A novel pathway of HXK1-mediated cell death in which two MIPS enzymes act cooperatively highlights a novel checkpoint of inositol homeostasis in plants.
Abstract
Programmed cell death (PCD) is essential for several aspects of plant life, including development and stress responses. We recently identified the mips1 mutant of Arabidopsis thaliana, which is deficient for the enzyme catalyzing the limiting step of myo-inositol (MI) synthesis. One of the most striking features of mips1 is the light-dependent formation of lesions on leaves due to salicylic acid (SA)-dependent PCD. Here, we identified a suppressor of PCD by screening for mutations that abolish the mips1 cell death phenotype. Our screen identified the hxk1 mutant, mutated in the gene encoding the hexokinase1 (HXK1) enzyme that catalyzes sugar phosphorylation and acts as a genuine glucose sensor. We show that HXK1 is required for lesion formation in mips1 due to alterations in MI content, via SA-dependant signaling. Using two catalytically inactive HXK1 mutants, we also show that hexokinase catalytic activity is necessary for the establishment of lesions in mips1. Gas chromatography-mass spectrometry analyses revealed a restoration of the MI content in mips1 hxk1 that it is due to the activity of the MIPS2 isoform, while MIPS3 is not involved. Our work defines a pathway of HXK1-mediated cell death in plants and demonstrates that two MIPS enzymes act cooperatively under a particular metabolic status, highlighting a novel checkpoint of MI homeostasis in plants.
INTRODUCTION
Myo-inositol (MI) is a ubiquitous molecule and the precursor of many inositol-containing compounds that play critical and diverse roles in signal transduction, membrane biogenesis, vesicle trafficking, and chromatin remodeling (Michell, 2008; Gillaspy, 2011, 2013). In higher plants, inositol and inositol derivatives such as inositol phosphates, phosphatidylinositide, and sphingolipids are involved in gene expression regulation (Alcázar-Román and Wente, 2008), hormonal signaling (Tan et al., 2007), biotic and abiotic stress responses (Taji et al., 2006), and the control of cell death (Liang et al., 2003; Wang et al., 2008; Berkey et al., 2012). MI is also a precursor of the osmoprotectants galactinol and raffinose (Van den Ende, 2013). Coupling of MI with UDP-galactose to produce galactinol is catalyzed by galactinol synthase. Galactinol can be further reacted with sucrose to produce raffinose, and this reaction recycles MI.
Thus, MI synthesis and catabolism impact metabolites involved in many different and critical plant biochemical pathways. MI is synthesized from d-glucose in three steps: First, glucose is phosphorylated by the hexokinase (HXK); second, glucose-6-P (G6P) is converted by the myo-inositol 1-phosphate synthase (MIPS) (Eisenberg et al., 1964); and third, myo-inositol-1-phosphate is dephosphorylated to MI, in a reaction that is catalyzed by myo-inositol monophosphatase. This series of reactions is known as the Loewus pathway, which was first studied in plants and is the only known route for MI biosynthesis (Loewus and Murthy, 2000).
Although yeast and animal genomes contain a single gene encoding the MIPS protein (GhoshDastidar et al., 2006), plants have multiple MIPS genes (Torabinejad and Gillaspy, 2006). The Arabidopsis thaliana genome encompasses three MIPS isoforms that share 89 to 93% identity at the amino acid level (GhoshDastidar et al., 2006; Torabinejad and Gillaspy, 2006). Microarray databases, and previous works, indicate that the transcript level of MIPS1 is predominant over that of MIPS2 and MIPS3 at all developmental stages except seed maturation and early germination stages, when the transcript level of MIPS2 are comparable with that of MIPS1. The transcript level of MIPS3 is much lower than that of MIPS1 and MIPS2 at all developmental stages (Zimmermann et al., 2004; Mitsuhashi et al., 2008; Chen and Xiong, 2010). Furthermore, MIPS1 is expressed in most Arabidopsis tissues, whereas MIPS2 and MIPS3 are specifically expressed in vascular or related tissues (Donahue et al., 2010).
