Abstract
Polymorphisms rs6232 and rs6234/rs6235 in PCSK1 have been associated with extreme obesity [e.g. body mass index (BMI) ≥ 40 kg/m2], but their contribution to common obesity (BMI ≥ 30 kg/m2) and BMI variation in a multi-ethnic context is unclear. To fill this gap, we collected phenotypic and genetic data in up to 331 175 individuals from diverse ethnic groups. This process involved a systematic review of the literature in PubMed, Web of Science, Embase and the NIH GWAS catalog complemented by data extraction from pre-existing GWAS or custom-arrays in consortia and single studies. We employed recently developed global meta-analytic random-effects methods to calculate summary odds ratios (OR) and 95% confidence intervals (CIs) or beta estimates and standard errors (SE) for the obesity status and BMI analyses, respectively. Significant associations were found with binary obesity status for rs6232 (OR = 1.15, 95% CI 1.06–1.24, P = 6.08 × 10−6) and rs6234/rs6235 (OR = 1.07, 95% CI 1.04–1.10, P = 3.00 × 10−7). Similarly, significant associations were found with continuous BMI for rs6232 (β = 0.03, 95% CI 0.00–0.07; P = 0.047) and rs6234/rs6235 (β = 0.02, 95% CI 0.00–0.03; P = 5.57 × 10−4). Ethnicity, age and study ascertainment significantly modulated the association of PCSK1 polymorphisms with obesity. In summary, we demonstrate evidence that common gene variation in PCSK1 contributes to BMI variation and susceptibility to common obesity in the largest known meta-analysis published to date in genetic epidemiology.
Introduction
The prevalence of obesity has reached epidemic proportions throughout the world (1). In addition to being the main risk predictor for the rapid increase of type 2 diabetes (T2D) (2), obesity also significantly increases the global disease burden of cardiovascular disease and cancer (3,4). Rising rates of childhood obesity, combined with an increasing prevalence of obesity in aging adult populations, suggest that the impact of this disease on human health will continue to grow in the future (5). Therefore, there is an urgent need to improve understanding of the etiology of obesity to help curb the obesity epidemic (6).
Genetic factors have been shown to play a substantial role in the etiology of obesity (7) and accordingly research has focused on identifying specific underlying genetic determinants of body weight regulation. Candidate-gene, gene-centric and genome-wide association (GWAS) studies have identified 42 loci with single-nucleotide polymorphisms (SNPs) that significantly associate (P < 5 × 10−8) with body mass index (BMI, as a continuous variable) (8–17). Additionally, case–control candidate gene and GWAS approaches have been used to examine the genetics of childhood and adult obesity (as a binary variable) (13,18–28). These studies have identified 46 loci with alleles associated with obesity at the genome-wide significance level. The majority of alleles (N = 24) influence both BMI variation and the risk for obesity, but 18 and 22 loci have been shown to contribute more specifically to BMI variation and the genetic risk for obesity, respectively (13,18–27). These data indicate that the genetic architecture of BMI variation and obesity may not be totally overlapping. Obesity may not only represents the extreme of the phenotypic spectrum of BMI (18), but perhaps a partially distinct inherited condition (29).
The PCSK1 gene may be illustrative of this paradigm. Mutations in PCSK1 lead to PC1/3 enzyme deficiency in neuroendocrine cells, which is characterized by monogenic obesity in mice and humans resulting from the abnormal maturation of hormones involved in energy and glucose metabolism (30–32). In a positional candidate-gene study, Benzinou et al. showed convincing evidence for the association of coding variants rs6232 and rs6234/rs6235 (pooled given perfect linkage disequilibrium between the two SNPs among diverse ethnic backgrounds) with childhood and adult severe obesity (27). The candidacy of these common variants is further strengthened in that they have been shown to reduce PC1/3 enzymatic activity through altered protein secretion, biosynthesis and catalytic activity (27,30,33,34) and determine glucose-stimulated proinsulin conversion (35,36), which is also a characteristic of complete human PCSK1 deficiency (31).
However, replication of the association of PCSK1 variants with obesity has provided conflicting results (27,37–45) with the lack of statistical power and genetic/phenotypic/ethnic heterogeneity being likely contributors to this variability. Additionally, conflicting evidence for the association of PCSK1 variants with BMI variation has been observed in individual studies (35,37–40,43,44,46,47) and only nominal evidence of association with BMI variation has been found for rs6232 and rs6235 in large GWAS meta-analyses (12,36). To give a more conclusive answer regarding whether PCSK1 variants differ in their contribution to extreme obesity (e.g. BMI ≥ 40 kg/m2), common obesity (BMI ≥ 30 kg/m2) and continuous BMI variation, we have systematically collected data from the literature and unpublished sources to perform a meta-analysis of the association of variants rs6232 and rs6234/rs6235 with quantitative BMI variation and common obesity risk in up to 331 175 subjects from diverse ethnic groups.
Results
Study selection
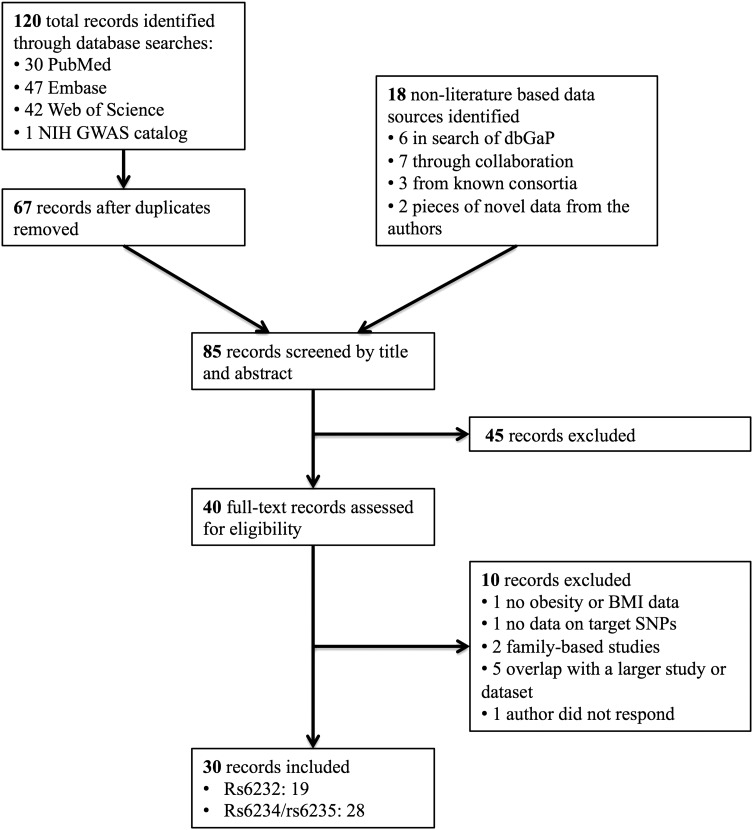
Results of the systematic search and data collection are presented in Figure 1. In total, 85 unique records were screened by title and abstract and 40 records were reviewed in full text, of which 10 were excluded. Reasons for exclusion after full text review included overlap with larger studies from the literature review, family-based studies (where clustering was not accounted for in the analysis), candidate-gene studies not examining the variants of interest, neither obesity nor BMI variation having been evaluated, lack of response from the authors and overlap with larger datasets. In total, 30 records were included from the literature, consortia, the Database of Genotypes and Phenotypes (dbGap), direct collaboration and novel contributions by the authors, as detailed in Supplementary Material, Table S1. Of these, 19 contained data on rs6232 and 28 contained data on rs6234/rs6235. Details and characteristics of all data sources included in the analysis can be found in Supplementary Material, Table S1. The minor allele frequency for each variant for rs6232, rs6234 and rs6235 are provided for the EpiDream cohort and from the 1000 Genomes Project in Supplementary Material, Tables S2 and S3, respectively.
Figure 1.
