Summary
WNT10A is upregulated in invasive esophageal tumor cells versus non-invasive cells in 3D-organotypic cultures. Functionally, WNT10A promotes proliferation, migration, invasion and self-renewal. WNT10A is expressed during development, downregulated postnatally and re-expressed during carcinogenesis, where it correlates with poor prognosis.
Abstract
Esophageal cells overexpressing epidermal growth factor receptor (EGFR) and TP53 mutation can invade into the extracellular matrix when grown in 3D-organotypic cultures (OTC) and mimic early invasion in esophageal squamous cell carcinoma (ESCC). We have performed laser capture microdissection with RNA microarray analysis on the invasive and non-invasive tumor cells of p53R175H-overexpressing OTC samples to determine candidate genes facilitating tumor invasion. WNT10A was found to be >4-fold upregulated in the invasive front. Since WNT10A is also prominently upregulated during placode promotion in hair follicle development, a process that requires epithelial cells to thicken and elongate, in order to allow downward growth, we hypothesized that WNT10A may be important in mediating a similar mechanism of tumor cell invasion in ESCC. We have found that WNT10A expression is significantly upregulated in human ESCC, when compared with normal adjacent tissue. Furthermore, high WNT10A expression levels correlate with poor survival. Interestingly, we observe that WNT10A is expressed early in embryogenesis, but is reduced dramatically postnatally. We demonstrate that overexpression of WNT10a promotes migration and invasion, and proliferation of transformed esophageal cells. Lastly, we show that WNT10A overexpression induces a greater CD44High/CD24Low population, which are putative markers of cancer stem cells, and increases self-renewal capability. Taken together, we propose that WNT10A acts as an oncofetal factor that is highly expressed and may promote proper development of the esophagus. During tumorigenesis, it is aberrantly overexpressed in order to promote ESCC migration and invasion, and may be linked to self-renewal of a subset of ESCC cells.
Introduction
Esophageal squamous cell carcinoma (ESCC) is the sixth leading cause of cancer-related death amongst American men, and sixth overall cause worldwide (1). Common genetic alterations of ESCC include overexpression of epidermal growth factor receptor (EGFR) and CYCLIND1, as well as mutation in TP53 and either loss or mislocalization of P120 CATENIN (2–5). A recent genome sequencing effort from a cohort of 158 Chinese ESCC samples revealed that WNT and NOTCH signaling pathways are also highly deregulated (4).
We have shown previously that overexpression of mutant p53 (R175H) along with EGFR in primary immortalized esophageal epithelial cells induces transformation (6). Furthermore, when these engineered cells were grown in 3D-organotypic culture (OTC), which mimics the stratified epithelium and its crosstalk with the underlying stroma, they invade into surrounding stroma, similar to early invasion observed in ESCC (7). To understand what molecular mechanisms may be responsible for invasion, we dissected out the invasive and non-invasive regions from these 3D cultures and performed comparative microarray analysis (8). PERIOSTIN, was found to be the highest upregulated gene, and has been described previously as a potential biomarker for ESCC (9,10). Strikingly, we observed additional dysregulation of a variety of WNT pathway genes, such as upregulation of WNT ligands, WNT10A, WNT5B, and downregulation of the WNT pathway inhibitor DKK1 (8).
Wnt signaling is critical in the embryonic development of different invertebrate and vertebrate organisms. In particular, Wnt signaling is critical in the regulation of axis patterning, cell fate specification, cell proliferation and cell migration during development (11,12). Wnt ligands are secreted glycoproteins that are cysteine-rich and comprise a short N-terminal signal sequence with a mature segment that has variable length (13). There are nearly 20 different Wnt proteins, the expression of which is spatially and temporally regulated during development, and maintain homeostasis and drive cancers in a context dependent manner (14,15). Mouse WNT10A is synthesized as a 417 amino acid precursor that contains a 382 amino acid mature region, the latter of which contains two potential glycosylation sites. Mouse, rat and human WNT10A are highly conserved and WNT10A’s amino acid sequence is 64% identical to WNT10B (16,17).
Developmentally, WNT10A is best studied in the context of ectodermal lineages. It is studied primarily in the deregulation of ectodermal tissues resulting in a variety of disorders classified as: odonto-onychal dermal dysplasia (18,19). Manifestation of WNT10A mutations in humans can result in defects in dentinogenesis, tooth morphogenesis, odontoblast differentiation, hair follicle development, nail formation, papillae of the tongue and sweat gland, and regeneration of the epidermis (19–21). Contributing to this developmental phenotype, WNT10A messenger RNA has been shown to strongly localize to the dermal condensates during the earliest stages of embryonic hair follicle formation and postnatal anagen (22). This process requires coordinated cross-talk between epithelial cells and underlying dermal cells in order to facilitate elongated epithelial cell down growths (22,23). Moreover, publically available in situ hybridization data indicates that in addition to localization in ectodermal tissues, WNT10A messenger RNA also strongly localizes to the embryonic esophagus at embryonic day 14.5, suggesting a role in esophageal development (24).
WNT10A has been previously implicated in a variety of cancers and has been shown to promote proliferation, migration and chemoresistance in renal cell carcinoma cell lines by regulation of β-CATENIN (25–28). Other reports suggest that WNT10A is also upregulated in esophageal cancer, gastric and colon cancer cells and tumors (27). By contrast, the WNT10A promoter has been suggested to be hypermethylated in head/neck squamous cell carcinoma and oligodendroglioma cell lines (29,30). Herein, we show that WNT10A is upregulated both in early development, as well as in early and late stages of ESCC, due its ability to promote proliferation, migration, invasion and self-renewal. Taken together, we suggest that WNT10A may act as an oncofetal factor in the context of both esophageal development and tumorigenesis.
