Abstract
Abnormally expanded DNA repeats are associated with several neurodegenerative diseases. In Friedreich's ataxia (FRDA), expanded GAA repeats in intron 1 of the frataxin gene (FXN) reduce FXN mRNA levels in averaged cell samples through a poorly understood mechanism. By visualizing FXN expression and nuclear localization in single cells, we show that GAA-expanded repeats decrease the number of FXN mRNA molecules, slow transcription, and increase FXN localization at the nuclear lamina (NL). Restoring histone acetylation reverses NL positioning. Expanded GAA-FXN loci in FRDA patient cells show increased NL localization with increased silencing of alleles and reduced transcription from alleles positioned peripherally. We also demonstrate inefficiencies in transcription initiation and elongation from the expanded GAA-FXN locus at single-cell resolution. We suggest that repressive epigenetic modifications at the expanded GAA-FXN locus may lead to NL relocation, where further repression may occur.
Introduction
An abnormal GAA trinucleotide repeat expansion in intron 1 of the frataxin gene (FXN) causes Friedreich's ataxia (FRDA; OMIM 229300), a progressive neurodegenerative disease and the most common form of recessive ataxia (1). FXN alleles in healthy individuals contain <36 GAA repeats, whereas in FRDA patients GAA expansions ranging from 70 to 1700 GAA repeats lead to FXN mRNA deficiency and subsequent reduced levels of frataxin, a nuclear-encoded mitochondrial protein essential for life (1,2). GAA expansion-mediated transcriptional dysregulation occurs due to the generation of unusual DNA structures such as triplexes or sticky DNA (3,4), R-loops (5,6) and heterochromatin (7,8), which lead to increased DNA methylation at specific CpG sites (9–11), reduced histone acetylation (H3/H4ac) and increased levels of methylated histones H3K9me2 and H3K9me3 (8,10). It has been suggested that these epigenetic changes surrounding the GAA expansion impair RNA polymerase II (RNAPII) elongation (12), but also spread upstream towards the FXN promoter, inducing a non-permissive chromatin configuration for transcription initiation, altering nucleosome positioning and preventing binding of insulator CCCTC-binding factor (CTCF) (6,13–15). However, these studies only provide the probable state of the FXN gene, as these observations come from experiments in which the outputs of bulk cell cultures are averaged. A dissection of the silencing mechanism in FRDA in situ, in which FXN localization and expression are quantified at single-cell level, is crucial to improve our understanding of the underlying pathogenesis and ultimately to design effective therapies for FRDA.
Studies indicate that the radial positioning of a gene within the nucleus correlates with its transcriptional output, but whether a gene is transcribed due to its position or its position is determined by its transcriptional state is the subject of current research (16–19). In particular, genomic DNA interactions with the nuclear periphery (NP) can actively contribute to gene repression (20–22). However, this is not a general phenomenon (20,23), rather is gene-specific, and may depend on multiple parameters such as transcription factor accessibility, promoter strength, existence of insulator elements and pre-existing chromatin marks, which may counteract the mechanisms underlying transcription repression. The nuclear lamina (NL) tends to be in contact with heterochromatin and is associated with markers of gene repression, such as enrichment in histone modifications H3K9me2 and H3K27me3 and depletion of activating histone marks and RNAPII occupancy (reviewed in ref. 24). Given that the expanded FXN heterochromatic state is coupled with gene repression, we asked where GAA-expanded FXN alleles are found in the nucleus, and how their location impacts on repression.
Here, we report a single-cell analysis of FXN repression in which we identify the NL as a novel and key player in FXN transcriptional impairment and silencing. Using a multidisciplinary approach including analysis in both fixed and living single cells, we show that expanded GAA repeats increase FXN positioning at the NL, leading to decreased numbers of FXN mRNA molecules and slower transcription kinetics in an FXN-GAA-MS2 cell model. We observe the same abnormal repositioning to the NL in carrier and FRDA patient cells and show that this tightly correlates with a marked decrease in the number of actively expressing FXN alleles. Furthermore, we show that those few active expanded FXN alleles located at the NL express at a significantly lower level than the alleles located in the interior of the nucleus. Finally, we demonstrate that expanded GAA repeats predominantly disrupt FXN transcription initiation. The mechanisms we describe may extend to other genetic diseases mediated by repeat expansions within regions of non-coding DNA.
Results
GAA repeat expansion increases FXN-GAA-MS2 positioning at the NL
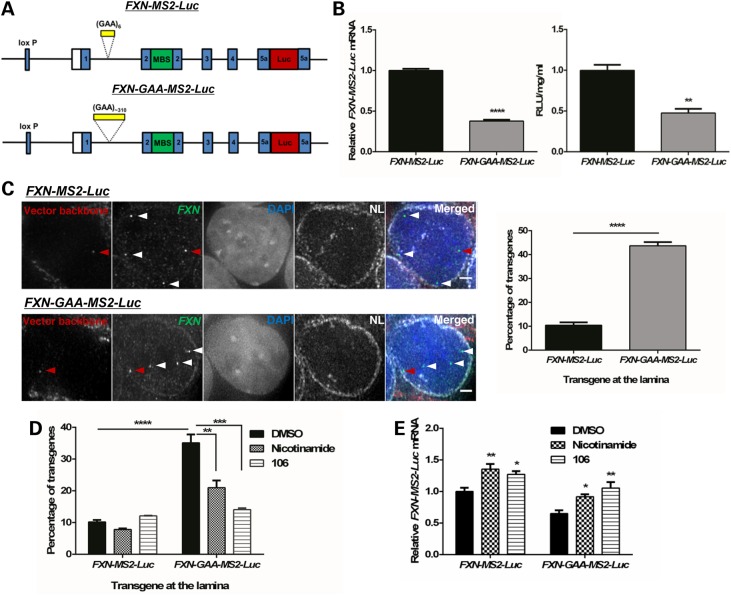
To investigate the link between FXN localization and expression at the single-cell level, we modified our previously described reporter model (25), which carries the whole 80 kb FXN locus with its native promoter, including exons 1–5b and all regulatory elements necessary to achieve physiologically relevant FXN expression, a luciferase reporter gene in exon 5a and the presence of either six GAA repeats or a ∼310 GAA repeat expansion in intron 1. To fluorescently label the transgenic FXN mRNA and quantify the effect of the expansion on FXN transcription, we inserted 24 repeats of MS2 protein-binding sites (MBS) into exon 2 by homologous recombination, generating pBAC-FXN-MS2-Luc and pBAC-FXN-GAA-MS2-Luc (Fig. 1A). We used site-specific integration to generate several stable human clones of FXN-MS2-Luc and FXN-GAA-MS2-Luc cell lines carrying a single copy of each vector in the same integration site in chromosome 1 (25) (Supplementary Material, Fig. S1A–D), because this allows a direct single-allele comparison of normal and expanded FXN loci. We selected FXN-MS2-Luc clone 2 and FXN-GAA-MS2-Luc clone 22 (referred to throughout the text as FXN-MS2-Luc and FXN-GAA-MS2-Luc, respectively) for the following studies, as these best represented the average reduction in FXN mRNA (Supplementary Material, Fig. S1E) and protein levels (Supplementary Material, Fig. S1F) in screened clones. The ∼310 GAA repeat expansion in the FXN-GAA-MS2-Luc cell line recapitulates the characteristic FXN gene repression seen in FRDA by altering the FXN epigenetic landscape (25) and reducing FXN mRNA levels by 62% and frataxin protein levels by 52% (Fig. 1B).
Figure 1.
The expanded GAA repeat FXN transgene associates with the NL more frequently in an FXN-GAA-MS2-Luc cell model. (A) Schematic representation of the pBAC-FXN-MS2-Luc and pBAC-FXN-GAA-MS2-Luc vectors. Each vector carries either six or ∼310 GAA repeats in intron 1, an array of 24 MBS in exon 2 and expresses an FXN-luciferase fusion protein. (B) FXN-MS2-Luc clone 2 and FXN-GAA-MS2-Luc clone 22 mRNA expression (left) was determined by qRT-PCR using qFXN-Luc primers and showed a significant reduction in FXN-GAA-MS2-Luc clone 22 mRNA levels, as determined by unpaired two-tailed Student's t-test (****P < 0.0001). Data are mean ± SEM (n = 3). Luciferase assay (right) showed a significant reduction in FXN-MS2-luciferase protein levels in FXN-GAA-MS2-Luc clone 22 cells, as determined by unpaired two-tailed Student's t-test (**P < 0.01). Data are mean ± SEM (n = 3). (C) 3D Immuno-FISH detection of FXN-MS2-Luc and FXN-GAA-MS2-Luc association with the NL (pseudocoloured white in merged image). Transgenes were identified by co-localization of FISH signals from FXN loci (green) and the vector backbone (red) in FXN-MS2-Luc and FXN-GAA-MS2-Luc cells (left). Nuclei were identified by DAPI staining (blue). Arrows show endogenous (white) and transgene (red) alleles. Comparison of the proportion of transgenes contacting the NL showed a significant difference in FXN-MS2-Luc and FXN-GAA-MS2-Luc cells (right) (****P < 0.0001, as determined by Fisher's two-tailed exact test, n = 140 cells per condition). Data are mean ± SEM from two independent FISH experiments. Association of FISH signals with the NL was analysed using ImageJ: image stacks were analysed manually, and association was determined by eye when FXN FISH signals were touching or superimposing the NL signal at a given z-section. Images are maximum projections of z-stacks covering the signals. Scale bar: 2 µm. (D) Treatment with nicotinamide (10 mm) or compound 106 (10 µm) for 48 h significantly decreased FXN-GAA-MS2-Luc interaction with NL (**P < 0.01, ***P < 0.001, ****P < 0.0001, as determined by Fisher's two-tailed exact test, n = 140 cells per condition). Data are mean ± SEM from 2–3 independent FISH experiments. (E) FXN-MS2-Luc mRNA expression after incubation with HDACi for 48 h was determined by qRT-PCR using qFXN-Luc primers (25). Data were normalized to GAPDH mRNA levels. Results are relative to FXN-MS2-Luc DMSO. Two-way analysis of variance (ANOVA) with Sidak's correction test showed a significant difference in the two cell lines when comparing with the DMSO control of each line (*P < 0.05; **P < 0.01). Data are mean ± SEM from three independent experiments.
