The cytidine analog zebularine triggers DNA damage at replication forks, which is signaled by ATR and ATM and detoxified by synthesis-dependent strand-annealing homologous recombination.
Abstract
DNA damage repair is an essential cellular mechanism that maintains genome stability. Here, we show that the nonmethylable cytidine analog zebularine induces a DNA damage response in Arabidopsis thaliana, independent of changes in DNA methylation. In contrast to genotoxic agents that induce damage in a cell cycle stage-independent manner, zebularine induces damage specifically during strand synthesis in DNA replication. The signaling of this damage is mediated by additive activity of ATAXIA TELANGIECTASIA MUTATED AND RAD3-RELATED and ATAXIA TELANGIECTASIA MUTATED kinases, which cause postreplicative cell cycle arrest and increased endoreplication. The repair requires a functional STRUCTURAL MAINTENANCE OF CHROMOSOMES5 (SMC5)-SMC6 complex and is accomplished predominantly by synthesis-dependent strand-annealing homologous recombination. Here, we provide insight into the response mechanism for coping with the genotoxic effects of zebularine and identify several components of the zebularine-induced DNA damage repair pathway.
INTRODUCTION
Genome stability is frequently challenged by internal and external damaging factors, leading to formation of aberrant bonds, breakage, or cleavage of DNA (Britt, 1996). Genome damage is opposed by diverse surveillance mechanisms, with the DNA damage repair machinery playing the central role (Kolodner et al., 2002). Depending on the type of DNA damage, the plant induces different repair pathways, with evolutionarily conserved kinases activating specific repair processes. ATAXIA TELANGIECTASIA MUTATED (ATM) signals the existence of DNA double-strand breaks, and ATAXIA TELANGIECTASIA MUTATED AND RAD3-RELATED (ATR) signals the presence of single-stranded DNA, mostly at stalled replication forks (Cimprich and Cortez, 2008). This induces a cascade of responses affecting cell cycle progression (De Schutter et al., 2007) and activates the corresponding DNA damage repair effectors (Garcia et al., 2003; Culligan et al., 2006).
Recent studies have demonstrated the connections between DNA damage repair, genome integrity, and chromatin control (Downey and Durocher, 2006). Functional chromatin is important for genome stability, as loss of DNA methylation or defective nucleosome assembly increases sensitivity to genotoxic stress and alters homologous recombination (HR) frequencies in Arabidopsis thaliana (Kirik et al., 2006; Melamed-Bessudo and Levy, 2012; Rosa et al., 2013). However, higher frequency of somatic HR can be induced by zebularine, the nonmethylable cytidine analog used for interference with transcriptional gene silencing (TGS) of various genetic elements (Zhou et al., 2002; Egger et al., 2004; Baubec et al., 2009, 2014; Pecinka et al., 2009). In addition, zebularine and 5-azacytidine (a less stable cytidine analog) treatments affect plant growth more severely than mutations in the genes responsible for DNA methylation, e.g., the SWI2/SNF2 class chromatin remodeling factor DECREASED DNA METHYLATION1 (DDM1) (Baubec et al., 2009). This contrasts with the weaker DNA demethylation induced by zebularine treatment compared with that in the ddm1 mutants (Baubec et al., 2009) and suggests that toxicity of nonmethylable cytidine analogs, and not DNA demethylation, could cause the reduction of plant growth in the presence of zebularine.
Zebularine and 5-azacytidine have been described as suppressors of tumor growth and are frequently used in cancer treatment, where zebularine is preferred, in some cases, over 5-azacytidine because of its lower toxicity (Dote et al., 2005; Yang et al., 2013). This is most likely due to the extensive metabolism of zebularine into zebularine-deoxyphosphate-cholines and diphosphoethanolamine, which may reduce the amount of biologically active drug (Ben-Kasus et al., 2005). Up to 5% of total cytosines can be replaced by 5-azacytidine, but the rate of zebularine incorporation into genomic DNA seems to be much lower (Jones and Taylor, 1980; Ben-Kasus et al., 2005). Both drugs are bound by DNA METHYLTRANSFERASEs (DNMTs) and form nucleoprotein adducts (NPAs), which effectively deplete the DNMT pool (Egger et al., 2004). In vitro studies using synthetic oligonucleotides containing 5-azacytidine or zebularine revealed higher stability of NPAs when compared with DNMT bound to 5-methyl-deoxycytosine (Champion et al., 2010; Kiianitsa and Maizels, 2013). The data generated using 5-azacytidine and 5-azadeoxicytidine suggest that NPAs represent a physical barrier for enzymes sliding along the DNA molecule and are repaired by HR coupled with replication restart and nucleotide excision repair (Kuo et al., 2007; Salem et al., 2009).
Nucleoside analogs are frequently used in basic and medical research. However, their mode of action and spectrum of effects is not well understood. Using Arabidopsis as a model system, we show that administration of zebularine triggers a specific type of DNA damage response, which dominates over DNA methylation changes. Reduced DNA damage response in the DNMT triple mutant suggests zebularine-DNMT NPAs as the possible causal aberrations. Zebularine treatment extends the G2 phase of the cell cycle and promotes endoreplication. Activation of DNA damage repair of zebularine-induced lesions is additively mediated by ATR and ATM kinases, and the damage is repaired by HR with only a minor contribution of nucleotide excision repair (NER). Absence of higher level of DNA strand breaks upon zebularine treatment differentiated its effects from those of 5-azacytidine inducing large amount of DNA single-strand breaks. The STRUCTURAL MAINTENANCE OF CHROMOSOMES5 (SMC5)-SMC6 complex plays an essential role in the repair of zebularine-induced DNA damage.
RESULTS
Transcriptional Activation of DNA Damage Repair Genes by Zebularine Treatment
To understand the effects of zebularine treatment, we used RNA-sequencing to perform genome-wide transcriptome analysis of dissected shoot apices of 12-d-old wild-type Arabidopsis plants treated with 20 μM zebularine for 24 h (short) and 5 d (long). Short and long zebularine treatment caused significant (adjusted P value < 0.05) upregulation of 31 and 678 genes and downregulation of 12 and 392 genes, respectively (Figure 1A, Table 1; Supplemental Data Set 1). The RNA-sequencing results for 12 significantly up- or downregulated genes were validated by reverse transcription-quantitative PCR (RT-qPCR) and revealed >75% agreement between both methods, including for key DNA damage repair genes (Supplemental Table 1). Only 38.7% of up- and 50% of downregulated genes after short zebularine treatment overlapped with the set of genes differentially transcribed after long exposure (Figure 1A, Table 1). This indicated duration-dependent contrasting effects of zebularine treatment on the Arabidopsis transcriptome.
Figure 1.
Zebularine Treatment Activates DNA Damage Repair Genes.
(A) Genes significantly up- or downregulated in response to 24 h (blue) and 5 d (pink) 20 μM zebularine (zeb) treatment of wild-type plants.
(B) Significantly up- and downregulated genes in response to 24 h zebularine (blue) and 24 h 10 μM MMC treatment (green).
(C) Histochemical staining of pGMI1:GUS reporter line after the specified hours of treatment with 20 μM zebularine, 10 μM MMC, and 100 nM bleocin. Representative rosettes and root tips are shown.
(D) RT-qPCR analysis of DNA damage repair marker genes GMI1, RAD51, PARP2, and BRCA1 in dissected shoot apices after given hours of treatment with 20 μM zebularine, 10 μM MMC, and 100 nM bleocin. The bars represent a mean of mRNA levels from a pool of 5 to 10 seedlings in one biological replicate.
Table 1. Genes Significantly Up- and Downregulated after Short (24 h) 20 μM Zebularine Treatment.