The enzyme activity of MIPS1 in inositol biosynthesis has been demonstrated by both in vitro assay and yeast complementation (Johnson and Sussex, 1995). Consistent with gene expression data, MIPS1 appears to be responsible for most of the MI biosynthesis in leaves since MI levels are reduced in mips1 but not in mips2 and mips3 null mutants (Meng et al., 2009; Donahue et al., 2010). Consequently, mips1 mutants are dramatically impacted during growth and development, while mips2 and mips3 mutants appear phenotypically normal. The mips1 mutant displays pleiotropic developmental defects, such as reduced root growth or altered venation in cotyledons (Meng et al., 2009; Chen and Xiong, 2010; Donahue et al., 2010). Moreover, a striking feature of the mips1 mutant is the light-dependent formation of leaf lesions due to programmed cell death (PCD) (Meng et al., 2009; Donahue et al., 2010). PCD is essential for several aspects of plant life, including development and stress responses. Incompatible plant-pathogen interactions are well known to induce the hypersensitive response, which is often accompanied by localized cell death due to PCD. How MI levels regulate such PCD is not clear. Salicylic acid (SA) production in response to low MI content is a key component of this pathway, since disruption of SA biosynthesis by the sid2 mutation is sufficient to prevent lesion formation in mips1 (Meng et al., 2009). Furthermore, it has been shown that peroxisomal H2O2 induces the formation of SA-dependent lesions in the catalase2 (cat2) Arabidopsis mutant and the expression of disease resistance genes only under lowered MI levels (Chaouch and Noctor, 2010). We have also shown that MIPS1 positively regulates its own expression by inhibiting the activity of histone methyltransferases and that this positive feedback loop is abolished by flagellin treatment (Latrasse et al., 2013). Hence, the tissue content of MI could be a key factor in defense responses, but the underlying molecular mechanisms remain to be unraveled.
Using a candidate gene approach, we recently identified a polyadenylation factor subunit, CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR30 (CPSF30), as a regulator of MI-dependent plant PCD. We showed that CPSF30 activity was required for lesion formation in mips1 via SA-dependant signaling (Bruggeman et al., 2014). Furthermore, CPSF30 activity was also necessary for PCD establishment in cat2, lesion-simulating disease1, constitutive expressor of pathogenesis-related genes5 (cpr5), and mitogen-activated protein kinase4 Arabidopsis mutants, indicating that posttranscriptional regulation plays a key role in the control of PCD.
To gain insight into the molecular mechanisms involved in MI-dependent cell death, we performed a screen for extragenic secondary mutations that abolish lesion formation in mips1. Here, we report that a mutation in the HXK1 enzyme suppresses mips1 light-dependent PCD.
In plants, HXKs are the only enzymes that can phosphorylate glucose. Multiple isoforms of HXK are found in most organisms and six HXK genes have been found in Arabidopsis. Type B HXK (HXK1) was the most studied over recent years and was clearly identified as a core component in plant sugar sensing and signaling (Granot et al., 2014). Indeed, in bacteria, yeast, animals, and plants, hexokinases not only catalyze sugar phosphorylation as the first step of hexose metabolism but also sense glucose levels and transmit the sugar signal to the nucleus (Rolland et al., 2006). The role of HXK1 has been investigated by exposing plant cells and seedlings to HXK substrates or analogs and by overexpressing and downregulating HXK1 in planta (Jang et al., 1997). Isolation and characterization of the Arabidopsis HXK1 mutants, called glucose-insensitive2 (gin2), show that HXK1 is a genuine glucose sensor that mediates sugar sensing independently of its glucose phosphorylation activity, thereby coordinating sugar availability with plant physiology and development (Moore et al., 2003). Uncoupling of metabolic and signaling activity was confirmed by constructing and analyzing two catalytically inactive HXK1 alleles. Although these forms of HXK1 are deficient in ATP binding (G104D) and phosphoryl transfer (S177A), respectively, they restore wild-type growth, repression of photosynthetic gene expression, and hormone responsiveness when expressed in a gin2 background (Moore et al., 2003).
Here, we show, by genetic approaches, an epistatic relationship between mips1 and hxk1 mutants and that PCD triggered by a decrease in MI content and SA accumulation can be reverted by a hxk1 mutation.
RESULTS
Suppression of Cell Death and Defense Responses of mips1 by somi1
The most striking aspect of the mips1 mutant phenotype is the spontaneous lesion formation observed when plants were transferred to long-day (LD) conditions, whereas when grown under short-day (SD) conditions, mature mips1 mutants are indistinguishable from the wild type (Meng et al., 2009). To understand how MIPS1 negatively regulates cell death, we performed a screen for suppressors of mips1-1 using ethyl methanesulfonate mutagenesis. A total of ∼50 suppressors of mips1 (somi1) mutants were identified. The somi1 mutant was further characterized in detail in this study.