Flow diagram of literature search and study selection for meta-analysis of the association of rs6232 and rs6234/rs6235 with obesity and BMI. GWAS, genome-wide association study; dbGaP, Database of Genotypes and Phenotypes; BMI, body mass index; SNPs, single-nucleotide polymorphisms.
Study quality
Study characteristics, genotyping and analysis methods of the included studies are described in the Supplementary Material, Table S1. As many studies were not initially designed to evaluate obese cases compared with non-obese controls, population structure was variable with many studies containing few obese cases and therefore likely underpowered to evaluate genetic associations with obesity. Hardy–Weinberg equilibrium was either reported or obtained via correspondence with P > 0.05 for all studies included. Similarly, all studies included were found to have SNP-wise call rates of >95% and all study estimates were adjusted for age and/or sex as covariates.
Binary obesity analysis
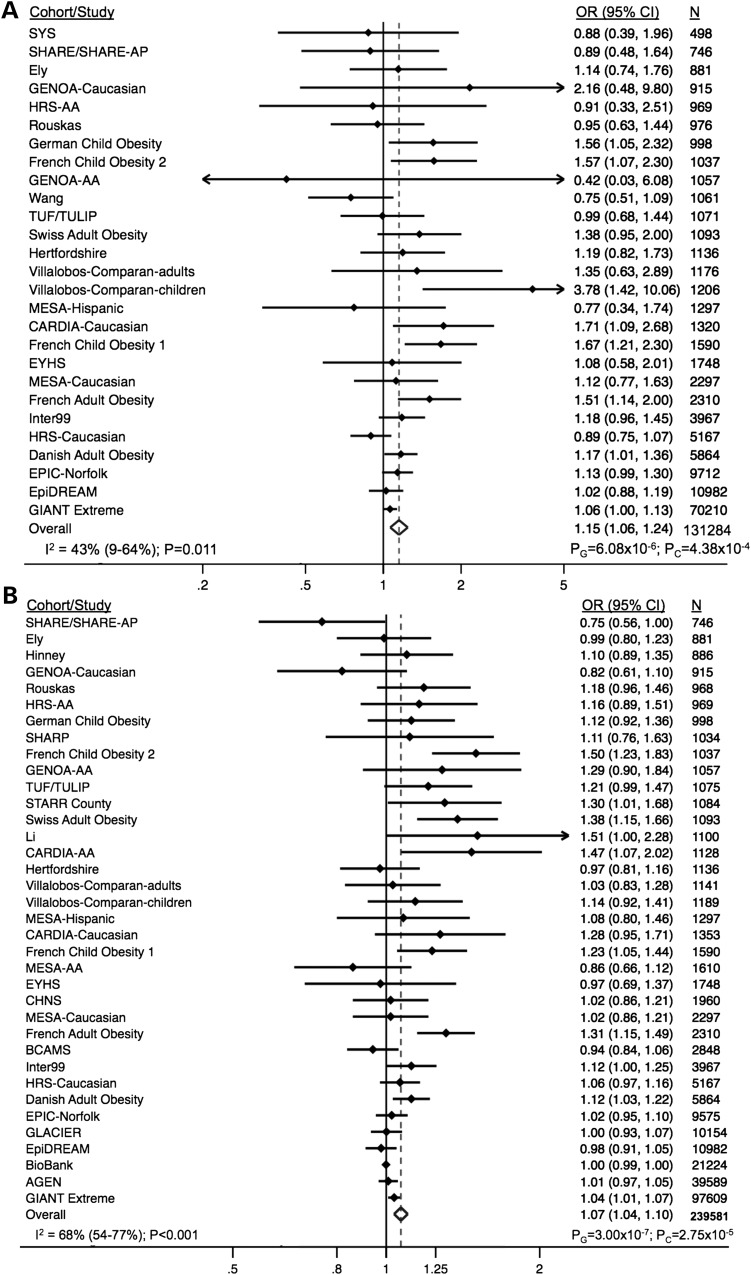
The results for the meta-analyses of rs6232 and rs6234/rs6235 with obesity are presented in Figure 2 using both classical random-effects meta-analytic techniques and a global random-effects meta-analytic method, designed to detect genetic associations among multi-cohort studies with high heterogeneity (48). The combined analysis of 131 284 individuals demonstrated a significant association of the G allele of rs6232 with obesity status [odds ratio (OR) = 1.15; 95% confidence interval (CI), 1.06–1.24] with a global method P-value (PG) = 6.08 × 10−6 and a classical random-effects method P-value (PC) = 4.38 × 10−4. Similarly, the analysis of 239 581 individuals demonstrated an OR of 1.07 (95% CI 1.04–1.10; PG = 3.00 × 10−7; PC = 2.75 × 10−5) for the association of rs6234/rs6235 with obesity status. Low-to-moderate between-study heterogeneity was observed for the association of both rs6232 (I2 = 43%; 95% CI, 9–64%, P = 0.011) and rs6234/rs6235 (I2 = 68%; 95% CI, 54–77%, P = 2.18 × 10−9) with binary obesity. Finally, exclusion of the initial PCSK1 discovery cohort (27) consisting of 1045 obese French adults and 1265 non-obese controls did not significantly impact the significance of our analysis for rs6232 (OR = 1.13; 95% CI 1.05–1.22; PG = 8.31 × 10−5; PC = 0.002) or rs6234/rs6235 (OR = 1.06; 95% CI 1.03–1.09; PG = 4.45 × 10−5; PC = 2.05 × 10−4).
Figure 2.
Association of rs6232 (A) and rs6234/rs6235 (B) with obesity status. OR, odds ratio; CI, confidence interval.
Continuous BMI variation analysis
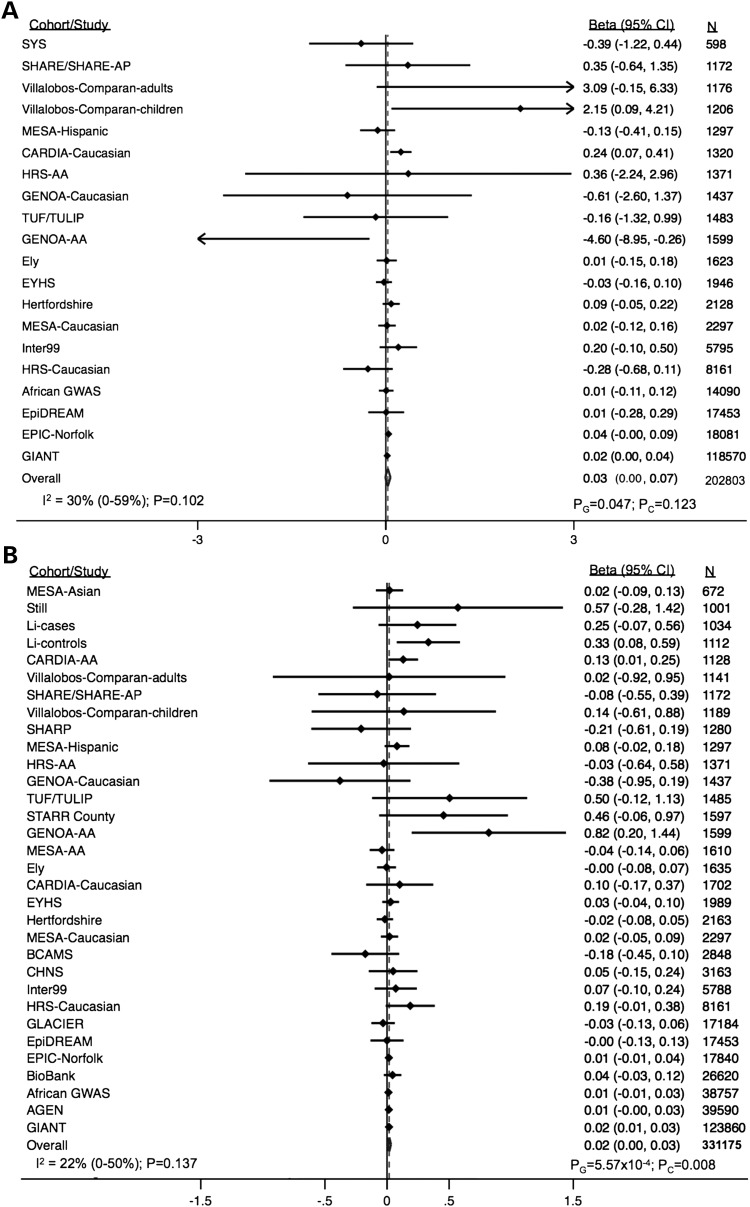
The results for the meta-analysis of rs6232 and rs6234/rs6235 with BMI variation are presented in Figure 3. In the analysis of 202 803 individuals, the average BMI increase for each G allele at rs6232 was 0.03 (95% CI 0.00–0.07; PG = 0.047; PC = 0.123). In the analysis of rs6234/rs6235, each effect-allele conferred a 0.02 (95% CI 0.00–0.03; PG = 5.57 × 10−4; PC = 0.008) unit increase in BMI among 331 175 individuals. The between-study heterogeneity was non-significant for the associations of rs6232 (I2 = 30%; 95% CI, 0–59%, P = 0.102) and rs6234/rs6235 (I2 = 22%; 95% CI, 0–50%, P = 0.137).