| AGI Locus | Gene Annotation | Mock | Zeb | Log2 Fold Change | Adjusted P Value | DDR | ||
|---|---|---|---|---|---|---|---|---|
| RPKM | ±sd | RPKM | ±sd | |||||
| Upregulated genes | ||||||||
| At1g11580 | METHYLESTERASE PCR A (PMEPCRA) | 8.3 | 0.1 | 15.1 | 0.4 | 0.84 | 0.008 | |
| At1g20750 | RAD3-like | 0.0 | 0.0 | 0.4 | 0.1 | Infinite | 0.004 | + |
| At1g48460 | Unknown protein | 12.5 | 0.5 | 20.2 | 1.2 | 0.68 | 0.049 | |
| At1g63660 | GMP synthase | 16.1 | 1.7 | 26.7 | 1.8 | 0.72 | 0.049 | + |
| At1g65310 | XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE17 (XTH17) | 0.8 | 0.1 | 3.2 | 0.1 | 1.95 | 0.043 | |
| At1g70260 | USUALLY MULTIPLE ACIDS MOVE IN AND OUT TRANSPORTERS36 (UMAMIT36) | 2.7 | 0.4 | 6.5 | 0.4 | 1.23 | 0.009 | |
| At1g72440 | SLOW WALKER2 (SWA2) | 10.0 | 1.5 | 16.5 | 0.2 | 0.70 | 0.048 | |
| At1g75780 | TUBULIN β-1 CHAIN (TUB1) | 8.5 | 0.2 | 14.3 | 0.8 | 0.73 | 0.037 | |
| At1g78370 | GLUTATHIONE S-TRANSFERASE TAU 20 (GSTU20) | 368.0 | 12.6 | 721.3 | 74.7 | 0.95 | 0.000 | |
| At2g21790 | RIBONUCLEOTIDE REDUCTASE1 (RNR1) | 23.7 | 2.3 | 40.5 | 1.4 | 0.76 | 0.001 | + |
| At2g40360 | ARABIDOPSIS THALIANA PESCADILLO ORTHOLOG1 (ATPEP1) | 18.9 | 2.7 | 31.9 | 0.4 | 0.74 | 0.021 | |
| At2g43100 | ISOPROPYLMALATE ISOMERASE2 (IPMI2) | 60.8 | 6.7 | 113.8 | 12.4 | 0.88 | 0.000 | |
| At3g03780 | METHIONINE SYNTHASE2 (MS2) | 77.3 | 10.3 | 137.3 | 0.9 | 0.81 | 0.011 | |
| At3g07800 | THYMIDINE KINASE 1A (TK1A) | 13.9 | 2.6 | 29.7 | 4.1 | 1.07 | 0.000 | + |
| At3g13470 | CHAPERONIN-60BETA2 (CPN60BETA2) | 99.1 | 8.3 | 158.8 | 16.1 | 0.67 | 0.005 | |
| At3g15950 | NAI2 | 32.2 | 1.4 | 49.1 | 2.0 | 0.59 | 0.009 | |
| At3g16150 | ASPARAGINASE B1 (ASPGB1) | 2.9 | 0.1 | 8.3 | 0.3 | 1.48 | 0.007 | |
| At3g19680 | Protein of unknown function (DUF1005) | 14.0 | 2.8 | 30.1 | 3.1 | 1.07 | 0.008 | |
| At3g27060 | TSO2 | 63.2 | 3.7 | 127.2 | 9.7 | 0.99 | 0.005 | + |
| At3g27630 | SIAMESE-RELATED7 (SMR7) | 0.6 | 0.5 | 5.0 | 0.1 | 3.08 | 0.049 | + |
| At3g54810 | BLUE MICROPYLAR END3 (BME3) | 19.6 | 0.8 | 30.5 | 1.0 | 0.62 | 0.024 | |
| At3g59670 | Unknown protein | 4.4 | 0.5 | 9.9 | 0.5 | 1.17 | 0.000 | + |
| At4g21070 | BREAST CANCER SUSCEPTIBILITY1 (BRCA1) | 3.4 | 0.5 | 9.3 | 1.1 | 1.43 | 0.000 | + |
| At4g22410 | Ubiquitin C-terminal hydrolase protein | 0.0 | 0.0 | 1.8 | 0.1 | Infinite | 0.048 | |
| At4g22880 | LEUCOANTHOCYANIDIN DIOXYGENASE (LDOX) | 7.2 | 0.2 | 15.7 | 2.3 | 1.10 | 0.003 | |
| At4g31210 | DNA topoisomerase | 10.7 | 0.9 | 16.4 | 0.2 | 0.61 | 0.035 | + |
| At5g14200 | ISOPROPYLMALATE DEHYDROGENASE1 (IMD1) | 76.1 | 2.0 | 146.9 | 23.2 | 0.92 | 0.000 | |
| At5g20850 | RAS ASSOCIATED WITH DIABETES51 (RAD51) | 3.3 | 0.3 | 7.3 | 0.8 | 1.11 | 0.043 | + |
| At5g42800 | DIHYDROFLAVONOL 4-REDUCTASE (DFR) | 5.3 | 0.4 | 11.8 | 0.9 | 1.14 | 0.000 | |
| At5g52470 | FIBRILLARIN1 (FIB1) | 83.8 | 1.4 | 128.8 | 4.7 | 0.60 | 0.049 | |
| At5g55920 | OLIGOCELLULA2 (OLI2) | 12.0 | 2.8 | 22.9 | 0.1 | 0.92 | 0.011 | |
| Downregulated genes | ||||||||
| At1g28330 | DORMANCY-ASSOCIATED PROTEIN1 (DYL1) | 170.6 | 5.9 | 106.3 | 14.3 | −0.71 | 0.022 | |
| At1g35612 | Transposable element gene | 40.9 | 1.3 | 26.7 | 4.8 | −0.64 | 0.037 | |
| At1g68050 | FLAVIN-BINDING, KELCH REPEAT, F BOX1 (FKF1) | 4.8 | 0.7 | 1.8 | 0.6 | −1.46 | 0.003 | |
| At2g21210 | SMALL AUXIN UPREGULATED RNA6 (SAUR6) | 63.1 | 0.3 | 31.8 | 5.7 | −1.02 | 0.049 | |
| At2g33830 | DORMANCY ASSOCIATED GENE2 (DRM2) | 317.4 | 62.1 | 90.6 | 13.0 | −1.83 | 0.000 | |
| At2g42530 | COLD REGULATED 15B (COR15B) | 50.7 | 2.6 | 16.9 | 2.7 | −1.59 | 0.005 | |
| At3g05880 | RARE-COLD-INDUCIBLE 2A (RCI2A) | 144.0 | 4.0 | 89.7 | 9.6 | −0.71 | 0.005 | |
| At3g62550 | Adenine nucleotide α-hydrolase-like | 80.3 | 2.3 | 48.1 | 5.6 | −0.75 | 0.003 | |
| At4g04330 | HOMOLOG OF CYANOBACTERIAL RBCX1 (RBCX1) | 55.2 | 4.9 | 33.8 | 5.1 | −0.72 | 0.049 | |
| At4g39090 | RESPONSIVE TO DEHYDRATION19 (RD19) | 241.8 | 1.9 | 163.8 | 7.4 | −0.58 | 0.008 | |
| At5g14780 | FORMATE DEHYDROGENASE (FDH) | 84.4 | 3.5 | 57.9 | 3.5 | −0.56 | 0.010 | |
| At5g54190 | PROTOCHLOROPHYLLIDE OXIDOREDUCTASE A (PORA) | 10.4 | 0.2 | 5.3 | 0.2 | −1.00 | 0.009 | |
Reads per kilobase per million reads (RPKM) are an average of two biological replicates ± sd. Adjusted P values were calculated using DESeq statistics in R. DNA damage repair (DDR) genes (TAIR10) are marked with a “+.” Genes in bold were significantly up- or downregulated after a long (5 d) zebularine treatment.
To identify how many of the zebularine up- or downregulated genes are targets of TGS, we compared our data to the RNA-sequencing data set of ddm1 plants (Zemach et al., 2013). No overlap was found for short zebularine treatment and only four out of 908 genetic elements upregulated in ddm1 were also significantly upregulated after the long zebularine treatment (TE gene AT1G42050; MuDr AT2G15810, LINE1-6 AT3G28915, and Gypsy-like AT5G35057; Figure 1A). Therefore, <1% of the zebularine upregulated genes in shoot apices are TGS targets. A functional annotation analysis (TAIR10) of the 31 genes induced by the short zebularine treatment revealed that 32.3% are linked to DNA metabolism and DNA damage repair, e.g., the genes encoding the RIBONUCLEOTIDE REDUCTASE (RNR) complex subunits RNR1 and TSO2, and the genes BREAST CANCER SUSCEPTIBLE1 (BRCA1), RAS ASSOCIATED WITH DIABETES51 (RAD51), or SIAMESE-RELATED7 (Table 1). Several additional DNA damage repair genes, including GAMMA-IRRADIATION AND MITOMYCIN C INDUCED1 (GMI1), were significantly upregulated after the long zebularine treatment (Supplemental Data Set 1). To test whether these mRNA changes represent a bona fide response to a DNA damage stimulus, we exposed plants to mitomycin C (MMC), a drug that induces DNA interstrand cross-links (Iyer and Szybalski, 1963; Tomasz, 1995). Short (24 h) 10 μM MMC treatment significantly up- and downregulated 815 and 579 genes, respectively, including numerous DNA damage repair genes (adjusted P value < 0.05; Figure 1B; Supplemental Data Set 2). Importantly, the sets of genes up- and downregulated in response to 24 h of zebularine exposure overlapped 93.1% (29 out of 31) and 91.7% (11 out of 12), respectively, with the MMC treatment (Figure 1B).
Prior to incorporation into DNA, zebularine undergoes modification in several steps (Ben-Kasus et al., 2005). This raises the question of the kinetics of the DNA damage response and its tissue specificity. To examine this, we used a pGMI1::GUS (β-glucuronidase) reporter line that allows the visualization of tissues with ongoing DNA damage repair (Böhmdorfer et al., 2011). The reporter lines were exposed to zebularine, MMC, and the radiomimetic drug bleocin. GUS was not detected in mock-treated plants, while 3 h of bleocin and 6 h of MMC or zebularine treatment were sufficient to obtain GUS staining in the shoot apices, petioles of the youngest leaves, and in the cotyledon vasculature (Figure 1C). Over time, the staining became more prominent in the entire true leaves and cotyledon vasculature. GUS was also detected in root apical meristems of MMC- and bleocin-treated, but not of zebularine-treated, samples. These results suggest a rapid induction of GMI1 by zebularine and its different drug processing or stability in root and shoot apical meristem tissues. To assess the kinetics of transcriptional activation in more detail, we dissected shoot apices of mock- and drug-treated plants over the 24-h time series and validated GMI1 activation by RT-qPCR (Figure 1D). However, the amount of transcript did not simply accumulate over time as observed in histochemical staining (Figure 1C), probably reflecting the higher stability of the GUS protein compared with GMI1 mRNA. Other tested DNA damage repair genes, including those detected in our RNA-sequencing (RAD51, BRCA1, and PARP2) were also upregulated in response to zebularine with kinetics and amplitudes similar to the MMC and bleocin treatments (Figure 1D). Hence, zebularine treatment leads to transcriptional upregulation of a specific set of DNA damage repair genes in shoot apical tissues, in a rapid and high amplitude manner.