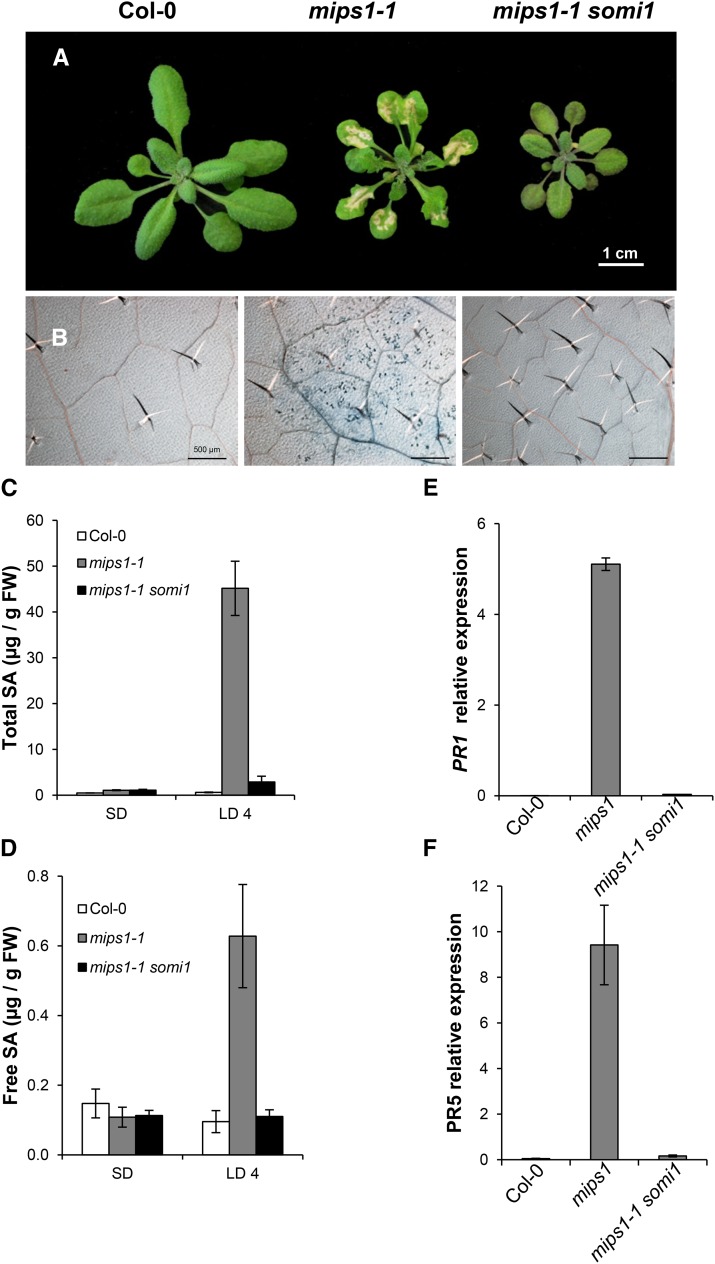
Four days after transfer in restrictive LD conditions, cell death was obvious in mips1-1, whereas the leaves of the mips1-1 somi1 double mutant did not show any cell death (Figure 1A). Trypan blue staining (Figure 1B) and ion leakage measurements (Supplemental Figure 1A) confirmed this observation.
Figure 1.
mips1-Dependent Cell Death Suppression by the somi1 Mutation.
(A) Suppression of mips1-mediated cell death in the mips1-1 somi1 background. Plants were grown for 1 week in vitro on growth medium, then for 14 d under SD conditions in soil and photographed 5 d after transfer to LD conditions. Bar = 1 cm.
(B) Trypan blue staining confirmed the absence of cell death in the suppressor mips1 somi1 mutant. Bar = 500 µm.
(C) and (D) Total (C) and free SA (D) levels in the indicated genotypes, with means and standard deviations calculated from four biological replicates. Plants were grown under SD conditions and transferred to LD conditions for 4 d (LD 4). FW, fresh weight.
(E) and (F) Real-time RT-PCR analysis of PR1 (AT2G14610) and PR5 (AT1G75040) expression in rosette leaves of plants with the indicated genotypes, 4 d after transfer to LD conditions. Transcript abundance is expressed relative to UBQ10 (AT4G05320) transcript abundance. Average relative quantities ± sd (n = 3).
Since lesion formation in mips1-1 is SA dependent, we measured the effect of the somi1 mutation on SA content in the mips1-1 background. As previously reported (Meng et al., 2009), free and total SA contents were similar in the different backgrounds under permissive conditions, but dramatically increased in the mips1-1 mutant 4 d after transfer to restrictive conditions (Figures 1C and 1D). This increase in SA was abolished in mips1-1 somi1 double mutants. In addition, the somi1 mutation led to a dramatic downregulation of the SA pathogenesis-related marker transcripts PATHOGENESIS-RELATED1 (PR1) and PR5 (Figures 1E and 1F) in mips1-1 somi1 compared with their constitutive expression in mips1-1.
Two others striking phenotypes of the mips1 mutant were defects in primary root development and alteration in cotyledon morphology, which was in part due to mislocalization of the auxin carrier PIN (Meng et al., 2009; Chen and Xiong, 2010; Donahue et al., 2010; Luo et al., 2011). Nevertheless, the shorter primary root observed in mips1-1 was not suppressed in the mips1-1 somi1 mutant (Supplemental Figure 1B). To investigate whether the alteration in cotyledons is suppressed by the somi1 mutation, we classified cotyledon abnormalities into four classes according to the severity of the phenotype (Supplemental Figure 1C). The mips1-1 somi1 mutant displayed a similar proportion of plants with severe cotyledon defects as the mips1-1 mutant.
