Genome-wide scanning of large size sequence changes revealed a tandem duplication of a DNA segment that gives rise to cucumbers bearing only female flowers.
Abstract
Structural variations (SVs) represent a major source of genetic diversity. However, the functional impact and formation mechanisms of SVs in plant genomes remain largely unexplored. Here, we report a nucleotide-resolution SV map of cucumber (Cucumis sativas) that comprises 26,788 SVs based on deep resequencing of 115 diverse accessions. The largest proportion of cucumber SVs was formed through nonhomologous end-joining rearrangements, and the occurrence of SVs is closely associated with regions of high nucleotide diversity. These SVs affect the coding regions of 1676 genes, some of which are associated with cucumber domestication. Based on the map, we discovered a copy number variation (CNV) involving four genes that defines the Female (F) locus and gives rise to gynoecious cucumber plants, which bear only female flowers and set fruit at almost every node. The CNV arose from a recent 30.2-kb duplication at a meiotically unstable region, likely via microhomology-mediated break-induced replication. The SV set provides a snapshot of structural variations in plants and will serve as an important resource for exploring genes underlying key traits and for facilitating practical breeding in cucumber.
INTRODUCTION
Genomic structural variations (SVs), including deletions, insertions, inversions, and duplications, represent an important source of genetic diversity (Alkan et al., 2011; Baker, 2012). SVs have been associated with a range of human disorders such as autism (Sebat et al., 2007; Pinto et al., 2010), schizophrenia (Stefansson et al., 2008; McCarthy et al., 2009), and neuroblastoma (Diskin et al., 2009). In plants, SVs are related to numerous phenotypic variations such as leaf size (Horiguchi et al., 2009), fruit shape (Xiao et al., 2008), and aluminum tolerance (Maron et al., 2013). Initially, for different species such as human (Homo sapiens) (Iafrate et al., 2004), rice (Oryza sativa) (Yu et al., 2013), soybean (Glycine max) (McHale et al., 2012), and barley (Hordeum vulgare) (Muñoz-Amatriaín et al., 2013), the majority of SVs were detected by microarray-based comparative genomic hybridization. However, array-based technology can only detect SVs with sequences that are homologous to probes and cannot determine the exact copy number or breakpoint. These disadvantages have hampered the detection of new SVs, as well as the exploration of the mechanism and functional impacts of SVs. Recent advances in next-generation sequencing (NGS) technologies have allowed nucleotide resolution mapping of SVs on a large scale, which further provides necessary information that can be used to explore SV formation mechanisms and to investigate their functional impact, as has been shown in human (Mills et al., 2011; Yang et al., 2013), mouse (Mus musculus) (Yalcin et al., 2011, 2012), and fruit fly (Drosophila melanogaster) (Zichner et al., 2013) studies. In plants, several reports have described SV mapping at the population scale using NGS technologies in species such as Arabidopsis thaliana (Cao et al., 2011) and maize (Zea mays) (Chia et al., 2012). In Arabidopsis, SV deletions were found to affect more than 2000 protein coding genes (Cao et al., 2011), and in maize, SVs were found to be enriched at loci associated with important agronomical traits including leaf development and disease resistance (Chia et al., 2012). Despite these findings, the formation mechanisms of plant SVs and their functional impact still remain largely unexplored (Saxena et al., 2014).
Cucumber (Cucumis sativus) is a major vegetable crop consumed worldwide and has served as a model system for sex determination studies (Tanurdzic and Banks, 2004). We previously reported the genome sequence of cucumber (Huang et al., 2009; Li et al., 2011) as well as a variome map comprising ∼3.3 M single nucleotide polymorphisms (SNPs) (Qi et al., 2013), based on the resequencing data from a core collection of 115 cucumber accessions with an average depth of 18.3X. The core collection was estimated to capture 77.2% of the total genetic diversity of 3342 accessions from a wide geographic distribution (Lv et al., 2012) and could be divided into four groups including one wild group (Indian) and three cultivated groups (East Asian, Eurasian, and Xishuangbanna) (Qi et al., 2013). In this study, we exploited this population-scale genome sequence resource to systematically investigate the occurrence, functional impact, and formation mechanisms of SVs in cucumber genomes. This resulted in the construction of a nucleotide resolution SV map that was used to explore the genetic basis of the Female (F) locus, which is involved in the determination of cucumber sex types.
RESULTS AND DISCUSSION
A Sequence-Based SV Map of Cucumber
The previously described deep sequencing data set (Qi et al., 2013) was used to generate a cucumber SV map by applying read pair (RP; Zeitouni et al., 2010) and split-read (SR; Ye et al., 2009) analyses. A set of filters was then developed to remove false SVs (Supplemental Figure 1), as the false discovery rate (FDR) in SV identification using NGS data is often high for current computational algorithms (Handsaker, et al., 2011; Mills, et al., 2011). Recent findings from the 1000 Genomes Project indicated that among 36 SV call-sets compiled from 15 SV discovery programs, only eight call-sets showed FDR less than 10%, whereas the other 28 call-sets yielded FDR ranging from 13 to 89% (Mills, et al., 2011). The filters we developed can identify potential false SVs caused by library construction artifacts, artifacts of short read mapping algorithms, incomplete reference genome assemblies, or genome assembly errors (see Methods for details) and thus substantially improve the accuracy of our SV detection.
Finally, relative to the reference genome sequence of the ‘Chinese long’ inbred line 9930, we identified a total of 19,168 deletions ranging from 50 to 16,600 bp, 7337 insertions from 50 to 400 bp, 205 tandem duplications from 340 to 155,800 bp, and 78 inversions from 180 to 40,500 bp, which jointly affect ∼12 Mb of sequences (Table 1; Supplemental Figure 2 and Supplemental Data Set 1). The genotypes of more than 70% of these SVs can be determined in over 80 of the 115 accessions (Supplemental Figure 3). Relative to the reference genome, more than 65% of the SVs occurred in at least two cucumber accessions (Supplemental Figure 4), and more than 60% of the identified SVs had inferred breakpoints, resulting in a high-resolution sequence-based map of SVs in cucumber genomes (Supplemental Data Set 1).