Zebularine-Triggered DNA Damage Response Is Independent of DNA Methylation Changes
Zebularine has been shown to reduce DNA methylation in a dose-dependent manner (Baubec et al., 2009). Therefore, the activation of DNA damage repair genes observed after 20 μM zebularine treatment may be caused by DNA demethylation. We identified methylated DNA regions <1 kb upstream of TSO2 and RAD51, two DNA damage repair genes activated by zebularine treatment (Supplemental Figures 1 and 2). Analysis of these regions by bisulfite sequencing in dissected shoot apices of mock, short, and long zebularine-treated plants revealed <5% reduction of DNA methylation (Figure 2A; Supplemental Data Sources 1 to 4). Similarly, we observed normal levels of DNA methylation at the LINE1-6 retrotransposon (AT3G28915/AT3TE45385) identified as a common target of zebularine and activation in ddm1 mutants. DNA methylation was also maintained in the repetitive region upstream of the SUPPRESSOR OF drm1 drm2 cmt3 (SDC) gene (Henderson and Jacobsen, 2008) upregulated by long zebularine treatment (Figures 2A and 2B; Supplemental Data Set 1). Hence, zebularine-induced upregulation of several genetic elements occurred without loss of DNA methylation. Recently, it has been shown that SDC can be activated by disturbed higher chromatin order structure in MORC6 ATPase mutants (Moissiard et al., 2012). Because zebularine treatment leads to heterochromatin decondensation in Arabidopsis (Baubec et al., 2009), we tested whether disturbed chromatin structure in morc6 is sufficient for induction of DNA damage repair genes. However, SDC but not TSO2 and RAD51 were activated in dissected apices of morc6 plants (Supplemental Figure 3A). This suggests that disturbed heterochromatin structure alone is not sufficient to induce DNA damage repair response and that zebularine treatment interferes with at least two independent genome maintenance pathways. Furthermore, zebularine-induced transcriptional activation of DNA damage repair genes and TGS targets may occur without stable changes in DNA methylation.
Figure 2.
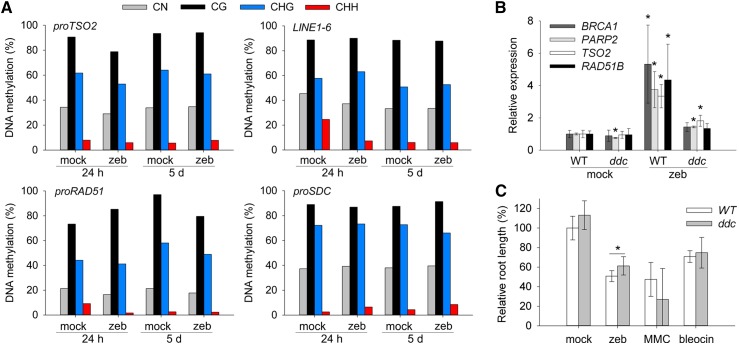
Zebularine Effects on DNA Methylation and Nucleoprotein Adduct Formation.
(A) Percentage of DNA methylation in dissected shoot apices based on bisulfite sequencing of 24 h and 5 d mock- and 20 μM zebularine (zeb)-treated samples. A minimum number of 12 reads per experimental point has been analyzed. Schematic view of the analyzed genomic regions is provided in Supplemental Figure 1.
(B) RT-qPCR measurement of DNA damage marker gene induction in the wild type (WT) and drm1 drm2 cmt3 (ddc) triple mutant after 24 h treatment with mock and 20 μM zebularine normalized to ACTIN7. Error bars represent sd of three biological replicates and asterisks P < 0.05 in t test.
(C) Relative root length of wild-type and ddc plants in response to 20 μM zebularine, 15 μM MMC, or 50 nM bleocin treatment. Error bars represent sd of three biological replicates and asterisk P < 0.05 in t test.
Next, we tested the frequency of zebularine incorporation into plant genomic DNA. We grew Arabidopsis plants in medium containing 20 μM zebularine, which we refreshed every 3 d, for 14 d, and analyzed the amount of deoxyzebularine in genomic DNA using reverse-phase HPLC (RP-HPLC). Even with a detection limit at ∼1 deoxyzebularine per 5000 deoxycytosines (Supplemental Figure 4), we could not detect deoxyzebularine incorporated into plant DNA. Although surprising, these data are generally in line with the low rate (∼0.1 pmol per μg DNA = ∼1 deoxyzebularine per ∼8000 deoxycytosines) of zebularine incorporation into DNA of mammalian cell lines (Ben-Kasus et al., 2005). This suggests that zebularine may not be efficiently and/or stably incorporated into DNA, in particular in Arabidopsis, a plant with very small meristems.
In vitro experiments with synthetic oligonucleotides revealed that DNMTs covalently bind to zebularine-containing DNA molecules (Champion et al., 2010). Since we could not detect zebularine directly in DNA, we tested whether the NPAs could cause DNA damaging effects by reducing the amount of available DNMTs. Due to strongly reduced fitness and pleiotropic effects of mutants in DNA METHYLTRANSFERASE1 (Mathieu et al., 2007), we used CHROMOMETHYLASE3 (CMT3), DOMAINS REARRANGED METHYLTRANSFERASE1 (DRM1), and DRM2 triple homozygous mutant (ddc) plants (Henderson and Jacobsen, 2008). We exposed ddc plants to mock treatment and 20 μM zebularine for 24 h and used RT-qPCR to measure mRNA levels of DNA damage repair genes. TSO2, BRCA1, PARP2, and RAD51B were 3.5- to 5.5-fold upregulated in response to zebularine in the wild type, whereas we observed <2-fold upregulation in zebularine-treated ddc plants (Figure 2B). Furthermore, zebularine-induced inhibition of root growth was significantly reduced in ddc compared with wild-type plants (t test, P < 0.05; Figure 2C; Supplemental Figure 3B). Therefore, the DNMT-zebularine NPAs seem to be at least partly responsible for the DNA damage phenotypes and zebularine toxicity.
This indicates that zebularine incorporation into DNA is rare or unstable, the transcriptional activation of zebularine-induced targets occurs without stable DNA demethylation, and the DNA damage response is triggered at least partially by the zebularine-DNMT NPAs.
ATR and ATM Redundantly Signal Repair of Zebularine-Induced DNA Damage
The >90% overlap between MMC and zebularine-induced mRNA changes suggests that the damage they induce is repaired by a pathway with at least some components in common. Interstrand DNA cross-linking activity of MMC causes stalled replication forks that are repaired by the ATR pathway (Culligan et al., 2004). Therefore, we performed RNA-sequencing of the shoot apices of atr mutant plants exposed to mock, 20 μM zebularine, and 10 μM MMC for 24 h and compared this with their effects on the wild type. In mock-treated atr, 227 and 119 genes were significantly up- and downregulated, respectively (Supplemental Figure 5A and Supplemental Data Set 3). This corresponded to 70 and 20 significantly enriched Gene Ontology term categories, respectively, pointing toward stress and immune responses (Supplemental Data Set 4). As atr plants were grown under conditions that did not induce stress in the wild type, this indicates that ATR prevents a hypersensitive reaction to the environment in Arabidopsis. A 24-h zebularine and MMC treatment of atr led to significant upregulation of 62 and 78 genes (29 common), respectively (Supplemental Figure 5A). In total, 363 and 421 genes (225 overlapping) were significantly downregulated in atr in response to zebularine and MMC treatment, respectively (Supplemental Figure 5A). This confirms the role of ATR as a positive regulator of transcription in response to stress. Importantly, only four genes were commonly upregulated and two downregulated in zebularine-treated wild type and atr, suggesting that most of the transcriptional response to zebularine treatment is ATR dependent (Figure 3A). This was less pronounced for the MMC treatment, where 50% of upregulation (408 out of 815) and 61% of downregulation (353 out of 579) occurred in an ATR-independent manner (Figure 3B).
Figure 3.
Both ATR and ATM Signal Repair of Zebularine-Induced Damage.
(A) Effects of zebularine-atr on gene transcript levels. Blue ovals in Venn diagrams show genes significantly up- or downregulated in response to short zebularine (zeb) exposure. Pink depicts genes significantly up- or downregulated in zebularine-treated relative to mock-treated atr. The genes in overlap are upregulated in response to zebularine independent of ATR.
(B) MMC-atr effects on gene transcript amounts analyzed as described in (B).
(C) Representative phenotypes of wild-type, atr, atm, and atm atr double mutant root elongation on 20 μM zebularine, 15 μM MMC, and 50 nM bleocin. The graph shows quantitative root length data for individual genotypes. Asterisks indicate statistically significant (t test, P < 0.05), and error bars denote sd of three biological replicates. n.a., not analyzed.