Figure 3.
Association of rs6232 (A) and rs6234/rs6235 (B) with BMI. OR, odds ratio; CI, confidence interval.
Heterogeneity and subgroup analysis
As low-to-moderate between-study heterogeneity was observed for the association of both rs6232 and rs6234/rs6235 with binary obesity, causes of heterogeneity were explored through pre-specified subgroup analyses (Table 1). Stratification by ethnicity, cohort size (≤1000 or >1000) and study ascertainment did not have any significant impact on the association of rs6232 with obesity. Stratification by cohort age-group (child/adolescent versus adult) resulted in a significantly different association between rs6232 and obesity in children/adolescents (OR = 1.53, 95% CI 1.22–1.93; PG = 3.68 × 10−5; PC = 2.84 × 10−4) and in adults (OR = 1.10, 95% CI 1.03–1.17; PG = 0.001; PC = 0.006; Pdifference = 3.00 × 10−4). Between-study heterogeneity still remained significant for the association between rs6232 and obesity, after stratifying by ethnicity, cohort size or study ascertainment (Table 1). However, no more between-study heterogeneity was observed when children/adolescent and adult subgroups were analyzed apart (P = 0.250 and 0.182, respectively).
Table 1.
Subgroup analysis for the association of rs6232 and rs6234/rs6235 with obesity
| rs6232 | Random-effects OR (95% CI) |
P-value |
Heterogeneity |
χ2-test for difference | No. of studies/cohorts | Sample size | ||
|---|---|---|---|---|---|---|---|---|
| Classical | Global | I2 (95% CI) | P-value | |||||
| Ethnicity | ||||||||
| White Caucasian | 1.14 (1.06–1.24) | 0.001 | 1.08 × 10−5 | 44 (7–66) | 0.015 | 0.485 | 22 | 125 579 |
| East Asian | 0 | 0 | ||||||
| Hispanic | 1.52 (0.65–3.55) | 0.335 | 0.182 | 67 (0–90) | 0.049 | 3 | 3679 | |
| African | 0.83 (0.32–2.13) | 0.694 | 0.810 | a | 0.597 | 2 | 2026 | |
| Cohort age-group | ||||||||
| Child/adolescent | 1.53 (1.22–1.93) | 2.84 × 10−4 | 3.68 × 10−5 | 25 (0–68) | 0.250 | 3.00 × 10−4 | 6 | 7077 |
| Adult | 1.10 (1.03–1.17) | 0.006 | 0.001 | 22 (0–54) | 0.182 | 21 | 124 207 | |
| Cohort size | ||||||||
| ≤1000 | 1.14 (0.92–1.40) | 0.223 | 0.351 | 0 (0–71) | 0.536 | 0.756 | 7 | 5983 |
| >1000 | 1.16 (1.06–1.26) | 0.003 | 1.17 × 10−5 | 53 (21–72) | 0.003 | 20 | 125 301 | |
| Population-based recruitment | ||||||||
| No | 1.19 (1.02–1.38) | 0.030 | 7.15 × 10−4 | 53 (13–75) | 0.012 | 0.222 | 13 | 29 799 |
| Yes | 1.12 (1.03–1.22) | 0.010 | 0.004 | 28 (0–62) | 0.157 | 14 | 101 485 | |
| rs6234/rs6235 | ||||||||
| Ethnicity | ||||||||
| White Caucasian | 1.09 (1.04–1.14) | 2.04 × 10−4 | 4.28 × 10−9 | 64 (44–77) | 1.25 × 10−5 | 5.13 × 10−7 | 23 | 163 385 |
| East Asian | 1.00 (0.98–1.03) | 0.961 | 1.00 | 22 (0–67) | 0.274 | 5 | 66 721 | |
| Hispanic | 1.13 (0.99–1.27) | 0.050 | 0.103 | 0 (0–85) | 0.579 | 4 | 4711 | |
| African | 1.16 (0.92–1.46) | 0.222 | 0.115 | 59 (0–86) | 0.064 | 4 | 4764 | |
| Cohort age-group | ||||||||
| Child/adolescent | 1.13 (1.00–1.29) | 0.053 | 5.61 × 10−4 | 68 (29–86) | 0.004 | 0.005 | 7 | 10 296 |
| Adult | 1.06 (1.02–1.09) | 0.001 | 6.35 × 10−5 | 66 (49–77) | 4.19 × 10−7 | 29 | 229 285 | |
| Cohort size | ||||||||
| ≤1000 | 1.02 (0.91–1.15) | 0.69 | 0.576 | 44 (0–77) | 0.096 | 0.439 | 7 | 6363 |
| >1000 | 1.07 (1.04–1.11) | 2.56 × 10−5 | 3.48 × 10−7 | 71 (58–80) | 1.71 × 10−9 | 29 | 23 3218 | |
| Population-based recruitment | ||||||||
| No | 1.11 (1.05–1.17) | 1.60 × 10−4 | 3.65 × 10−6 | 76 (61–85) | 1.35 × 10−7 | 0.001 | 16 | 92 067 |
| Yes | 1.05 (1.00–1.09) | 0.040 | 0.005 | 46 (8–68) | 0.014 | 20 | 14 7514 | |
OR, odds ratio; CI, confidence interval.
aAnalysis not possible if <2 degrees of freedom.
Stratification by cohort size also did not have a significant impact on the association of rs6234/rs6235 with obesity. However, stratification by ethnicity, cohort age-group (child/adolescent versus adult) or study ascertainment resulted in a significantly different association between rs6234/rs6235 and obesity (Table 1). The OR estimate of rs6234/rs6235 for obesity was comparable in white Caucasian (OR = 1.09, 95% CI 1.04–1.14; PG = 4.28 × 10−9; PC = 2.04 × 10−4), Hispanic (OR = 1.13, 95% CI 0.99–1.27; PG = 0.103; PC = 0.050) and African (OR = 1.16, 95% CI 0.92–1.46; PG = 0.115; PC = 0.222) ethnic groups, but the rs6234/rs6235 variant conferred no evidence for an increase in the odds for obesity in East Asian populations (OR = 1.00, 95% CI 0.98–1.03; PG = 1.00; PC = 0.961; Pdifference = 5.13 × 10−7), which included Chinese, Japanese, Korean and Malay individuals. A significantly different association between rs6234/rs6235 and obesity was observed in children/adolescents (OR = 1.13, 95% CI 1.00–1.29; PG = 5.61 × 10−4; PC = 0.053) and in adults (OR = 1.06, 95% CI 1.02–1.09; PG = 6.35 × 10−5; PC = 0.001; Pdifference = 0.005). The association of rs6234/rs6235 with obesity was significantly different depending on the type of recruitment [population-based or other recruitment (e.g. hospital): OR = 1.05, 95% CI 1.00–1.09; PG = 0.005; PC = 0.040 and OR = 1.11, 95% CI 1.05–1.17; PG = 3.65 × 10−6; PC = 1.60 × 10−4, respectively; Pdifference = 0.001]. Between-study heterogeneity still remained significant for the association between rs6234/rs6235 and obesity, after stratifying by ethnicity, cohort age-group, cohort size or study ascertainment (Table 1).
