbart M., Salamon P., Edwards R. A., Carlson C., et al. (2006). The marine viromes of four oceanic regions. PLoS Biol. 4:e368 10.1371/journal.pbio.0040368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M., Gascuel O. (2006). Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55 539–552. 10.1080/10635150600755453 [DOI] [PubMed] [Google Scholar]
- Bellay J., Han S., Michaut M., Kim T., Costanzo M., Andrews B. J., et al. (2011). Bringing order to protein disorder through comparative genomics and genetic interactions. Genome Biol. 12 R14 10.1186/gb-2011-12-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinkova O., Victoria J., Li Y., Keele B. F., Sanz C., Ndjango J. B., et al. (2010). Novel circular DNA viruses in stool samples of wild-living chimpanzees. J. Gen. Virol. 91 74–86. 10.1099/vir.0.015446-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M., Salamon P., Andresen B., Mahaffy J. M., Segall A. M., Mead D., et al. (2002). Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U.S.A. 99 14250–14255. 10.1073/pnas.202488399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briddon R. W., Stanley J. (2006). Subviral agents associated with plant single-stranded DNA viruses. Virology 344 198–210. 10.1016/j.virol.2005.09.042 [DOI] [PubMed] [Google Scholar]
- Brown C. J., Johnson A. K., Dunker A. K., Daughdrill G. W. (2011). Evolution and disorder. Curr. Opin. Struct. Biol. 21 441–446. 10.1016/j.sbi.2011.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. J., Takayama S., Campen A. M., Vise P., Marshall T. W., Oldfield C. J., et al. (2002). Evolutionary rate heterogeneity in proteins with long disordered regions. J. Mol. Evol. 55 104–110. 10.1007/s00239-001-2309-6 [DOI] [PubMed] [Google Scholar]
- Burge C., Karlin S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268 78–94. 10.1006/jmbi.1997.0951 [DOI] [PubMed] [Google Scholar]
- Chang C. K., Hsu Y. L., Chang Y. H., Chao F. A., Wu M. C., Huang Y. S., et al. (2009). Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J. Virol. 83 2255–2264. 10.1128/JVI.02001-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charif D., Lobry J. R. (2007). Seqin{R} 1.0-2: a Contributed Package to the {R} Project for Statistical Computing Devoted to Biological Sequences Retrieval and Analysis. New York: Springer Verlag. [Google Scholar]
- Chen J. W., Romero P., Uversky V. N., Dunker A. K. (2006). Conservation of intrinsic disorder in protein domains and families: I. A database of conserved predicted disordered regions. J. Proteome Res. 5 879–887. 10.1021/pr060048x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A. K., Ng T. F., Lager K. M., Alt D. P., Delwart E. L., Pogranichniy R. M. (2014). Unique circovirus-like genome detected in pig feces. Genome Announc. 2:e00251-14 10.1128/genomeA.00251-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A. K., Ng T. F., Lager K. M., Bayles D. O., Alt D. P., Delwart E. L., et al. (2013). A divergent clade of circular single-stranded DNA viruses from pig feces. Arch. Virol. 158 2157–2162. 10.1007/s00705-013-1701-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayaram A., Galatowitsch M., Harding J. S., Arguello-Astorga G. R., Varsani A. (2014). Novel circular DNA viruses identified in Procordulia grayi and Xanthocnemis zealandica larvae using metagenomic approaches. Infect. Genet. Evol. 22 134–141. 10.1016/j.meegid.2014.01.013 [DOI] [PubMed] [Google Scholar]
- Dayaram A., Goldstien S., Arguello-Astorga G. R., Zawar-Reza P., Gomez C., Harding J. S., et al. (2015a). Diverse small circular DNA viruses circulating amongst estuarine molluscs. Infect. Genet. Evol. 31 284–295. 10.1016/j.meegid.2015.02.010 [DOI] [PubMed] [Google Scholar]
- Dayaram A., Potter K. A., Pailes R., Marinov M., Rosenstein D. D., Varsani A. (2015b). Identification of diverse circular single-stranded DNA viruses in adult dragonflies and damselflies (Insecta: Odonata) of Arizona and Oklahoma, USA. Infect. Genet. Evol. 30 278–287. 10.1016/j.meegid.2014.12.037 [DOI] [PubMed] [Google Scholar]