However, several genes upregulated in response to the zebularine treatment were also previously identified as ATM targets (Culligan et al., 2006). Therefore, we performed genetic studies to test for the involvement of both kinases in detoxifying zebularine-induced damage. Besides the reduced root length of atr, phenotypes of atr and atm were similar to the wild type on medium without zebularine. But both mutants had partially reduced growth on 20 μM zebularine (Figure 3C; Supplemental Figures 5B and 5C and Supplemental Tables 2 and 3). This resembled the phenotype obtained after bleocin treatment and contrasted with the MMC treatment, which caused an extreme hypersensitivity in atr and only weak sensitivity in atm. Next, we tested for potential functional redundancy of ATM and ATR in repair of zebularine-induced damage. Because the atm atr double mutants are sterile (Culligan et al., 2006), we phenotyped and genotyped a population of plants homozygous for atr (ATR−/−) and segregating for atm alleles (ATM−/+). In total 27.6% (16 out of 58) of plants were atm atr homozygous double mutants and corresponded to individuals with extreme hypersensitivity to the zebularine treatment (Figure 3C; Supplemental Figures 5B and 5C). All ATR−/− ATM−/+ plants were fully sensitive to MMC treatment due to atr single mutant hypersensitivity, and no fully sensitive ATR−/− ATM−/+ plants were observed upon bleocin treatment (Figure 3C). These experiments provide molecular and genetic evidence for the additive role of ATR and ATM in signaling repair of zebularine-induced DNA damage.
Zebularine-Induced DNA Damage Is Detoxified Predominantly by Intermolecular HR
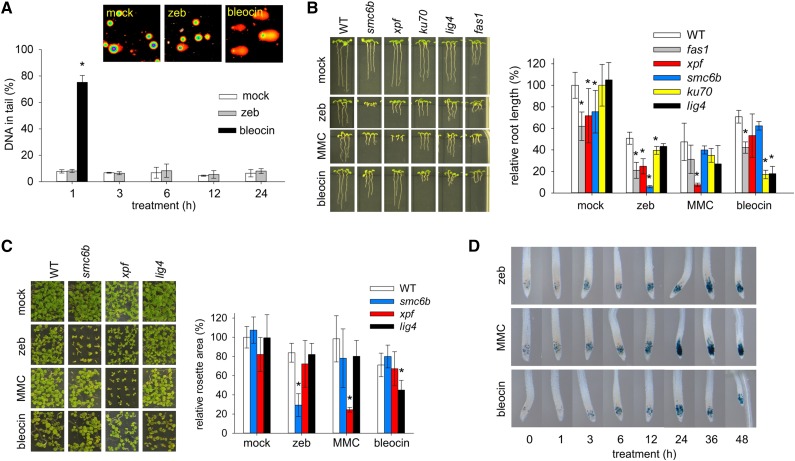
Metazoan data suggest that activation of ATM may be triggered by both DNA strand breaks and disturbed chromatin structure (Bakkenist and Kastan, 2003). To test for the former, we performed single cell electrophoresis (comet assays) using the alkaline/neutral protocol to detect both DNA single- and double-strand breaks. One-hour treatment of wild-type plants with 25 μg/mL bleocin resulted in 70% of DNA in comet tails, while only 10% of DNA was in the tails in the mock-treated sample (Figure 4A). However, the amount of DNA in the tail did not increase beyond mock levels during 24-h treatment with 800 μM zebularine (Figure 4A). This strongly suggests that even high zebularine concentrations over long time periods do not cause substantial fragmentation of the nuclear genome. This was further supported by the lack of zebularine hypersensitivity in mutants of nonhomologous end joining components KU70 and LIGASE (LIG4), which were hypersensitive to bleocin treatment (Figures 4B and 4C; Supplemental Figure 6 and Supplemental Tables 2 and 3). We also tested effects of treatment with 5-azacytidine, another nonmethylable cytidine analog, on DNA integrity (Supplemental Figure 7). We observed significantly (t test, P < 0.01) more DNA in comet tails after 1 h of 100 and 200 μM 5-azacytidine treatment followed by alkaline/neutral comet assays. However, no increased tail DNA was found in neutral/neutral comet assays, indicative of DNA double-strand breaks. This suggests that 5-azacytidine treatment is associated with extensive DNA single-strand breakage, in contrast with zebularine treatment where no large amount of DNA strand breaks could be detected.
Figure 4.
Zebularine Treatment Blocks Cell Cycle and Is Lethal for smc6b Plants.
(A) Analysis of DNA fragmentation in response to genotoxic treatment. Images of representative comet assays based on nuclei isolated from plants treated with mock, 800 μM zebularine (zeb), and 25 μg/mL bleocin for 1 h. The graph shows percentage of DNA in comet tail. Error bars indicate sd of means from three biological replicates, and asterisk marks statistically significantly different groups relative to mock control (t test; P < 0.05).
(B) and (C) Images show representative root length (B) and rosettes (C) of the wild type (WT) and mutants grown on mock, 20 μM zebularine, 15 μM MMC, and 100 nM bleocin for 7 and 15 d, respectively. Quantitative data presented in graphs are based on three to five biological replicates with the sd indicated by error bars. Statistically significant (P < 0.05) differences in t test are labeled by asterisk.
(D) Representative GUS-stained root tips of the cyclin-GUS reporter line after treatment with 20 μM zebularine, 10 μM MMC, and 100 nM bleocin for the given number of hours.
Strongly reduced growth of mutants in the genes encoding the CHROMATIN ASSEMBLY FACTOR1 components FASCIATA1 (FAS1) and FAS2 on zebularine suggested an additive effect of chemical and genetic interference with chromatin structure (Figure 4B; Supplemental Figures 6A to 6C and Supplemental Tables 2 and 3). Hence, ATM activation in response to zebularine treatment might occur via disturbed chromatin or DNA double helix structure.
ATR is activated by the presence of single-stranded DNA, typically at stalled replication forks (Cimprich and Cortez, 2008). Interference with the ATR pathway frequently leads to cell cycle prolongation or arrest (Culligan et al., 2004, 2006). We tested for zebularine-induced effects on the cell cycle using a cyclin-GUS (pCYCB1;1::CYCB1;1:GUS) reporter line (Colón-Carmona et al., 1999). This reporter protein is synthesized in G2 and degraded at the onset of mitosis. Under mock treatment conditions, the accumulation of cyclin-GUS can be observed in few root apical meristem cells (Figure 4D). Application of 10 μM MMC, 100 nM bleocin, or 20 μM zebularine led to time-dependent accumulation of GUS positive cells in root apical meristems. However, the strongest interference with the cell cycle occurred after MMC treatment followed by zebularine and bleocin treatments. Hence, zebularine-induced damage blocks progression of G2 to M phase. This block is weaker than MMC cross-links, but stronger than DNA double-strand breaks induced by bleocin, with the latter proposed to be repaired in a cell cycle stage-independent manner (Schubert et al., 2004)
To explore the detoxification mechanism of zebularine-induced DNA damage further, we analyzed the sensitivity of mutants of several DNA repair pathways. In bacteria, mutants defective in NER were hypersensitive to 5-azacytidine (Betham et al., 2010). Therefore, we exposed plants mutated in the XERODERMA PIGMENTOSUM GROUP F (XPF) gene, the endonuclease involved in NER and removal of nonhomologous overhangs in intramolecular homologous recombination events (Gaillard and Wood, 2001; Dubest et al., 2002; Molinier et al., 2008; Yoshiyama et al., 2009), to zebularine and other drugs (Figures 4B and 4C; Supplemental Figures 6A and 6B and Supplemental Table 2). While the xpf plants were hypersensitive to MMC treatment, they showed much weaker sensitivity to zebularine. This suggests a minor role of NER and intramolecular homologous recombination in the repair of zebularine-induced DNA damage in Arabidopsis. Similar weak zebularine sensitivity was observed for rad5a plants (Supplemental Figure 8), indicating that repair of zebularine-induced damage does not occur via replication fork regression (Heyer et al., 2010). An opposite pattern was found for the mutants of SMC6B, which were hypersensitive to zebularine and only moderately sensitive to MMC treatment (Figures 4B and 4C; Supplemental Figures 6A, 6B, and 6D). SMC6B is the core component of the SMC5-SMC6 complex (Yan et al., 2013), which has been implicated in DNA damage repair processes in both animals and plants (Mengiste et al., 1999; Chiolo et al., 2011). In Arabidopsis, SMC6B (and presumably the entire SMC5-SMC6 complex) is required for the normal speed of lesion removal and frequency of HR (Mengiste et al., 1999; Hanin et al., 2000; Kozak et al., 2009; Watanabe et al., 2009).
We previously observed that zebularine strongly increases the frequency of somatic HR in Arabidopsis (Pecinka et al., 2009). However, a detailed analysis of this phenotype and comparison to other types of DNA damage was missing. We selected HR reporter lines 651 and IC9C with a similar basal recombination frequency, but differing as to the recombination mechanism (Puchta et al., 1995; Molinier et al., 2004). Line 651 contains a direct repeat of the recombination substrate and allows scoring of intramolecular HR by single strand annealing (SSA). In contrast, an inverted repeat reporter region in the IC9C line is repaired by intermolecular recombination mechanism of synthesis-dependent strand annealing (SDSA). All drug treatments increased HR of both lines (Figure 5A; Supplemental Table 4). However, the damage induced by MMC and bleocin treatments was repaired predominantly by SSA, which was also the preferred HR pathway under non-stress conditions (Figure 5B). However, zebularine-induced damage was repaired significantly more frequently by SDSA than SSA when compared with other treatments (Fisher’s exact test, P < 0.001), suggesting that intermolecular HR by SDSA is the favored HR mechanism to remove zebularine-induced damage. To test whether this SDSA occurs between sister chromatids or homologous chromosomes, we analyzed plants homozygous and hemizygous for the IC9C reporter construct as described (Molinier et al., 2004). The homozygous and hemizygous IC9C plants contained on average 0.83 and 0.41 GUS spots per plant, respectively (Figure 5C; Supplemental Table 5). The number of GUS spots in hemizygous plants was ∼49.1% of that in homozygous ones, suggesting that virtually all zebularine-induced SDSA events occurred between sister chromatids.