Taken together, these results suggest that the somi1 mutation specifically suppresses mips1-1-dependent lesions by blocking events that lead to cell death upstream of SA accumulation without impacting other mips1 phenotypes.
SOMI1 Encodes HXK1
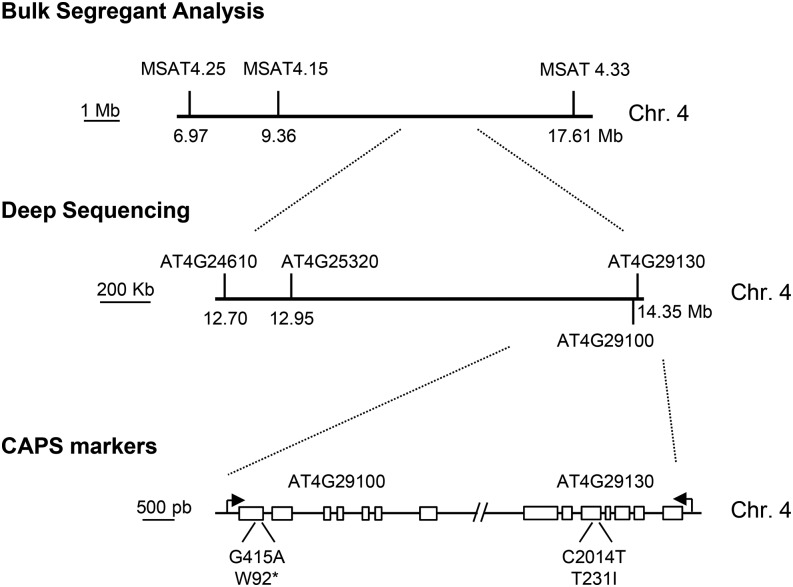
To map the somi1 mutation, mips1-1 somi1 plants in the Columbia-0 (Col-0) background were crossed with an allelic mutant of mips1-1, the mips1-2 allele in the Wassilewskija (Ws) background. We performed bulk segregant analysis (BSA) using F2 plants without (n = 22) or with lesions (n = 22) in LD conditions and revealed that somi1 was flanked by microsatellite markers MSAT4.15 and MSAT4.33, on a region of ∼8 Mb on chromosome 4 (Figure 2). Further mapping was performed by high-throughput sequencing of nuclear DNA obtained from a bulk of 156 F2 plants without lesions in LD. Sequence analysis using the Col-0 genome as reference revealed that in the region previously mapped by BSA, four candidate genes (AT4G24610, AT4G25320, AT4G29100, and AT4G29130) presented a single-nucleotide polymorphism (SNP) in 100% of paired-end reads (Figure 2). After two backcrosses of mips1-2 somi1 with the mips1-2 single mutant to clean background mutations, cleaved amplified polymorphic sequence (CAPS) markers were used on 30 mips1-2 somi1 plants to eliminate SNP mutations that did not account for lesion suppression. Two genes remained after this step: AT4G29100, encoding a putative DNA binding protein, and AT4G29130, encoding Hexokinase1 (At-HXK1, hereafter called HXK1). Mutations introduced a stop codon or an amino acid change of Thr to Ile at position 231 (T231I) in the AT4G29100 and HXK1 genes, respectively. In HXK1, this mutation induced the replacement of a polar amino acid by a nonpolar residue, within a region highly conserved in animals, plants, and fungi directly involved in glucose fixation (Supplemental Figures 2A and 2B).
Figure 2.
Mapping of the somi1 Mutation.
A BSA was performed using F2 from the cross between mips1-1 somi1, in the Col-0 background, and mips1-2, in the Ws background, and revealed that somi1 was flanked by markers MSAT4.15 and MSAT4.33 on chromosome 4 (Chr. 4). High-throughput sequencing of nuclear DNA obtained from a bulk of 156 F2 plants without lesions in LD identified four candidate genes (AT4G24610, AT4G25320, AT4G29100, and AT4G29130). CAPS markers were used on 30 mips1-1 somi1 plants to removed SNP mutations not linked to the somi1 locus. Two genes remained after this step: AT4G29100, encoding a putative DNA binding protein, and AT4G29130, encoding HXK1. Mutations introduced an early stop or an amino acid change of Thr to Ile, in AT4G29100 and HXK1 genes, respectively.