Table 1. Summary of Cucumber SVs and Their Functional Impact.
| Deletions | Insertions | Duplications | Inversions | Total | |
|---|---|---|---|---|---|
| No. | 19,168 | 7,337 | 205 | 78 | 26,788 |
| Size (bp) | 6,501,467 | 679,902 | 4,212,813 | 325,747 | 11,751,414 |
| Number of SVs overlapping with protein coding genesa | |||||
| Full CDS overlap | 112 (130)b | 0 (0) | 134 (509) | 15 (27) | 261 (666) |
| Partial CDS overlap | 619 (622) | 226 (217) | 117 (149) | 17 (22) | 979 (1,010) |
| UTR overlapc | 266 (262) | 90 (90) | 7 (7) | 1 (1) | 364 (360) |
| Intron overlapc | 2,409 (1,979) | 863 (775) | 3 (3) | 2 (3) | 3,277 (2,760) |
| Total | 3,379 (2,952) | 1,175 (1,076) | 171 (668) | 27 (53) | |
For insertions, we further extracted the inserted sequences relative to the reference genome by performing de novo assembly of paired-end reads from each cucumber accession and comparing the assembled contigs to the reference sequences. Approximately 40% of insertion events had inserted sequences extracted in at least one cucumber line. All of these sequences are provided in Supplemental Data Set 2. It is worth noting that there were much fewer SVs identified in cucumber than in Arabidopsis (Cao et al., 2011) and maize (Chia et al., 2012). Such a difference was also observed between human and animals, with ∼28,000 SVs identified in human genomes (Mills et al., 2011) and more than 280,000 identified in mouse (Yalcin et al., 2011). The major factors contributing to this difference include the degree of genetic diversity within the population and the frequency of active transposable elements in the genomes. Cucumber has a lower level of genetic diversity compared with Arabidopsis and maize (Cao et al., 2011; Chia et al., 2012; Qi et al., 2013) and a much lower frequency of active transposable elements compared with maize (Huang et al., 2009; Schnable et al., 2009). Another reason that much fewer SVs were identified in cucumber than in Arabidopsis is that in Arabidopsis, SVs >20 bp in length were kept, while in this study, only SVs with length >50 bp were kept.
To evaluate the quality of our SV calling, we performed PCR analyses of 500 randomly selected SVs, including 400 deletions and 100 insertions, in four cucumber lines (the reference line 9930 and the nonreference lines CG6663, CG9207, and CG0002). Based on the PCR data, we estimated that the FDRs for deletion and insertion calling were 5.2 and 9.4%, respectively, and the FDRs for deletion and insertion genotyping were 4 and 3.9%, respectively (Supplemental Data Set 3). This SV data set is of sufficient quality for downstream population genetic analyses and genome-wide association studies (GWASs).
Mechanisms of SV Formation
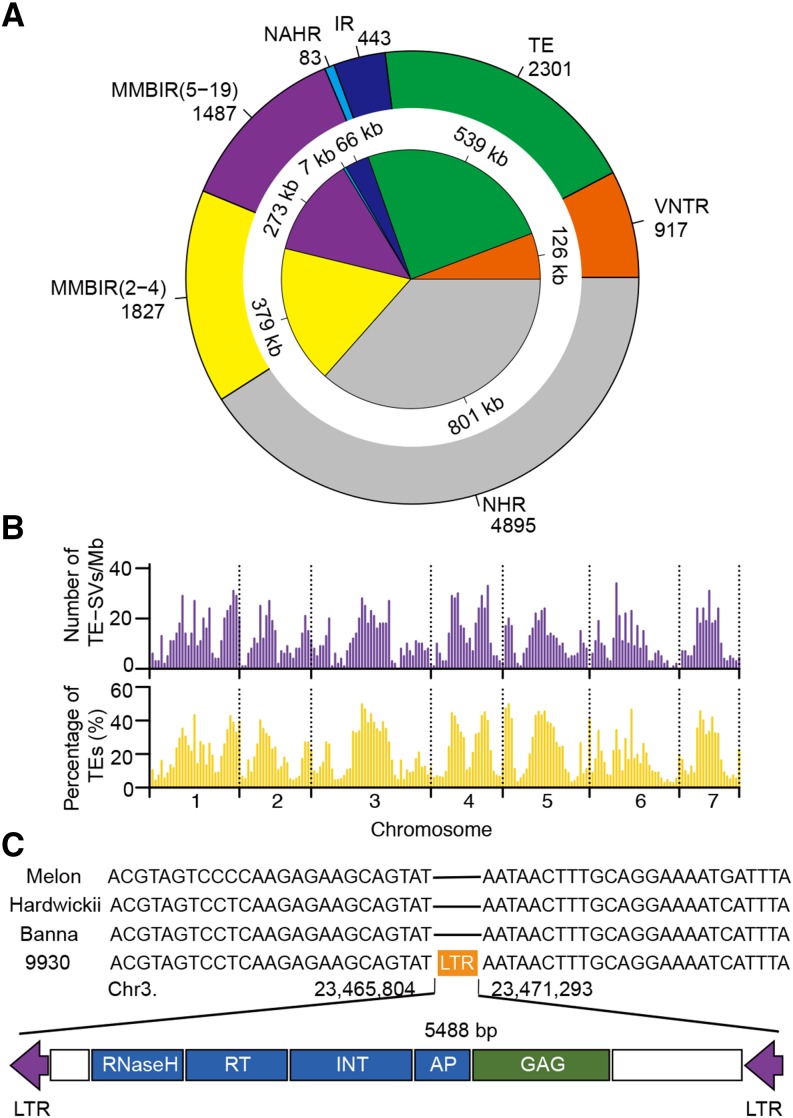
A total of 11,891 deletions with base-pair resolution breakpoint information were used to infer their formation mechanisms (Figure 1A). As in human (Mills et al., 2011; Zichner et al., 2013), the largest category of deletions arose from nonhomologous rearrangement events, which are considered to be associated with DNA double-strand repair by nonhomologous end joining. However, the nonallelic homologous recombination events that are abundant (23%) in the human genome (Mills et al., 2011) contribute a very small portion of the deletions in the cucumber genome (0.7%). This may be due to the fact that the cucumber genome lacks recent bursts of interspersed repeats (e.g., Alu elements) and segmental duplications (Kim et al., 2008). In addition, the contribution of transposable element (TE)-related events in cucumber (19.2%) is substantially more than that found in human (4.2%) and fruit fly (9.0%) genomes (Mills et al., 2011; Zichner et al., 2013), possibly due to the more active transposable elements in the cucumber genome. The distribution of TE-derived deletions across the cucumber genome was highly consistent with the occurrence of TEs (Figure 1B), suggesting that TE enriched regions represent an important source of SVs. For example, we predicted 12 full-length long terminal repeat (LTR) retrotransposons for deletions with lengths >1 kb (Supplemental Table 1); one of these (SV3G00315300) is absent in the wild cucumber and melon genomes but is present in East Asian and Eurasian cucumbers, representing a recent transposition event (Figure 1C).
Figure 1.
SV Formation Mechanisms in the Cucumber Genome.
(A) Relative contribution of different SV formation mechanisms inferred from deletions with nucleotide resolution. Outer circle: number of deletions derived from different mechanisms. Inner circle: cumulative size of the deletions derived from different mechanisms. NAHR, nonallelic homologous recombination; MMBIR (5-19), MMBIR with homologous fragments from 5 to 19 bp; MMBIR (2-4), 2- to 4-bp identical flanking nucleotides at the two sides of the breakpoint; IR, inverted repeat; NHR, nonhomologous rearrangement.
(B) Distribution of TE-related deletions and TEs across the seven cucumber chromosomes.
(C) Example of a deleted full-length LTR retrotransposon in the genomes of Indian and Xishuangbanna cucumbers, as well as melon, compared with the 9930 reference genome.