Figure 5.
Zebularine Treatment Induces Endoreplication and Requires Repair by HR.
(A) HR assays. Left: representative cotyledons of mock and zebularine (zeb)-treated line 651. HR events, visible as blue dots, are indicated by red arrows. Right: HR frequency of SSA reporter line 651 and SDSA reporter line IC9C after 20 μM zebularine, 15 μM MMC, and 100 nM bleocin stress relative to mock treatment. Error bars denote sd of three biological replicates.
(B) The ratio of SDSA versus SSA after different treatments. Asterisk indicates significant differences (P < 0.001) relative to mock treatment in Fisher’s exact test.
(C) Average number of GUS spots in homozygous and hemizygous IC9C line after treatment with 20 μM zebularine. Error bars show sd of four biological replicates.
(D) Mean cycle values of nuclei isolated from cotyledons of wild-type and mutant plants after 15 d of treatment with 10 μM zebularine, 10 μM MMC, and 50 nM bleocin. Error bars indicate sd of three to five biological replicates, and asterisks denote statistically significant differences (t test, P < 0.05).
To prevent mitosis with potentially aberrant chromosomes, some cells may undergo endoreplication (De Veylder et al., 2011). We used flow cytometry to measure the endoreplication frequency in cotyledons of drug-treated seedlings (Figure 5D; Supplemental Table 6). The cycle value (CV) of mock-treated plants was 1.36 and increased significantly to 1.52 in response to 10 μM zebularine treatment (relative CV = 111%; t test, P < 0.05). Control treatments with 10 μM MMC and 50 nM bleocin yielded CVs of 1.51 (relative CV = 111%, P < 0.05) and 1.32 (relative CV = 97%), respectively. Hence, zebularine treatment increased the endoreplication level similarly to MMC, while DNA strand break induction did not. Next, we extended the analysis to zebularine and MMC hypersensitive mutants (Figure 5D; Supplemental Table 4). Mock-treated mutants were similar to the wild type, except for atr and fas1, which reached 93% (CV 1.26) and 113% (CV 1.54) of the wild type endoreplication level, respectively. The CV of fas1 was further enhanced by zebularine and MMC treatments (CV 1.81 and 1.74, respectively, both P < 0.05 in t test). For atm and atm atr plants, zebularine treatment increased endoreplication to 123.0 and 130.7% (CV 1.6 and 1.77; P = 0.386 and 0.024, respectively), while treatments with bleocin and MMC significantly increased endoreplication in both genotypes (Figure 5D). In contrast, response to either treatment did not increase significantly in atr, probably owing to large variation between biological replicates. The endoreplication levels of smc6b did not change significantly upon zebularine treatment (Figure 5D), despite its hypersensitivity. This contrasted with the effect of nonfunctional XPF, where hypersensitivity to MMC correlated with strongly increased cycle value (168%, CV 2.24, P < 0.05).
Collectively, this provides evidence that zebularine induces a complex type of lesion that affect the cell cycle, leading to significantly increased frequency of endoreplication. These lesions are repaired by HR with a crucial role of the SMC5-SMC6 complex.
DISCUSSION
Chromatin mediates the proper regulation of transcription and maintains the stability of genetic information. Nonmethylable cytidine analogs are widely used in epigenetic and cancer research (Ben-Kasus et al., 2005; Yang et al., 2013; Baubec et al., 2014). However, their biological effects and the mechanism(s) of their action are not well understood (Pecinka and Liu, 2014). Here, we showed that exposure of Arabidopsis to zebularine induces a DNA damage response that is signaled additively by ATR and ATM and repaired through SDSA.
Approximately 32% of the genes upregulated by short zebularine treatment were associated with DNA damage repair and additional genes were induced after longer zebularine treatment. This contrasts with transcriptome analysis after 16 d of 5-azacytidine treatment in Arabidopsis, which revealed upregulation of a functionally diverse set of genes with no association to DNA damage repair (Chang and Pikaard, 2005). This is most likely due to differences in treatment length, stability, and biological effects of both drugs. In contrast, mRNA level changes induced by short zebularine treatment overlapped >90% with those induced by the alkylating agent MMC.
Because zebularine has been proposed to be an inhibitor of DNA methylation (Egger et al., 2004), we analyzed its genome-wide effects on the transcription of DNA methylation targets and also used bisulfite sequencing to analyze its effects on DNA methylation. Only four zebularine-activated genetic elements (<1%) were among the genes controlled by key DNA methylation factor DDM1 (Zemach et al., 2013). Another zebularine-activated TGS target included SDC, a gene under surveillance of DRM2 and CMT3 DNA methyltransferases and higher chromatin order established by MORC6 (Henderson and Jacobsen, 2008; Moissiard et al., 2012). However, SDC and the other three analyzed genes did not show DNA demethylation after the zebularine treatment. We cannot exclude DNA methylation changes in some specific genomic regions, but transcriptional activation of all analyzed genes occurred without loss of DNA methylation. This may be due to fast removal of zebularine, rapid DNA remethylation in apical meristems, or activation by reduced heterochromatin compaction (Baubec et al., 2009, 2014).
We were not able to detect deoxyzebularine in genomic DNA of treated plants with sensitivity of 1 deoxyzebularine per ∼5000 deoxycytosines. Hence, the exact nature of zebularine-induced damage remains unknown. As a ribonucleotide, zebularine might be incorporated into RNA primers of Okazaki fragments and interfere with their removal. However, this model could not be experimentally tested owing to its technical difficulties. The reduced DNA damage response in ddc suggested that the damage is triggered at least partially by deoxy-zebularine-DNMT NPAs (Champion et al., 2010). NPAs (or DNA protein cross-links) are formed by the action of specific chemicals, UV radiation, or compromised activity of topoisomerases (Sheridan and Bishop, 2006; Stingele et al., 2014). Covalent binding of proteins to DNA is a common characteristic of NPAs that differentiates them from many other types of damage and requires specific repair components (Stingele et al., 2014). NPAs most likely represent a heterogeneous group due to different chemical or physical properties of their inducers and share some common features with other damaging agents. Our data also suggest fundamental differences in the nature of DNA damage induced by zebularine and 5-aza-cytidine, two structurally similar cytidine analogs.
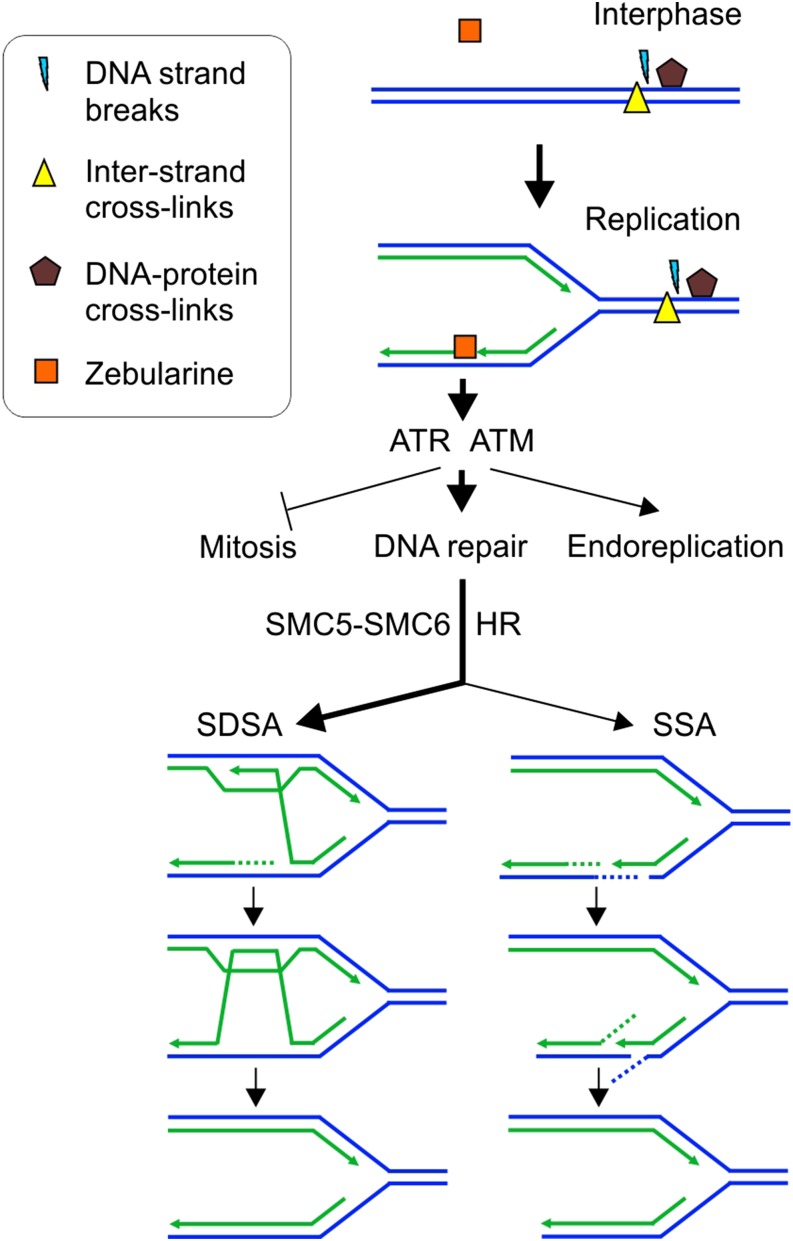
Presumably, the nucleobase-like nature of zebularine allows its interference with genome stability only in a narrow window during DNA replication (Figure 5). As outlined above, this can be by incorporation into either newly synthesized DNA strands and/or RNA primers of Okazaki fragments. This contrasts with effect of MMC-induced interstrand cross-links, where damage is sensed before the replication fork; zebularine-induced damage most likely occurs later, during new strand synthesis. Hence, zebularine-induced DNA damage most likely occurs specifically after DNA strand separation. This activates the DNA damage repair machinery by additive functions of the kinases ATR and ATM. Previously, an additive role of ATM and ATR has been observed for the repair of DNA damage induced by ionizing radiation and in the course of meiosis (Culligan et al., 2006). However, our comparison to radiomimetic treatments revealed that zebularine treatment interferes more strongly with DNA replication and does not cause extensive DNA strand break formation. Furthermore, zebularine treatment had a much stronger potential to increase endoreplication, which was similar to the replication-blocking agent. This creates a unique set of phenotypes that are not observed upon induction of DNA damage with other genotoxic agents and allows us to address the mechanism that repairs this damage.