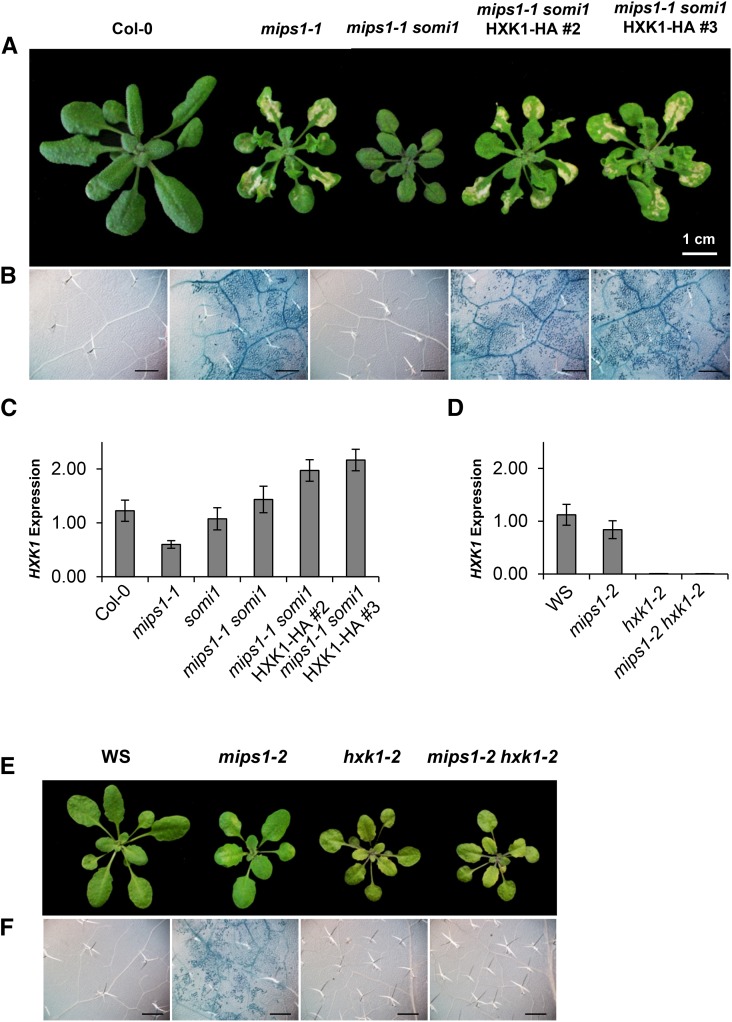
To identify the somi1 locus, complementation of the mips1-1 somi1 mutant with wild-type forms of AT4G29100 and HXK1 was performed. While complementation with AT4G29100 did not restore lesion formation, independent mips1-1 somi1 transgenic lines expressing wild-type HXK1 fused to a hemagglutinin tag (HXK1-HA) showed identical lesions to mips1-1 in LD conditions (Figures 3A and 3B). Quantitative RT-PCR (RT-qPCR) analysis of two independent complemented lines, mips1-1 somi1 HXK1-HA#2 and mips1-1 somi1 HXK1-HA#3, showed that HXK1 transcripts were slightly overaccumulated in these lines, while mips1-1, somi1, and mips1-1 somi1 mutants presented a similar level of HXK1 expression to the wild type (Figure 3C). To confirm the involvement of HXK1 in mips1-1-dependent cell death, we crossed the mips1-2 mutant with the hxk1-2 T-DNA mutant (Flag_346H03). The T-DNA insertion was located in the promoter of HXK1, inducing a very low expression of this gene in hxk1-2 and mips1-2 hxk1-2 mutants (Figure 3D). mips1-2 hxk1-2 did not show any lesions in LD (Figures 3E and 3F), confirming that disruption of HXK1 suppresses MI-dependent cell death. Expression analysis showed that the overexpression of the immune markers PR1 and PR5, which were suppressed in mips1-1 somi1, was restored by complementation with wild-type HXK1 in the mips1-1 somi1 HXK1-HA#2 and mips1-1 somi1 HXK1-HA#3 lines (Supplemental Figure 3A). Overexpression of PR1 and PR5 was also observed in mips1-2, in LD conditions, but was suppressed in mips1-2 hxk1-2 double mutants (Supplemental Figure 3B). According to these results, it was obvious that the mutation located in HXK1 was responsible for the suppression of mips1-dependent cell death and PR gene expression in the somi1 suppressor, hereafter referred to as the hxk1-1 allele.
Figure 3.
somi1 Locus Corresponds to HXK1.
(A) Transformation of the suppressor mips1-1 somi1 with a cDNA encoded wild-type form of HXK1 fused to the HA tag restores lesions in two independent transgenic lines (mips1-1 somi1 HXK1-HA #2 and #3).
(B) and (F) Trypan blue staining confirms the cell death phenotype of the different indicated lines. Bar = 500 µm.
(C) and (D) RT-qPCR analysis of the HXK1 expression in indicated lines in LD conditions. Transcript abundance is expressed relative to UBQ10 (AT4G05320) transcripts abundance.