Materials and methods
Network analysis
Ingenuity Pathway Analysis was used to identify potential upstream regulators that control gene expression patterns enriched in invasive TP53 mutant cell lines. The analysis is based on prior knowledge of expected effects between transcriptional factors and their target genes stored in the Ingenuity Knowledge Base (31). Briefly, the analysis examines the known targets of each transcription factor in the signature and compares their direction of change (expression in invasive relative to the non-invasive gene signature) to what is expected from the literature. If the direction of change is consistent with the literature across the majority of targets, then the regulator is predicted to be active in invasive cells, whereas if the direction of change is mostly inconsistent (anti-correlated) with the literature, then the regulator is predicted to be inactive in invasive cells. Regulation z-score was used to estimate the activation state of the regulators. The overlap P values generated by Fisher’s exact test were used to estimate the statistical significance of overlap between the dataset genes and the genes regulated by a regulator.
Cell culture
EPC2-hTERT cells and derivatives, established and extensively characterized by us, were grown in keratinocyte serum-free media (Invitrogen, Carlsbad, CA), supplemented with 40 µg/ml of bovine pituitary extract, 1ng/ml epidermal growth factor and 1% penicillin/streptomycin, as described previously (6,7,32). The TE series human ESCC cells were a gift from Dr Nishihira who established the cell lines and were grown in Dulbecco's modified Eagle's medium supplemented by 10% fetal calf serum (Sigma–Aldrich, St. Louis, MO) and 1% penicillin/streptomycin (Invitrogen) as described previously (33,34). The earliest frozen stocks of all cell lines have been stored at the Cell Culture Core of the University of Pennsylvania. We have propagated cells from frozen stocks of the original vials that were authenticated by short tandem repeat analysis for highly polymorphic microsatellites FES/FPS, vWA31, D22S417, D10S526 and D5S592 as performed by the Cell Culture Core to validate the identity of cells by comparing cells at the earliest stocks we have and those grown >8–12 passages. All cell lines have been tested for mycoplasma contamination on a regular basis.
Wnt10A was a gift from Marian Waterman (Addgene plasmid # 35920), and was subcloned into pBabe-Blast empty vector (35). Retroviral spinfection was performed as described previously (8).
3D-OTCs
Epithelial cells were grown as described (7), in order to recreate the microenvironement of ESCC, by supplying extracellular membrane components, including collagen and fetal esophageal fibroblasts.
Mouse treatments
4-nitroquinoline 1-oxide (4-NQO) experiments were performed as described previously and under Institutional Animal Care and Use Committee (IACUC) approval (36). In brief, 6-week-old C57Bl/6 and mixed background mice were given 10mg/ml of 4-NQO diluted in 10% propylene glycol in their drinking water ad libitum. Mice were randomized amongst littermates into control versus treatment groups. Mice were euthanized either directly after treatment or after a 12-week wash out period. Three to five mice, per condition, were euthanized and tissues were processed for histology and immunohistochemistry (IHC).
Tumor microarrays and IHC/immunofluorescence
ESCC tissue along with adjacent non-cancerous mucosa were obtained as surgical biopsies from Kagoshima University Hospital, as described previously (37). The clinical materials were obtained from informed-consent patients according to the Institutional Review Board standards and guidelines. IHC and immunofluorescence were performed as described previously (37). About 5 μM paraffin embedded sections were incubated with WNT10A (Ab-106522, Abcam, Cambridge, MA 1:200) or WNT10A (SC-376028, Santa Cruz Biotechnologies, Santa Cruz, CA 1:100). Immunofluorescence signal was quantified in at least four independent high-powered fields per slide for mean intensity using ImageJ (38).
RNA isolation and quantitative PCR
Total RNA was isolated using GeneJet RNA Purification Kit (Thermo Fisher Scientific, Waltham, MA) and cDNA was synthesized utilizing the Taqman Reverse Transcription Reagents kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Quantitative PCR was performed with primer sequences for WNT10A Forward: 5′-ATCCACGAATGCCAACACCA-3′, Reverse: 5′-CTCTCTCGGAAACCTCTGCT-3′ and ACTB Forward: 5′-CCTGGCACCC AGGACAAT-3′, Reverse: 5′-GCCGAGCCACACGGAGTACT-3′ using Power SYBR Green PCR Master Mix (PE, Applied Biosystems), according to manufacturer’s instructions.
Western blotting
Proteins were separated through 4–12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis Bis-Tris gel (Invitrogen) and then transferred to a poly membrane (Immobilon-P; Millipore, Billerica, MA). Membranes were then blocked with 5% non-fat milk in TBS-T (1× TBS and 0.01% Tween-20), probed with primary antibody (1:500 WNT10A SC-376028, Santa Cruz, 1:1000 β-CATENIN D10A8, Cell Signaling, and 1:1000 GAPDH, Millipore) diluted in blocking buffer overnight at 4°C, washed with TBST-T and incubated with horseradish peroxidase-coupled secondary antibodies (GE Healthcare, Pittsburgh, PA). Signal was visualized with Amersham ECL-Prime (GE Healthcare) as per manufacturer’s instructions.
Migration and invasion assays
Boyden chambers (8-μm pore size, FluoroBlok-HTS inserts; BD Biosciences, San Jose, CA) were used for migration and invasion assays, as described previously (6). In brief, Boyden chambers were coated with 200 μg/ml of Matrigel Matrix (BD Biosciences) 2h before cell plating. Inserts were placed in a 24-well plate containing full keratinocyte serum-free medium to stimulate cell migration and invasion. Cells (5×104) in keratinocyte basal media (Lonza, Basel, Switzerland) were placed in each insert. Twenty-four hours later, migrating or invading cells were labeled with 4 μg/ml Calcein AM dye (Invitrogen) in Hanks’ buffered salt solution buffer (Invitrogen) for 1h. Labeled cells were then read on a Synergy HT multidetection microplate reader (BioTek, Winooski, VT) at 485-nm excitation and 528-nm detection. Data represents the average of three independent experiments performed in triplicate for each genotype.