There was no evidence of small study effects in the analysis of rs6232 for obesity (Supplementary Material, Figure S1). However, a funnel plot for rs6234/rs6235 demonstrated asymmetry for the obesity analysis (Supplementary Material, Figure S2), which was supported by the Egger's (P < 0.05), but not the Begg's (P > 0.05) tests. Additionally, the association of rs6234/rs6235 with obesity status did not significantly differ in studies with greater than 1000 individuals compared with those with 1000 participants or less (P = 0.439).
Finally, published data from the Genetic Investigation of Anthropometric Traits (GIANT) consortium for the association of rs6232 and rs6235 with different classes of obesity are presented in Table 2. In all analyses, the control group consisted of normal weight individuals (BMI < 25 kg/m2). For the rs6232 variant, there was an OR of 1.04 (95% CI 1.00–1.08), 1.06 (95% CI 1.00–1.13), 1.08 (95% CI 0.98–1.20) and 1.13 (95% CI 0.95–1.34), across the overweight and obese classes I, II and III groups, respectively (0.057 ≤ PC ≤ 0.18). Similarly, the rs6235 analysis showed a similar increasing risk pattern with an OR of 1.03 (95% CI 1.01–1.05), 1.04 (95% CI 1.01–1.07), 1.07 (95% CI 1.02–1.11) and 1.11 (95% CI 1.02–1.20), across the overweight and obese classes I, II and III groups, respectively (0.003 ≤ PC ≤ 0.011). However, the ORs of rs6232 and rs6235 SNPs for different classes of obesity were not significantly different (P > 0.05).
Table 2.
Association of rs6232 and rs6235 with obesity class in GIANT
| OR (95% CI) | P-value | Cases | Controls | |
|---|---|---|---|---|
| rs6232 | ||||
| Overweight | 1.04 (1.00–1.08) | 0.078 | 78 671 | 60 578 |
| Obesity class I | 1.06 (1.00–1.13) | 0.057 | 22 947 | 47 263 |
| Obesity class II | 1.08 (0.98–1.20) | 0.120 | 5983 | 35 721 |
| Obesity class III | 1.13 (0.95–1.34) | 0.180 | 1534 | 23 221 |
| rs6235 | ||||
| Overweight | 1.03 (1.01–1.05) | 0.005 | 92 808 | 65 660 |
| Obesity class I | 1.04 (1.01–1.07) | 0.010 | 32 746 | 64 864 |
| Obesity class II | 1.07 (1.02–1.11) | 0.003 | 9723 | 61 085 |
| Obesity class III | 1.11 (1.02–1.20) | 0.011 | 2550 | 35 900 |
OR, odds ratio; CI, confidence interval.
Discussion
In this study, we provide evidence that common variants in PCSK1 contribute to BMI variation and common obesity. The proprotein convertase 1 encoded by the PCSK1 gene belongs to the subtilisin-like proprotein convertase family. Proprotein convertase 1 is known to cleave key peptides in the regulation of energy balance such as proinsulin or proopiomelanocortin (49). Rare loss-of-function coding mutations in PCSK1 have been associated with a Mendelian form of hyperphagic obesity in mice and humans (30,32,50). Benzinou and colleagues provided evidence of association between the frequent coding variants N221D (rs6232) and Q665E/S690T (rs6234/rs6235) and childhood and adult severe obesity in European populations (27). However, conflicting results have been reported regarding the association of the rs6232 and rs6234/rs6235 polymorphisms with common obesity (27,37–45) or BMI variation (12,35–40,43,44,46,47) in diverse ethnic backgrounds. This prompted us to assess the contribution of rs6232 and rs6234/rs6235 with common obesity and BMI variation using a meta-analytic approach. We enhanced the power of our analyses, and therefore the ability to detect associations, using both a classic meta-analytic approach in addition to data extraction from pre-existing GWAS or custom-arrays from consortia and single studies as well as internal data and collaboration. In total, we collected phenotypic and genetic data in up to 331 175 individuals from diverse ethnic groups, which represents to our knowledge the largest meta-analysis published to date in the field of genetic epidemiology.
Overall, our data demonstrate that the common functional variants rs6232 and rs6234/rs6235 in PCSK1 not only predispose to severe obesity but also modestly increase the risk of common obesity and increased BMI within populations. Our results are in line with the conclusions of a large-scale GWAS meta-analysis for BMI and different clinical classes of obesity performed in up to 263 407 subjects (20). In this study, Berndt et al. found a large overlap of SNPs contributing to both quantitative BMI and different thresholds of obesity and concluded that there was little etiological heterogeneity between these traits at least at the level of common SNP variation (20). Our data are only partially concordant with a recent meta-analysis published by Stijnen and colleagues (51). However, the 65% lower sample size in this study in comparison with ours (N = 200 000 versus 331 175), added to methodological considerations lead us to interpret the conclusions of this study with caution (52).
Children with congenital proprotein convertase one-third deficiency display early growth abnormalities and reduced height as a consequence of growth hormone deficiency (53). Therefore, we cannot totally exclude that the association of rs6232 and rs6234/rs6235 SNPs with BMI may be in part confounded by an additional genetic effect on height variation. We consulted the publically available data released in 2014 by the GIANT consortium (54) and did not find an association between the rs6232 SNP and height (B = −0.0086 ± 0.0068, P = 0.21, N = 233 697). On the contrary, and in line with the observations made in monogenic patients, the rs6235 SNP BMI-increasing allele evidenced a strong association with decreased height (B = −0.0224 ± 0.0033, P = 5.4 × 10−11, N = 251 342).
If the genetic architecture of BMI variation in general populations and the risk of common obesity includes many overlapping genetic variants, these variants may be more prevalent in severe and/or familial forms of childhood and adult obesity. Benzinou et al. indeed reported odds ratios (OR) for obesity of 1.34 (95% CI 1.20–1.49; P = 7.27 × 10−8) and 1.22 (95% CI 1.15–1.29; P = 2.31 × 10−12) for rs6232 and rs6234/rs6235 in a European sample enriched in familial forms of childhood and adult extreme obesity in European populations (27). Lower OR for common obesity were observed for rs6232 (OR = 1.15) and rs6234/rs6235 (OR = 1.07) in our meta-analysis. Importantly, the larger ORs for obesity observed in the Benzinou et al. original study are less likely to result from initial overestimation of the true effect (i.e. winner's curse), as they were estimated from a meta-analysis of seven independent cohorts (27). Additionally, exclusion of the initial PCSK1 discovery cohort in sensitivity analyses did not impact the significance of our results. A more plausible explanation is that extreme familial forms of early-onset and adult obesity are enriched for susceptibility variants. Consistent with this hypothesis, a non-significant progressive enrichment of rs6232 and rs6234/rs6235 effect variants was observed for increasing degrees of obesity in the GIANT sample (Supplementary Material, Table S3). Our results indicate that an enrichment sampling strategy (selection of obese individuals having a strong family background of the disease, an early age of onset and/or a more severe phenotype) is a cost-effective and efficient approach to identify loci that also contribute to BMI variation and risk for common obesity in general populations (20,55).