- Dayaram A., Potter K. A., Moline A. B., Rosenstein D. D., Marinov M., Thomas J. E., et al. (2013). High global diversity of cycloviruses amongst dragonflies. J. Gen. Virol. 94 1827–1840. 10.1099/vir.0.052654-0 [DOI] [PubMed] [Google Scholar]
- Delwart E. L. (2007). Viral metagenomics. Rev. Med. Virol. 17 115–131. 10.1002/rmv.532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwart E., Li L. (2012). Rapidly expanding genetic diversity and host range of the Circoviridae viral family and other Rep encoding small circular ssDNA genomes. Virus Res. 164 114–121. 10.1016/j.virusres.2011.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diemer G. S., Stedman K. M. (2012). A novel virus genome discovered in an extreme environment suggests recombination between unrelated groups of RNA and DNA viruses. Bio. Dir. 7 1–14. 10.1186/1745-6150-7-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmic M. W., Rest J. S., Mindell D. P., Goldstein R. A. (2002). rtRev: An amino acid substition matrix for inference of retrovirus and reverse transcriptase phylogeny. J. Mol. Evol. 55 65–73. 10.1007/s00239-001-2304-y [DOI] [PubMed] [Google Scholar]
- Du Z., Tang Y., Zhang S., She X., Lan G., Varsani A., et al. (2014). Identification and molecular characterization of a single-stranded circular DNA virus with similarities to Sclerotinia sclerotiorum hypovirulence-associated DNA virus 1. Arch. Virol. 159 1527–1531. 10.1007/s00705-013-1890-5 [DOI] [PubMed] [Google Scholar]
- Duffy S., Holmes E. C. (2009). Validation of high rates of nucleotide substitution in geminiviruses: phylogenetic evidence from East African cassava mosaic viruses. J. Gen. Virol. 90 1539–1547. 10.1099/vir.0.009266-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S., Shackelton L. A., Holmes E. C. (2008). Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9 267–276. 10.1038/nrg2323 [DOI] [PubMed] [Google Scholar]
- Dunker A. K., Lawson J. D., Brown C. J., Williams R. M., Romero P., Jeong S. O., et al. (2001). Intrinsically disordered protein. J. Mol. Graph. Model. 19 26–59. 10.1016/S1093-3263(00)00138-8 [DOI] [PubMed] [Google Scholar]
- Dunlap D. S., Ng T. F., Rosario K., Barbosa J. G., Greco A. M., Breitbart M., et al. (2013). Molecular and microscopic evidence of viruses in marine copepods. Proc. Natl. Acad. Sci. U.S.A. 110 1375–1380. 10.1073/pnas.1216595110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R. A., Rohwer F. (2005). Viral metagenomics. Nat. Rev. Microbiol. 3 504–510. 10.1038/nrmicro1163 [DOI] [PubMed] [Google Scholar]
- Garigliany M. M., Borstler J., Jost H., Badusche M., Desmecht D., Schmidt-Chanasit J., et al. (2015). Characterization of a novel circo-like virus in Aedes vexans mosquitoes from Germany: evidence for a new genus within the family Circoviridae. J. Gen. Virol. 96 915–920. 10.1099/vir.0.000036 [DOI] [PubMed] [Google Scholar]
- Garigliany M. M., Hagen R. M., Frickmann H., May J., Schwarz N. G., Perse A., et al. (2014). Cyclovirus CyCV-VN species distribution is not limited to Vietnam and extends to Africa. Sci. Rep. 4 7552 10.1038/srep07552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Li Y., Yang X., Zhang H., Zhou P., Zhang Y., et al. (2012). Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. J. Virol. 86 4620–4630. 10.1128/JVI.06671-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh G. K., Dunker A. K., Uversky V. N. (2008a). Protein intrinsic disorder toolbox for comparative analysis of viral proteins. BMC Genomics 9(Suppl. 2):S4 10.1186/1471-2164-9-S2-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh G. K., Dunker A. K., Uversky V. N. (2008b). A comparative analysis of viral matrix proteins using disorder predictors. Virol. J. 5 126 10.1186/1743-422X-5-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V. (1993). Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3 419–429. 10.1016/S0959-440X(05)80116-2 [DOI] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Wolf Y. I. (1990). A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 262 145–148. 10.1016/0014-5793(90)80175-I [DOI] [PubMed] [Google Scholar]