To dissect repair pathways, we tested XPF, a component involved in NER and to some extent also in the SSA type of HR (Dubest et al., 2002; Molinier et al., 2008). The partial sensitivity of xpf shows that a minor fraction of zebularine-induced damage is repaired by NER or SSA, which is also consistent with our HR data. This contrasts with the effects of 5-azacytidine, where NER is the dominant repair pathway in bacteria and humans (Salem et al., 2009; Orta et al., 2014). Because the smc6b mutant was more sensitive to zebularine than to other tested drugs, we suggest that the SMC5-SMC6 complex plays an essential role in the repair of zebularine-induced NPAs in Arabidopsis. We hypothesize that this could be either due to transcriptional deregulation of specific genes in smc6b or lack of DNA damage repair competence. It has been demonstrated that the SMC5-SMC6 complex functions as a facilitator of HR (Mengiste et al., 1999; Hanin et al., 2000; Watanabe et al., 2009) and its absence affects the speed of repair in Arabidopsis (Kozak et al., 2009). This is consistent with the proposed function of SMC5-SMC6 in controlling HR timing in DNA damage repair in Drosophila melanogaster (Chiolo et al., 2011) and also matches with the elevated frequency of somatic HR upon zebularine treatment (Pecinka et al., 2009).
The analysis of specific HR pathways revealed that SSA is a preferred HR pathway for repair of bleocin- and MMC-induced damage, while SDSA seems to be more important for repair of zebularine-induced damage. This is genetically supported by a minor role of XPF, an enzyme involved in HR by removing nonhomologous overhangs in SSA events (Dubest et al., 2002; Molinier et al., 2008). SSA can occur at both nonreplicated and replicated chromosomes, but SDSA only occurs at replicated chromosomes. By comparing plants allowing HR between sister chromatids and/or homologous chromosomes, we showed that zebularine-induced SDSA occurs strictly between sister chromatids. The lack of zebularine sensitivity of rad5a plants indicated the absence of replication fork regression (Heyer et al., 2010). Collectively, this suggests that zebularine-induced damage is removed after strand separation, during or shortly after the new strand synthesis (Figure 6). This further differentiates the zebularine effects from other DNA damaging agents and supports the presence of a specific repair strategy (Figure 6).
Figure 6.
The Model of Zebularine-Induced Damage and Its Repair.
Most types of DNA damage, including DNA-protein cross-links, DNA strand breaks, or interstrand cross-links, can occur irrespectively of the cell cycle phase. In contrast, zebularine damage occurs during DNA replication in course of new DNA strand synthesis. This causes DNA damage stress, which suppresses cell division, promotes endoreplication, and activates DNA damage repair signaling by ATR and ATM activity. The repair depends strongly on SMC5-SMC6 activity and is pursued primarily by SDSA and to a smaller extent also SSA homologous recombination pathways.
Zebularine is an anticancer agent that effectively suppresses growth of several types of tumors (Egger et al., 2004; Ben-Kasus et al., 2005; Yang et al., 2013). Mechanistically, this has been proposed to be due to its interference with DNA methylation and p53-dependent endoplasmic reticulum stress. Our data indicate an alternative mechanism based on the induction of specific DNA damage. Furthermore, Arabidopsis data suggest that this interference may be particularly effective for treatment of cells with deficient ATM and ATR functions.
METHODS
Plant Material
Arabidopsis thaliana wild type and mutants were in Columbia-0 background: atm-1 (SALK_040423C), atr-2 (SALK_032841C), fas1 (Sail_662.D10), fas2 (SALK_033228), ku70 (SALK_123114C), lig4 (SALK_044027C), rad5a-2 (SALK_047150), smc6b-1 (SALK_101968C), smc6b-2 (SALK_135638), smc6b-3 (Mengiste et al., 1999), and xpf-3 (SALK_096156C). The atr-2 atm-2 plants were identified in the atr-2−/− (SALK_032841C) and atm-2+/− (SALK_006953) segregating population. We also used cyclin-GUS containing the pCYCB1;1::CYCB1;1:GUS construct (Colón-Carmona et al., 1999) and pGMI1:GUS (Böhmdorfer et al., 2011). All mutants and reporter lines were used as homozygous lines unless stated otherwise. smc6b-1 was used for experiments unless specified otherwise.
Drug Treatments
The seeds were sterilized, evenly spread on sterile half-strength Murashige and Skoog (1/2 MS) medium with or without zebularine (Sigma-Aldrich), MMC (Duchefa Biochemie), and bleocin (Calbiochem) in concentrations specified in the text and grown at 16 h light:8 h dark at 21°C. For RNA-sequencing, RT-qPCR, and reporter analysis, plants were grown for 7 d on solid 1/2 MS medium and then transferred to control 1/2 MS plates or freshly prepared drug plates (Figures 1A, 1B, 2, 3, 4B, 4C, and 5; Supplemental Figures 3, 5, 6, and 8) or liquid media (Figures 1C, 1D, 4A, and 4D, Supplemental Figures 4 and 7) for specified times. For root elongation assays, 7-d-old plants grown continuously on mock and drug containing solid media were used. Fifteen-day-old plants grown under the same conditions were used for rosette area measurements and endoreplication analysis. RNA-sequencing was performed on dissected shoot apices of 12-d-old plants grown on solid media.
Nucleic Acid Isolation, cDNA Synthesis, and RNA-Sequencing
DNA was extracted using the DNeasy kit (Qiagen) or Nucleon Phytopure kit (GE Healthcare). RNA was extracted using the RNeasy kit (Qiagen) with on-column DNase I (Roche) treatment. cDNA for quantitative PCR experiments was synthesized from 1 μg RNA per sample with Revert Aid H-Minus First Strand cDNA synthesis kit using the oligo-d(T) primer (Thermo Scientific). The purity of cDNA was monitored by PCR with an intron-spanning primer pair.
RNA sequencing was performed with two biological replicates per experimental point. The libraries were prepared from 1 μg total RNA with RNA integrity number >7.8 (Bioanalyzer; Agilent) using TruSeq RNA kit (Illumina) and sequenced as 100-bp single-end reads on HiSeq2500 (Illumina). Reads were trimmed and low-quality reads filtered with FAST-X tools (http://hannonlab.cshl.edu/fastx_toolkit/) using custom made scripts. This yielded an average of 15 million high-quality reads per library. The reads were mapped to the TAIR10 Arabidopsis reference genome using Tophat2 (Kim et al., 2013) with default settings. The coverage of individual genes was retrieved with the Qualimap from the set of uniquely mapped reads and significance (adjusted P value < 0.05) of mRNA level changes estimated with the DEseq package (Anders and Huber, 2010) in R. Venn diagrams were drawn using the venneuler package in R. Publicly available ddm1 transcriptional data from the Gene Expression Omnibus data set GSE41302 (Zemach et al., 2013) were analyzed in the same way.
Primers
Primers used in this study are provided in Supplemental Table 6.
DNA Methylation Analysis
Approximately 120 ng of genomic DNA extracted from shoot apices of 15 seedlings was bisulfite treated using the EZ DNA methylation-Gold kit (Zymo Research). Desired fragments were PCR amplified from 1 μL of converted DNA and cloned into the pJET1.2 vector using the CloneJET PCR cloning kit (Thermo Scientific). At least 12 clones were analyzed per condition. Individual bisulfite sequencing reads used for analysis of DNA methylation are provided as Supplemental Data Sources 1 to 4.
Comet Assays
Ten-day-old plants were transferred from 1/2 MS solid to liquid media containing no drug (mock), 25 μg/mL bleocin, 800 μM zebularine, and 100 or 200 μM 5-azacytidine for the specified times. Afterward, nuclei were isolated from entire seedlings and alkaline/neutral or neutral/neutral comet assays were performed using the CometAssay kit (Trevigen) with the following modifications: The nuclei lysis was reduced to 5 min, unwinding to 10 min and electrophoresis to 6 min. Preparations were stained with Sybr Gold, and images were captured with a Zeiss Axio Imager A2 epifluorescence microscope equipped with Axiocam HRc camera. A total of 100 to 150 comets per experimental point were analyzed with CometScore (Tritek).