Our data show a substantial degree of between-study heterogeneity in the association of PCSK1 SNPs rs6232 and rs6234/rs6235 with obesity. Benzinou et al. similarly reported significant between-study heterogeneity while analyzing the association of the SNP rs6235 with severe obesity in seven independent cohorts (27). We investigated the potential causes of this heterogeneity and made several observations. Ethnicity significantly modulated the association between rs6234/rs6235 and obesity. Whereas similar effect sizes for the association of rs6234/rs6235 with obesity were found in white Caucasian, African and Hispanic ethnic groups, no evidence for association was found in East Asian populations. As the rs6234/rs6235 coding non-synonymous SNPs exhibit functional effects on the proprotein convertase 1 activity (34), and therefore do not represent proxy SNPs, the absence of evidence for an association restricted to only East Asian populations is very unlikely to be explained by differential linkage disequilibrium structure at the PCSK1 locus in certain ethnic groups. Notably, another SNP in PCSK1, independent of the rs6234/rs6235 signal, has been recently identified as an important contributor to BMI variation in East Asian populations (15). Therefore, the lack of association of rs6234/rs6235 with obesity in East Asians in the current study is unlikely to be explained by an interaction between the PCSK1 locus and ethnic-specific lifestyle factors that may inhibit the genetic effect on obesity at this specific locus. Ethnic-specific epistasis effects may account for this intriguing pattern of association. In contrast to rs6234/rs6235, ethnicity did not modulate the association between rs6232 and obesity. However, this may have been secondary to decreased power and no available data examining the association of rs6232 and obesity or BMI in East Asian populations.
Study ascertainment in the overall meta-analysis significantly modulated the association of rs6234/rs6235 with obesity. Not surprisingly, obese cases recruited in hospitals, which tended to be more obese, or obtained from pedigrees enriched in obese cases displayed 2-fold higher ORs for obesity in comparison with cases issued from the general population. Additionally, there is a suggestion of possible small study effects for the rs6234/rs6235, due to a lower than predicted representation of smaller negative studies in our analysis, which may also explain a degree of the observed heterogeneity.
Cohort age-group significantly modulated the association between rs6232, rs6234/rs6235 and obesity with the effect sizes for both SNPs being stronger in children/adolescents than in adults (Table 1). Similar trends have been observed for the association of rs6232 with BMI and obesity in younger versus older adult Europeans in two independent reports (38,44). As prohormone convertase 1 cleaves proopiomelanocortin, a key peptide in the regulation of energy balance and appetite, it is tempting to speculate that the genetic effect of common functional variants in PCSK1 may be more pronounced in the context of the more recent ‘obesogenic’ environment with unlimited access to high-caloric food (56).
Additional environmental or biological factors (e.g. physical activity) are likely to modulate the association between rs6234/rs6235 and obesity in the current study. Between-study heterogeneity indeed remained significant after stratifying the genetic association test by ethnicity, cohort age-group or population recruitment. Our data, in line with previous reports, confirm that heterogeneity may be a common feature of genetic association studies involving obesity predisposing variants (57–59). In that context, the global meta-analytic random-effects method recently developed by Lebrec et al. is especially relevant as it is designed to detect genetic associations among multi-cohort studies that convey a high level of between-study heterogeneity (48). Neupane and colleagues recently demonstrated that the global method achieves higher power and lower rates of false positives compared with classic methods in the presence of high between-study heterogeneity, using both simulated and real datasets (60). We observed the same trends in our study with the global method providing greater precision of estimates than the classic random-effects inverse variance weighted method in the setting of high between-study heterogeneity. We therefore support the wider use of global meta-analytic random-effects model for the genetic dissection of complex traits.
The strengths of this meta-analysis include a comprehensive data collection strategy, an exceptionally large sample size representative of high ethnic diversity, the use of the most up-to-date meta-analytic methods and the selection of functionally relevant coding polymorphisms. Limitations of this study include the modest statistical power of subgroup analyses and the limited access to a broad range of environmental exposure information that may modulate the association of PCSK1 SNPs with obesity.
In summary, we provide evidence that SNPs rs6232 and rs6234/rs6235 in PCSK1 contribute to BMI variation as well as increased susceptibility to common obesity. Our data confirm the power of gene identification strategies based on extreme forms of obesity to identify loci that also contribute to BMI variation and risk for common obesity in general populations.
Methods
The PRISMA statement guidelines were utilized for this systematic review and meta-analyses (61).
Eligibility criteria
Individual studies, meta-analyses and GWAS consortia examining obesity status or BMI variation with respect to exposure to the previously defined effect alleles of rs6232 (G), rs6234 (G) or rs6235 (C) were eligible for inclusion (27,41). As rs6234 and rs6235 are in perfect linkage disequilibrium among diverse ethnic backgrounds (r2 = 1.0), they are considered as identical and are analyzed together (62,63). Additionally, studies in East Asian, South Asian and white Caucasian populations reporting on the SNP rs7713317 (effect allele: G) were eligible, as this variant is in perfect linkage disequilibrium (r2 = 1.0) with rs6234 and rs6235 among these groups in both the HapMap (62) and 1000 Genomes projects (63). Only analyses of variants under the additive model were eligible for inclusion as this has been demonstrated to be the most likely inheritance model (27). In addition, studies that recorded BMI or obesity status and used genotyping platforms known to contain these variants were also eligible and corresponding authors were contacted to share the unpublished data in a collaborative manner. Studies examining obesity status were eligible if they compared an obese group with a non-obese group as defined according to the study population. Our exclusion criteria were clustered datasets such as family-based studies (unless clustering was accounted for in the analysis), data from the analysis of variants not shown to be in Hardy–Weinberg equilibrium of P ≥ 0.05 and data with an imputation quality of <0.9.
Information sources
The search strategy was designed to identify all sources of published and unpublished data both in the literature and in available databases. Electronic searches without language or date restriction were carried out in PubMed (1966–present), Web of Science (1899–present), Embase (1974–present) and the NIH GWAS catalog (64). Search terms used in all databases to identify relevant studies, along with a representative search strategy used for the PubMed query, can be found in Supplementary Material, Table S4. All articles identified through the search were evaluated based on the title and abstract. Clearly, irrelevant studies were excluded from further consideration. The remaining articles received a full text review. The last search was undertaken on 9 June 2013.
Studies recording data on study participant BMI and/or obesity status and utilizing genotyping arrays known to include rs6232, rs6234, rs6235 or rs7713317 were searched for in dbGaP (65). Relevant genotyping arrays and proxy SNPs were identified using the SNP Annotation and Proxy Search (66). Additional data were obtained through collaboration with other investigators and through consortia (Supplementary Material, Table S1).
Descriptive information was extracted from each study including: (i) SNPs available, (ii) study design, (iii) participant selection, (iv) eligible phenotype (obesity or BMI), (v) genotyping method, (vi) sample size, (vii) number of obese and non-obese individuals (for obesity analyses), (viii) obese and non-obese definitions, (ix) ethnicity, (x) included age groups and (xi) model adjustments. As data were gathered from individual studies, meta-analyses and GWAS consortia, particular care was taken to avoid the inclusion of duplicate data. Detailed cohort information was requested from participating consortia and key study characteristics were compared across all eligible studies to determine if multiple publications with data from the same study were present. If study duplication existed, data from the publication with the most complete analysis of the duplicated study was used. Where possible, corresponding authors were contacted to provide study data that did not overlap with other data sources. The literature search and article review were carried out independently by two reviewers (K.N. and D.M.) with consensus reached by discussion. Details of study selection can be found in Figure 1.
Unpublished data from the EpiDREAM (Epidemiological arm of the Diabetes Reduction Assessment with Ramipril and Rosiglitazone Medication) (67), SHARE (Study of Health Assessment and Risk in Ethnic groups) (68) and SHARE-AP (Study of Health Assessment and Risk Evaluation in Aboriginal Peoples) (69) cohorts were also included. Briefly, the EpiDREAM multi-ethnic longitudinal cohort is the epidemiological arm of the DREAM study comprised of individuals with an increased risk for T2D who were screened for trial eligibility. The SHARE and SHARE-AP studies are cross-sectional cohorts investigating atherosclerosis and cardiovascular diseases among different ethnic groups living in Canada. Genetic and clinical data were available from 1172 participants from the SHARE and SHARE-AP population. In the EpiDREAM study, 17 453 subjects from six ethnic groups (South Asian, East Asian, European, African, Latin American and Native North American) and having both genetic and baseline clinical information have been included here. Self-reported ethnicity has been validated in the SHARE, SHARE-AP and EpiDREAM studies using the eigensoft software (http://genepath.med.harvard.edu/~reich/Software.htm). The SHARE, SHARE-AP and EpiDREAM cohorts were genotyped using the cardiovascular gene-centric 50 K SNP ITMAT-Broad-CARe (IBC) array (70). Subjects in the SHARE and SHARE-AP cohorts were genotyped using the Illumina HumanCVD BeadChip. SNPs rs6232, rs6234 and rs6235 in PCSK1 were part of the 50K array SNP list and were extracted for further study.