FXN transgene localization in FXN-MS2-Luc and FXN-GAA-MS2-Luc lines was determined by immunofluorescence in situ hybridization (Immuno-FISH), using co-localized probes for the vector backbone and FXN loci to distinguish the transgene from the three additional endogenous FXN loci (25) present in each clone (Fig. 1C). We found FXN at the NL in ∼44% of FXN-GAA-MS2-Luc cells compared to only ∼10% of FXN-MS2-Luc cells. Treatment with the histone deacetylase inhibitors (HDACi) nicotinamide and compound 106—previously shown to upregulate FXN expression and increase histone acetylation levels and DNAse I accessibility at the expanded FXN locus in FRDA patient cells (26,27)—repositioned only FXN-GAA-MS2-Luc away from the NL (Fig. 1D), but increased transgene FXN mRNA levels in both cell lines (Fig. 1E), suggesting a complex interplay between FXN repression and intranuclear localization.
GAA-expanded repeats decrease the number of FXN mRNA molecules and slow transcription kinetics in single FXN-GAA-MS2 cells
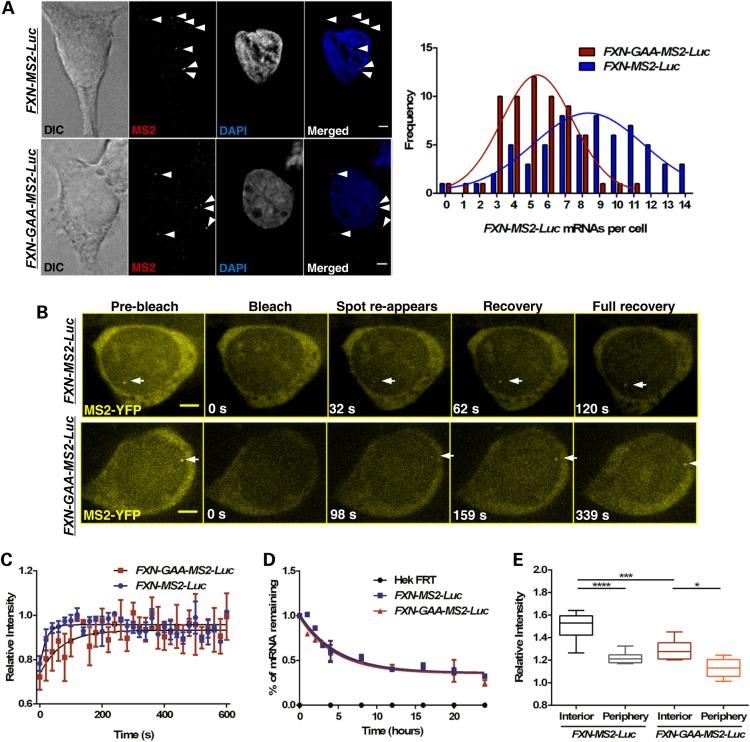
To further understand FXN repression, we analysed the transcriptional output of individual transgenic FXN alleles in FXN-MS2-Luc and FXN-GAA-MS2-Luc cells by RNA FISH using a probe hybridizing to the MBS sequence. Single FXN-MS2-Luc mRNA molecules were identified as sharp signals above non-specific fluorescence HEK cell background in accordance with previous studies demonstrating single-molecule resolution of this technique (Supplementary Material, Fig. S2) (28–30). The insertion of the MBS in exon 2 allowed us to identify all fluorescence spots as mature FXN mRNA molecules, considering transcription sites were not distinguishable. FXN-GAA-MS2-Luc cells contained ∼5 ± 2 mRNAs per cell, and FXN-MS2-Luc cells contained ∼9 ± 4 mRNA per cell (Fig. 2A), therefore ∼310 GAA repeats reduce the number of mature mRNA molecules by 44% at single-cell level. This quantification is in agreement with oligonucleotide array assays previously showing the majority of mRNAs are present at <10 copies per cell (31).
Figure 2.
The GAA repeat expansion reduces FXN-GAA-MS2-Luc transcriptional output and impedes transcription in fixed and living FXN-GAA-Luc cells. (A) Quantification of FXN-MS2-Luc mRNA molecules in FXN-MS2-Luc and FXN-GAA-MS2-Luc cells by RNA FISH. FXN-MS2-Luc mRNA molecules were identified as sharp dots (arrows) above background of non-specific fluorescence using an MS2-Cy3 probe (red in merged images) in FXN-MS2-Luc (top images) and FXN-GAA-MS2-Luc (bottom images) cells. Nonlinear regression curve-fit comparison showed a significant difference between the distribution of FXN-MS2-Luc mRNA molecules in FXN-MS2-Luc and FXN-GAA-MS2-Luc cells (P < 0.0001, F = 33.31, d.f. = 3, 21, n = 63 cells per line) (right). Images are maximum projections of z-stacks covering the signals. (B) Live-cell fluorescence video frames taken before and at indicated times after photobleaching of the transcription site. Scale bar: 2 µm. (C) FRAP recovery curves and fitted exponential curves show a difference in transcription kinetics (P = 0.0048, F = 4.434, d.f. = 3, 210, n = 5 cells per line). (D) Degradation of FXN-MS2-Luc mRNA was analysed by qRT-PCR using qFXN-Luc primers in FXN-MS2-Luc and FXN-GAA-MS2-Luc cells after treatment with Actinomycin D for 24 h. Data were normalized to β-actin mRNA. Comparison of fitted exponential decay curves shows no difference between the two cell lines (P = 0.5979, F = 0.6351, d.f. = 3, 32). FXN-MS2-Luc mRNA half-life was ∼4 h. Data are mean ± SEM (n = 3). (E) Quantification of fluorescence intensities of active transgenes in living FXN-MS2-Luc and FXN-GAA-MS2-Luc cells. Spot intensity of active FXN transgenes above MS2-YFP background was significantly lower in the nuclear interior in FXN-GAA-MS2-Luc cells when compared with FXN-MS2-Luc cells (***P < 0.001), and significantly lower when alleles were expressing at the periphery than in the nuclear interior in the two cell lines, as determined by one-way ANOVA with Sidak's multiple comparisons test (****P < 0.0001, *P < 0.05, n = 15 cells per line).
We then followed the kinetic behaviour of FXN-MS2-Luc and FXN-GAA-MS2-Luc transcription using fluorescence recovery after photobleaching (FRAP) (30). Nascent transcripts were detected as fluorescent spots as they were bound by MS2-YFP fusion protein and visualized in real time (Supplementary Material, Video S1 and S2). Recovery of fluorescence indicates new transcripts are actively synthesized at the transcription site (Fig. 2B). FRAP curves showed that the time required to reach steady state after photobleaching was different between the two cell lines. FXN-MS2-Luc cells showed half-time of recovery (t1/2) = 12.90 s with full recovery = 120 s (Fig. 2C and Supplementary Material, Video S3), whereas FXN-GAA-MS2-Luc cells showed slower kinetics t1/2 = 48.10 s with full recovery = 260 s (Fig. 2C and Supplementary Material, Video S4). Since we found no difference in FXN mRNA stability in FXN-GAA-MS2-Luc and FXN-GAA-MS2-Luc cells (Fig. 2D) (12), RNA FISH and FRAP data indicate that the GAA expansion decreases the amount of FXN-GAA-MS2-Luc mRNA molecules by impeding RNAPII transcription initiation and/or elongation. We could not distinguish between the two events given the placement of the MBS in exon 2, where the FRAP readout is obtained.
To elucidate the link between FXN localization and repression, we analysed the fluorescence intensities of active transgenes in living FXN-MS2-Luc and FXN-GAA-MS2-Luc cells. The fluorescence intensity of transcription sites was significantly lower when transgenes were expressed at the NP compared with the interior in both FXN-MS2-Luc and FXN-GAA-MS2-Luc cells (Fig. 2E). This indicates that both FXN transgenes express when localized at NP, although they do so at lower levels. When compared to active FXN-MS2-Luc transgenes, spot intensity of FXN-MS2-GAA-Luc transgenes was significantly lower only in the nuclear interior. Taken together with data from Figure 1C, these data suggest that expanded GAA repeats increase FXN-GAA-MS2-Luc positioning at the NL, where expression levels are reduced when compared to the nuclear interior or with FXN-MS2-Luc expression levels in the nucleoplasm.
Expanded FXN alleles localize more frequently at the NL in carrier cells
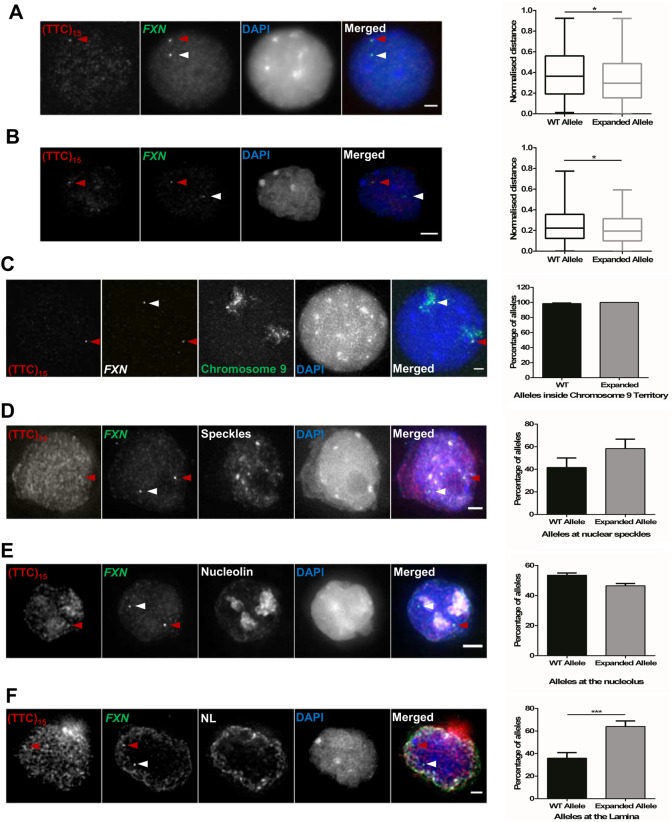
Reporter transgenes are a powerful tool to reveal mechanisms of gene expression, however their expression may be influenced by the local environment of the integration site (32). FXN-MS2-Luc and FXN-GAA-MS2-Luc transgenes are integrated in chromosome 1, and given that the native FXN is located in chromosome 9, we asked whether our findings from chromosome 1 would replicate in a more physiologically relevant context. We first analysed FXN localization in its native genomic environment in carrier-derived lymphoblastoid cells (GM14519) carrying 9 and 1285 GAA repeats in each FXN allele, respectively. Each carrier cell exhibited a 55% reduction in mRNA compared with healthy cells (Supplementary Material, Fig. S3). Co-detection of a DNA probe for FXN with a (GAA)15 or (TTC)15 oligo signal allowed discrimination of the expanded allele and a direct intracellular comparison of the normal and the expanded FXN allele, with distance measurements for each allele to the NP by 2D and 3D FISH (33,34). We found that the expanded FXN allele localized preferentially closer to the NP than the normal allele (Fig. 3A and B). To further analyse the observed difference in nuclear positioning, we then assessed FXN association with specific nuclear subdomains known to either (i) suggest the transcriptional status of a gene, such as nuclear SC35-enriched speckles and chromosome territories (CT), or (ii) interact with heterochromatin, such as the nucleolus and the NL (21,35–39). FXN never looped out of chromosome 9 nuclear territory in 2D FISH experiments (Fig. 3C), demonstrating that this does not account for the observed difference in nuclear localization. We found no evidence suggesting the expanded and normal FXN alleles associated differently with nuclear speckles (Fig. 3D) or with heterochromatin surrounding nucleoli (Fig. 3E). However, when we analysed FXN positioning at the NL, we found an increased number of expanded FXN alleles contacting with the NL when compared with the normal allele (Fig. 3F). Taken together, these data suggest that GAA repeat expansions increase FXN positioning at the NL in carrier-derived cells.