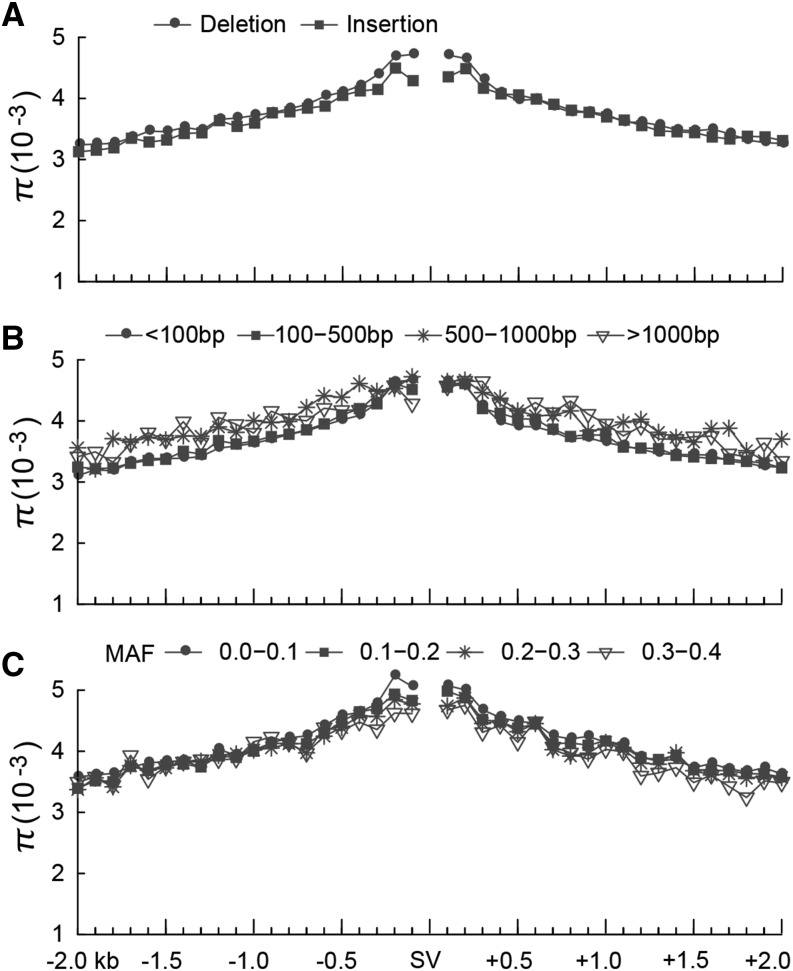
Next, we investigated the relationship between nucleotide diversity and SV occurrence. The nucleotide diversity was markedly elevated in the vicinity of indel events and decreased with increasing distance from the SVs (Figure 2A). This observation is consistent with the indel-associated mutation hypothesis (Tian et al., 2008; Hollister et al., 2010; Hodgkinson and Eyre-Walker, 2011; Jovelin and Cutter, 2013). Previous studies on indel-associated mutation have been limited to relatively small indels (<300 bp) (Tian et al., 2008; Hollister et al., 2010; Hodgkinson and Eyre-Walker, 2011; Jovelin and Cutter, 2013). Here, we classified deletions into four categories according to their length (<100 bp, 100 to 500 bp, 500 to 1000 bp, and >1000 bp). We found that nucleotide diversity patterns surrounding these four different size classes of deletions were similar (Figure 2B), suggesting that the diversity is independent of the length of deletions. It has been proposed that indels are mutagenic and therefore promote higher nucleotide diversity in their vicinity (Tian et al., 2008; Hollister et al., 2010; Jovelin and Cutter, 2013), a proposition that assumes that the heterozygosity of an indel increases the rate of point mutations. Were this to be the case, we would expect that SVs with higher minor allele frequency (MAF) in the population would result in higher nucleotide diversity in their vicinity. However, in this study, we found that the nucleotide diversity was similar in the flanking regions of SVs with different MAFs (Figure 2C), implying that SVs in cucumber might not be mutagenic agents, which is similar to a previous report (McDonald et al., 2011). It is worth noting that the higher nucleotide diversity in the vicinity of SVs might be caused to some extent by the potentially high rate of false SNP calls due to inaccurate read mapping near SVs.
Figure 2.
Relationship between SVs and Nucleotide Diversity (π).
Distribution of nucleotide diversity of the 2-kb upstream and the 2-kb downstream regions (bin size: 100 bp) of deletions, insertions, inversions, and duplications (A), SVs of different sizes (<100 bp, 100 to 500 bp, 500 to 1000 bp, and >1000 bp) (B), and SVs with different MAF (<0.1, 0.1 to 0.2, 0.2 to 0.3, and 0.3 to 0.4) (C).
Functional Impact of Cucumber SVs
To assess the functional impact of cucumber SVs, we investigated their relationship with annotated protein coding genes. We found that 1240 SVs affected at least a portion of the coding sequences (CDS) of 1676 genes (752 deletions, 217 insertions, 658 tandem duplications, and 49 inversions; Table 1; Supplemental Data Sets 4 to 7). Of these, the CDSs of 130 genes were entirely eliminated by deletions, 509 were completely duplicated, and 27 were located entirely within the inverted genomic regions. Functional category enrichment analyses indicated that of the genes affected by SVs through deletions in their CDS, those related to histone methylation and abiotic stress responses were significantly enriched, whereas of the insertion-affected genes, those related to histone acetylation were highly enriched (Supplemental Data Set 8). In addition, we found that tandemly duplicated genes were significantly enriched in those involved in the reproductive process and plant responses to biotic and abiotic stresses, while genes located within the inversed genomic regions were highly enriched in those playing a role in plant responses to chemical stimulus (e.g., zinc ion) (Supplemental Data Set 8).
The three groups of cultivated cucumbers (East Asian, Eurasian, and Xishuangbanna) were domesticated from the Indian group (Qi et al., 2013). To identify potential SVs associated with domestication, we searched for those that were highly differentiated between the Indian group (n = 30) and the three cultivated groups (n = 85) (Supplemental Data Set 9). A total of 943 SVs, including 44 affecting the CDS of 52 genes, were found to be highly divergent between the three cultivated groups and the wild Indian group. More than 70% (675) of these SVs were largely fixed in cultivated groups, an indication that they might have undergone selection during domestication. Among them, 560 do not reside in the previously identified regions that have undergone selective sweeps during domestication (Qi et al., 2013), 31.4% (176) of which are located in regions with higher ratios (≥3) of genetic diversity between wild and cultivated groups and may serve as complementary candidates for the exploration of domestication. For example, a 183-bp deletion (SV2G00185800) was only found in six wild accessions (CG0001, CG0002, CG0004, CG0005, CG0017, and CG0020) that form a most distant cluster relative to the cultivated cucumbers in the phylogenetic tree (Qi et al., 2013). This deletion affects the untranslated region of a kinesin gene, Csa2G237130. Kinesin has been associated with organ size through regulating cell division (Müller et al., 2006). Interestingly, Csa2G237130 is highly expressed in roots, stems, leaves, and fruits of cultivar 9930 (Li et al., 2011) and is not expressed in counterparts of the wild accession CG0002 (Supplemental Figure 5), implying that this deletion might be associated with the larger sizes of leaves and fruits in cultivated cucumbers. Therefore, it is likely that this deletion changed the expression pattern of Csa2G237130, a candidate domestication gene.