Cell proliferation assay
A total of 3×103 cells were cultured in 96-well plates. Cell proliferation was quantified using the WST-1 colorimetric assay (Roche, Penzberg, Germany) according to manufacture’s instructions. Absorbance was measured on a microplate reader (Tecan Sunrise, Männedorf, Switzerland) at a wavelength of 450nm. Data represents the average of five independent experiments performed in quintuplicate.
Flow cytometry and fluorescence-activated cell sorting
Flow cytometry was performed using LSRII (BD Biosciences) and FlowJo (Tree Star, Ashland, OR). Cells were suspended in phosphate-buffered saline (Invitrogen) containing 1% BSA (Sigma–Aldrich) and stained with PE/Cy7-anti-CD24 (1:40; BioLegend, San Diego, CA) and APC-anti-CD44 (1:20; BD Biosciences) on ice for 30min. Flow cytometry data represent at least three independent experiments.
Self-renewal assays
About 100 cells were plated in single suspension in an ultra-low attachment 96 well-plate in 100 μl of mammary epithelial cell growth medium (Invitrogen). After 7 days, spheres were visualized and counted. All spheres consisted of greater than three cells. Data represents three independent experiments performed in 96 replicates for each cell type.
Statistics
Each experiment is presented as mean ± standard error (at least n = 3) and was analysed by paired or non-paired two-tailed Student’s t-test. P < 0.05 was considered significant. Overall survival curves were plotted by the Kaplan–Meier method and subjected to the Log-rank and Gehan–Wilcoxon test. Chi-square test was used to determined differences in proportion of WNT10A scoring in normal versus tumor samples.
Results
WNT10A is preferentially expressed in the invasive compartment of transformed epithelial cells in 3D-OTC
We have identified WNT10A as a prominent upregulated gene in invasive versus non-invasive EPC2-hTERT-EGFR-p53R175H cells grown in 3D-OTC, an in vivo like model of ESCC (8). WNT10A was a strong candidate for further study, as it was upregulated significantly in the invasive region of another TP53 mutant (R248W) (unpublished data). Furthermore, based on pathway analysis of all TP53 mutants analysed (R175H, V143A, R248W and R273H), there was a significant enrichment for WNT/β-catenin signaling genes (Figure 1). Interestingly, WNT10A was included in other significantly upregulated pathways, such as regulation of epithelial to mesenchymal transition and human embryonic stem cell pluoripotency (Supplementary Table 1 is available at Carcinogenesis Online).
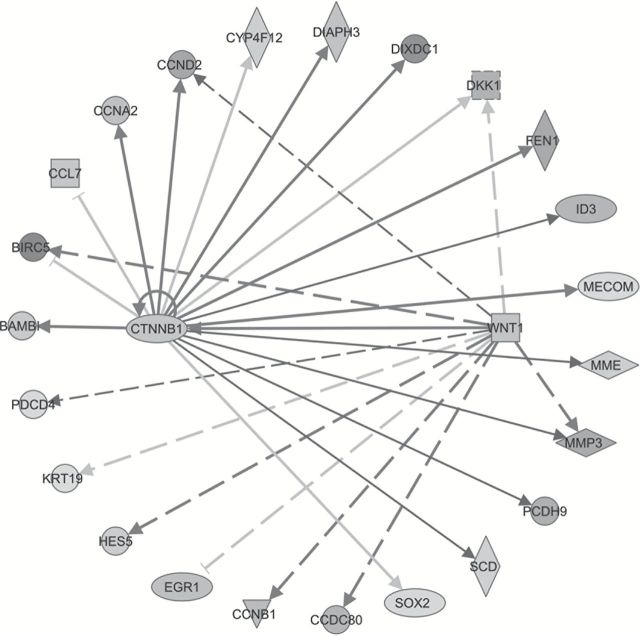
Figure 1.
WNT pathway is significantly upregulated in invasive mutant TP53 cells. Ingenuity Pathway analysis shows networks of genes, known to be regulated by WNT signaling in gene expression data, are significantly associated with TP53 mutations. Upregulated and downregulated genes are indicated by dark shade and light shade, respectively. The lines and arrows represent functional and physical interactions and the directions of regulation, as indicated in the literature.
We sought to confirm this expression of WNT10A through immunofluorescent staining of 3D-OTCs. WNT10A stained predominately in the cytoplasm of epithelial cells. Strikingly, we saw stronger staining within the invasive regions (I), as compared with the basal non-invasive (NI) regions (Figure 2A). We quantified the mean intensity of the staining, and saw that invasive regions had an ~4-fold increase in fluorescent level as compared with the basal layer (Figure 2B). We next confirmed that WNT10A expression was increased through quantitative PCR. Using a non-invasive cell line, EPC2-hTERT-EGFR-empty vector cells, we confirmed that the more invasive EPC2-hTERT-EGFR-p53R175H cells expressed significantly higher levels of WNT10A messenger RNA (Figure 2C). These results were replicated with several primer sets for WNT10A as compared with GAPDH and HPRT (data not shown).