Figure 3.
Expanded FXN allele localizes closer to the NP and sits at the NL in carrier cells (GM14519). Each allele was detected with a DNA probe for FXN (green in merged image) and the expanded allele was identified by co-localization with a (GAA)15 or (TTC)15 oligo signal (red) (left). Nuclei were identified by DAPI staining (blue). Arrows indicate normal (white) and expanded (red) alleles. (A and B) Distance measurements between FISH signals, and the NP were measured and normalized by the local ray between the nuclear centre and periphery passing through the FISH signal (right). The expanded FXN allele (1285 + 50 GAA repeats) localized preferentially closer to the NP than the WT (9 GAA repeats) (*P = 0.0153, as determined by Wilcoxon matched-paired two-tailed test; data are mean ± SEM from three independent hybridizations, n = 147) in 2D (A) and 3D DNA FISH analysis (B) (*P = 0.0254, as determined by Wilcoxon matched-paired two-tailed test; data are mean ± SEM from three independent hybridizations, n = 107). (C) FXN (white) did not loop out of chromosome 9 territories (green) in 2D FISH experiments. Data are mean ± SEM, n = 136 cells from three independent hybridizations. (D and E) 3D Immuno-FISH detection of FXN association with nuclear SC35-enriched speckles (D) and nucleoli (E) showed no significant difference between alleles, as determined by Fisher's two-tailed exact test (Speckles: P = 0.4942, n = 86 cells from two independent hybridizations; nucleoli: P = 0.3607, n = 156 cells from two independent hybridizations) (F) 3D Immuno-FISH detection of FXN association with the NL (pseudocoloured white). Comparison of the proportion of cells with only one allele contacting with the NL revealed the mutation induced a significant increase in co-localization with the NL, as determined by Fisher's two-tailed exact test (***P = 0.0005). Data are mean ± SEM from two independent hybridizations (n = 166). Images are maximum projections of z-stacks covering the signals. Association of FISH signals with subnuclear domains or chromosome territories was analysed using ImageJ: image stacks were analysed manually and the association was determined by eye when FXN FISH signals were touching or superimposing the signal from a specific subnuclear domain at a given z-section. Scale bar: 2 µm.
NL is a key player in FXN transcriptional impairment and silencing
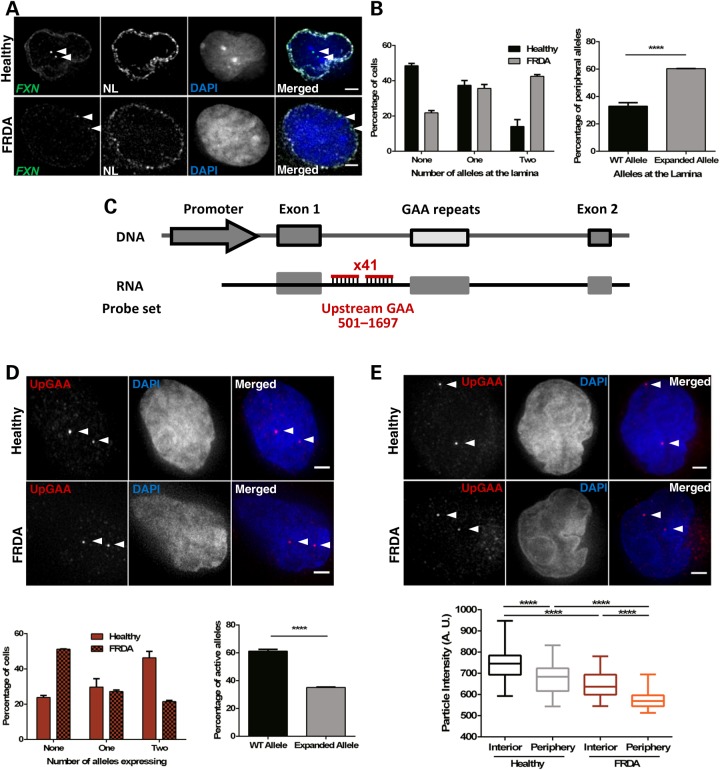
We next analysed FXN localization at the NL in healthy (GM15851) and patient-derived cells (GM16209) using Immuno-FISH (Fig. 4A). The patient cell line contains two FXN alleles carrying 800 GAA repeats each and was chosen to avoid differing behaviour of each locus due to different repeat size. Eight-hundred GAA repeats decreased mRNA levels by ∼83% (Supplementary Material, Fig. S3). The distribution of cells with FXN FISH signals contacting with the NL was significantly different in the two cell lines (Fig. 4B). We found one or both FXN alleles localized at the NL in a small proportion of healthy cells (∼37 and ∼14%, respectively), with ∼49% of cells containing both FXN alleles in the nucleoplasm. In contrast, the majority of FRDA cells presented both or one allele contacting with the NL (∼43 and ∼36%, respectively), with only 21% of FRDA cells containing both FXN alleles in the nucleoplasm. When data were analysed at allele level instead of cell level, ∼60% of expanded FXN alleles were found at the NL compared with only ∼33% of normal FXN alleles (Fig. 4B). These data suggest that the GAA expansion increases the probability of an allele to be found at the NL in FRDA cells, in agreement with data presented in Figures 1C and 3F.
Figure 4.
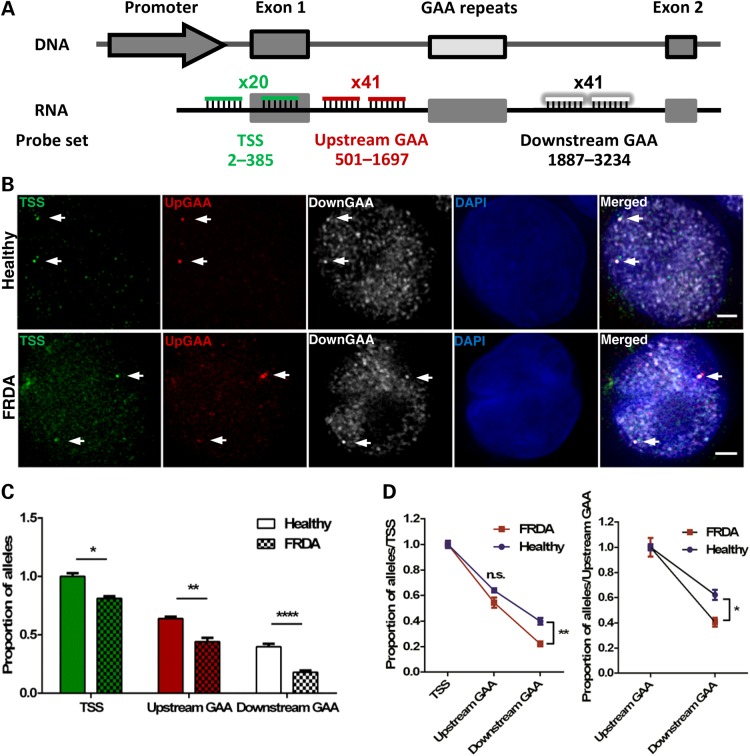
GAA repeat expansion increases FXN localization at the NL, reduces the number of active FXN alleles and downregulates transcription from active FXN in FRDA cells (GM16209). (A) 3D Immuno-FISH detection of FXN loci association with the NL in healthy (top images) and patient-derived (bottom images) cells. Each allele was detected with a DNA probe for FXN (green in merged image), and the NL was detected with an antibody for Lamin B1 (white). Nuclei were identified by DAPI staining (blue). Arrows indicate alleles. Association of FISH signals with the NL was analysed using ImageJ: image stacks were analysed manually and the association was determined by eye when FXN FISH signals were touching or superimposing the signal from the NL at a given z-section. (B) Histogram (left) showing the distribution of cells with FXN FISH signals contacting with the NL was significantly different in the two cell lines, as determined by Chi-squared test (P < 0.0001, χ2 = 63.056, d.f. = 2). When data were analysed at allele level instead of cell level (right), the proportion of FXN alleles positioned at the NL is significantly different when comparing the localization of the expanded and normal FXN alleles (****P < 0.0001, as determined by Fisher's two-tailed exact test). (C) Schematic representation showing the relative position to TSS and labelling of the DNA oligo probe set used in nascent RNA FISH experiments. (D) Nascent RNA FISH detection of FXN expression in healthy and patient-derived cells (top). Each allele was detected with a mix of 41 DNA oligos hybridizing with the region upstream of the GAA repeats (pseudocoloured red in merged image). Histogram (bottom left) showing the distribution of cells with RNA FXN FISH signals was significantly different in the two cell lines, as determined by Chi-squared test (P < 0.0001, χ2 = 75.424, d.f. = 2). When data were analysed at allele level instead of cell level (bottom right), the proportion of active FXN alleles is significantly different when comparing the expanded and normal FXN alleles (****P < 0.0001, as determined by Fisher's two-tailed exact test). Data are mean ± SEM from three independent hybridizations (n = 187–202 cells). (E) Nascent RNA FISH detection of FXN expression in healthy and patient-derived cells (top) showing different signal intensities of internal versus peripheral nascent RNA loci. Detection was performed as described in (D). Box plot (bottom) showing the RNA FISH spot intensity of active FXN alleles above the threshold was significantly lower in the interior and at the periphery in FRDA versus healthy nuclei (****P < 0.0001) and significantly lower when alleles were expressing at the periphery than in the nuclear interior in the two cell lines, as determined by one-way ANOVA with Tukey's multiple comparisons test (****P < 0.0001, n = 245 signals). Images are maximum projections of z-stacks covering the signals. Scale bar: 2 µm.