GUS Staining and Endoploidy Analysis
GUS histochemical staining was performed as described (Baubec et al., 2009). Images were acquired using MZ16 FA stereomicroscope equipped with DFC490 CCD camera (both Leica). For endoploidy analysis, cotyledons were dissected, chopped with a razor blade in 300 μL extraction buffer (Partec), filtered through 30-μm nylon mesh, stained with 900 to 1800 μL CyStain dye (Partec), and analyzed with PAS I ploidy analyzer (Partec). The endopolyploidy cycle value was calculated using the formula: CV = ((n 2C*0) + (n 4C*1) + (n 8C+2) + (n 16C*3) + (n 32C*4)) / (n 2C + n 4C + n 8C + n 16C + n 32C), where n = number of counts per given C-value content.
Quantitative PCR
The RT-qPCR was performed using 1 μL cDNA per 10-μL reaction with the SensiMix kit (PeqLab) on an CFX384 instrument (Bio-Rad). Fold changes were calculated relative to mock-treated controls using the standard curve method.
Root Elongation and Rosette Area Measurements
For root length assay, plants were grown for 7 d on control and drug containing media, then carefully taken out using forceps without breaking roots and stretched on agar plates. Rosette area measurements were performed in independent experiments with 15-d-old plants. Plants were photographed with a D90 digital camera (Nikon). For rosette area measurements, color photographs were converted into binary mode using ImageJ (http://rsbweb.nih.gov/ij/). Both types of traits were then measured using ImageJ calibrated with an internal size control. Sensitivity to the DNA damaging agent in individual replicates was determined by calculating mean(treatment)/mean(mock). The roots and rosettes of at least 10 plants per genotype and treatment were measured per each of the three biological replicates.
HR Assays
The 651 and IC9C reporter lines (Puchta et al., 1995; Molinier et al., 2004) were grown in liquid 1/2 MS media with or without drug treatment for 14 d, with the medium being replenished every 3 to 4 d. GUS staining was performed as described (Pecinka et al., 2009), and the number of GUS spots was examined under a stereomicroscope (Leica).
RP-HPLC
DNA samples of zebularine- and mock-treated plants were prepared using the Plant DNA MaxiPrep kit (Qiagen). Two to six micrograms of DNA per sample was treated with DNase I and Nuclease P1 and subsequently with alkaline phosphatase to obtain the free dNs as described previously (Rozhon et al., 2008). The dNs composition was subsequently analyzed by RP-HPLC using a Nucleodur C18ec 100-5 125 × 4.6 mm column and a gradient starting with 98% eluent A (20 mM HCOOH set with NaOH to pH 4.0 in water) and 2% eluent B (20 mM HCOOH set with NaOH to pH 4.0 in 30% acetonitrile) at a flow rate of 0.8 mL/min. The concentration of eluent B was linearly increased to 5% within 7 min and subsequently to 50% within another 13 min. Finally, the initial settings were applied and the column equilibrated for 9.5 min prior injection of the next sample. Fluorescence of deoxyzebularine was detected at an excitation wavelength of 300 nm and an emission wavelength of 370 nm. UV absorbance was recorded at 277 nm.
Accession Numbers
Illumina reads and read counts per gene for all 16 samples are deposited at the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) with the code GSE63355. The following genes names and symbols are associated with this article: ATM (AT3G48190), ATR (AT5G40820), BRCA1 (AT4G21070), CMT3 (AT1G69770), DDM1 (AT5G66750), DRM1 (AT5G15380), DRM2 (AT5G14620), FAS1 (AT1G65470), FAS2 (AT5G64630), GMI1 (AT5G24280), Gypsy-like (AT5G35057), KU70 (AT1G16970), LIG4 (AT5G57160), MORC6 (AT1G19100), MuDr (AT2G15810), LINE1-6 (AT3G28915/AT3TE45385), PARP2 (AT4G02390), RAD3-LIKE (AT1G20750), RAD51 (AT5G20850), RNR1 (AT2G21790), SDC (AT2G17690), SMC6B (AT5G61460), SMR7 (AT3G27630), TE gene (AT1G42050), TSO2 (AT3G27060), and XPF (AT5G41150).
Supplemental Data
Supplemental Figure 1. DNA methylation analyzed regions.
Supplemental Figure 2. DNA sequences of genomic regions analyzed by bisulfite sequencing.
Supplemental Figure 3. Comparison of zebularine, morc6, and ddc phenotypes.
Supplemental Figure 4. Reverse-phase high performance liquid chromatography analysis of zebularine incorporation into genomic DNA.
Supplemental Figure 5. Rosette area of atm, atr, and atm atr under genotoxic stress.
Supplemental Figure 6. Mutant growth under genotoxic stress.
Supplemental Figure 7. 5-Azacytidine treatment causes DNA single-strand breaks.
Supplemental Figure 8. Phenotype of rad5a in zebularine root assay.
Supplemental Table 1. Validation of RNA-sequencing.
Supplemental Table 2. Relative root length (%) of mutants and the wild type treated by mock, zebularine, MMC, and bleocin.
Supplemental Table 3. Relative rosette area (%) of mutants and the wild type treated by mock, zebularine, MMC, and bleocin.
Supplemental Table 4. GUS spot numbers in HR reporter lines 651 and IC9C.
Supplemental Table 5. Number of GUS spots in IC9C homozygous and hemizygous plants after 20 μM zebularine treatment.
Supplemental Table 6. Cycle values after treatment with genotoxic stress.
Supplemental Table 7. PCR primers used in this study.
Supplemental Data Set 1. Genes significantly up- and downregulated after long (5 d) 20 μM zebularine treatment.
Supplemental Data Set 2. Genes significantly up- and downregulated after short (24 h) 10 μM mitomycin C treatment.
Supplemental Data Set 3. mRNA level changes in mock-, zebularine-, and mitomycin C-treated atr mutant.
Supplemental Data Set 4. Gene Ontology terms significantly enriched for the sets of genes up- and downregulated in mock-treated atr plants.
Supplemental Data Source 1. Bisulfite sequencing reads of TSO2 promoter.
Supplemental Data Source 2. Bisulfite sequencing reads of RAD51 promoter.
Supplemental Data Source 3. Bisulfite sequencing reads of LINE1-6.
Supplemental Data Source 4. Bisulfite sequencing reads of SDC promoter.
Acknowledgments
We thank G. Böhmdorfer for GMI1, P. Doerner for cyclin-GUS, H. Puchta for xpf-3, rad5a-2, and atm-1, K. Culligan for atr-2 atm-2, and V. Cavrak and O. Mittelsten Scheid for ddc. We thank B. Eilts, P. Pecinkova, and R. Gentges for technical assistance, M. Koornneef for critical reading of the article, and T. Harrop for language editing. This work was supported by funding from the Max Planck Society to A.P., C.-H.L., and A.F., by DAAD scholarship A/12/77772 to M.D., and by FWF Grant P22734 to B.P.
AUTHOR CONTRIBUTIONS
A.P, A.F., and C.-H.L. designed the research. C.-H.L., A.F., M.D., W.R., A.P., T.B., and B.P. performed experiments and analyzed data. A.P. wrote the article with contribution from all authors.
Glossary
- HR
homologous recombination
- TGS
transcriptional gene silencing
- NPA
nucleoprotein adduct
- NER
nucleotide excision repair
- RT-qPCR
reverse transcription-quantitative PCR
- MMC
mitomycin C
- RP-HPLC
reverse-phase HPLC
- SDSA
synthesis-dependent strand annealing
- SSA
single strand annealing
- CV
cycle value
- 1/2 MS
half-strength Murashige and Skoog
Footnotes
Articles can be viewed online without a subscription.