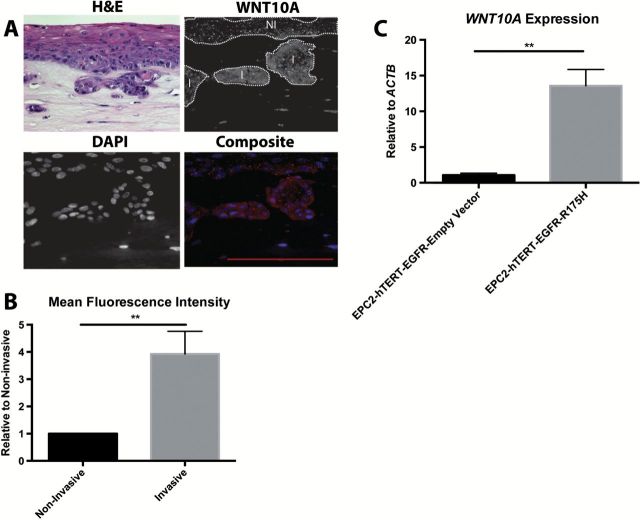
Figure 2.
WNT10A is upregulated in invasive esophageal cells. (A) Representative Hematoxylin and Eosin (H&E), WNT10A and DAPI staining of EPC2-hTERT-EGFR-p53R175H cells grown in 3-D OTC. Non-invasive (NI) and invasive (I) regions are delineated by white dots (B) Quantification utilizing ImageJ for mean fluorescent intensity of WNT10A staining in the non-invasive versus invasive front (n = 4 independent experiments). Fluorescent intensity of invasive front is relative to non-invasive regions. Bar graphs represent relative mean ± SEM. **P < 0.005 (paired Student’s t-test) (C) Quantitative reverse transcription–PCR of WNT10A expression relative to ACTB comparing non-invasive EPC2-hTERT-EGFR cells versus invasive EPC2-hTERT-EGFR-p53R175H cells (n = 3 experiments). Bar graph represents fold change ± SEM. **P < 0.005 (unpaired Student’s t-test).
WNT10A overexpression promotes migration and invasion in transformed esophageal keratinocytes
Since WNT10A was upregulated in the invasive front of transformed esophageal cells grown in 3D OTC, we next wanted to determine the role of WNT10A in mediating migration and invasion. To test this hypothesis, we first generated EPC2-hTERT-EGFR-R175H cells that either overexpressed WNT10A or an empty vector control (herin known as WNT10A and empty vector, respectively). We confirmed WNT10A expression by western blot (Figure 3A and Supplementary Figure 1). Several bands were visualized for WNT10A, indicative of post-translational modifications, such as glycosylation and lipidataion, mainly at the 40 and the 50 kD molecular masses (13). Interestingly, overexpression of WNT10A resulted in a slight increase in β-catenin protein (Figure 3A), suggesting increased stabilization of total β-catenin. WNT10A overexpressing cells exhibited increased migratory and invasive capability, as compared with empty vector controls in Boyden chamber assays (Figure 3B and C). WNT10A cells exhibited also a significant increase in proliferative ability (Figure 3D).
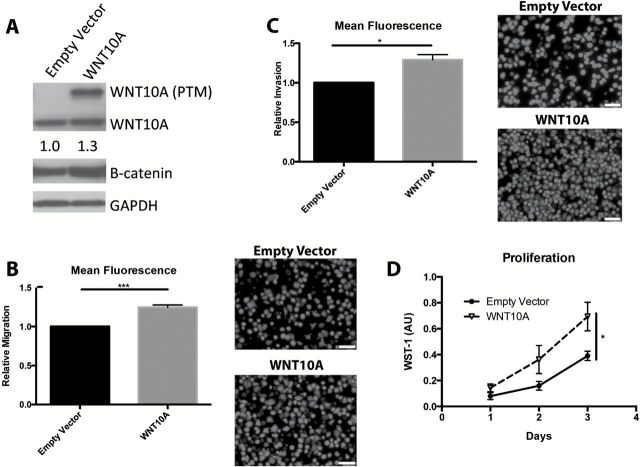
Figure 3.
WNT10A promotes migration and invasion. (A) Western blot (cropped image) of EPC2-hTERT-EGFR-p53R175H either overexpressing empty vector or WNT10A. Multiple bands indicate post-translational modifications of WNT10A. B-catenin densitometry indicates a modest 1.3-fold increase in total β-catenin relative to GAPDH, with WNT10A overexpression (n = 3 experiments). (B) Transwell Boyden Chamber migration assay of empty vector compared with WNT10A cells along with representative images of migratory cells stained with Calcein AM. (C) Transwell Boyden Chamber invasion assay of empty vector compared with WNT10A cells with representative images of invasive cells stained with Calcein AM. Scale bars =100 μM. n = 3 in triplicate. Bar graphs represent fold changes ± SEM. *P < 0.05, **P < 0.005 (paired Student’s t-test). (D) Proliferation assay utilizing a WST-1 colorimetric assay. (n = 5 experiments in quintuplicate). *P < 0.05 (paired Student’s t-test).
WNT10A promotes a self-renewal phenotype
Based upon our microarray results, WNT10A was suggested to be important in a category of genes that promote stem cell pluoripotency, along with genes such as WNT5B and SOX2. We therefore wanted to investigate the role of WNT10A in stemness marker expression, as well as its ability to promote self-renewal through single sphere assays. CD44, a cell surface marker for hyaluronic acid, has been previously implicated as a putative stem cell marker in ESCC, and has been suggested to be regulated by the WNT/β-catenin signaling pathway (39,40). Additionally, CD44High and CD24Low expressing cells have also been described to represent a tumor-initiating population with mesenchymal-like properties (41,42). By fluorescence-activated cell sorting-analysis, WNT10A overexpressing cells exhibited a marked increase in CD44High and decrease in CD24Low cells (Figure 4A), resulting in a nearly 2-fold increase in CD44High/CD24Low cells with WNT10A overexpression (Figure 4B). We next performed single cell sphere formation assays in ultra-low attachment plates in the presence of sphere-promoting media. WNT10A overexpressing and empty vector control cells were plated in a single cell suspension of 100 cells per well. After 7 days, there was a significant increase in the average number of spheres formed per well in the WNT10A overexpressing cells as compared with control cells (Figure 4C), although there was no difference in the size of the spheres that formed. Together, these data demonstrate that WNT10A increases migratory and invasive capabilities and promotes a shift toward CD44High cells as well as self-renewal.