To evaluate the utility of the SV data set in identifying agronomic traits, we performed GWAS using the SV set for the tuberculate fruit trait, which is characterized by epidermal spines and tubercules on the fruits. Seven signals were significantly associated with the trait (P < 1e−8) (Supplemental Figure 6), including one that resided within the genetically mapped tu region on chromosome 5, which has previously been reported to determine the formation of fruit tubercules (Zhang et al., 2010a). This SV involves the deletion of the entire tu gene (Csa5G577350, encoding a C2H2 type zinc finger transcription factor) (Yang et al., 2014) in five Eurasian cucumber lines (Supplemental Figure 6). In addition, our analysis indicated that this deletion is caused by a microhomolog (AATT) at the breakpoints (Supplemental Figure 6), elucidating the formation of tu, a null allele explored by breeders for developing cultivars for European and US supermarkets where cucumbers are generally packed in plastic for transportation and longer shelf-life (and thus must be spineless).
Genesis of the F Locus
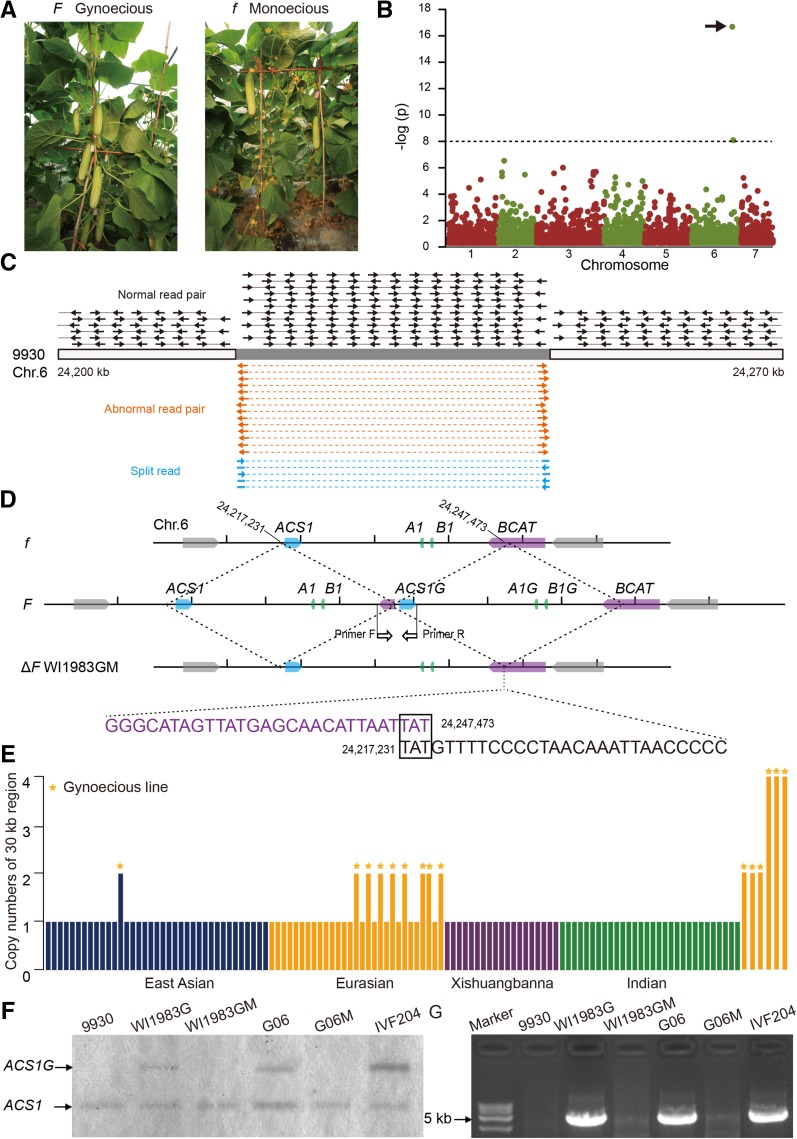
Most cucumbers are monoecious, bearing male and female flowers on a single plant, with the female flowers developing into fruits (Figure 3A). In contrast, gynoecious cucumbers bear only female flowers and can set fruit at nearly each node. This trait was exploited by Dutch breeders to develop gynoecious cultivars specifically suited for resource-rich soil-less greenhouse cultivation supporting continuous fruit set. As a result, the fruit number per square meter in the greenhouse has increased from ∼53 in 1973 (Anonymous, 1973), when monoecious cucumbers were cultivated, to 200 in 2012 (Vermeulen, 2012). It was previously reported that gynoecy is conferred by the Mendelian locus F (Shifriss, 1961) and that the F locus is genetically associated with a new copy (ACS1G) of an aminocyclopropane-1-carboxylic acid synthase gene, ACS1 (Trebitsh et al., 1997; Knopf and Trebitsh, 2006), an ethylene synthesis gene. However, no functional evidence was provided and its exact genomic location and formation mechanism were previously unclear.
Figure 3.
A 30.2-kb Duplicated Sequence Is Responsible for the Gynoecious Cucumber Phenotype.
(A) Phenotypes of gynoecious (WI1983G) and monoecious (WI1983GM) cucumbers. WI1983GM is a mutant of WI1983G with a predominance of male flowers.
(B) Manhattan plot of GWAS using the SV set for the gynoecy phenotype. The arrow points to the SV that is significantly associated with gynoecy.
(C) Schematic illustration of the mapped reads from gynoecious accession (CG5278) on the 9930 reference genome. Read depth is represented by reads marked by black arrows, abnormally mapped RPs are indicated in orange, and SRs are indicated in light blue.
(D) Structural organization of the 30.2-kb duplicated region in gynoecious and monoecious lines. ACS1, Csa6G496450; A1, Csa6G496950; B1, Csa6G496960; BCAT, Csa6G496970.
(E) Copy numbers of the 30.2-kb region and geographical distribution of the core collection and six additional gynoecious lines (GCA07, GCA09, HAU106, IVF204, HAU107, and SJ08). The 115 accessions were ordered as listed in Supplemental Data Set 1. The copy numbers of the duplicated 30.2-kb region for the cucumbers from Indian, Xishuangbanna, Eurasian, and East Asian groups are denoted as green, red, orange, and blue bars, respectively. The six additional lines (GCA07, GCA09, HAU106, IVF204, HAU107, and SJ08) from Europe are represented by the orange bars on the far right of the histogram.
(F) DNA gel blot analysis of the duplicated region of 9930 (monoecious), WI1983G (gynoecious), WI1983GM (mutant of WI1983G, monoecious), G06 (gynoecious), G06M (mutant of G06, monoecious), and IVF204 (gynoecious).
(G) PCR validation of the duplication.