(E) T-DNA mutant hxk1-2 (Flag_346H03), with an insertion in the promoter of the HXK1 gene, abolishes lesions that normally occur in mips1-2. These lines are in the Ws background. In (A) and (E), plants were photographed 5 d after transfer in LD conditions. Bar = 1 cm.
The hxk1-1 Mutation Affects Both the Metabolic Activity and the Glucose Signaling Function of HXK1
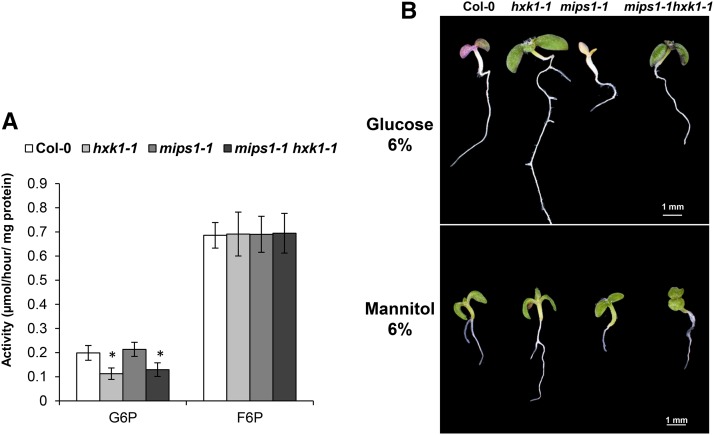
HXK1 encodes a bifunctional enzyme that mediates sugar sensing in addition to its involvement in hexose phosphorylation, thereby coordinating sugar availability with plant physiology and development. Hence, the consequences of hxk1-1 mutation could be due to an effect on HXK1 protein catalytic activity and/or HXK1 signaling functions. To analyze whether the hxk1-1 T231I mutation resulted in reduced hexokinase activity, we measured glucose and fructose phosphorylation activity using protein extracts isolated from wild-type, hxk1-1, mips1-1, and mips1-1 hxk1-1 lines. Leaves of hxk1-1 and mips1-1 hxk1-1 mutants displayed ∼56 and 60% of the glucose phosphorylation activity, respectively, of the wild-type control and mips1-1 leaves, whereas the fructose phosphorylation activity was similar to that of the control (Figure 4A). The mips1-1 mutation alone did not affect either phosphorylation activity. A decrease in glucose phosphorylation was also observed in hxk1-2 and mips1-2 hxk1-2 lines (Supplemental Figure 4A). Because hexokinases have a preferential affinity for glucose, these results suggested that the hxk1-1 mutation affects hexokinase activity.
Figure 4.
Effect of the hxk1-1 Mutation on HXK1 Functions.
(A) Glucokinase and fructokinase activity measurements show the effect of the indicated mutations on the synthesis of G6P and fructose-6-phosphate (F6P) compounds. Samples were harvested 2 d after transfer in LD conditions, and means and se were calculated from four biological replicates. *Student’s t test: P < 0.05.
(B) Glucose sensitivity assays. Plants were grown on 6% glucose (Glc) or 6% mannitol (Man) 0.5× MS medium for 10 d.
To test the glucose sensitivity and signaling of the seedlings, a widely used test is based on the ability to develop normally on otherwise inhibitory concentrations of exogenous glucose in which wild-type plants display inhibited cotyledon expansion, chlorophyll accumulation, and shoot growth at the seedling stage of development (Rolland et al., 2006). Therefore, seedling growth of the various mutants was assessed in the presence of 6% glucose. As shown in Figure 4B, hxk1-1 and mips1-1 hxk1-1 mutants were able to overcome the developmental arrest displayed by wild-type control plants, whereas wild-type and mips1-1 single mutants showed glucose-dependent developmental arrest and repression of chlorophyll accumulation. The same response was observed for lines in the Ws background (Supplemental Figure 4B). As an osmotic control, all lines were shown to have a similar phenotype on Murashige and Skoog (MS) plates with 6% mannitol (Figure 4C). These results indicate that the hxk1-1 mutant was affected in both its metabolic activity and signaling function.