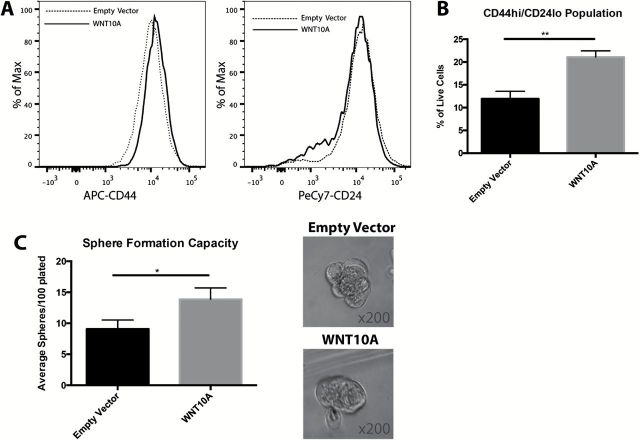
Figure 4.
WNT10A increases CD44High population and promotes self-renewal. (A) Representative histograms of CD44 and CD24 levels between empty vector and WNT10A cells, as expressed in a logarithmic scale. (B) Quantification of the CD44High/CD24Low population between empty vector and WNT10A cells (n = 3 experiments). Bar graphs represent fold change ± SEM within three different passages of cells within one representative experiment. **P < 0.005 (unpaired Student’s t-test). (C) Quantification of the average number of spheres formed per well between empty vector and WNT10A cells (n = 3 experiments in 96 replicates). Bar graph represents fold change ± SEM. *P < 0.05 (paired Student’s t-test). Representative images of the spheres from each cell type indicate no difference in the size of the spheres formed. Each image was taken at a magnification of ×200.
WNT10A expression in esophageal development and ESCC
WNT10A has been implicated previously to be upregulated during early follicle morphogenesis as well as in a variety of other ectodermal tissue types (22,43). WNT10A is rarely expressed in the normal adult mouse esophagus. Interestingly though, RNA in situ hybridization data of whole mount embryos suggest that in addition to ectodermal lineages, WNT10A is also expressed in the developing esophagus (24). To confirm this finding, we conducted WNT10A IHC in the esophagus at embryonic days (E) 12.5, 18.5, postnatal day (P) 1 and adult tissue. We found that WNT10A is expressed as early as E12.5, but exhibits the strongest staining at E18.5 (Figure 5A). Interestingly, by P1, WNT10A staining is mostly abrogated, although there may be some residual low levels. In normal adult stages, WNT10A is expressed minimally. This expression pattern suggests that WNT10A may play a role in the development of the early mouse esophagus.
Figure 5.
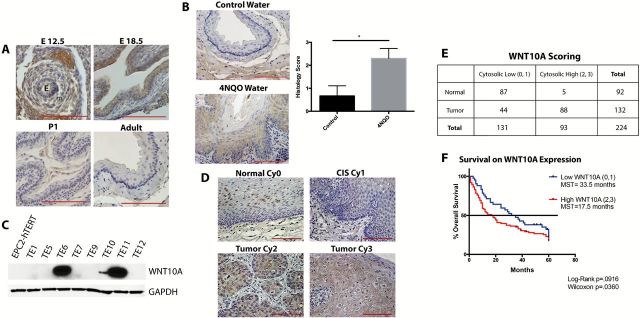
WNT10A is upregulated during esophageal development and in cancer. (A) WNT10A IHC of embryonic (E12.5, E18.5), postnatal day 1 (P1), and adult esophagus (E). Epithelial (e) and mesenchyme (m) are as indicated. (B) Representative WNT10A staining in esophagus of control mice versus mice treated with 10mg/ml of 4-NQO for 16 weeks. Downward growths and hyperplasia/dysplasia are indicative of early ESCC. Bar graph represents average scoring of esophagi ± SEM. *P < 0.05 (unpaired Student’s t-test) of three control and five experimental mice. (C) Western blot (cropped image) for WNT10A on a panel of ESCC human cell lines, as compared with normal, immortalized epithelial cells (EPC2-hTERT). (D) Representative images of WNT10A scoring classifications of adjacent normal (upper left panel) versus carcinoma in situ (CIS), and tumor tissue. Scoring was based on cytoplasmic intensity (Cy0, Cy1, Cy2 and Cy3). All scale bars are 100 μM. (E) Scoring stratified between low staining intensity (Cy0 and Cy1) and high staining intensity (Cy2 and Cy3) WNT10A staining between normal adjacent tissue and tumor sections, respectively P < 0.0001 (chi-squared test). (F) Kaplan–Meyer survival curve of tumor staining stratified between low WNT10A staining intensity versus high WNT10A staining intensity, with MST. P < 0.05 (Wilcoxon).
In order to examine WNT10A expression in pre-malignant and malignant stages of ESCC, we utilized a mouse model of ESCC. The 4-NQO is a quinolone derivative that causes DNA-adduct formation similar to carcinogens in tobacco, leading to oral squamous and esophageal dysplastic lesions, similar to that of human head neck squamous cell and esophageal carcinomas (36). In mice treated with 4-NQO ad libitum in drinking water, we saw increased hyperplastic and dysplastic lesions in the esophagi, with downward growths into the submucosa, consistent with invasive cancer, as compared with mice on water (control) (Figure 5B). Interestingly, 4-NQO-treated mice exhibited greater cytoplasmic WNT10A staining in the esophagi, especially in the invasive regions, as compared with control mice (Figure 5B). We additionally screened a panel of ESCC cell lines via western blot analysis for expression of WNT10A. As compared with immortalized primary epithelial cells (EPC2-hTERT), we saw an increase in WNT10A expression in three out of the eight ESCC cell lines screened (Figure 5C), namely TE6, TE10 and TE11.