The cucumber population used for SV discovery and genotyping consists of nine gynoecious and 106 monoecious accessions, which enabled us to perform GWAS to search for the underlying variant responsible for cucumber gynoecy. We observed a significant association (P < 1e−16) of a 30.2-kb duplication with gynoecy (Figure 3B). This signal was also supported by the nearby strongly associated SNPs identified from the GWAS analysis using the SNP data set (Supplemental Figure 7). Evidence derived from the analysis of abnormally mapped RPs, SRs, and read depths (RDs) strongly supports this copy number variation (CNV) (Table 2, Figure 3C). The duplicated region contains the ACS1 (Csa6G496450) gene as well as Csa6G496950 (a gene of unknown function), Csa6G496960 (encoding a truncated MYB transcription factor), and part of Csa6G496970 (encoding a branched-chain amino acid aminotransferase) (Figure 3D). The previously reported differentially expressed genes between female and male flowers, such as auxin-related and short-chain dehydrogenase or reductase genes related to the development of unisexual flowers (Huang et al., 2009; Guo et al., 2010), were not found in this region; thus, they are likely downstream flowering-related genes. To further confirm the association between the CNV and gynoecy, we resequenced six additional gynoecious lines (GCA07, GCA09, HAU106, IVF204, HAU107, and SJ08). All six lines possess the duplication, and three of them (IVF204, HAU107, and SJ08) were estimated to contain four copies of the 30.2-kb region (Figure 3E) based on RD analysis. DNA gel blot analysis also supported the higher copy number of the 30.2-kb region in IVF204 than in G06 and WI1983G, which contain two copies (Figure 3F).
Table 2. Summary of Sequence Evidence Supporting the CNV (F) and the Variation between Copies.
| Accession | RPsa | SRsa | RDsa | CNb | Heteroc | |
|---|---|---|---|---|---|---|
| Monoecious | CG1322 | 0 | 0 | 1 | 1 | – |
| CG8724 | 0 | 0 | 1 | 1 | – | |
| CG4357 | 0 | 0 | 1 | 1 | – | |
| CG4182 | 0 | 0 | 1 | 1 | – | |
| CG1031d | 0 | 0 | 1 | 1 | – | |
| Gynoecious | CG3007 | 16 | 7 | 2 | 2 | 1 |
| CG5071 | 17 | 4 | 2 | 2 | 13 | |
| CG5278 | 21 | 5 | 2 | 2 | 2 | |
| CG5420 | 8 | 3 | 1 | 2 | 2 | |
| CG5786 | 19 | 3 | 2 | 2 | 16 | |
| CG5790 | 6 | 3 | 1 | 2 | 0 | |
| CG6542 | 8 | 7 | 1 | 2 | 6 | |
| CG6578 | 6 | 4 | 1 | 2 | 1 | |
| CG6663 | 7 | 1 | 2 | 2 | 13 | |
| SJ08 | 79 | 15 | 4 | 4 | 5 | |
| HAU107 | 113 | 7 | 4 | 4 | 8 | |
| IVF204 | 59 | 31 | 4 | 4 | 7 | |
| HAU106 | 12 | 0 | 2 | 2 | 2 | |
| GCA09 | 18 | 1 | 2 | 2 | 5 | |
| GCA07 | 17 | 1 | 2 | 2 | 4 |
Number of RPs or SRs that support the tandem duplication of the 30.2-kb region. RDs: the estimated copy numbers based on RDs.
Number of heterozygous SNP sites within the 30.2-kb region. Assuming each accession is homozygous, the heterozygous sites should have resulted from the new copy.
CG1031 was clustered together with all gynoecious cucumbers in Supplemental Figure 8.
Two naturally occurring mutants, WI1983GM and G06M, derived from the gynoecious lines WI1983G and G06, respectively, display monoecious phenotypes. DNA gel blot and PCR analyses indicated that both mutants have lost the 30.2-kb duplicated region (Figures 3F and 3G), providing convincing evidence that this duplication is responsible for gynoecy. In addition, resequencing the line WI1983GM further confirmed the loss of the entire duplicated region (Figure 3D). Taken together, our data strongly support the conclusion that the CNV defines the F locus. It appears that this genomic region is meiotically unstable and prone to generate or eliminate additional copies. We further explored the formation mechanism of the CNV and found a 3-bp microhomolog (TAT) at the breakpoint of this duplication (Figure 3C). This suggests that, as with Pelizaeus-Merzbacher disease, a central nervous system disorder in humans (Lee et al., 2007), microhomology-mediated break-induced replication (MMBIR) likely gave rise to the F locus.
It was previously reported that the source of cucumber gynoecium was a group of Japanese and Korean accessions (Shifriss, 1961). In the core collection, one gynoecious accession (CG3007) is from Japan, and so to better define the origin of this locus, we performed a phylogenetic analysis of the 115 cucumber accessions based on the alignments of the variants within the 30.2-kb duplicated region (Supplemental File 1). Nine gynoecious cucumber accessions were clustered together with one monoecious accession (CG1031) from East Asia (Table 2; Supplemental Figure 8). This indicates that the gynoecy trait may have originated from the same cucumber (likely an Asian cultivar) and then radiated during cultivation to other parts of the world. This also indicates that CG1031 might be a close relative of the ancestor of gynoecious cucumbers. In addition, to determine the identity between the new and original copies, we explored the heterozygous SNPs within the 30.2-kb region by mapping reads from gynoecious accessions. Very small numbers of heterozygous sites (0 to 16) were observed in nine gynoecious accessions (Table 2), suggesting that the different copies of the duplication are almost identical. Therefore, the genesis of the F locus is likely a very recent event.
Notably, eight of nine gynoecious accessions in the core collection, as well as the six additional resequenced gynoecious lines, were from the Eurasian group. We note that this phenomenon appears to be associated with resource-rich soil-less greenhouse cultivation in Europe, which supports fruit setting at each node; therefore, the gynoecy trait was selected to ensure high yield. In contrast, in most of East Asia, gynoecious cucumbers are not cultivated because the resource-limited cultivation practice does not support fruit set at each node.