Hexokinase Activity Is Necessary for the Establishment of Lesions in mips1
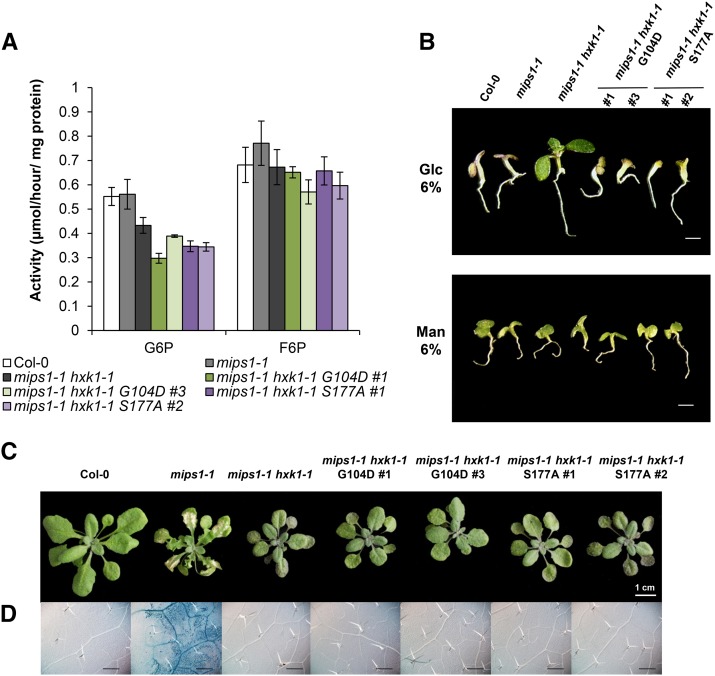
To test whether the involvement of HXK1 in cell death relies on its signaling function or on its glucose phosphorylation activity, we used targeted mutagenesis to produce HXK1 mutants that retain signaling function but not catalytic activity. We generated the two catalytically inactive HXK1 mutants described by Moore et al. (2003): one that eliminated adenosine triphosphate binding [Gly-104 → Asp-104 (G104D)] and another that prevented phosphoryl transfer [Ser-177 → Ala-177 (S177A)], but both maintained the glucose binding site (Ma et al., 1989; Kraakman et al., 1999). It has been previously shown that these two catalytically inactive mutants exhibited a similar glucose signaling function as the wild-type HXK1 in repressing two promoters of photosynthesis genes (Moore et al., 2003). Hence, to further evaluate the respective roles of glucose signaling and metabolism during lesion formation, we generated independent transgenic plants, mips1-1 hxk1-1 S177A and mips1-1 hxk1-1 G104D, which expressed the two catalytically inactive HXK1 mutant alleles in the mips1-1 hxk1-1 background. Measurements of glucose phosphorylation activity (Figure 5A) and of glucose sensitivity (Figure 5B) confirmed that complementation with catalytically inactive forms was able to restore the response to glucose, whereas production of G6P was still lower than in the wild-type control.
Figure 5.
Uncoupling Glucose Signaling and Metabolism on Cell Death.
(A) Glucokinase and fructokinase activity measurement in mips1-1 hxk1-1 complemented with metabolically inactive forms of HXK1. Amino acid changes, such as Gly-104 to Asp (G104D) and Ser-177 to Ala (S177A), deactivate the kinase activity of HXK1. The suppressor mips1-1 hxk1-1 was transformed with cDNA encoding HXK1 with the G104D or with the S177A amino acid changes fused to the HA tag, and hexokinase activity was measured in two independent transgenic lines (mips1-1 hxk1-1 G104D#1 and #3, and mips1-1 hxk1-1 S177A#1 and #2). Means and standard errors were calculated from four biological replicates. F6P, fructose-6-phosphate.
(B) Glucose sensitivity assays. Plants were grown on 6% glucose (Glc) or 6% mannitol (Man) 0.5× MS medium for 10 d. Bar = 500 µm.
(C) Cell death phenotypes of the indicated lines after 5 d in LD conditions. Bar = 1 cm.
(D) Trypan blue staining confirms the cell death phenotype of the different indicated lines. Bar = 500 µm.
As shown on Figures 5C and 5D 4 d after transfer to restrictive LD conditions, cell death was obvious in mips1-1, as expected, whereas the leaves of the mips1-1 hxk1-1 and wild-type Col-0 did not show any cell death. Two independent transgenic lines for each HXK1 catalytically inactive form did not display lesions on leaves as the wild type and mips1-1 hxk1-1 double mutant.
Thus, these results suggest that HXK1 catalytic activity is necessary for the establishment of the lesions in the double mutant mips1 hxk1.