References
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist C.J., Kastan M.B. (2003). DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421: 499–506. [DOI] [PubMed] [Google Scholar]
- Baubec T., Finke A., Mittelsten Scheid O., Pecinka A. (2014). Meristem-specific expression of epigenetic regulators safeguards transposon silencing in Arabidopsis. EMBO Rep. 15: 446–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baubec T., Pecinka A., Rozhon W., Mittelsten Scheid O. (2009). Effective, homogeneous and transient interference with cytosine methylation in plant genomic DNA by zebularine. Plant J. 57: 542–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Kasus T., Ben-Zvi Z., Marquez V.E., Kelley J.A., Agbaria R. (2005). Metabolic activation of zebularine, a novel DNA methylation inhibitor, in human bladder carcinoma cells. Biochem. Pharmacol. 70: 121–133. [DOI] [PubMed] [Google Scholar]
- Betham B., Shalhout S., Marquez V.E., Bhagwat A.S. (2010). Use of Drosophila deoxynucleoside kinase to study mechanism of toxicity and mutagenicity of deoxycytidine analogs in Escherichia coli. DNA Repair (Amst.) 9: 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhmdorfer G., Schleiffer A., Brunmeir R., Ferscha S., Nizhynska V., Kozák J., Angelis K.J., Kreil D.P., Schweizer D. (2011). GMI1, a structural-maintenance-of-chromosomes-hinge domain-containing protein, is involved in somatic homologous recombination in Arabidopsis. Plant J. 67: 420–433. [DOI] [PubMed] [Google Scholar]
- Britt A.B. (1996). DNA damage and repair in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 75–100. [DOI] [PubMed] [Google Scholar]
- Champion C., Guianvarc’h D., Sénamaud-Beaufort C., Jurkowska R.Z., Jeltsch A., Ponger L., Arimondo P.B., Guieysse-Peugeot A.-L. (2010). Mechanistic insights on the inhibition of c5 DNA methyltransferases by zebularine. PLoS ONE 5: e12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Pikaard C.S. (2005). Transcript profiling in Arabidopsis reveals complex responses to global inhibition of DNA methylation and histone deacetylation. J. Biol. Chem. 280: 796–804. [DOI] [PubMed] [Google Scholar]
- Chiolo I., Minoda A., Colmenares S.U., Polyzos A., Costes S.V., Karpen G.H. (2011). Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144: 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich K.A., Cortez D. (2008). ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9: 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A., You R., Haimovitch-Gal T., Doerner P. (1999). Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20: 503–508. [DOI] [PubMed] [Google Scholar]
- Culligan K., Tissier A., Britt A. (2004). ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16: 1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan K.M., Robertson C.E., Foreman J., Doerner P., Britt A.B. (2006). ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 48: 947–961. [DOI] [PubMed] [Google Scholar]
- De Schutter K., Joubès J., Cools T., Verkest A., Corellou F., Babiychuk E., Van Der Schueren E., Beeckman T., Kushnir S., Inzé D., De Veylder L. (2007). Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19: 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Larkin J.C., Schnittger A. (2011). Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 16: 624–634. [DOI] [PubMed] [Google Scholar]
- Dote H., Cerna D., Burgan W.E., Carter D.J., Cerra M.A., Hollingshead M.G., Camphausen K., Tofilon P.J. (2005). Enhancement of in vitro and in vivo tumor cell radiosensitivity by the DNA methylation inhibitor zebularine. Clin. Cancer Res. 11: 4571–4579. [DOI] [PubMed] [Google Scholar]
- Downey M., Durocher D. (2006). Chromatin and DNA repair: the benefits of relaxation. Nat. Cell Biol. 8: 9–10. [DOI] [PubMed] [Google Scholar]
- Dubest S., Gallego M.E., White C.I. (2002). Role of the AtRad1p endonuclease in homologous recombination in plants. EMBO Rep. 3: 1049–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger G., Liang G., Aparicio A., Jones P.A. (2004). Epigenetics in human disease and prospects for epigenetic therapy. Nature 429: 457–463. [DOI] [PubMed] [Google Scholar]
- Gaillard P.-H.L., Wood R.D. (2001). Activity of individual ERCC1 and XPF subunits in DNA nucleotide excision repair. Nucleic Acids Res. 29: 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V., Bruchet H., Camescasse D., Granier F., Bouchez D., Tissier A. (2003). AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15: 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanin M., Mengiste T., Bogucki A., Paszkowski J. (2000). Elevated levels of intrachromosomal homologous recombination in Arabidopsis overexpressing the MIM gene. Plant J. 24: 183–189. [DOI] [PubMed] [Google Scholar]
- Henderson I.R., Jacobsen S.E. (2008). Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes Dev. 22: 1597–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W.-D., Ehmsen K.T., Liu J. (2010). Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44: 113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V.N., Szybalski W. (1963). A molecular mechanism of mitomycin action: linking of complementary DNA strands. Proc. Natl. Acad. Sci. USA 50: 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.A., Taylor S.M. (1980). Cellular differentiation, cytidine analogs and DNA methylation. Cell 20: 85–93. [DOI] [PubMed] [Google Scholar]
- Kiianitsa K., Maizels N. (2013). A rapid and sensitive assay for DNA-protein covalent complexes in living cells. Nucleic Acids Res. 41: e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik A., Pecinka A., Wendeler E., Reiss B. (2006). The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell 18: 2431–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R.D., Putnam C.D., Myung K. (2002). Maintenance of genome stability in Saccharomyces cerevisiae. Science 297: 552–557. [DOI] [PubMed] [Google Scholar]
- Kozak J., West C.E., White C., da Costa-Nunes J.A., Angelis K.J. (2009). Rapid repair of DNA double strand breaks in Arabidopsis thaliana is dependent on proteins involved in chromosome structure maintenance. DNA Repair (Amst.) 8: 413–419. [DOI] [PubMed] [Google Scholar]
- Kuo H.K., Griffith J.D., Kreuzer K.N. (2007). 5-Azacytidine induced methyltransferase-DNA adducts block DNA replication in vivo. Cancer Res. 67: 8248–8254. [DOI] [PubMed] [Google Scholar]
- Mathieu O., Reinders J., Čaikovski M., Smathajitt C., Paszkowski J. (2007). Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130: 851–862. [DOI] [PubMed] [Google Scholar]
- Melamed-Bessudo C., Levy A.A. (2012). Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: E981–E988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T., Revenkova E., Bechtold N., Paszkowski J. (1999). An SMC-like protein is required for efficient homologous recombination in Arabidopsis. EMBO J. 18: 4505–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissiard G., et al. (2012). MORC family ATPases required for heterochromatin condensation and gene silencing. Science 336: 1448–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J., Lechner E., Dumbliauskas E., Genschik P. (2008). Regulation and role of Arabidopsis CUL4-DDB1A-DDB2 in maintaining genome integrity upon UV stress. PLoS Genet. 4: e1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J., Ries G., Bonhoeffer S., Hohn B. (2004). Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell 16: 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orta M.L., Höglund A., Calderón-Montaño J.M., Domínguez I., Burgos-Morón E., Visnes T., Pastor N., Ström C., López-lázaro M., Helleday T. (2014). The PARP inhibitor Olaparib disrupts base excision repair of 5-aza-2′-deoxycytidine lesions. Nucleic Acids Res. 42: 9108–9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A., Liu C.-H. (2014). Drugs for plant chromosome and chromatin research. Cytogenet. Genome Res. 143: 51–59. [DOI] [PubMed] [Google Scholar]
- Pecinka A., Rosa M., Schikora A., Berlinger M., Hirt H., Luschnig C., Mittelsten Scheid O. (2009). Transgenerational stress memory is not a general response in Arabidopsis. PLoS ONE 4: e5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H., Swoboda P., Gal S., Blot M., Hohn B. (1995). Somatic intrachromosomal homologous recombination events in populations of plant siblings. Plant Mol. Biol. 28: 281–292. [DOI] [PubMed] [Google Scholar]
- Rosa M., Von Harder M., Cigliano R.A., Schlögelhofer P., Mittelsten Scheid O. (2013). The Arabidopsis SWR1 chromatin-remodeling complex is important for DNA repair, somatic recombination, and meiosis. Plant Cell 25: 1990–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhon W., Baubec T., Mayerhofer J., Mittelsten Scheid O., Jonak C. (2008). Rapid quantification of global DNA methylation by isocratic cation exchange high-performance liquid chromatography. Anal. Biochem. 375: 354–360. [DOI] [PubMed] [Google Scholar]
- Salem A.M.H., Nakano T., Takuwa M., Matoba N., Tsuboi T., Terato H., Yamamoto K., Yamada M., Nohmi T., Ide H. (2009). Genetic analysis of repair and damage tolerance mechanisms for DNA-protein cross-links in Escherichia coli. J. Bacteriol. 191: 5657–5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert I., Pecinka A., Meister A., Schubert V., Klatte M., Jovtchev G. (2004). DNA damage processing and aberration formation in plants. Cytogenet. Genome Res. 104: 104–108. [DOI] [PubMed] [Google Scholar]
- Sheridan S., Bishop D.K. (2006). Red-Hed regulation: recombinase Rad51, though capable of playing the leading role, may be relegated to supporting Dmc1 in budding yeast meiosis. Genes Dev. 20: 1685–1691. [DOI] [PubMed] [Google Scholar]
- Stingele J., Schwarz M.S., Bloemeke N., Wolf P.G., Jentsch S. (2014). A DNA-dependent protease involved in DNA-protein crosslink repair. Cell 158: 327–338. [DOI] [PubMed] [Google Scholar]
- Tomasz M. (1995). Mitomycin C: small, fast and deadly (but very selective). Chem. Biol. 2: 575–579. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Pacher M., Dukowic S., Schubert V., Puchta H., Schubert I. (2009). The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. Plant Cell 21: 2688–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Wang W., Marqués J., Mohan R., Saleh A., Durrant W.E., Song J., Dong X. (2013). Salicylic acid activates DNA damage responses to potentiate plant immunity. Mol. Cell 52: 602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P.-M., Lin Y.-T., Shun C.-T., Lin S.-H., Wei T.-T., Chuang S.-H., Wu M.-S., Chen C.-C. (2013). Zebularine inhibits tumorigenesis and stemness of colorectal cancer via p53-dependent endoplasmic reticulum stress. Sci. Rep. 3: 3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama K., Conklin P.A., Huefner N.D., Britt A.B. (2009). Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc. Natl. Acad. Sci. USA 106: 12843–12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., Kim M.Y., Hsieh P.-H., Coleman-Derr D., Eshed-Williams L., Thao K., Harmer S.L., Zilberman D. (2013). The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Cheng X., Connolly B.A., Dickman M.J., Hurd P.J., Hornby D.P. (2002). Zebularine: a novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J. Mol. Biol. 321: 591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]