METHODS
SV Discovery and Genotyping
The genome resequencing data from the 115 cucumber (Cucumis sativas) accessions (NCBI SRA accession: SRA056480) reported in our previous study (Qi et al., 2013) were used for SV detection. The paired-end reads were mapped to the ‘Chinese long’ 9930 cucumber reference genome using BWA (version 0.6.2) (Li and Durbin, 2009). Only one of the duplicated paired-end reads was kept to minimize the artifacts of PCR amplification, and only reads uniquely mapped (having one single best hit) to the genome were used. Based on the alignments, discordant paired-end reads were identified and used to detect deletions, insertions, inversions, and tandem duplications using the SVDetect program (Zeitouni et al., 2010) (Supplemental Figure 1A). In addition, SR information was also generated and used to identify deletion events with Pindel (Ye et al., 2009). For SVDetect, we set the minimum number of pairs in a cluster to three and the minimal number of sigma fold for the insert size filtering to six. For Pindel, we set the minimum number of reads to support the deletion to three and the minimum number of match bases of a read to 30. We then developed a set of filters (http://bioinfo.bti.cornell.edu/tool/SVFilter/) to further remove potential false SVs identified by SVDetect and/or Pindel (Supplemental Figure 1). First, we removed SVs spanning regions of the reference genome that contain gaps. Such SVs would span at least two contigs or even two scaffolds, and the estimated size of a gap between two contigs or scaffolds is usually not accurate. Second, we removed SVs that were contradictory to a substantial number (≥3) of normally mapped reads. These SVs might be derived from wrong read mapping or from abnormally mapped reads from chimeric inserts in the library. Third, we removed SVs that showed SNVs between normally and abnormally mapped reads within the anchoring windows, as those abnormal reads could be misaligned to the reference genome. Fourth, we removed deletions spanning regions on the reference genome of which a large portion (>80%) was covered by normally mapped reads. Finally, we used RD information to select tandem duplications derived from the analysis of the abnormally mapped RPs. The copy numbers of duplications were estimated based on RD information using the cn.MOPS package (Klambauer et al., 2012).
Subsequently, we combined SVs that were detected in each cucumber accession and assigned genotypes (presence, absence, and undetermined) of structural polymorphisms across all 115 accessions. The genotype of an SV was assigned as undetermined if insufficient reads covered the SV region. Finally, to remove redundancy, deletions found by the two approaches (RP and SR, which complement each other) (Supplemental Figure 9) were merged if they overlapped by at least 80%. In this study, only SVs longer than 50 bp were considered.
We then performed de novo assembly to identify sequences that were in the insertions relative to the reference genome. Paired-end reads from each of the 115 cucumber lines were assembled into contigs using SOAPdenovo (Li et al., 2010). The assembled contigs longer than 100 bp from each cucumber accession were compared with the reference sequences surrounding the insertion sites using BLAST. The corresponding inserted sequences relative to the reference genome were then extracted based on the BLAST results.
Experimental Validation of Cucumber SVs
To evaluate the accuracy of our SV calling by PCR, a total of 400 deletion and 100 insertion loci were randomly selected from four cucumber accessions (East Asian, 9930; Eurasian, CG6663; Xishuangbanna, CG9207; India, CG0002). The lengths of the selected deletions ranged from 50 to ∼13 kb, and the insertions from 50 to 400 bp. For SVs <2 kb, PCR was performed according to the user manual of Tiangen 2× Taq Master Mix (Tiangen KT201). For SVs ≥2 kb, PCR was performed according to the user manual of KOD-FX (Toyobo KFX-101). PCR products were fractionated on a 1.5% agarose gel containing ethidium bromide, and the sizes of the amplified fragments were determined and used to infer the presence/absence of insertions or deletions. To identify potential PCR errors such as those caused by nonspecific amplifications, we first compared genomic regions of the 400 deletions and 100 insertions between 9930 and CG0002 to infer their genotypes in CG0002 and then removed those (27 deletions and two insertions; Supplemental Data Set 3) that had different genotypes in CG0002 inferred from PCR analyses.
Inferring Mechanisms of SV Formation
In this study, we only used deletions with nucleotide resolution breakpoints to infer their formation mechanisms. First, these SVs were related to annotated TEs and variable number of tandem repeats (VNTRs) in the cucumber genome (Li et al., 2011). If more than 80% of a certain SV overlapped with a TE or VNTR, this SV was considered to be derived from the corresponding TE or VNTR. Second, we extracted the 100-bp flanking sequences from both sides of each breakpoint and then aligned the two sequences using BLAST. If the two flanking sequences contained homologous blocks with a minimum length of 20 bp and >85% identity, the SV was considered to be derived from a nonallelic homologous recombination event when the alignment was in the same orientation or an inverted repeat event when in the opposite orientation. If the homologous fragments were from 5 to 19 bp, they were designated as MMBIR 5-19 events. If the flanking nucleotides with the length from 2 to 4 bp of the two sides at the breakpoint were identical, they were designated as MMBIR 2-4 events. Finally, all of the remaining SVs were classified as nonhomologous rearrangement events.
Prediction of Full-Length LTRs for TE-Related Deletions
The underlying sequences of TE-related deletions with length >1 kb were extracted from the 9930 genome and their structures were analyzed with LTR_FINDER (Xu and Wang, 2007) to identify full-length LTRs. Output score threshold was set to 6.0, and the maximum LTR length was set to 20 kb. The ps_scan program was used to identify integrase and RNaseH. The tRNA database of Arabidopsis thaliana was used to predict primer binding sites.
Identification of Highly Divergent SVs between Cultivated and Wild Groups
To identify highly divergent SVs between the three cultivated groups and the wild group, the numbers of lines with or without the specific genotypes for each SV were compared between pairs of groups using Fisher's exact tests. The resulting raw P values were adjusted for multiple tests using FDR (Benjamini and Hochberg, 1995). SVs with FDR < 0.01 were identified as significantly divergent SVs. In addition, an SV was regarded as largely fixed in cultivated groups if more than 95% of the cultivated accessions had the same genotype for this SV.
GWAS Using the Cucumber SV Set
SVs with MAF > 0.05 were used to perform a GWAS for the tuberculate fruit and gynoecy traits in the 115 cucumber accessions. The analysis was performed using TASSEL 4.0 (Bradbury et al., 2007) with the inferred population structure based on SNPs (Qi et al., 2013) and the compressed mixture line model (Zhang et al., 2010b).
Experimental Validation of the Duplicated Region in Gynoecious Cucumbers
Unique primers (forward 5′-CCACGGTCAAGATTCCTCCTAC-3′ and reverse 5′-GCTGCGTCTGGATTTTTTGT-3′) were used to generate DIG labeled DNA probes for DNA gel blot analysis according to the manufacturer’s instructions (Roche). The probe hybridizes to the breakpoint region of the duplication. Genomic DNA was extracted from young cucumber leaves using the CTAB method and further purified using spin columns (Tiangen DP305). Five micrograms of genomic DNA was digested with 150 units of MspI or MboI (NEB) overnight at 37°C and then fractionated on 0.8% agarose gels in fresh 0.5× TBE buffer at 80 V for 5 to 8 h. Size-fractionated DNA was transferred to a Hybond-N+ nylon membrane in 20× SSC (3 M sodium chloride and 300 mM trisodium citrate, adjusted to pH 7.0 with HCl) for 6 h and immobilized by baking the Hybond-N+ nylon membrane at 80°C for 2 h. Membranes were prehybridized in the DIG EASY Hyb buffer (10 mL/100 cm2) of the DIG High Prime DNA Labeling and Detection Starter Kit I (Roche) at 52°C for 2 h. The DIG-labeled DNA probe was denatured by boiling for 10 min followed by rapid cooling on ice. The denatured DIG-labeled DNA probe was then added to preheated DIG EASY Hyb buffer to a concentration of 25 ng/mL and hybridized with the membrane overnight. After hybridization, the membrane was briefly rinsed in washing buffer, then in low-stringency buffer (2× SSC and 0.1% SDS) twice for 15 min each, and in high stringency buffer (0.5× SSC and 0.1% SDS) twice for 15 min each. Immunological detection using antidigoxigenin-AP was performed according to the instructions provided by the DIG High Prime DNA Labeling and Detection Starter Kit I.