To further dissect FXN repression, we analysed nascent FXN RNA synthesis at a single-cell level, using probes hybridizing with the upstream region of the GAA repeats (Fig. 4C). Figure 4D shows ∼46% of healthy cells contained two FXN alleles expressing, and ∼30 and ∼24% of cells with either one or no expressing FXN allele, respectively. We found a higher proportion of FRDA cells (51%) containing both FXN alleles not expressing, followed by ∼27 and ∼22% of cells with either one or both FXN alleles expressing, respectively. Analysing these data at allele level instead of cell level, we found that ∼61% of normal FXN alleles were active compared to only ∼35% of expanded FXN alleles (Fig. 4D). These data suggest that the mutation increases the probability of an allele being silenced. Collectively, data from Figures 1C, 2A, E, 3F, 4B and D indicate a clear link between expanded FXN positioning at the NL and GAA-mediated transcriptional repression. Furthermore, these results confirm the existence of biological variability within cell populations and highlight the importance of single-cell studies (40).
To further elucidate the interplay between the repressive environment at the NL and FXN repression, we analysed RNA FISH spot intensities from active FXN alleles in Figure 4D as an indication of the quantity of transcript being synthesized at each transcription site relative to its interior or peripheral nuclear position (Fig. 4E). The signal variability of the technique was assessed by comparing RNA FISH intensities between replicates. Spot intensities were comparable among replicates (Supplementary Material, Fig. S4A), and background intensity was not significantly different in nuclei of healthy and FRDA patient cells (Supplementary Material, Fig. S4B). We found that FXN RNA FISH spot intensity was significantly lower when FXN alleles were expressed in the NP compared with the interior in both healthy and FRDA cells. This indicates that the NP was not incompatible with FXN expression because FXN RNA FISH signals could be seen at the periphery in both healthy and FRDA cells, albeit at lower intensities. Strikingly, RNA FISH spot intensity in FRDA cells was significantly lower than in healthy cells both in the interior and at the periphery. When collectively considering with the data in Figures 3F, 4B and D, we conclude that several factors combine to diminish transcription from the expanded FXN gene. The expanded FXN gene has a significantly increased likelihood of being positioned at the periphery (Figs 3F and 4B) and markedly reduced probability of being transcribed at all (Fig. 4D). Crucially, when transcription does occur at the expanded GAA-FXN locus, it is at a significantly reduced level both in the nucleoplasm and especially at the periphery (Fig. 4E). The NP can therefore be seen as a generally less supportive environment for FXN transcription, but the combined effect of expansion with peripheral relocation results in a catastrophic reduction in transcriptional output.
Expanded GAA repeats impair preferentially FXN transcription initiation in single FRDA cells
To analyse the effect of the mutation at the initiation and elongation stages of FXN transcription at single-cell resolution, probes hybridizing with the promoter after the transcription start site (TSS) and regions upstream and downstream of GAA repeats were included in our RNA FISH analysis (Fig. 5A and B). Healthy FXN alleles showed the expected decrease in transcription elongation throughout the analysed regions (Fig. 5C and D), confirming previous real-time reverse transcription-polymerase chain reaction (qRT-PCR) analysis (6,41). However, when compared with healthy alleles, expanded FXN alleles showed a ∼19% decrease in RNA levels at the TSS site, demonstrating a marked reduction in FXN transcription initiation (Fig. 5C). We also observed a ∼20% decrease at the region upstream of GAA repeats and a ∼22% decrease at the downstream region of GAA repeats. We then normalized the data in Figure 5C to exclude the effect of (i) transcription initiation at the TSS and (ii) elongation through the regions upstream of GAA repeats and found that expanded FXN alleles showed a small but significant decrease in FXN mRNA synthesis only downstream and not upstream of the GAA repeat expansion when compared with the same regions in healthy alleles (Fig. 5D), demonstrating that transcription elongation through expanded GAA repeats is also affected. Taken together, these data indicate that GAA repeat expansions diminish FXN mRNA synthesis predominantly by impairing FXN transcription initiation, but also by impairing elongation through the expansion.
Figure 5.
GAA repeat expansion disrupts transcription initiation and elongation in FRDA cells (GM16209). (A) Schematic representation showing the relative position to TSS and labelling of the three DNA oligo probe sets used to detect transcription initiation and elongation in RNA FISH experiments. (B) FXN RNA FISH detection of transcription initiation after the transcription starting site (TSS probe set in green) and elongation through the regions upstream (pseudocoloured red) and downstream (pseudocoloured white) of the GAA repeats. Nuclei were identified by DAPI staining (blue). Arrows indicate alleles. Images are maximum projections of z-stacks covering the signals. Scale bar: 2 µm. (C) Data were normalized to the TSS of healthy cells. When compared with the same region in healthy alleles, expanded FXN alleles showed a ∼19% decrease at the TSS, a ∼20% and a ∼22% decrease at the regions upstream and downstream of GAA repeats, respectively. This data demonstrates GAA repeat expansions reduce FXN transcription initiation by 19% and elongation downstream of expanded GAA repeats by 3% (*P = 0.0119, **P = 0.003, ****P < 0.0001, as determined by Fisher’'s two-tailed exact test, n = 218–226 cells per line). Data are mean ± SEM from three independent FISH experiments. (D) Data of each line in (C) were normalized to their TSS, thus excluding the effect of transcription initiation (left) and elongation through the region upstream of GAA repeats (right). Expanded FXN alleles show a significant difference in the region downstream of the GAA repeats when comparing with the same region in healthy alleles (*P < 0.05, **P < 0.01, as determined by two-way ANOVA with Sidak's multiple comparisons test). Data are mean ± SEM from three independent FISH experiments.
Discussion
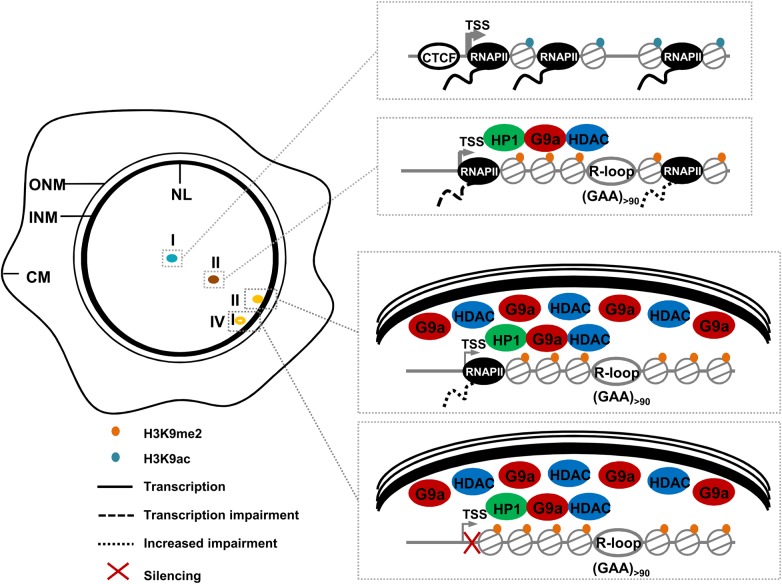
Gene relocation can be driven by chromatin remodelling coupled with or without transcription changes (19). Through these experiments, we demonstrate that the GAA expansion-mediated impairment of FXN expression is the result of a complex interplay of multiple factors contributing to different extents to decreased FXN transcriptional activity (Fig. 6). Among these factors, we identify the NL, a location known to be a generally repressive environment, as a novel major player in FXN silencing in FRDA. Using a variety of models, we found a marked increase in expanded FXN positioning at the NL and a concomitant reduction in the number of active FXN alleles, suggesting that FXN localized at the NL leads to FXN silencing. We have also identified a small proportion of expanded FXN alleles sitting at the NL which are transcriptionally active, however these show significantly reduced expression when compared with alleles in the nuclear interior. We observed two distinct events affecting FXN expression. First, we observed that the GAA repeat expansion impairs FXN transcription regardless of FXN localization in the nuclear interior or periphery. Second, although the proportion of normal and expanded FXN alleles positioned at the NL is remarkably different, we found that the NL reduces the expression of normal and expanded FXN at the NP to a similar extent, demonstrating that the repressive action of the NL is irrespective of a GAA expansion. This leads us to a consideration of the order of molecular events occurring at the expanded FXN locus. One of the initial steps in FXN repression is the binding of nascent FXN mRNA to the GAA repeat expansion as the gene is being transcribed, a feature shared with the CGG repeat expansion disease Fragile X Syndrome (6,42). These RNA/DNA hybrids (R-loops) have been previously shown to form on both transgenic and endogenous expanded FXN loci in cell lines used in this study (6). R-loops impair RNAPII elongation and trigger epigenetic changes, such as H3K9me2 repressive marks through recruitment of G9a methyltransferase (6). The effects of inhibition of histone deacetylases were interesting in this regard. FXN transcription from both normal and expanded alleles is upregulated by enforced acetylation by similar amounts, yet results in significant movement from the NL of only the expanded alleles. The mechanism of retention at the NL of these expanded repeat structures is therefore highly sensitive to HDAC inhibition. However, the expanded alleles are unable to achieve the same level of upregulation as their normal counterparts. This could be the result of a different extent of histone acetylation achieved by normal and expanded alleles following treatment with HDAC inhibitors, which could be caused by reduced accessibility of compacted FXN alleles. However, the observed difference in FXN upregulation could also imply that other structural changes such as R-loop formation contribute to transcriptional dysregulation and that the trigger event is induced by structural and epigenetic changes to the DNA followed by an increased likelihood of retention at the NL.