- Goss J., Burch D., Rickson R. E. (2000). Agri-food restructuring and third world transnationals: Thailand, the CP Group and the global shrimp industry. World Dev. 28 513–530. 10.1016/S0305-750X(99)00140-0 [DOI] [Google Scholar]
- Gronenborn B. (2004). Nanoviruses: genome organisation and protein function. Vet. Microbiol. 98 103–109. 10.1016/j.vetmic.2003.10.015 [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Gutierrez C. (1999). Geminivirus DNA replication. Cell. Mol. Life Sci. 56 313–329. 10.1007/s000180050433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewson I., Eaglesham J. B., Höök T. O., Labarre B. A., Sepúlveda M. S., Thompson P. D., et al. (2013a). Investigation of viruses in Diporeia spp. from the Laurentian Great Lakes and Owasco Lake as potential stressors of declining populations. J. Great Lakes Res. 39 499–506. 10.1016/j.jglr.2013.06.006 [DOI] [Google Scholar]
- Hewson I., Ng G., Li W., Labarre B. A., Aguirre I., Barbosa J. G., et al. (2013b). Metagenomic identification, seasonal dynamics, and potential transmission mechanisms of a Daphnia-associated single-stranded DNA virus in two temperate lakes. Limnol. Oceanogr. 58 1605–1620. 10.4319/lo.2013.58.5.1605 [DOI] [Google Scholar]
- Ilyina T. V., Koonin E. V. (1992). Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 20 3279–3285. 10.1093/nar/20.13.3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M. R., Communie G., Ribeiro E. A., Jr., Martinez N., Desfosses A., Salmon L., et al. (2011). Intrinsic disorder in measles virus nucleocapsids. Proc. Natl. Acad. Sci. U.S.A. 108 9839–9844. 10.1073/pnas.1103270108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. H., Bae J. W. (2011). Amplification methods bias metagenomic libraries of uncultured single-stranded and double-stranded DNA viruses. Appl. Environ. Microbiol. 77 7663–7668. 10.1128/AEM.00289-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. H., Chang H. W., Nam Y. D., Roh S. W., Kim M. S., Sung Y., et al. (2008). Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl. Environ. Microbiol. 74 5975–5985. 10.1128/AEM.01275-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleppel G. S., Burkart C. A., Carter K., Tomas C. (1996). Diets of calanoid copepods on the West Florida continental shelf: relationships between food concentration, food composition and feeding activity. Mar. Biol. 127 209–217. 10.1007/BF00942105 [DOI] [Google Scholar]
- Kraberger S., Arguello-Astorga G. R., Greenfield L. G., Galilee C., Law D., Martin D. P., et al. (2015). Characterisation of a diverse range of circular replication-associated protein encoding DNA viruses recovered from a sewage treatment oxidation pond. Infect. Genet. Evol. 31 73–86. 10.1016/j.meegid.2015.01.001 [DOI] [PubMed] [Google Scholar]
- Labonte J. M., Suttle C. A. (2013). Previously unknown and highly divergent ssDNA viruses populate the oceans. ISME J. 7 2169–2177. 10.1038/ismej.2013.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefeuvre P., Lett J. M., Varsani A., Martin D. P. (2009). Widely conserved recombination patterns among single-stranded DNA viruses. J. Virol. 83 2697–2707. 10.1128/JVI.02152-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Kapoor A., Slikas B., Bamidele O. S., Wang C., Shaukat S., et al. (2010a). Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J. Virol. 84 1674–1682. 10.1128/JVI.02109-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Victoria J. G., Wang C., Jones M., Fellers G. M., Kunz T. H., et al. (2010b). Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J. Virol. 84 6955–6965. 10.1128/JVI.00501-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H., Liu Y., Li N., Wang Y., Zhang S., Hu R. (2014). Novel circovirus from mink, China. Emerging Infect. Dis. 20 1548–1550. 10.3201/eid2009.140015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bueno A., Tamames J., Velázquez D., Moya A., Quesada A., Alcamí A. (2009). High diversity of the viral community from an Antarctic lake. Science 326 858–861. 10.1126/science.1179287 [DOI] [PubMed] [Google Scholar]