For PCR validation, primer sequences spanning the breakpoint of the duplicated region were designed. The positions of primer sequences in the duplicated region (primer F, 5′-TTACTTCTCTTCAAACACACACATCTAATT-3′; primer R, 5′-GAAAAATGTTAATGATTTGGGGGTT-3′) are shown in Figure 3D. Genomic DNA was extracted from young cucumber leaves with the DNasecure Plant Kit (Tiangen DP320). PCR reactions were performed according to the user manual of KOD-FX (Toyobo KFX-101).
Phylogenetic Analysis of the 30.2-kb CNV Region
SNP genotyping data within the 30.2-kb CNV region of the 115 accessions were extracted from the previously released SNP data set (Qi et al., 2013). The alignments were generated based on the positions of these SNPs on the chromosome using our own script and used to construct the phylogenetic tree. SNPs with missing genotype data in more than 10 accessions were excluded in the analysis. The tree was constructed with PhyML (Guindon and Gascuel, 2003) using the maximum likelihood method. Support for each branch was calculated based on 1000 bootstrap replicates.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries or Cucurbit Genomics Database (icugi.org) under the following accession numbers: NCBI SRA accession (SRA056480), Csa2G237130, Csa5G577350, Cs-ACS1 (DQ839410.1 or Csa6G496450), Csa6G496950, Csa6G496960, and Csa6G496970.
Supplemental Data
Supplemental Figure 1. SV discovery, filtering, and genotyping.
Supplemental Figure 2. Size distribution of identified SVs.
Supplemental Figure 3. Genotyping summary of cucumber SVs in the 115 accessions.
Supplemental Figure 4. Distribution of different numbers of accessions for which a structural variation was detected compared with the reference genome.
Supplemental Figure 5. Expression profile of Csa2G237130 in cultivated cucumber line 9930 and wild line CG0002.
Supplemental Figure 6. SVs associated with tuberculate fruit in cucumber.
Supplemental Figure 7. Manhattan plot of GWAS using the SNP set for the gynoecy phenotype.
Supplemental Figure 8. Phylogenetic relationships of the 115 cucumber accessions based on the SNPs within the 30.2-kb duplicated region (F locus).
Supplemental Figure 9. Size distribution of deletions detected by read pair and split read approaches.
Supplemental Table 1. SVs predicted to be full-length LTR retrotransposons.
Supplemental Data Set 1. List of cucumber SVs and SV genotyping of 115 cucumber accessions.
Supplemental Data Set 2. Inserted sequences extracted by de novo assembly relative to the reference genome.
Supplemental Data Set 3. PCR validation of selected SVs.
Supplemental Data Set 4. Genes affected by deletions.
Supplemental Data Set 5. Genes affected by insertions.
Supplemental Data Set 6. Genes affected by duplications.
Supplemental Data Set 7. Genes affected by inversions.
Supplemental Data Set 8. Enriched Gene Ontology terms for genes with coding regions affected by SVs.
Supplemental Data Set 9. Highly divergent SVs between the three cultivated groups and the wild Indian group.
Supplemental File 1. Alignments of the SNPs within the 30.2-kb duplicated region for 115 cucumber lines in PHYLIP format.
Supplementary Material
Acknowledgments
We thank Jim Giovannoni and Jocelyn Rose for critical reading of the article and Shunong Bai and Zhihong Xu (Peking University) for useful discussion. This work was supported by funding from the National Natural Science Foundation of China (31225025 to S.H., 31471871 to H.C., and 31322047 to Z.Z.), the National Program on Key Basic Research Projects in China (the 973 Program; 2012CB113900), the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-IVFCAAS), the United States National Science Foundation (IOS-0923312 and IOS-1313887 to Z.F.), and Multidisciplinary Research Partnership “Bioinformatics: from nucleotides to networks” of Ghent University (01MR0310W to Y.V.d.P.).
AUTHOR CONTRIBUTIONS
S.H., Z.F., and Z.Z. conceived and designed the research. Z.Z., L.M., F.B., G.L., J.S., H.S., C.J., R.B., R.L., Y.V.d.P., E.J., Z.F., and S.H. conducted the data analysis. H.C., F.B., G.L., J.S., S.L., J.P., and R.C. performed biology experiments. Z.Z., S.H., and Z.F. wrote the article. Y.V.d.P., Z.F., L.M., and S.H. revised the article.
Glossary
- SV
structural variation
- NGS
next-generation sequencing
- SNP
single nucleotide polymorphism
- FDR
false discovery rate
- GWAS
genome-wide association study
- TE
transposable element
- LTR
long terminal repeat
- MAF
minor allele frequency
- CDS
coding sequences
- RP
read pair
- RD
read depth
- CNV
copy number variation
- MMBIR
microhomology-mediated break-induced replication
- VNTR
variable number of tandem repeat
- SR
split read
References
- Alkan C., Coe B.P., Eichler E.E. (2011). Genome structural variation discovery and genotyping. Nat. Rev. Genet. 12: 363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous (1973). Vademecum voor de Glastuinbouw: Glasgroenten, Glasbloemen, Champignons. (The Hague, The Netherlands: Landbouw-Economisch Instituut; ). [Google Scholar]
- Baker M. (2012). Structural variation: the genome’s hidden architecture. Nat. Methods 9: 133–137. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate-a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Bradbury P.J., Zhang Z., Kroon D.E., Casstevens T.M., Ramdoss Y., Buckler E.S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. [DOI] [PubMed] [Google Scholar]
- Cao J., et al. (2011). Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43: 956–963. [DOI] [PubMed] [Google Scholar]
- Chia J.-M., et al. (2012). Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 44: 803–807. [DOI] [PubMed] [Google Scholar]
- Diskin S.J., et al. (2009). Copy number variation at 1q21.1 associated with neuroblastoma. Nature 459: 987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Guo S., Zheng Y., Joung J.G., Liu S., Zhang Z., Crasta O.R., Sobral B.W., Xu Y., Huang S., Fei Z. (2010). Transcriptome sequencing and comparative analysis of cucumber flowers with different sex types. BMC Genomics 11: 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handsaker R.E., Korn J.M., Nemesh J., McCarroll S.A. (2011). Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat. Genet. 43: 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson A., Eyre-Walker A. (2011). Variation in the mutation rate across mammalian genomes. Nat. Rev. Genet. 12: 756–766. [DOI] [PubMed] [Google Scholar]
- Hollister J.D., Ross-Ibarra J., Gaut B.S. (2010). Indel-associated mutation rate varies with mating system in flowering plants. Mol. Biol. Evol. 27: 409–416. [DOI] [PubMed] [Google Scholar]
- Horiguchi G., Gonzalez N., Beemster G.T., Inzé D., Tsukaya H. (2009). Impact of segmental chromosomal duplications on leaf size in the grandifolia-D mutants of Arabidopsis thaliana. Plant J. 60: 122–133. [DOI] [PubMed] [Google Scholar]
- Huang S., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41: 1275–1281. [DOI] [PubMed] [Google Scholar]
- Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. (2004). Detection of large-scale variation in the human genome. Nat. Genet. 36: 949–951. [DOI] [PubMed] [Google Scholar]