Figure 6.
Model of GAA repeat-mediated FXN repression and the repressive role of NL in FXN transcriptional impairment. The euchromatic state of normal FXN alleles allows physiological levels of expression (I). Expanded GAA repeats trigger R-loop formation and locally recruit HP1 (13), G9a methyltransferase (6) and class I (8) and III (27) HDACs, causing epigenetic changes that contribute to transcriptional dysregulation (II). These GAA expansion-mediated structural and epigenetic changes may lead to FXN relocation to the NL—a location known to be a generally repressive environment and enriched in HDAC and histone methyltransferase activity—where further repression (III) and silencing (IV) occur. ONM, outer nuclear membrane; INM, inner nuclear membrane; CM, cell membrane.
It seems plausible that, once a locus adopts an abnormal DNA structure and becomes modified by specific markers, it is more likely to be retained at the NL after mitosis (43). There are several mechanisms that could be involved. Genome contacts with the NL during interphase are generally dynamic, but confined to a narrow zone near the NL. Peripheral siting of chromatin after mitosis is largely determined by a stochastic mechanism and directly linked to cellular or allelic variation in H3K9me2 levels where G9a methyltransferase acts as a regulator of NL contacts (44). The NP is enriched in heterochromatin and the presence of HP1, whose binding is promoted by methylation at lysine 9, may help to regulate the post-mitotic binding of inner nuclear membrane proteins (43). There are also spatial considerations. The FXN locus is located between two lamina-associated domains (LADs)—NL-interacting chromosome regions demarcated by the insulator protein CTCF that present heterochromatic histone modifications and low expression levels—in lung fibroblasts (45) and various neural cell types (46). LAD topology has yet to be assessed in the lymphoblastoid cell lines used in this study, but as different cell types present remarkable LAD similarities (46), it is tempting to speculate that FXN is also in an interLAD in lymphoblastoid cells. Given the severe depletion of CTCF in the 5′ UTR of the expanded FXN in FRDA (13), we speculate that the boundary between FXN and the LAD located upstream is inefficient, leading to the inclusion of expanded FXN inside the LADs and increased proximity to the NL, possibly through recruitment of NL-associated HDACs and G9a methyltransferase (6). It would be interesting to explore the LAD layout over this region in FRDA versus healthy individuals, and whether interruption of the boundary CTCF elements can allow consolidation of the two flanking LADs. A further spatial contribution could be that even normal FXN can stochastically contact the NL, considering the intranuclear mobility of chromatin (47) and the FXN native location on the usually peripheral chromosome 9. Indeed, we observe that transcription from the normal allele is downregulated to an equivalent degree when located at the NP. FXN retention may result in a molecular interaction with the NL, as lamina-associating sequences are enriched in a GAGA motif which is bound by the transcriptional repressor cKrox associated with HDAC3 and inner nuclear membrane Lap2β (22). It would be interesting to knockdown cKrox in FRDA patient cells and analyse whether expanded FXN alleles at the NL relocalize to the nuclear interior. These events in turn could act through a positive feedback mechanism by mediating further impairment or silencing since the NL is enriched in HDAC and histone methyltransferase activity. Given the shared features of DNA repeat expansions, we hypothesize that the NL may play a role in heterochromatin-mediated transcriptional dysregulation in other repeat-expansion diseases and it would be interesting to interrogate the nuclear location of other repeat expansion loci. Although we show a marked degree of repression at the NP, one striking finding is that of transcriptionally active FXN alleles sitting at the NL, confirming what others have shown that the NP creates a repressive environment, but is not incompatible with expression (20,23). Our findings support the existence of NL heterogeneous microdomains composed of nuclear pore complexes and different lamin subtypes (24) able to impact differently on gene expression.
Non-coding triplet repeat expansions can mediate gene repression of other genes in cis (48,49). There is evidence suggesting that PIP5K1B is downregulated in FRDA lymphocytes and fibroblasts (50). The PIP5K1B locus is located ∼26 kb upstream of FXN and encodes phosphatidylinositol 4-phosphate 5-kinase β type I, an enzyme functionally linked to actin cytoskeleton dynamics. This gene is highly expressed in the brain, but also in the heart and lungs to a lesser extent (51), thus partially overlapping with tissues where FXN is expressed (1). Two other genes, PRKACG and TJP2, are located immediately upstream the 5′ end and downstream the 3′ end of the FXN locus, respectively, and show no differences in expression in FRDA patient cells (50). Information on the FXN locus interaction in trans is not yet available and is beyond the scope of our study. However, it would be interesting to analyse the expanded FXN localization in FRDA-relevant tissues, such as cardiomyocytes and sensory neurons, and assess expression levels of potential spatial FXN-interacting genes.
Here, we have focused on the nuclear location of and transcription from the expanded FXN gene at the single-cell level. We find an increased retention of the expanded FXN gene at the NL. We show that a higher proportion of expanded FXN alleles are inactive when compared with normal alleles and we have demonstrated reduced transcription from the expanded alleles that are active. We provide a single-cell quantification showing that expanded GAA repeats impair expression predominantly at the initiation stage of transcription, but also block RNAPII elongation. It is the combination of these factors which together reduce the levels of frataxin protein to cause disease. Our work provides new mechanistic insights into the molecular causes of FRDA which may extend to other genetic diseases mediated by repeat expansions within regions of non-coding DNA.
Materials and Methods
Cell culture
Epstein Barr virus-transformed lymphoblastoid cell lines GM14519 (from an FRDA clinically unaffected carrier, alleles with 1285 + 50 and 9 GAA repeats), GM15851 (from an unaffected individual with normal range of GAA repeats) and GM16209 (from an FRDA clinically affected individual, both alleles with 800 GAA repeats) were obtained from the Human Genetic Cell Repository of the Coriell Institute (USA) and propagated in RPMI 1640 medium supplemented with 15% fetal bovine serum (FBS) and 2 mm l-glutamine. HEK FRT cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, 2 mm l-glutamine, 100 U/ml penicillin/streptomycin and 100 µg/ml Zeocin (Life Technologies). FXN-MS2-Luc and FXN-GAA-MS2-Luc clonal cell lines were propagated in complete DMEM medium (see above) supplemented with 100 µg/ml Hygromycin B (Life Technologies). Actinomycin D (ActD) time course was performed by adding dimethyl sulphoxide (DMSO) or 8 µg/ml ActD to the media of FXN-MS2-Luc and FXN-GAA-MS2-Luc clonal cell lines before lysing cells every 4 h up to 24 h for RNA extraction. FXN-MS2-Luc and FXN-GAA-MS2-Luc cells were treated with DMSO, nicotinamide (10 mm) or compound 106 (10 µm) for 48 h before RNA extraction and Immuno-FISH.
Vector construction and stable cell line generation
pBAC-FXN-GAA-MS2-Luc was generated from the pBAC-FXN-GAA-Luc vector (25) which carries the whole 80 kb FXN locus with exons 1–5b of the FXN gene, an insertion of the luciferase gene in exon 5a and an ∼310 GAA repeat expansion in intron 1. A selection/counter-selection homologous recombination protocol based on RedET recombination (GeneBridges) was used. The recombination using the rpsL-neo and the pSC101-BAD plasmids was carried out in two steps in E. coli according to the manufacturer's instructions. In the first step, a PCR product containing the rpsL-neo cassette flanked by 55 bp homology arms to either side of exon 2 was used to insert the cassette. The replacement of the rpsL-neo cassette in the second step of recombination by 24 MS2 Binding Sites in exon 2 was performed as follows. A plasmid containing exon 2 with an engineered BamHI restriction site and flanking homology arms (147 bp upstream and 172 bp downstream of exon 2) was purchased from GeneArt (Life Technologies). The 24 MS2 Binding Sites were excised by BamHI/BglII digestion from pCR4-24XMS2SL-stable (52) (Addgene plasmid 31865) and inserted in exon 2 at the BamHI site (Exon2-MBS). The rpsL-neo cassette was replaced with a PCR product containing the Exon2-MBS sequence flanked by homology arms, generating pBAC-FXN-GAA-MS2-Luc. To generate pBAC-FXN-MS2-Luc, the ∼310 GAA repeats in intron 1 in pBAC-FXN-MS2-Luc were replaced by the rpsL-neo cassette amplified with primers carrying 58 bp homology arms to sequences immediately upstream and downstream of GAA repeats. The rpsL-neo was then replaced with a PCR product containing six GAA repeats amplified from the genomic DNA of GM15851 using GAA-F and GAA-R primers (1). Successful construction was confirmed by PCR, restriction enzyme digestion followed by pulsed-field gel electrophoresis and sequencing. Cre-loxP-mediated retrofitting of pBAC-FXN-MS2-Luc and pBAC-FXN-GAA-MS2-Luc vectors to pH-FRT-Hy was performed as previously described (53). Stable clonal cell lines were obtained by transfecting HEK FRT cells with either pBAC-FXN-MS2-Luc or pBAC-FXN-GAA-MS2-Luc together with Flp recombinase-encoding plasmid pOG44 (Life Technologies), generating the lines FXN-MS2-Luc and FXN-GAA-MS2-Luc, respectively. Stable selection was performed with 100 µg/ml hygromycin (Life Technologies). Stable clones were expanded and characterized as previously described (25).
Amplification of GAA repeats
Genomic DNA from FXN-GAA-MS2-Luc and FXN-MS2-Luc clonal cell lines was isolated using Illustra Tissue and Cells GenomicPrep Mini Spin Kit (GE Healthcare), according to the manufacturer's instructions. PCR amplification of the GAA repeat sequence was carried out on 200 ng of genomic DNA using primers 147F and 602R (54) and Expand Long Template DNA Polymerase (Roche) as previously described (1,55). PCR products were visualized on a 1% agarose gel. The GAA repeat sequences were sequenced to check for interruptions.