- Martin D. P., Biagini P., Lefeuvre P., Golden M., Roumagnac P., Varsani A. (2011). Recombination in eukaryotic single stranded DNA viruses. Viruses 3 1699–1738. 10.3390/v3091699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel L. D., Rosario K., Breitbart M., Paul J. H. (2014). Comparative metagenomics: natural populations of induced prophages demonstrate highly unique, lower diversity viral sequences. Environ. Microbiol. 16 570–585. 10.1111/1462-2920.12184 [DOI] [PubMed] [Google Scholar]
- Muhire B. M., Varsani A., Martin D. P. (2014). SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 9:e108277 10.1371/journal.pone.0108277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawagitgul P., Morozov I., Bolin S. R., Harms P. A., Sorden S. D., Paul P. S. (2000). Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 81 2281–2287. 10.1099/0022-1317-81-9-2281 [DOI] [PubMed] [Google Scholar]
- Ng T. F., Alavandi S., Varsani A., Burghart S., Breitbart M. (2013). Metagenomic identification of a nodavirus and a circular ssDNA virus in semi-purified viral nucleic acids from the hepatopancreas of healthy Farfantepenaeus duorarum shrimp. Dis. Aquat. Org. 105 237–242. 10.3354/dao02628 [DOI] [PubMed] [Google Scholar]
- Ng T. F., Chen L. F., Zhou Y., Shapiro B., Stiller M., Heintzman P. D., et al. (2014). Preservation of viral genomes in 700-y-old caribou feces from a subarctic ice patch. Proc. Natl. Acad. Sci. U.S.A. 111 16842–16847. 10.1073/pnas.1410429111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T. F., Marine R., Wang C., Simmonds P., Kapusinszky B., Bodhidatta L., et al. (2012). High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J. Virol. 86 12161–12175. 10.1128/JVI.00869-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T. F., Willner D. L., Lim Y. W., Schmieder R., Chau B., Nilsson C., et al. (2011). Broad surveys of DNA viral diversity obtained through viral metagenomics of mosquitoes. PLoS ONE 6:e20579 10.1371/journal.pone.0020579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niagro F. D., Forsthoefel A. N., Lawther R. P., Kamalanathan L., Ritchie B. W., Latimer K. S., et al. (1998). Beak and feather disease virus and porcine circovirus genomes: intermediates between the geminiviruses and plant circoviruses. Arch. Virol. 143 1723–1744. 10.1007/s007050050412 [DOI] [PubMed] [Google Scholar]
- Nilsson J., Grahn M., Wright A. P. (2011). Proteome-wide evidence for enhanced positive Darwinian selection within intrinsically disordered regions in proteins. Genome Biol. 12 R65. 10.1186/gb-2011-12-7-r65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obradovic Z., Peng K., Vucetic S., Radivojac P., Brown C. J., Dunker A. K. (2003). Predicting intrinsic disorder from amino acid sequence. Proteins 53(Suppl. 6), 566–572. 10.1002/prot.10532 [DOI] [PubMed] [Google Scholar]
- Padilla-Rodriguez M., Rosario K., Breitbart M. (2013). Novel cyclovirus discovered in the Florida woods cockroach Eurycotis floridana (Walker). Arch. Virol. 158 1389–1392. 10.1007/s00705-013-1606-x [DOI] [PubMed] [Google Scholar]
- Paezosuna F. (2003). Shrimp aquaculture development and the environment in the Gulf of California ecoregion. Mar. Pollut. Bull. 46 806–815. 10.1016/S0025-326X(03)00107-3 [DOI] [PubMed] [Google Scholar]
- Patterson E. I., Swarbrick C. M., Roman N., Forwood J. K., Raidal S. R. (2013). Differential expression of two isolates of beak and feather disease virus capsid protein in Escherichia coli. J. Virol. Methods 189 118–124. 10.1016/j.jviromet.2013.01.020 [DOI] [PubMed] [Google Scholar]