Statistical analysis
The primary analyses examined the effect of exposure to the effect alleles of rs6232 (G) or rs6234/rs6235/ rs7713317 (G/C/C) on obesity status and BMI variation. The OR and 95% CI were collected to examine the effects of the variants on obesity status. The beta estimates and SE were collected to examine the effects of the variants on BMI. If eligible studies did not report data necessary for inclusion in the meta-analysis, the corresponding authors were contacted directly.
We implemented a global random-effects meta-analytic method, designed to detect genetic associations among multi-cohort studies with high heterogeneity (48), to calculate a summary OR and 95% CI or summary beta estimate and SE for the obesity status and BMI variation analyses, respectively. Compared with classical random-effects meta-analytic techniques, which consider each study to be a random sample of the true effect distribution and calculates the combined effect estimate as the mean of this distribution, the global random-effects method tests if the overall association or between-cohort variance of associations is non-zero. This approach was implemented because it has demonstrated improved power and lower rates of false positives compared with classical methods in the setting of heterogeneity (60). The meta-analyses were additionally performed using classical random-effects inverse variance weighted methods.
Between-study heterogeneity was evaluated using Cochran's Q-statistic and the proportion of heterogeneity due to study variation was quantified using the I2 statistic (71). If substantial heterogeneity was detected (P < 0.1), sources of this heterogeneity were explored. The presence of small study effects was evaluated using funnel plots, by calculating Begg and Egger statistics and by comparing subgroup analysis of small studies (n ≤ 1000) versus large studies (n > 1000) as previously described (72). Subgroup analysis was further examined according to pre-specified categories including ethnicity (white Caucasian, East Asian and Hispanic, African), studies using population-based recruitment compared with studies using other recruitment methods (e.g. hospital based) and by age category. Within subgroup heterogeneity was examined using a chi-squared test for difference.
Additionally, using summary statistics from the GIANT consortium (20), we examined the risk of obesity conferred by the PCSK1 variants with increasing classes of obesity including overweight (BMI ≥ 25 kg/m2) and obesity classes I (BMI ≥ 30 kg/m2), II (BMI ≥ 35 kg/m2) and III (BMI ≥ 40 kg/m2) compared with a lean control group (BMI < 25 kg/m2).
All analyses were carried out using Stata version 12 (StataCorp, College Station, TX, USA) or the R software.
Supplementary Material
Funding
K.T.N. was supported by the Gates Cambridge Trust. The EpiDREAM study was funded by a grant from the Canadian Institutes of Health Research University Industry competition with partner funding from Glaxo-SmithKline and Sanofi Aventis Global, Sanofi Aventis Canada, the McGill University and Genome Quebec Innovation Centre, Heart and Stroke Foundation of Canada. The SHARE/SHARE-AP study was funded by the Canadian Institutes of Health Research, the Heart and Stroke Foundation of Ontario and Genome Canada through the Ontario Genomics Institute. D.M. holds a Canada Research Chair in Genetic Epidemiology at McMaster University. S.S.A. holds a Canada Research Chair in Ethnic Diversity and Cardiovascular Disease, and is the Michael G. DeGroote and Heart and Stroke Foundation of Ontario Chair in Population Health, McMaster University. H.G. holds the Population Health Research Institute Chair in Diabetes Research sponsored by Aventis. S.Y. holds a Heart and Stroke Foundation Chair in Cardiovascular Research, McMaster University. R.A. Hegele is a Career Investigator of the Heart and Stroke Foundation of Ontario and holds the Edith Schulich Vinet Canada Research Chair (Tier I) in Human Genetics, the Martha G. Blackburn Chair in Cardiovascular Research, and the Jacob J. Wolfe Distinguished Medical Research Chair at the University of Western Ontario. G.S.G. and C.D.S. were supported by National Institute of Health grants DK072488 and DK088231. The CAMP Genetics Ancillary Study is supported by contracts U01 HL076419, U01 HL65899, P01 HL083069 and T32 HL07427 from the National Heart, Lung, and Blood Institute, National Institutes of Health. The GLACIER Study was funded primarily by grants from the Swedish Research Council, The Swedish Heart-Lung Foundation, The Swedish Diabetes Association, and Novo Nordisk (all to P.W.F.). A.S. is supported by the Federal Ministry of Education and Research (BMBF), Germany (FKZ: 01EO1002). T.O.K. was supported by grant no. DFF-1333-00124 from The Danish Council for Independent Research.
Supplementary Material
Acknowledgements
We thank all staff of the Laboratory for Statistical Analysis, Medical Informatics and Genotyping Development at the Center for Genomic Medicine, RIKEN and the BioBank Japan Project for the support of this the study. BioBank Japan Project was supported by Ministry of Education, Culture, Sports, Science and Technology, Japan.
Conflict of Interest statement. None declared.
References
- 1.Finucane M.M., Stevens G.A., Cowan M.J., Danaei G., Lin J.K., Paciorek C.J., Singh G.M., Gutierrez H.R., Lu Y., Bahalim A.N., et al. (2011) National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9.1 million participants. Lancet, 377, 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen L., Magliano D.J., Zimmet P.Z. (2012) The worldwide epidemiology of type 2 diabetes mellitus – present and future perspectives. Nat. Rev. Endocrinol., 8, 228–236. [DOI] [PubMed] [Google Scholar]
- 3.Calle E.E., Rodriguez C., Walker-Thurmond K., Thun M.J. (2003) Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N. Engl. J. Med., 348, 1625–1638. [DOI] [PubMed] [Google Scholar]
- 4.Hubert H.B., Feinleib M., McNamara P.M., Castelli W.P. (1983) Obesity as an independent risk factor for cardiovascular disease: a 26-year follow-up of participants in the Framingham Heart Study. Circulation, 67, 968–977. [DOI] [PubMed] [Google Scholar]
- 5.James P.T., Leach R., Kalamara E., Shayeghi M. (2001) The worldwide obesity epidemic. Obes. Res., 9(Suppl. 4), 228S–233S. [DOI] [PubMed] [Google Scholar]
- 6.Wright S.M., Aronne L.J. (2011) Obesity in 2010: the future of obesity medicine: where do we go from here? Nat. Rev. Endocrinol., 7, 69–70. [DOI] [PubMed] [Google Scholar]
- 7.Elks C.E., den Hoed M., Zhao J.H., Sharp S.J., Wareham N.J., Loos R.J., Ong K.K. (2012) Variability in the heritability of body mass index: a systematic review and meta-regression. Front. Endocrinol. (Lausanne), 3, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scuteri A., Sanna S., Chen W.M., Uda M., Albai G., Strait J., Najjar S., Nagaraja R., Orru M., Usala G., et al. (2007) Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet., 3, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loos R.J., Lindgren C.M., Li S., Wheeler E., Zhao J.H., Prokopenko I., Inouye M., Freathy R.M., Attwood A.P., Beckmann J.S., et al. (2008) Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet., 40, 768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Lango Allen H., Lindgren C.M., Luan J., Magi R., et al. (2010) Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet., 42, 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorleifsson G., Walters G.B., Gudbjartsson D.F., Steinthorsdottir V., Sulem P., Helgadottir A., Styrkarsdottir U., Gretarsdottir S., Thorlacius S., Jonsdottir I., et al. (2009) Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet., 41, 18–24. [DOI] [PubMed] [Google Scholar]