PCR, qPCR and copy number assay
Vector integration and copy number determination were performed as previously described (25). The integration of pBAC-FXN-MS2-Luc and pBAC-FXN-GAA-MS2-Luc at the docking site was assessed by PCR analysis using primers pSV40-F and Hygro-R. Copy number was determined by real-time PCR, using the relative standard curve method. The upstream region of the GAA repeats in intron 1 of the FXN gene was amplified and normalized by GAPDH. The number of transgene copies was determined by comparison with the acceptor cell line HEK FRT as reference sample, which carries three endogenous FXN loci (25).
qRT-PCR
Total RNA from HEK FRT, FXN-MS2-Luc and FXN-GAA-MS2-Luc clonal cell lines and from the lymphoblastoid-derived cell lines GM15851, GM14519 and GM16209 was extracted using RNeasy Mini Kit (Qiagen) and treated with RNase-free DNase (Qiagen). cDNA was synthesized from 1 µg of total RNA using random primers (Life Technologies) and SuperScript III Reverse Transcriptase (Life Technologies) as previously described (25). Three independent cDNA samples per cell line were quantified in triplicate by real-time PCR using the SYBR Green PCR Master Mix (Applied Biosystems). FXN-MS2-Luc mRNA expression was determined using primers qFXN-Luc-F and qFXN-Luc-R (25) normalized by GAPDH. FXN mRNA expression was detected using primers FXN-F and FXN-R (8) normalized by β-actin.
Luciferase assay
To assay luciferase expression, 1.5 × 106 cells of FXN-MS2-Luc and FXN-GAA-MS2-Luc clonal cell lines were used as previously described (25). Cells were seeded in 6-well plates, washed in phosphate-buffered saline (PBS) and lysed in lysis buffer (25 mm Tris–PO4, pH 7.8, 2 mm CDTA, 10% glycerol and 1% Triton-X 100) for 20 min at 4°C. Seventy-five microlitres of lysates were mixed with 100 μl of luciferase assay buffer (15 mm MgSO4, 15 mm KPO4, pH 7.8, 4 mm ethylene glycol tetraacetic acid, pH 7.8, 2 mm adenosine 5′-triphosphate and 2 mm dithiothreitol) and 50 μl of d-luciferin (0.3 mg/ml). The relative light units of luciferase of each cell line were determined using the Dynex MLX 96 Well Plate Luminometer and were normalized by total protein concentration, determined using bicinchoninic acid solution (BCA, Sigma).
Probes for DNA FISH
Human FXN gene was detected using either G248P87590H8 or G248P89980G8 fosmid clones obtained from BACPAC Resources Center (Children's Hospital, Oakland Research Institute). Probes were labelled by nick translation with digoxigenin (Roche). Briefly, 1 µg of DNA from each fosmid clone was treated with 200 ng RNase (Sigma) for 30 min at 37°C. After RNA removal, each probe was mixed with 5 µl of nick translation buffer [(0.5 M Tris–HCl, pH 8.0, 50 mm MgCl2, 0.5 mg/ml bovine serum albumin (BSA)], 5 µl of β-mercaptoethanol, 5 µl of deoxynucleotide triphosphate mix (0.5 mm deoxyadenosine triphosphate, 0.5 mm deoxyguanosine triphosphate, 0.5 mm deoxycytidine triphosphate), 1 nmol of digoxigenin, 6 units of DNAse I (Roche) and 10 units of DNA polymerase I (Life Technologies) and incubated for 2 h at 16°C. Unincorporated nucleotides were removed using Illustra G-50 columns (GE Healthcare), according to the manufacturer's instructions. In carrier cells, the expanded allele was distinguished from the WT allele using biotinylated (GAA)15 or (TTC)15 oligos (Biomers). Chromosome 9 was detected using a green XCyting 9 whole chromosome paint for HSA9 (MetaSystems).
2D. DNA FISH
DNA FISH was performed on GM14519 carrier cell line as described previously with slight modifications (56). Cells were fixed in 3:1 methanol–acetic acid and treated with 100 μg/ml RNase for 30 min at 37°C, washed in 2× SSC and dehydrated. Cells were denatured in 70% formamide/2× SSC at 72°C for 2 min and dehydrated. Labelled oligos (100 ng) were denatured in FISH hybridization buffer (Kreatech) at 90°C for 8 min and immediately allowed to cool on ice. Labelled fosmid clones (100 ng) were precipitated with 3 µg of human Cot-1 DNA (Life Technologies) and 20 µg of salmon sperm DNA (Sigma), denatured in FISH hybridization buffer at 90°C for 8 min and were preannealed at 37°C for 8 min. Whole chromosome 9 paint (4 µl) was denatured at 75°C for 2 min and combined with 8 μl of denatured oligo and fosmid and applied onto the slide. Slides were hybridized overnight at 37°C. After hybridization, slides were washed in 0.4× SSC/0.3% Igepal for 2 min and then 1 min at room temperature. Slides were blocked in 3% BSA/4× SSC for 30 min, biotin was detected with streptavidin Cy3 (Jackson ImmunoResearch Laboratories, 1/200 dilution) and digoxygenin was detected with sheep anti-digoxigenin FITC (Roche, 1/50 dilution) followed by rabbit anti-sheep FITC (Vector Laboratories, 1/100 dilution) or mouse anti-digoxigenin (Jackson ImmunoResearch Laboratories, 1/100 dilution) followed by Alexa Fluor 647 goat anti-mouse IgG (Life Technologies, A-21236, 1/200 dilution) when the green whole chromosome 9 paint was used. Cells were mounted in Vectashield (Vector Laboratories) with 1 µg/ml 4′,6-diamidino-2-phenylindole (DAPI) counterstain.
3D. DNA FISH
3D DNA FISH was performed on GM14519 carrier cell line as described previously with slight modifications (56). Cells were washed in PBS and allowed to settle on poly-l-lysine-treated coverslips for 5 min. Cells were fixed in 4% paraformaldehyde (PFA)/25 mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) for 15 min and permeabilized in 0.5% Triton X-100 (Calbiochem) in PBS for 10 min. RNA was removed with 100 μg/ml RNase in 2× SSC for 1 h at 37°C. Cells were denatured in 3 N HCl for 20 min and neutralized in ice-cold PBS. Probes were prepared as in the previous section, and slides were hybridized overnight at 37°C. Cells were washed twice in 2× SSC at 37°C, once in 1× SSC at RT for 30 min, and blocked in 3% BSA/4× SSC for 1 h. Probes were detected as in the previous section. Cells were washed between layers in 4× SSC with 0.05% Tween-20. Coverslips were mounted in Vectashield with 1 µg/ml DAPI counterstain.
Immuno-FISH
Immuno-FISH was performed on FXN-MS2-Luc and FXN-GAA-MS2-Luc cells and on the lymphoblastoid cell lines GM14519, GM15851, and GM16209 as described previously with slight modifications (56). Adherent cells were seeded on poly-l-lysine-treated coverslips and allowed to grow overnight. Suspension cells were washed in PBS and allowed to settle on poly-l-lysine-treated coverslips for 5 min. Cells were fixed in 4% PFA/25 mm HEPES for 15 min and permeabilized in 0.5% Triton X-100 for 10 min. Non-specific sites were blocked with 10% FBS. Antibodies were prepared in blocking solution. The NL was detected with goat anti-lamin B1 (Santa Cruz, sc-6217, 1/200 dilution). Nuclear speckles were detected with mouse anti-SC35 (Sigma, S4045, 1/1000 dilution). The nucleolus was detected with mouse anti-nucleolin (Abcam, ab13541, 1/25 dilution). Secondary antibodies used were Alexa Fluor 647 goat anti-mouse IgG (Life Technologies, A-21236, 1/1000 dilution) and Alexa Fluor 647 rabbit anti-goat IgG (Life Technologies, A-21446, 1/500 dilution). After protein detection, the slides were post-fixed in 4% PFA/25 mm HEPES at RT for 10 min. Cells were treated with 100 μg/ml RNase for 1 h at 37°C, denatured in 3 N HCl for 20 min and neutralized in ice-cold PBS. Probes were prepared as in the 2D DNA FISH section, and slides were hybridized overnight at 37°C. Cells were washed twice in 2× SSC at 37°C, once in 1× SSC at RT and blocked in 3% BSA/4× SSC. Probes were detected as in the 3D DNA-FISH section.
RNA FISH
RNA FISH was performed on HEK FRT and FXN-MS2-Luc cell lines as previously described (57) with minor modifications. Briefly, cells were allowed to grow on poly-l-lysine-treated coverslips overnight, washed in PBS and fixed in 4% formaldehyde (Electron Microscopy Science) for 10 min at RT. Cells were permeabilized in 0.5% Triton X-100 for 6 min and washed in 50% formamide/2× SSC for 5 min. Cells were hybridized overnight at 37°C with 10 ng Cy3-MS2 DNA probe (58) in 40 µl hybridization mix containing 2× SSC, 3% BSA, 10% dextran sulphate (Sigma), 2 mm vanadyl-ribonucleoside complex (New England Biolabs), 40 µg tRNA (Roche) and 50% formamide. Cells were washed once in 50% formamide/2× SSC at 37°C for 10 min, once in 2× SSC for 10 min at 37°C and once in 2× SSC for 5 min at RT. Cells were washed briefly in PBS and mounted in Vectashield with 1 µg/ml DAPI counterstain.
RNA FISH on GM15851 and GM16209 cell lines was performed as described previously (59) with minor modifications. Custom Stellaris FISH probes TSS (20 oligos), UpGAA (41 oligos) and DownGAA (41 oligos) were designed to hybridize with the regions TSS-exon1, upstream and downstream of the GAA repeats, respectively (Supplementary Material, Table S1), according to the manufacturer's instructions. Cells were washed in PBS and allowed to settle on poly-l-lysine-treated coverslips for 5 min. Cells were fixed in 3.7% formaldehyde/1× PBS for 10 min, permeabilized in 70% ethanol at 4°C for 1 h and washed in 10% formamide/2× SSC at RT for 5 min. Hybridization was performed overnight at 42°C with probes TSS (50 nm), UpGAA (50 nm) and DownGAA (25 nm) in 100 µl hybridization buffer containing 100 mg/ml dextran sulphate, 10% formamide in 2× SSC. Cells were washed in 10% formamide/2× SSC at 42°C for 10 min. Cells were washed briefly in PBS and mounted in Vectashield with 1 µg/ml DAPI counterstain.