- Pham H. T., Bergoin M., Tijssen P. (2013a). Acheta domesticus volvovirus, a novel single-stranded circular DNA virus of the house cricket. Genome Announc. 1:e00079-13 10.1128/genomeA.00079-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham H. T., Iwao H., Bergoin M., Tijssen P. (2013b). New volvovirus isolates from Acheta domesticus (Japan) and Gryllus assimilis (United States). Genome Announc. 1:e00328-13 10.1128/genomeA.00328-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham H. T., Yu Q., Boisvert M., Van H. T., Bergoin M., Tijssen P. (2014). A circo-like virus isolated from Penaeus monodon shrimps. Genome Announc. 2:e01172-13 10.1128/genomeA.01172-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. G., Kapusinszky B., Wang C., Rose R. K., Lipton H. L., Delwart E. L. (2011). The fecal flora of wild rodents. PLoS Pathog. 7:e1002218 10.1371/journal.ppat.1002218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. G., Mori D., Deng X., Rajindrajith S., Ranawaka U., Fan Ng T. F., et al. (2015). Small circular single stranded DNA viral genomes in unexplained cases of human encephalitis, diarrhea, and in untreated sewage. Virology 482 98–104. 10.1016/j.virol.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushker R., Mooney C., Davey N. E., Jacque J. M., Shields D. C. (2013). Marked variability in the extent of protein disorder within and between viral families. PLoS ONE 8:e60724 10.1371/journal.pone.0060724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2014). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Reavy B., Swanson M. M., Cock P., Dawson L., Freitag T. E., Singh B. K., et al. (2015). Distinct circular ssDNA viruses exist in different soil types. Appl. Environ. Microbiol. 81 3934–3945. 10.1128/AEM.03878-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario K., Breitbart M. (2011). Exploring the viral world through metagenomics. Curr. Opin. Virol. 1 289–297. 10.1016/j.coviro.2011.06.004 [DOI] [PubMed] [Google Scholar]
- Rosario K., Duffy S., Breitbart M. (2009a). Diverse circovirus-like genome architectures revealed by environmental metagenomics. J. Gen. Virol. 90 2418–2424. 10.1099/vir.0.012955-0 [DOI] [PubMed] [Google Scholar]
- Rosario K., Nilsson C., Lim Y. W., Ruan Y., Breitbart M. (2009b). Metagenomic analysis of viruses in reclaimed water. Environ. Microbiol. 11 2806–2820. 10.1111/j.1462-2920.2009.01964.x [DOI] [PubMed] [Google Scholar]
- Rosario K., Duffy S., Breitbart M. (2012a). A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics. Arch. Virol. 157 1851–1871. 10.1007/s00705-012-1391-y [DOI] [PubMed] [Google Scholar]
- Rosario K., Dayaram A., Marinov M., Ware J., Kraberger S., Stainton D., et al. (2012b). Diverse circular single-stranded DNA viruses discovered in dragonflies (Odonata: Epiprocta). J. Gen. Virol. 93 2668–2681. 10.1099/vir.0.045948-0 [DOI] [PubMed] [Google Scholar]
- Rosario K., Marinov M., Stainton D., Kraberger S., Wiltshire E. J., Collings D. A., et al. (2011). Dragonfly cyclovirus, a novel single-stranded DNA virus discovered in dragonflies (Odonata: Anisoptera). J. Gen. Virol. 92 1302–1308. 10.1099/vir.0.030338-0 [DOI] [PubMed] [Google Scholar]
- Roux S., Enault F., Bronner G., Vaulot D., Forterre P., Krupovic M. (2013). Chimeric viruses blur the borders between the major groups of eukaryotic single-stranded DNA viruses. Nat. Commun. 4 2700 10.1038/ncomms3700 [DOI] [PubMed] [Google Scholar]
- Roux S., Enault F., Robin A., Ravet V., Personnic S., Theil S., et al. (2012). Assessing the diversity and specificity of two freshwater viral communities through metagenomics. PLoS ONE 7:e33641 10.1371/journal.pone.0033641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachsenroder J., Twardziok S., Hammerl J. A., Janczyk P., Wrede P., Hertwig S., et al. (2012). Simultaneous identification of DNA and RNA viruses present in pig faeces using process-controlled deep sequencing. PLoS ONE 7:e34631 10.1371/journal.pone.0034631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Orba Y., Ueno K., Ishii A., Moonga L., Hang’ombe B. M., et al. (2015). Metagenomic analysis of the shrew enteric virome reveals novel viruses related to human stool-associated viruses. J. Gen. Virol. 96 440–452. 10.1099/vir.0.071209-0 [DOI] [PubMed] [Google Scholar]