- 12.Willer C.J., Speliotes E.K., Loos R.J., Li S., Lindgren C.M., Heid I.M., Berndt S.I., Elliott A.L., Jackson A.U., Lamina C., et al. (2009) Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet., 41, 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paternoster L., Evans D.M., Nohr E.A., Holst C., Gaborieau V., Brennan P., Gjesing A.P., Grarup N., Witte D.R., Jorgensen T., et al. (2011) Genome-wide population-based association study of extremely overweight young adults – the GOYA study. PLoS ONE, 6, e24303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada Y., Kubo M., Ohmiya H., Takahashi A., Kumasaka N., Hosono N., Maeda S., Wen W., Dorajoo R., Go M.J., et al. (2012) Common variants at CDKAL1 and KLF9 are associated with body mass index in east Asian populations. Nat. Genet., 44, 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen W., Cho Y.S., Zheng W., Dorajoo R., Kato N., Qi L., Chen C.H., Delahanty R.J., Okada Y., Tabara Y., et al. (2012) Meta-analysis identifies common variants associated with body mass index in east Asians. Nat. Genet., 44, 307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Y., Lanktree M.B., Taylor K.C., Hakonarson H., Lange L.A., Keating B.J. (2012) Gene-centric meta-analyses of 108 912 individuals confirm known body mass index loci and reveal three novel signals. Hum. Mol. Genet., 22, 184–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monda K.L., Chen G.K., Taylor K.C., Palmer C., Edwards T.L., Lange L.A., Ng M.C., Adeyemo A.A., Allison M.A., Bielak L.F., et al. (2013) A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat. Genet., 45, 690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotsapas C., Speliotes E.K., Hatoum I.J., Greenawalt D.M., Dobrin R., Lum P.Y., Suver C., Chudin E., Kemp D., Reitman M., et al. (2009) Common body mass index-associated variants confer risk of extreme obesity. Hum. Mol. Genet., 18, 3502–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinney A., Nguyen T.T., Scherag A., Friedel S., Bronner G., Muller T.D., Grallert H., Illig T., Wichmann H.E., Rief W., et al. (2007) Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS ONE, 2, e1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berndt S.I., Gustafsson S., Magi R., Ganna A., Wheeler E., Feitosa M.F., Justice A.E., Monda K.L., Croteau-Chonka D.C., Day F.R., et al. (2013) Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet., 45, 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyre D., Delplanque J., Chevre J.C., Lecoeur C., Lobbens S., Gallina S., Durand E., Vatin V., Degraeve F., Proenca C., et al. (2009) Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat. Genet., 41, 157–159. [DOI] [PubMed] [Google Scholar]
- 22.Scherag A., Dina C., Hinney A., Vatin V., Scherag S., Vogel C.I., Muller T.D., Grallert H., Wichmann H.E., Balkau B., et al. (2010) Two new loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and German study groups. PLoS Genet., 6, e1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bradfield J.P., Taal H.R., Timpson N.J., Scherag A., Lecoeur C., Warrington N.M., Hypponen E., Holst C., Valcarcel B., Thiering E., et al. (2012) A genome-wide association meta-analysis identifies new childhood obesity loci. Nat. Genet., 44, 526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K., Li W.D., Zhang C.K., Wang Z., Glessner J.T., Grant S.F., Zhao H., Hakonarson H., Price R.A. (2011) A genome-wide association study on obesity and obesity-related traits. PLoS ONE, 6, e18939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiao H., Arner P., Hoffstedt J., Brodin D., Dubern B., Czernichow S., van't Hooft F., Axelsson T., Pedersen O., Hansen T., et al. (2011) Genome wide association study identifies KCNMA1 contributing to human obesity. BMC Med. Genomics, 4, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ichimura A., Hirasawa A., Poulain-Godefroy O., Bonnefond A., Hara T., Yengo L., Kimura I., Leloire A., Liu N., Iida K., et al. (2012) Dysfunction of lipid sensor GPR120 leads to obesity in both mouse and human. Nature, 483, 350–354. [DOI] [PubMed] [Google Scholar]
- 27.Benzinou M., Creemers J.W., Choquet H., Lobbens S., Dina C., Durand E., Guerardel A., Boutin P., Jouret B., Heude B., et al. (2008) Common nonsynonymous variants in PCSK1 confer risk of obesity. Nat. Genet., 40, 943–945. [DOI] [PubMed] [Google Scholar]
- 28.Wheeler E., Huang N., Bochukova E.G., Keogh J.M., Lindsay S., Garg S., Henning E., Blackburn H., Loos R.J., Wareham N.J., et al. (2013) Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nat. Genet., 45, 513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Putter C., Pechlivanis S., Nothen M.M., Jockel K.H., Wichmann H.E., Scherag A. (2011) Missing heritability in the tails of quantitative traits? A simulation study on the impact of slightly altered true genetic models. Hum. Hered., 72, 173–181. [DOI] [PubMed] [Google Scholar]
- 30.Creemers J.W., Choquet H., Stijnen P., Vatin V., Pigeyre M., Beckers S., Meulemans S., Than M.E., Yengo L., Tauber M., et al. (2012) Heterozygous mutations causing partial prohormone convertase 1 deficiency contribute to human obesity. Diabetes, 61, 383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson R.S., Creemers J.W., Ohagi S., Raffin-Sanson M.L., Sanders L., Montague C.T., Hutton J.C., O'Rahilly S. (1997) Obesity and impaired prohormone processing associated with mutations in the human prohormone convertase 1 gene. Nat. Genet., 16, 303–306. [DOI] [PubMed] [Google Scholar]
- 32.Lloyd D.J., Bohan S., Gekakis N. (2006) Obesity, hyperphagia and increased metabolic efficiency in Pc1 mutant mice. Hum. Mol. Genet., 15, 1884–1893. [DOI] [PubMed] [Google Scholar]
- 33.Mbikay M., Sirois F., Nkongolo K.K., Basak A., Chretien M. (2011) Effects of rs6234/rs6235 and rs6232/rs6234/rs6235 PCSK1 single-nucleotide polymorphism clusters on proprotein convertase 1/3 biosynthesis and activity. Mol. Genet. Metab., 104, 682–687. [DOI] [PubMed] [Google Scholar]
- 34.Pickett L.A., Yourshaw M., Albornoz V., Chen Z., Solorzano-Vargas R.S., Nelson S.F., Martin M.G., Lindberg I. (2013) Functional consequences of a novel variant of PCSK1. PLoS ONE, 8, e55065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heni M., Haupt A., Schafer S.A., Ketterer C., Thamer C., Machicao F., Stefan N., Staiger H., Haring H.U., Fritsche A. (2010) Association of obesity risk SNPs in PCSK1 with insulin sensitivity and proinsulin conversion. BMC Med. Genet., 11, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strawbridge R.J., Dupuis J., Prokopenko I., Barker A., Ahlqvist E., Rybin D., Petrie J.R., Travers M.E., Bouatia-Naji N., Dimas A.S., et al. (2011) Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes, 60, 2624–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang Y.C., Chiu Y.F., Shih K.C., Lin M.W., Sheu W.H., Donlon T., Curb J.D., Jou Y.S., Chang T.J., Li H.Y., et al. (2010) Common PCSK1 haplotypes are associated with obesity in the Chinese population. Obesity (Silver Spring), 18, 1404–1409. [DOI] [PubMed] [Google Scholar]
- 38.Kilpelainen T.O., Bingham S.A., Khaw K.T., Wareham N.J., Loos R.J. (2009) Association of variants in the PCSK1 gene with obesity in the EPIC-Norfolk study. Hum. Mol. Genet., 18, 3496–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi Q., Li H., Loos R.J., Liu C., Hu F.B., Wu H., Yu Z., Lin X. (2010) Association of PCSK1 rs6234 with obesity and related traits in a Chinese Han population. PLoS ONE, 5, e10590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renstrom F., Payne F., Nordstrom A., Brito E.C., Rolandsson O., Hallmans G., Barroso I., Nordstrom P., Franks P.W. (2009) Replication and extension of genome-wide association study results for obesity in 4923 adults from northern Sweden. Hum. Mol. Genet., 18, 1489–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rouskas K., Kouvatsi A., Paletas K., Papazoglou D., Tsapas A., Lobbens S., Vatin V., Durand E., Labrune Y., Delplanque J., et al. (2012) Common variants in FTO, MC4R, TMEM18, PRL, AIF1, and PCSK1 show evidence of association with adult obesity in the Greek population. Obesity (Silver Spring), 20, 389–395. [DOI] [PubMed] [Google Scholar]