Fluorescence recovery after photobleaching
FRAP experiments were performed using the UltraView Spinning Disk system (PerkinElmer) mounted on an IX81 microscope (Olympus) equipped with a 100× 1.3 NA objective, a FRAP unit and an electron multiplying charge-coupled device (EMCCD) camera (Hamamatsu Photonics) and driven by Volocity software (PerkinElmer). HEK FRT and FXN-MS2-Luc cell lines were seeded on collagen-coated glass-bottom dishes (MatTek) in complete DMEM medium (see above) and transiently transfected with NES-YFP-MS2-NLS (60) (a kind gift from Y. Shav-Tal). For nuclear detection, NucBlue Live reagent was added to the cells 20 min before imaging. Cells were maintained at 37°C in a 5% CO2 humidified atmosphere using an on-scope incubator and an objective heater (Tokai). Fluorescent spots inside the nucleus indicating active FXN-MS2-Luc transcription sites were detected with the YFP channel (514 nm laser). The nucleus was detected with the DAPI channel (405 nm laser). z-stacks were acquired at 0.25 µm intervals to cover the entire nuclear volume of each cell. Before bleaching, three z-stacks of each cell were taken. The active transcription site was bleached for 200 ms with the 514 nm laser power set to 100%. Post-bleach recovery z-stacks images were acquired with an exposure time of 50 ms every 20 s for 20 min.
Imaging acquisition and analysis
Images of fixed cells were obtained using the DV Elite system based on an Olympus IX71 fully motorized wide-field deconvolution inverted microscope with a 100× objective 1.40 numerical aperture (NA) fitted with a CoolSNAP HQ2 cooled charge-coupled device (CCD) camera (Photometrics) driven by SoftWoRx 5.0 software (Applied Precision). Several cell positions were chosen randomly and recorded using the motorized stage. z-stacks were acquired at 0.2 µm intervals to cover the entire volume of each cell. Distance measurements between FISH signals and the NP were calculated with NEMO software (61). Briefly, a region of interest (ROI) was defined around each nucleus using an image stack from the DAPI channel. The software applied several segmentation filters (Median 3D, TopHat 3D, mathematical morphology 3D) on raw image stacks for object detection. Objects were detected by computing seeds as the brightest maximum local pixels above a global threshold, and a local threshold was then computed to aggregate neighbouring pixels to the seed. For each cell, the distance of each allele to the NP was normalized by the local ray between the nuclear centre and periphery passing through the FISH signal. For relative positional analysis, image stacks were deconvolved using the SoftWoRx 5.0 software. Association of FISH signals with subnuclear domains or chromosome territories was analysed using ImageJ (http://imagej.nih.gov/ij/): image stacks were analysed manually, and association was determined by eye when FXN FISH signals were touching or superimposing the signal from a specific subnuclear domain at a given z-section. Images are maximum projections of z-stacks covering the signals. Contrast-stretch and gamma adjustments were made using Adobe Photoshop CS5 only for display in Figures 3B, F and 4A.
RNA MS2 FISH signals were detected as described previously (29) using StarSearch software (http://rajlab.seas.upenn.edu/StarSearch/launch.html). Briefly, the program applied Laplacian of Gaussian filters in three dimensions, taking into consideration the size of the fluorescent spots to remove the non-uniform background while enhancing particles. After filtering, the program calculated the number of spots detected for all possible thresholds. After plotting this data, the graph showed a plateau region corresponding to a broad range of thresholds over which the spot count did not vary significantly. Spot counts calculated with these thresholds matched the number of spots identified by eye. In HEK FRT cells and cells from the FXN-MS2-Luc cell lines that did not express the transgene, the plateau region was not observed. The DIC channel was used to determine the cell edges.
In RNA FISH experiments on GM15851 and GM16209 cell lines, several cell positions were chosen randomly. z-stacks were acquired at 0.2 µm intervals to cover the entire volume of each nucleus. Images were analysed using ImageJ. For the analysis of nascent RNA FISH signals using the UpGAA probe only, a threshold was defined for both cell lines to eliminate non-specific background. RNA fluorescent spots were identified either at the nuclear rim or interior using the DAPI channel and their intensity measured using the 3D Object Counter plugin (62). Identification of RNA FISH signals using probes TSS, UpGAA and DownGAA was performed as described above. In addition to threshold determination, to distinguish true signals from background, the downstream signals were scored when co-localized with the upstream signals. Because the TSS probe hybridizes to exon 1, it could potentially label mature FXN mRNA. However, due to its short length spanning, the TSS probe did not generate more than two RNA FISH signals per cell. Therefore, all TSS RNA FISH signals were identified as nascent transcript accumulating at the transcription sites.
For live-cell experiments, HEK FRT and FXN-MS2-Luc cell lines were seeded on collagen-coated glass-bottom dishes (MatTek) in complete DMEM medium (see above) and transiently transfected with NES-YFP-MS2-NLS (60). For live-cell imaging of transcription sites, cells were imaged in four dimensions (three dimensions over time) using the DV Elite system based on an Olympus IX71 microscope with a 100× objective 1.40 NA fitted with an Evolve EMCCD camera (Photometrics) driven by SoftWoRx 5.0 software under physiological conditions inside an incubator at 37°C and 5% CO2.
FRAP image sequences were analysed with Volocity software (PerkinElmer). Tracking of fluorescence spots was performed using the manual tracking function on Volocity at each time point. Relative intensity of each time point was calculated using
where T0 and Tt are the average intensity of an arbitrary area in the nucleus before and after bleaching, respectively, and I0 and It are the average intensity of the ROI before and after bleaching, respectively. The background was subtracted from all measured values. The formula accounted for fluorescence loss due to bleaching monitoring, and finally fluorescence intensity was normalized to 1 (63). Data from five cells per lines were collected in three independent experiments, and the averaged FRAP measurements were fitted with exponential curves. The diffusion of MS2-YFP protein during FRAP analysis was ignored as the diffusion rate of free MS2-YFP is very rapid, whereas the bound MS2-YFP is associated with high affinity to the mRNA and does not detach, diffuse and bind again at the transcription site (30).
Supplementary Material
Funding
This work was supported by Ataxia UK (fellowship no. 7125 to M.M.L.), the Friedreich's Ataxia Research Alliance (FARA) (to M.M.L. and R.W.-M.), BabelFAmily (to M.M.L.), Associazione Italiana per la lotta alle Sindromi Atassiche (AISA) (to M.M.L.), the European Union 7th Framework Program EFACTS (grant agreement no. 242193) (to R.W.-M.). M.M.L. is an Ataxia UK research fellow co-funded by FARA and AISA. A.M.S. was supported by Fundação para a Ciência e a Tecnologia PhD studentship (SFRH/BD/61048/2009). Advanced microscopy at Micron Oxford was supported by a Wellcome Trust Strategic Award (091911).
Supplementary Material
Acknowledgements
We thank Y. Shav-Tal for the gift of NES-YFP-MS2-NLS construct, J.R. Chubb, R. Parton and T. Weil for helpful discussions.
Conflict of Interest statement. None declared.
References
- 1.Campuzano V., Montermini L., Moltò M.D., Pianese L., Cossée M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., et al. (1996) Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science, 271, 1423–1427. [DOI] [PubMed] [Google Scholar]
- 2.Cossée M., Puccio H., Gansmuller A., Koutnikova H., Dierich A., LeMeur M., Fischbeck K., Dollé P., Kœnig M. (2000) Inactivation of the Friedreich ataxia mouse gene leads to early embryonic lethality without iron accumulation. Hum. Mol. Genet., 9, 1219–1226. [DOI] [PubMed] [Google Scholar]
- 3.Sakamoto N., Chastain P.D., Parniewski P., Ohshima K., Pandolfo M., Griffith J.D., Wells R.D. (1999) Sticky DNA: self-association properties of long GAA·TTC repeats in R·R·Y triplex structures from Friedreich's ataxia. Mol. Cell, 3, 465–475. [DOI] [PubMed] [Google Scholar]
- 4.Bidichandani S.I., Ashizawa T., Patel P.I. (2014) The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. Am. J. Hum. Genet., 62, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grabczyk E., Mancuso M., Sammarco M.C. (2007) A persistent RNA·DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res., 35, 5351–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groh M., Lufino M.M.P., Wade-Martins R., Gromak N. (2014) R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and Fragile X Syndrome. PLoS Genet., 10, e1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saveliev A., Everett C., Sharpe T., Webster Z., Festenstein R. (2003) DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature, 422, 909–913. [DOI] [PubMed] [Google Scholar]
- 8.Herman D., Jenssen K., Burnett R., Soragni E., Perlman S.L., Gottesfeld J.M. (2006) Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat. Chem. Biol., 2, 551–558. [DOI] [PubMed] [Google Scholar]
- 9.Greene E., Mahishi L., Entezam A., Kumari D., Usdin K. (2007) Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res., 35, 3383–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Mahdawi S., Pinto R.M., Ismail O., Varshney D., Lymperi S., Sandi C., Trabzuni D., Pook M. (2008) The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum. Mol. Genet., 17, 735–746. [DOI] [PubMed] [Google Scholar]
- 11.Evans-Galea M.V., Carrodus N., Rowley S.M., Corben L.A., Tai G., Saffery R., Galati J.C., Wong N.C., Craig J.M., Lynch D.R., et al. (2012) FXN methylation predicts expression and clinical outcome in Friedreich ataxia. Ann. Neurol., 71, 487–497. [DOI] [PubMed] [Google Scholar]