- Sickmeier M., Hamilton J. A., Legall T., Vacic V., Cortese M. S., Tantos A., et al. (2007). DisProt: the database of disordered proteins. Nucleic Acids Res. 35 D786–D793. 10.1093/nar/gkl893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski A., Dayaram A., Varsani A. (2013a). Identification of a novel circular DNA virus in New Zealand fur seal (Arctocephalus forsteri) fecal matter. Genome Announc. 1:e00558-13 10.1128/genomeA.00558-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski A., Massaro M., Kraberger S., Young L. M., Smalley D., Martin D. P., et al. (2013b). Novel myco-like DNA viruses discovered in the faecal matter of various animals. Virus Res. 177 209–216. 10.1016/j.virusres.2013.08.008 [DOI] [PubMed] [Google Scholar]
- Smits S. L., Schapendonk C. M., Van Beek J., Vennema H., Schurch A. C., Schipper D., et al. (2014). New viruses in idiopathic human diarrhea cases, the Netherlands. Emerging Infect. Dis. 20 1218–1222. 10.3201/eid2007.140190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S. L., Zijlstra E. E., Van Hellemond J. J., Schapendonk C. M., Bodewes R., Schurch A. C., et al. (2013). Novel cyclovirus in human cerebrospinal fluid, Malawi, 2010-2011. Emerging Infect. Dis. 19 1511 10.3201/eid1909.130404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffer N., Brandt M. E., Correa A. M., Smith T. B., Thurber R. V. (2014). Potential role of viruses in white plague coral disease. ISME J. 8 271–283. 10.1038/ismej.2013.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Le V., Van Doorn H. R., Nghia H. D., Chau T. T., Tu Le T. P., De Vries M., et al. (2013). Identification of a new cyclovirus in cerebrospinal fluid of patients with acute central nervous system infections. mBio 4:e00231-13 10.1128/mbio.00231-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brand J. M., Van Leeuwen M., Schapendonk C. M., Simon J. H., Haagmans B. L., Osterhaus A. D., et al. (2012). Metagenomic analysis of the viral flora of pine marten and European badger feces. J. Virol. 86 2360–2365. 10.1128/JVI.06373-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee R., Buljan M., Lang B., Weatheritt R. J., Daughdrill G. W., Dunker A. K., et al. (2014). Classification of intrinsically disordered regions and proteins. Chem. Rev. 114 6589–6631. 10.1021/cr400525m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whon T. W., Kim M. S., Roh S. W., Shin N. R., Lee H. W., Bae J. W. (2012). Metagenomic characterization of airborne viral DNA diversity in the near-surface atmosphere. J. Virol. 86 8221–8231. 10.1128/JVI.00293-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B., Blocquel D., Habchi J., Uversky A. V., Kurgan L., Uversky V. N., et al. (2014). Structural disorder in viral proteins. Chem. Rev. 114 6880–6911. 10.1021/cr4005692 [DOI] [PubMed] [Google Scholar]
- Xue B., Dunker A. K., Uversky V. N. (2012). Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J. Biomol. Struct. Dyn. 30 137–149. 10.1080/07391102.2012.675145 [DOI] [PubMed] [Google Scholar]
- Yoshida M., Takaki Y., Eitoku M., Nunoura T., Takai K. (2013). Metagenomic analysis of viral communities in (hado)pelagic sediments. PLoS ONE 8:e57271 10.1371/journal.pone.0057271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawar-Reza P., Arguello-Astorga G. R., Kraberger S., Julian L., Stainton D., Broady P. A., et al. (2014). Diverse small circular single-stranded DNA viruses identified in a freshwater pond on the McMurdo Ice Shelf (Antarctica). Infect. Genet. Evol. 26 132–138. 10.1016/j.meegid.2014.05.018 [DOI] [PubMed] [Google Scholar]
- Zhang W., Li L., Deng X., Kapusinszky B., Pesavento P. A., Delwart E. (2014). Faecal virome of cats in an animal shelter. J. Gen. Virol. 95 2553–2564. 10.1099/vir.0.069674-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.