In order to determine if WNT10A expression could also serve as a biomarker for ESCC prognosis, we utilized a tumor microarray (TMA) of 92 normal-matched ESCC tumors and 40 additional tumors, along with 5-year survival data on the patients. IHC indicated a faint nuclear pattern in samples from normal esophagus samples, while high-grade dysplasia and invasive tumors presented with overall stronger cytoplasmic staining (Figure 5D). An independent pathologist blinded to patient prognosis status scored the TMA for cytoplasmic signal (Cy0, Cy1, Cy2 and Cy3). Interestingly, most normal tissues expressed either very low (Cy1) or no (Cy0) staining, whereas tumor tissues were mostly scored as Cy2 or Cy3 (Figure 5E). When the tumor samples (n = 132) were stratified for 5-year survival, there was a decrease in median survival time (MST) between the high expressers (Cy2, Cy3) MST = 17.5 months versus the low expressers (Cy0, Cy1) MST = 33.5 (Figure 5F). Additionally, there was a significant (P = 0.0322) decrease in overall survival in the high expressers, based upon the Gehan–Breslow–Wilcoxon test, and was trending (P = 0.0916), based on Log-rank. Taken together, these findings suggest that WNT10A is over-expressed in ESCC tumors and cell lines, as well as in early esophageal development in the mouse, and that it may be an important biomarker for poor survival in ESCC.
Discussion
WNTs are critical signaling factors, required for the development of numerous tissue types, as well as maintenance of homeostasis in regenerative tissues, such as of the gastrointestinal tract and the skin. Hence, dysregulation of the WNT signaling cascade leads to a variety of cancers and diseases. Herein, we have demonstrated WNT10A to be expressed in mouse esophageal development, peaking at embryonic day 18.5 but diminished substantially postnatally. It is further re-expressed during esophageal carcinogenesis as observed in both a mouse model of esophageal epithelial dysplasia as well as in human ESCC. These findings suggest that WNT10A may serve as an oncofetal factor in the esophagus.
Interestingly, WNT10A embryonic expression has been described in another context of esophageal development. Notably, FOXA3cre;cdx2 conditional knockout mice, which exhibit esophagealization of the intestine, express increased levels of WNT10A in both the mutant intestines, as well as the embryonic esophagus, as these mice are embryonic lethal (44). In conjunction with our data, these findings would lend credence to the premise that differentiation of the esophagus during late development may involve WNT10A. SOX2 is a critical transcription factor, also highly expressed in embryonic development of the esophagus. It is vital in the division of the trachea and the esophagus, in order to promote a single esophageal tube, as well as fostering the stratified squamous epithelium within the esophagus (45). In adult life, SOX2 is limited to the basal compartment of esophageal epithelial cells, where it promotes proliferation and differentiation (45). In other squamous epithelial types, both SOX2 and WNT10A have been shown to be important in normal development. Interestingly, Sox2 hypomorphic mice and humans carrying WNT10A mutation both present with malformation of the fungiform papillae, and ultimately smooth tongues, as WNT signaling has been shown to drive SOX2 expression (18,46). These observations suggest a potential connection between WNT10A driving SOX2 expression in esophageal development. Indeed, SOX2 is amplified in ESCC (47), and it would be interesting to determine to what extent WNT10A and SOX2 are overexpressed concurrently in ESCC.
Employing a 3D-OTC model of ESCC, we found that WNT10A is upregulated in the invasive front compared with the non-invasive epithelium. We further demonstrate that overexpression of WNT10A enhances esophageal cells ability to proliferate, migrate and invade. Loss of function or germline mutation of WNT10A specifically leads to malformation of ectodermal appendages during development. These developmental processes require careful communication between epithelial and mesenchymal lineages, for proper migration of epithelial cells to form placodes and eventually ectodermal tissues (23). Specifically, during the hair follicle development, and then later in anagen, a period of growth during the hair follicle cycle, WNT10A is upregulated near the dermal condensate and later dermal root sheet, and may be required for the downward growth of the hair follicle (22). Therefore, we propose that WNT10A may be normally required for epithelial migration and proliferation during development, and that the oncogenic state is co-opting a developmental process to facilitate carcinogenesis. This pattern is frequently seen in the context of other oncofetal factors.
We also observe that WNT10A expression is also increased in an ESCC TMA when compared with adjacent normal esophagus, and correlates with decreased MST. Patients with ESCC that express higher WNT10A levels had an overall significant decrease in 5-year survival time, as well. These data suggest that WNT10A can annotate the invasive compartment of ESCC and could potentially be used as a biomarker for poorer prognosis in ESCC. Similarly, WNT10A has been shown to be upregulated in renal cell carcinoma biopsies and drives proliferation, migration and chemoresistance of renal carcinoma cell lines (25). Taken together, these data suggest that WNT10A can act as an oncogene in a variety of tissue types.
We additionally demonstrated that WNT10A could induce stem cell-like properties by promoting self-renewal. These data are supported by our pathway analysis of invasive mutant TP53 cells, which suggests that in addition to epithelial to mesenchymal transition, WNT10A may also play a role in maintaining embryonic stem cells. Indeed, WNT10A is secreted by mammary stem cells and it may be required for maintaining this population (48). WNT signaling is also required for the maintenance of a variety of stem cell niches, such as in intestinal crypts and hematopoietic systems (49,50). Interestingly, we have demonstrated previously that PERIOSTIN, a type of matricellular protein involved in remodelling the tumor stroma during tumor cell invasion, is upregulated in ESCC, mediates ESCC invasion and is critical in tumorigenesis in vivo (8–10,51). Indeed, metastatic tumor cells induce stromal PERIOSTIN expression in the secondary target organ (e.g. lung) to initiate colonization (52,53). PERIOSTIN is needed for cancer stem cell maintenance and recruits Wnt ligands for increased Wnt signaling in cancer stem cells (52). Whether PERIOSTIN and WNT10A interact in this context is an area for future investigation.