- 42.Sandholt C.H., Sparso T., Grarup N., Albrechtsen A., Almind K., Hansen L., Toft U., Jorgensen T., Hansen T., Pedersen O. (2010) Combined analyses of 20 common obesity susceptibility variants. Diabetes, 59, 1667–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi J., Long J., Gao Y.T., Lu W., Cai Q., Wen W., Zheng Y., Yu K., Xiang Y.B., Hu F.B., et al. (2010) Evaluation of genetic susceptibility loci for obesity in Chinese women. Am. J. Epidemiol., 172, 244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choquet H., Kasberger J., Hamidovic A., Jorgenson E. (2013) Contribution of common PCSK1 genetic variants to obesity in 8,359 subjects from multi-ethnic American population. PLoS ONE, 8, e57857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villalobos-Comparan M., Villamil-Ramirez H., Villarreal-Molina T., Larrieta-Carrasco E., Leon-Mimila P., Romero-Hidalgo S., Jacobo-Albavera L., Liceaga-Fuentes A.E., Campos-Perez F.J., Lopez-Contreras B.E., et al. (2012) PCSK1 rs6232 is associated with childhood and adult class III obesity in the Mexican population. PLoS ONE, 7, e39037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gjesing A.P., Vestmar M.A., Jorgensen T., Heni M., Holst J.J., Witte D.R., Hansen T., Pedersen O. (2011) The effect of PCSK1 variants on waist, waist-hip ratio and glucose metabolism is modified by sex and glucose tolerance status. PLoS ONE, 6, e23907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X.M., Ling Y., Lu D.R., Lu Z.Q., Liu Y., Chen H.Y., Gao X. (2012) The obesity-related polymorphism PCSK1 rs6235 is associated with essential hypertension in the Han Chinese population. Hypertens. Res., 35, 994–999. [DOI] [PubMed] [Google Scholar]
- 48.Lebrec J.J., Stijnen T., van Houwelingen H.C. (2010) Dealing with heterogeneity between cohorts in genomewide SNP association studies. Stat. Appl. Genet. Mol. Biol., 9, Article 8. [DOI] [PubMed] [Google Scholar]
- 49.Choquet H., Stijnen P., Creemers J.W. (2011) Genetic and functional characterization of PCSK1. Methods Mol. Biol., 768, 247–253. [DOI] [PubMed] [Google Scholar]
- 50.Farooqi I.S., Volders K., Stanhope R., Heuschkel R., White A., Lank E., Keogh J., O'Rahilly S., Creemers J.W. (2007) Hyperphagia and early-onset obesity due to a novel homozygous missense mutation in prohormone convertase 1/3. J. Clin. Endocrinol. Metab., 92, 3369–3373. [DOI] [PubMed] [Google Scholar]
- 51.Stijnen P., Tuand K., Varga T.V., Franks P.W., Aertgeerts B., Creemers J.W. (2014) The Association of Common Variants in PCSK1 With Obesity: a HuGE Review and Meta-Analysis. Am. J. Epidemiol., 180, 1051–1065. [DOI] [PubMed] [Google Scholar]
- 52.Meyre D. (2015) Re “The association of common variants in PCSK1 with obesity: a huge review and meta-analysis”. Am. J. Epidemiol., in press. [DOI] [PubMed] [Google Scholar]
- 53.Martin M.G., Lindberg I., Solorzano-Vargas R.S., Wang J., Avitzur Y., Bandsma R., Sokollik C., Lawrence S., Pickett L.A., Chen Z., et al. (2013) Congenital proprotein convertase 1/3 deficiency causes malabsorptive diarrhea and other endocrinopathies in a pediatric cohort. Gastroenterology, 145, 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z., et al. (2014) Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet., 46, 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li A., Meyre D. (2013) Challenges in reproducibility of genetic association studies: lessons learned from the obesity field. Int. J. Obes. (Lond), 37, 559–567. [DOI] [PubMed] [Google Scholar]
- 56.Ozen S., Aldemir O. (2012) Early-onset severe obesity with ACTH deficiency and red hair in a boy: the POMC deficiency. Genet. Couns., 23, 493–495. [PubMed] [Google Scholar]
- 57.Kurokawa N., Young E.H., Oka Y., Satoh H., Wareham N.J., Sandhu M.S., Loos R.J. (2008) The ADRB3 Trp64Arg variant and BMI: a meta-analysis of 44 833 individuals. Int. J. Obes. (Lond), 32, 1240–1249. [DOI] [PubMed] [Google Scholar]
- 58.Heid I.M., Huth C., Loos R.J., Kronenberg F., Adamkova V., Anand S.S., Ardlie K., Biebermann H., Bjerregaard P., Boeing H., et al. (2009) Meta-analysis of the INSIG2 association with obesity including 74,345 individuals: does heterogeneity of estimates relate to study design? PLoS Genet., 5, e1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng S., Zhu Y., Xu F., Ren X., Li X., Lai M. (2011) FTO gene polymorphisms and obesity risk: a meta-analysis. BMC Med., 9, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neupane B., Loeb M., Anand S.S., Beyene J. (2012) Meta-analysis of genetic association studies under heterogeneity. Eur. J. Hum. Genet., 20, 1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liberati A., Altman D.G., Tetzlaff J., Mulrow C., Gotzsche P.C., Ioannidis J.P., Clarke M., Devereaux P.J., Kleijnen J., Moher D. (2009) The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ, 339, b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.The International HapMap Consortium. (2005) A haplotype map of the human genome. Nature, 437, 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.1000 Genomes Project Consortium, Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A. (2010) A map of human genome variation from population-scale sequencing. Nature, 467, 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA, 106, 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mailman M.D., Feolo M., Jin Y., Kimura M., Tryka K., Bagoutdinov R., Hao L., Kiang A., Paschall J., Phan L., et al. (2007) The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet., 39, 1181–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson A.D., Handsaker R.E., Pulit S.L., Nizzari M.M., O'Donnell C.J., de Bakker P.I. (2008) SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics, 24, 2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Koning L., Gerstein H.C., Bosch J., Diaz R., Mohan V., Dagenais G., Yusuf S., Anand S.S. (2010) Anthropometric measures and glucose levels in a large multi-ethnic cohort of individuals at risk of developing type 2 diabetes. Diabetologia, 53, 1322–1330. [DOI] [PubMed] [Google Scholar]
- 68.Anand S.S., Yusuf S., Vuksan V., Devanesen S., Teo K.K., Montague P.A., Kelemen L., Yi C., Lonn E., Gerstein H., et al. (2000) Differences in risk factors, atherosclerosis, and cardiovascular disease between ethnic groups in Canada: the Study of Health Assessment and Risk in Ethnic groups (SHARE). Lancet, 356, 279–284. [DOI] [PubMed] [Google Scholar]
- 69.Anand S.S., Yusuf S., Jacobs R., Davis A.D., Yi Q., Gerstein H., Montague P.A., Lonn E. (2001) Risk factors, atherosclerosis, and cardiovascular disease among Aboriginal people in Canada: the Study of Health Assessment and Risk Evaluation in Aboriginal Peoples (SHARE-AP). Lancet, 358, 1147–1153. [DOI] [PubMed] [Google Scholar]
- 70.Keating B.J., Tischfield S., Murray S.S., Bhangale T., Price T.S., Glessner J.T., Galver L., Barrett J.C., Grant S.F., Farlow D.N., et al. (2008) Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE, 3, e3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. (2003) Measuring inconsistency in meta-analyses. BMJ, 327, 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ioannidis J.P., Trikalinos T.A., Ntzani E.E., Contopoulos-Ioannidis D.G. (2003) Genetic associations in large versus small studies: an empirical assessment. Lancet, 361, 567–571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.