- 12.Punga T., Bühler M. (2010) Long intronic GAA repeats causing Friedreich ataxia impede transcription elongation. EMBO Mol. Med., 2, 120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Biase I., Chutake Y.K., Rindler P.M., Bidichandani S.I. (2009) Epigenetic silencing in Friedreich ataxia is associated with depletion of CTCF (CCCTC-binding factor) and antisense transcription. PLoS ONE, 4, e7914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumari D., Biacsi R.E., Usdin K. (2011) Repeat expansion affects both transcription initiation and elongation in Friedreich ataxia cells. J. Biol. Chem., 286, 4209–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chutake Y.K., Costello W.N., Lam C., Bidichandani S.I. (2014) Altered nucleosome positioning at the transcription start site and deficient transcriptional initiation in Friedreich ataxia. J. Biol. Chem., 289, 15194–15202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dillon N. (2008) The impact of gene location in the nucleus on transcriptional regulation. Dev. Cell, 15, 182–186. [DOI] [PubMed] [Google Scholar]
- 17.Takizawa T., Meaburn K.J., Misteli T. (2008) The meaning of gene positioning. Cell, 135, 9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrai C., de Castro I.J., Lavitas L., Chotalia M., Pombo A. (2010) Gene positioning. Cold Spring Harb. Perspect. Biol., 2, a000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Therizols P., Illingworth R.S., Courilleau C., Boyle S., Wood A.J., Bickmore W.A. (2014) Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science, 346, 1238–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finlan L.E., Sproul D., Thomson I., Boyle S., Kerr E., Perry P., Ylstra B., Chubb J.R., Bickmore W.A. (2008) Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet., 4, e1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy K.L., Zullo J.M., Bertolino E., Singh H. (2008) Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature, 452, 243–247. [DOI] [PubMed] [Google Scholar]
- 22.Zullo J.M., Demarco I.A., Piqué-Regi R., Gaffney D.J., Epstein C.B., Spooner C.J., Luperchio T.R., Bernstein B.E., Pritchard J.K., Reddy K.L., et al. (2014) DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell, 149, 1474–1487. [DOI] [PubMed] [Google Scholar]
- 23.Kumaran R.I., Spector D.L. (2008) A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol., 180, 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kind J., van Steensel B. (2010) Genome–nuclear lamina interactions and gene regulation. Curr. Opin. Cell Biol., 22, 320–325. [DOI] [PubMed] [Google Scholar]
- 25.Lufino M.M.P., Silva A.M., Németh A.H., Alegre-Abarrategui J., Russell A.J., Wade-Martins R. (2013) A GAA repeat expansion reporter model of Friedreich's ataxia recapitulates the genomic context and allows rapid screening of therapeutic compounds. Hum. Mol. Genet., 22, 5173–5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu C., Soragni E., Chou C.J., Herman D., Plasterer H.L., Rusche J.R., Gottesfeld J.M. (2009) Chemical probes identify a role for histone deacetylase 3 in Friedreich's ataxia gene silencing. Chem. Biol., 16, 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan P.K., Torres R., Yandim C., Law P.P., Khadayate S., Mauri M., Grosan C., Chapman-Rothe N., Giunti P., Pook M., et al. (2013) Heterochromatinization induced by GAA-repeat hyperexpansion in Friedreich's ataxia can be reduced upon HDAC inhibition by vitamin B3. Hum. Mol. Genet., 22, 2662–2675. [DOI] [PubMed] [Google Scholar]
- 28.Fusco D., Accornero N., Lavoie B., Shenoy S.M., Blanchard J.M., Singer R.H., Bertrand E. (2003) Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol., 13, 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raj A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., Tyagi S. (2008) Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods, 5, 877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yunger S., Rosenfeld L., Garini Y., Shav-Tal Y. (2010) Single-allele analysis of transcription kinetics in living mammalian cells. Nat. Methods, 7, 631–633. [DOI] [PubMed] [Google Scholar]
- 31.Lockhart D.J., Winzeler E.A. (2000) Genomics, gene expression and DNA arrays. Nature, 405, 827–836. [DOI] [PubMed] [Google Scholar]
- 32.Akhtar W., De Jong J., Pindyurin A.V., Pagie L., Meuleman W., De Ridder J., Berns A., Wessels L.F.A., Van Lohuizen M., van Steensel B. (2013) Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell, 154, 914–927. [DOI] [PubMed] [Google Scholar]
- 33.Meaburn K.J., Misteli T. (2008) Locus-specific and activity-independent gene repositioning during early tumorigenesis. J. Cell Biol., 180, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takizawa T., Gudla P.R., Guo L., Lockett S., Misteli T. (2008) Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP. Genes Dev., 22, 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shopland L.S., Johnson C.V., Byron M., McNeil J., Lawrence J.B. (2003) Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J. Cell Biol., 162, 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chambeyron S., Bickmore W.A. (2004) Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev., 18, 1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown J.M., Leach J., Reittie J.E., Atzberger A., Lee-Prudhoe J., Wood W.G., Higgs D.R., Iborra F.J., Buckle V.J. (2006) Coregulated human globin genes are frequently in spatial proximity when active. J. Cell Biol., 172, 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L.-F., Huynh K.D., Lee J.T. (2007) Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell, 129, 693–706. [DOI] [PubMed] [Google Scholar]
- 39.Zhao R., Bodnar M.S., Spector D.L. (2009) Nuclear neighborhoods and gene expression. Curr. Opin. Genet. Dev., 19, 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levsky J.M., Singer R.H. (2003) Gene expression and the myth of the average cell. Trends Cell Biol., 13, 4–6. [DOI] [PubMed] [Google Scholar]
- 41.Kim E., Napierala M., Dent S.Y.R. (2011) Hyperexpansion of GAA repeats affects post-initiation steps of FXN transcription in Friedreich's ataxia. Nucleic Acids Res., 39, 8366–8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colak D., Zaninovic N., Cohen M.S., Rosenwaks Z., Yang W.-Y., Gerhardt J., Disney M.D., Jaffrey S.R. (2014) Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science, 343, 1002–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schooley A., Vollmer B., Antonin W. (2012) Building a nuclear envelope at the end of mitosis: coordinating membrane reorganization, nuclear pore complex assembly, and chromatin de-condensation. Chromosoma, 121, 539–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kind J., Pagie L., Ortabozkoyun H., Boyle S., de Vries S.S., Janssen H., Amendola M., Nolen L.D., Bickmore W.A., van Steensel B. (2013) Single-cell dynamics of genome-nuclear lamina interactions. Cell, 153, 178–192. [DOI] [PubMed] [Google Scholar]
- 45.Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., de Klein A., Wessels L., de Laat W., et al. (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature, 453, 948–951. [DOI] [PubMed] [Google Scholar]
- 46.Peric-Hupkes D., Meuleman W., Pagie L., Bruggeman S.W.M., Solovei I., Brugman W., Gräf S., Flicek P., Kerkhoven R.M., van Lohuizen M., et al. (2010) Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell, 38, 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chubb J.R., Boyle S., Perry P., Bickmore W.A. (2002) Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol., 12, 439–445. [DOI] [PubMed] [Google Scholar]
- 48.Klesert T., Otten A., Bird T., Tapscott S. (1997) Trinucleotide repeat expansion at the myotonic dystrophy locus reduces expression of DMAHP. Nat. Genet., 16, 402–406. [DOI] [PubMed] [Google Scholar]
- 49.Thornton C., Wymer J., Simmons Z., McClain C., Moxley R. (1997) Expansion of the myotonic dystrophy CTG repeat reduces expression of the flanking DMAHP gene. Nat. Genet., 16, 407–409. [DOI] [PubMed] [Google Scholar]
- 50.Bayot A., Reichman S., Lebon S., Csaba Z., Aubry L., Sterkers G., Husson I., Rak M., Rustin P. (2013) cis-silencing of PIP5K1B evidenced in Friedreich's ataxia patient cells results in cytoskeleton anomalies. Hum. Mol. Genet., 22, 2894–2904. [DOI] [PubMed] [Google Scholar]
- 51.Volpicelli-Daley L.A., Lucast L., Gong L.-W., Liu L., Sasaki J., Sasaki T., Abrams C.S., Kanaho Y., De Camilli P. (2010) Phosphatidylinositol-4-phosphate 5-kinases and phosphatidylinositol 4,5-bisphosphate synthesis in the brain. J. Biol. Chem., 285, 28708–28714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bertrand E., Chartrand P., Schaefer M., Shenoy S.M., Singer R.H., Long R.M. (1998) Localization of ASH1 mRNA particles in living yeast. Mol. Cell, 2, 437–445. [DOI] [PubMed] [Google Scholar]
- 53.Lufino M.M.P., Manservigi R., Wade-Martins R. (2007) An S/MAR-based infectious episomal genomic DNA expression vector provides long-term regulated functional complementation of LDLR deficiency. Nucleic Acids Res., 35, e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Montermini L., Andermann E., Labuda M., Richter A., Pandolfo M., Cavalcanti F., Pianese L., Iodice L., Farina G., Monticelli A., et al. (1997) The Friedreich ataxia GAA triplet repeat: premutation and normal alleles. Hum. Mol. Genet., 6, 1261–1266. [DOI] [PubMed] [Google Scholar]
- 55.Al-Mahdawi S., Pinto R.M., Ruddle P., Carroll C., Webster Z., Pook M. (2004) GAA repeat instability in Friedreich ataxia YAC transgenic mice. Genomics, 84, 301–310. [DOI] [PubMed] [Google Scholar]
- 56.Brown J.M., Green J., das Neves R.P., Wallace H.A.C., Smith A.J.H., Hughes J., Gray N., Taylor S., Wood W.G., Higgs D.R., et al. (2008) Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J. Cell Biol., 182, 1083–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chartrand P., Bertrand E., Singer R.H., Long R.M. (2000) Long, sensitive and high-resolution detection of RNA in situ. Methods Enzymol., 318, 493–506. [DOI] [PubMed] [Google Scholar]
- 58.Hocine S., Raymond P., Zenklusen D., Chao J.A., Singer R.H. (2013) Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat. Methods, 10, 119–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raj A., Tyagi S. (2010) Detection of individual endogenous RNA transcripts in situ using multiple singly labeled probes. In Nils, G.M. (ed), Methods in Enzymology. Academic Press, New York, USA, Vol. 472, pp. 365–382. [DOI] [PubMed] [Google Scholar]
- 60.Ben-Ari Y., Brody Y., Kinor N., Mor A., Tsukamoto T., Spector D.L., Singer R.H., Shav-Tal Y. (2010) The life of an mRNA in space and time. J. Cell Sci., 123, 1761–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iannuccelli E., Mompart F., Gellin J., Lahbib-Mansais Y., Yerle M., Boudier T. (2010) NEMO: a tool for analyzing gene and chromosome territory distributions from 3D-FISH experiments. Bioinformatics, 26, 696–697. [DOI] [PubMed] [Google Scholar]
- 62.Bolte S., Cordelières F.P. (2006) A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc., 224, 213–232. [DOI] [PubMed] [Google Scholar]
- 63.Dundr M., Misteli T. (2003) Measuring dynamics of nuclear proteins by photobleaching. Curr. Protoc. Cell Biol., 18, 13.5.1–13.5.18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.