Our study is the first report to demonstrate a role for WNT10A in both esophageal development and in tumorigenesis. Our data suggest that in a TP53 mutant background, WNT10A mediates tumor cell migration and invasion, while increasing a population of self-renewing CD44High cells. Interestingly, up to 83% of ESCC cases exhibit mutant in TP53, while altered genes in the WNT pathway represent almost 85% cases of ESCC (4). These mutations may work synergistically to promote more aggressive and invasive tumors in ESCC.
Supplementary material
Supplementary Table 1 and Figure 1 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health/National Cancer Institute P01 (CA098101 to A.K.R., A.L., V.G., K.W., A.J.K.-S., K.T., H.N.); National Institute of Health/National Cancer Institute U01 (CA143056); National Institute of Health/National Institute of Diabetes and Digestive and Kidney Diseases P30 Center for Molecular Studies in Digestive and Liver Diseases (Molecular Pathology and Imaging, Molecular Biology/Gene Expression, Cell Culture Core Facilities) (DK050306); American Cancer Society Grant (RP-10-033-01-CCE to A.K.R.]); National Institute of Health/National Cancer Institute F30 (CA175133 to A.L.); National Institute of Health/National Cancer Institute T32 (CA115299-06 to A.L.); National Fonds de recherche en sante du Quebec (P-Giroux-27692 to V.G.); National Institute of Health/National Institute of Diabetes and Digestive and Kidney Diseases K01 (DK100485 to K.E.H.).
Supplementary Material
Acknowledgements
We wish to thank members of the Rustgi lab for helpful discussions, and the staffs of the Molecular Pathology and Imaging Core, Molecular Biology/Gene Expression Core, Cell Culture Core and Transgenic and Chimeric Mouse Core Facilities.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- 4-NQO
4-nitroquinoline 1-oxide
- EGFR
epidermal growth factor receptor
- ESCC
esophageal squamous cell carcinoma
- IHC
immunohistochemistry
- MST
median survival time
- OTC
organotypic culture
- TMAs
tumor microarrays
References
- 1. Altekruse S.F. et al. (eds) SEER Cancer Statistics Review, 1975–2011 National Cancer Institute, Bethesda, MD. http://seer.cancer.gov/csr/1975_2011/. [Google Scholar]
- 2. Enzinger P.C., et al. (2003) Esophageal cancer. N. Engl. J. Med., 349, 2241–2252. [DOI] [PubMed] [Google Scholar]
- 3. Lu F., et al. (2003) An alternatively spliced cyclin D1 isoform, cyclin D1b, is a nuclear oncogene. Cancer Res., 63, 7056–7061. [PubMed] [Google Scholar]
- 4. Song Y., et al. (2014) Identification of genomic alterations in oesophageal squamous cell cancer. Nature, 509, 91–95. [DOI] [PubMed] [Google Scholar]
- 5. Stairs D.B., et al. (2011) Deletion of p120-catenin results in a tumor microenvironment with inflammation and cancer that establishes it as a tumor suppressor gene. Cancer Cell, 19, 470–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Okawa T., et al. (2007) The functional interplay between EGFR overexpression, hTERT activation, and p53 mutation in esophageal epithelial cells with activation of stromal fibroblasts induces tumor development, invasion, and differentiation. Genes Dev., 21, 2788–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kalabis J., et al. (2012) Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nat. Protoc., 7, 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Michaylira C.Z., et al. (2010) Periostin, a cell adhesion molecule, facilitates invasion in the tumor microenvironment and annotates a novel tumor-invasive signature in esophageal cancer. Cancer Res., 70, 5281–5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wong G.S., et al. (2013) Periostin cooperates with mutant p53 to mediate invasion through the induction of STAT1 signaling in the esophageal tumor microenvironment. Oncogenesis, 2, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wong G.S., et al. (2013) Optical imaging of periostin enables early endoscopic detection and characterization of esophageal cancer in mice. Gastroenterology, 144, 294–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clevers H., et al. (2012) Wnt/β-catenin signaling and disease. Cell, 149, 1192–1205. [DOI] [PubMed] [Google Scholar]
- 12. MacDonald B.T., et al. (2009) Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell, 17, 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Willert K., et al. (2012) Wnt proteins. Cold Spring Harb. Perspect. Biol., 4, a007864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Polakis P. (2012) Wnt signaling in cancer. Cold Spring Harb. Perspect. Biol., 4, a008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Staal F.J., et al. (2008) WNT signalling in the immune system: WNT is spreading its wings. Nat. Rev. Immunol., 8, 581–593. [DOI] [PubMed] [Google Scholar]
- 16. Wang J., et al. (1996) Murine Wnt10a and Wnt10b: cloning and expression in developing limbs, face and skin of embryos and in adults. Oncogene, 13, 1537–1544. [PubMed] [Google Scholar]
- 17. Kirikoshi H., et al. (2001) WNT10A and WNT6, clustered in human chromosome 2q35 region with head-to-tail manner, are strongly coexpressed in SW480 cells. Biochem. Biophys. Res. Commun., 283, 798–805. [DOI] [PubMed] [Google Scholar]
- 18. Adaimy L., et al. (2007) Mutation in WNT10A is associated with an autosomal recessive ectodermal dysplasia: the odonto-onycho-dermal dysplasia. Am. J. Hum. Genet., 81, 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bohring A., et al. (2009) WNT10A mutations are a frequent cause of a broad spectrum of ectodermal dysplasias with sex-biased manifestation pattern in heterozygotes. Am. J. Hum. Genet., 85, 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nawaz S., et al. (2009) WNT10A missense mutation associated with a complete odonto-onycho-dermal dysplasia syndrome. Eur. J. Hum. Genet., 17, 1600–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cluzeau C., et al. (2011) Only four genes (EDA1, EDAR, EDARADD, and WNT10A) account for 90% of hypohidrotic/anhidrotic ectodermal dysplasia cases. Hum. Mutat., 32, 70–72. [DOI] [PubMed] [Google Scholar]