- Jovelin R., Cutter A.D. (2013). Fine-scale signatures of molecular evolution reconcile models of indel-associated mutation. Genome Biol. Evol. 5: 978–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.M., Lam H.Y., Urban A.E., Korbel J.O., Affourtit J., Grubert F., Chen X., Weissman S., Snyder M., Gerstein M.B. (2008). Analysis of copy number variants and segmental duplications in the human genome: Evidence for a change in the process of formation in recent evolutionary history. Genome Res. 18: 1865–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klambauer G., Schwarzbauer K., Mayr A., Clevert D.A., Mitterecker A., Bodenhofer U., Hochreiter S. (2012). cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res. 40: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf R.R., Trebitsh T. (2006). The female-specific Cs-ACS1G gene of cucumber. A case of gene duplication and recombination between the non-sex-specific 1-aminocyclopropane-1-carboxylate synthase gene and a branched-chain amino acid transaminase gene. Plant Cell Physiol. 47: 1217–1228. [DOI] [PubMed] [Google Scholar]
- Lee J.A., Carvalho C.M., Lupski J.R. (2007). A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131: 1235–1247. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., et al. (2010). De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20: 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhang Z., Yan P., Huang S., Fei Z., Lin K. (2011). RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genomics 12: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J., et al. (2012). Genetic diversity and population structure of cucumber (Cucumis sativus L.). PLoS ONE 7: e46919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron L.G., et al. (2013). Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc. Natl. Acad. Sci. USA 110: 5241–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S.E., et al. ; Wellcome Trust Case Control Consortium (2009). Microduplications of 16p11.2 are associated with schizophrenia. Nat. Genet. 41: 1223–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald M.J., Wang W.C., Huang H.D., Leu J.Y. (2011). Clusters of nucleotide substitutions and insertion/deletion mutations are associated with repeat sequences. PLoS Biol. 9: e1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale L.K., Haun W.J., Xu W.W., Bhaskar P.B., Anderson J.E., Hyten D.L., Gerhardt D.J., Jeddeloh J.A., Stupar R.M. (2012). Structural variants in the soybean genome localize to clusters of biotic stress-response genes. Plant Physiol. 159: 1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R.E., et al. ; 1000 Genomes Project (2011). Mapping copy number variation by population-scale genome sequencing. Nature 470: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S., Han S., Smith L.G. (2006). Two kinesins are involved in the spatial control of cytokinesis in Arabidopsis thaliana. Curr. Biol. 16: 888–894. [DOI] [PubMed] [Google Scholar]
- Muñoz-Amatriaín M., et al. (2013). Distribution, functional impact, and origin mechanisms of copy number variation in the barley genome. Genome Biol. 14: R58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., et al. (2010). Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466: 368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., et al. (2013). A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat. Genet. 45: 1510–1515. [DOI] [PubMed] [Google Scholar]
- Saxena R.K., Edwards D., Varshney R.K. (2014). Structural variations in plant genomes. Brief Funct. Genomics 13: 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Sebat J., et al. (2007). Strong association of de novo copy number mutations with autism. Science 316: 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifriss O. (1961). Sex control in cucumbers. J. Hered. 51: 5–12. [Google Scholar]
- Stefansson H., et al. (2008). Large recurrent microdeletions associated with schizophrenia. Nature 455: 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanurdzic M., Banks J.A. (2004). Sex-determining mechanisms in land plants. Plant Cell 16 (suppl.): S61–S71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Wang Q., Zhang P., Araki H., Yang S., Kreitman M., Nagylaki T., Hudson R., Bergelson J., Chen J.Q. (2008). Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes. Nature 455: 105–108. [DOI] [PubMed] [Google Scholar]
- Trebitsh T., Staub J.E., O’Neill S.D. (1997). Identification of a 1-aminocyclopropane-1-carboxylic acid synthase gene linked to the female (F) locus that enhances female sex expression in cucumber. Plant Physiol. 113: 987–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen, P.C.M. (2012). Kwantitatieve informatie voor de Glastuinbouw 2012–2013: Kengetallen voor groenten, snijbloemen, potplanten tewelten. (Wageningen, The Netherlands: Wageningen UR Glastuinbouw). [Google Scholar]
- Xiao H., Jiang N., Schaffner E., Stockinger E.J., van der Knaap E. (2008). A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319: 1527–1530. [DOI] [PubMed] [Google Scholar]
- Xu Z., Wang H. (2007). LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35: W265–W268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B., Wong K., Bhomra A., Goodson M., Keane T.M., Adams D.J., Flint J. (2012). The fine-scale architecture of structural variants in 17 mouse genomes. Genome Biol. 13: R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B., et al. (2011). Sequence-based characterization of structural variation in the mouse genome. Nature 477: 326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., et al. (2013). Diverse mechanisms of somatic structural variations in human cancer genomes. Cell 153: 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Zhang W., He H., Nie J., Bie B., Zhao J., Ren G., Li Y., Zhang D., Pan J., Cai R. (2014). Tuberculate fruit gene Tu encodes a C2 H2 zinc finger protein that is required for the warty fruit phenotype in cucumber (Cucumis sativus L.). Plant J. 78: 1034–1046. [DOI] [PubMed] [Google Scholar]
- Ye K., Schulz M.H., Long Q., Apweiler R., Ning Z. (2009). Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Wang C.H., Xu Q., Feng Y., Yuan X.P., Yu H.Y., Wang Y.P., Tang S.X., Wei X.H. (2013). Genome-wide copy number variations in Oryza sativa L. BMC Genomics 14: 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitouni B., Boeva V., Janoueix-Lerosey I., Loeillet S., Legoix-né P., Nicolas A., Delattre O., Barillot E. (2010). SVDetect: a tool to identify genomic structural variations from paired-end and mate-pair sequencing data. Bioinformatics 26: 1895–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., He H., Guan Y., Du H., Yuan L., Li Z., Yao D., Pan J., Cai R. (2010a). Identification and mapping of molecular markers linked to the tuberculate fruit gene in the cucumber (Cucumis sativus L.). Theor. Appl. Genet. 120: 645–654. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Ersoz E., Lai C.Q., Todhunter R.J., Tiwari H.K., Gore M.A., Bradbury P.J., Yu J., Arnett D.K., Ordovas J.M., Buckler E.S. (2010b). Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42: 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zichner T., Garfield D.A., Rausch T., Stütz A.M., Cannavó E., Braun M., Furlong E.E., Korbel J.O. (2013). Impact of genomic structural variation in Drosophila melanogaster based on population-scale sequencing. Genome Res. 23: 568–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.