- 22. Reddy S., et al. (2001) Characterization of Wnt gene expression in developing and postnatal hair follicles and identification of Wnt5a as a target of Sonic hedgehog in hair follicle morphogenesis. Mech. Dev., 107,69–82. [DOI] [PubMed] [Google Scholar]
- 23. Itin P.H. (2014) Etiology and pathogenesis of ectodermal dysplasias. Am. J. Med. Genet. A, 164A, 2472–2477. [DOI] [PubMed] [Google Scholar]
- 24. Visel A., et al. (2004) GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res., 32, D552–D556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hsu R.J., et al. (2012) WNT10A plays an oncogenic role in renal cell carcinoma by activating WNT/β-catenin pathway. PLoS One, 7, e47649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li J., et al. (2001) Novel NEMO/IkappaB kinase and NF-kappa B target genes at the pre-B to immature B cell transition. J. Biol. Chem., 276, 18579–18590. [DOI] [PubMed] [Google Scholar]
- 27. Kirikoshi H., et al. (2001) Expression of WNT10A in human cancer. Int. J. Oncol., 19, 997–1001. [DOI] [PubMed] [Google Scholar]
- 28. Kirikoshi H., et al. (2001) Up-regulation of WNT10A by tumor necrosis factor alpha and Helicobacter pylori in gastric cancer. Int. J. Oncol., 19, 533–536. [PubMed] [Google Scholar]
- 29. Kurasawa Y., et al. (2012) Stabilization of phenotypic plasticity through mesenchymal-specific DNA hypermethylation in cancer cells. Oncogene, 31, 1963–1974. [DOI] [PubMed] [Google Scholar]
- 30. Ordway J.M., et al. (2006) Comprehensive DNA methylation profiling in a human cancer genome identifies novel epigenetic targets. Carcinogenesis, 27, 2409–2423. [DOI] [PubMed] [Google Scholar]
- 31. Krämer A., et al. (2014) Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics, 30, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harada H., et al. (2003) Telomerase induces immortalization of human esophageal keratinocytes without p16INK4a inactivation. Mol. Cancer Res., 1, 729–738. [PubMed] [Google Scholar]
- 33. Nishihira T., et al. (1993) Molecular and cellular features of esophageal cancer cells. J. Cancer Res. Clin. Oncol., 119, 441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakagawa H., et al. (1995) Human cyclin D1 oncogene and esophageal squamous cell carcinoma. Cancer, 76, 541–549. [DOI] [PubMed] [Google Scholar]
- 35. Najdi R., et al. (2012) A uniform human Wnt expression library reveals a shared secretory pathway and unique signaling activities. Differentiation, 84, 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tang X.H., et al. (2004) Oral cavity and esophageal carcinogenesis modeled in carcinogen-treated mice. Clin. Cancer Res., 10(1 Pt 1), 301–313. [DOI] [PubMed] [Google Scholar]
- 37. Natsuizaka M., et al. (2014) IGFBP3 promotes esophageal cancer growth by suppressing oxidative stress in hypoxic tumor microenvironment. Am. J. Cancer Res., 4, 29–41. [PMC free article] [PubMed] [Google Scholar]
- 38. Schneider C.A., et al. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wielenga V.J., et al. (1999) Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am. J. Pathol., 154, 515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao J.S., et al. (2011) Tumor initiating cells in esophageal squamous cell carcinomas express high levels of CD44. PLoS One, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Al-Hajj M., et al. (2003) Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U. S. A., 100, 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mani S.A., et al. (2008) The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell, 133, 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Narita T., et al. (2005) Wnt10a is involved in AER formation during chick limb development. Dev. Dyn., 233, 282–287. [DOI] [PubMed] [Google Scholar]
- 44. Gao N., et al. (2009) Establishment of intestinal identity and epithelial–mesenchymal signaling by Cdx2. Dev. Cell, 16, 588–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Que J., et al. (2007) Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development, 134, 2521–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Okubo T., et al. (2006) Sox2 is required for development of taste bud sensory cells. Genes Dev., 20, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bass A.J., et al. (2009) SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat. Genet., 41, 1238–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ji H., et al. (2011) Proteomic profiling of secretome and adherent plasma membranes from distinct mammary epithelial cell subpopulations. Proteomics, 11, 4029–4039. [DOI] [PubMed] [Google Scholar]
- 49. Korinek V., et al. (1998) Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet., 19, 379–383. [DOI] [PubMed] [Google Scholar]
- 50. Reya T., et al. (2003) A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature, 423, 409–414. [DOI] [PubMed] [Google Scholar]
- 51. Wong G.S., et al. (2013) Matricellular proteins: priming the tumour microenvironment for cancer development and metastasis. Br. J. Cancer, 108, 755–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Malanchi I., et al. (2012) Interactions between cancer stem cells and their niche govern metastatic colonization. Nature, 481, 85–89. [DOI] [PubMed] [Google Scholar]
- 53. Holland J.D., et al. (2013) Wnt signaling in stem and cancer stem cells. Curr. Opin. Cell Biol., 25, 254–264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.