Restoration of MI Content in mips1 Mutants by hxk1 Mutations
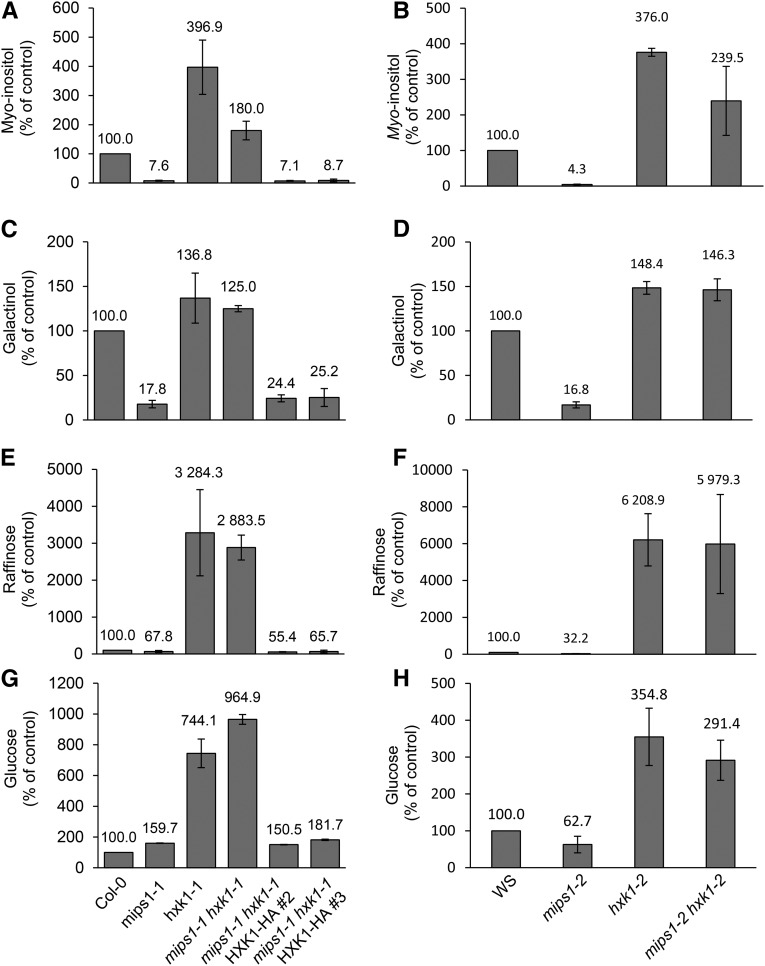
In the mips1 mutant, the induction of PCD was attributed to the reduced MI or galactinol accumulation, since treatment of mutants with either compound was able to suppress lesion formations (Meng et al., 2009). Using gas chromatography-mass spectrometry (GC-MS) analysis, we quantified MI in the different mutants and complemented lines. Surprisingly, while MI content was strongly decreased in mips1-1 and mips1-2 mutants compared with the corresponding wild type, as expected, this compound overaccumulated in mips1-1 hxk1-1 and mips1-2 hxk1-2 mutants and also in both single hxk1 mutants (Figures 6A and 6B). In mips1-1 hxk1-1 HXK1#2 and mips1-1 hxk1-1 HXK1#3 complemented lines, MI content was found again at a very low level, similar to what was observed in the mips1-1 mutant (Figure 6A). As shown in Figures 6C to 6F, measurements of galactinol and raffinose contents led to the same results: A restoration of their content by mutation of HXK1 in the mips1 mutant background could be responsible of the suppression of MIPS1-dependant cell death.
Figure 6.
Suppression of mips1-1-Dependent Cell Death Correlated with Changes in Metabolites.
GC-MS analysis was performed to determine the relative contents of myo-inositol ([A] and [B]), galactinol ([C] and [D]), raffinose ([E] and [F]), and glucose ([G] and [H]) in rosette leaves of the indicated lines. Samples were harvested 4 d after transfer in LD conditions. Data are expressed as the percentage of the wild-type control. Standard deviations were calculated from four biological replicates.
As expected, an overaccumulation of glucose, the immediate substrate of hexokinase, was also detected in both hxk1 allele backgrounds, which was abolished by complementation with the wild-type enzyme (Figures 6G and 6H).
MIPS2 Is Responsible for MI Production in mips1 hxk1 Mutants
Although organisms incorporate MI into various compounds, the Loewus pathway is the only biosynthetic route to produce MI de novo. Genetic analyses previously demonstrated that in Arabidopsis, the MIPS1 isoform is responsible for most MI biosynthesis in leaves. Thus, in this context, a restoration of MI content in the absence of functional MIPS1 enzyme was surprising. However, evidence exists that a minor pathway to ascorbic acid synthesis could proceed via MI synthesis (Radzio et al., 2003; Lorence et al., 2004; Torabinejad et al., 2009). It has been previously shown that the vitamin c4 (vtc4) mutant, impaired in l-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme (Conklin et al., 2006), has reduced MI levels, suggesting such a connection between these pathways (Torabinejad et al., 2009). Hence, to test whether MI accumulation observed in the mips1 hxk1 double mutant could originate from this pathway, this double mutant was crossed with the vtc4 mutant. Because no restoration of cell death was observed in hxk1 mips1 vtc4 triple mutants, this alternative pathway could be excluded as being responsible of restoration of the MI content in the mips1 hxk1 double mutant.
As the Arabidopsis genome contains three MIPS genes, we hypothesized that one or the two MIPS2 and MIPS3 enzymes could be responsible for restoring MI levels in the mips1 hxk1 suppressors. To test this, we crossed mips1-1 hxk1-1 with mips2-2 or mips3-2 mutants to obtain the corresponding triple mutants.
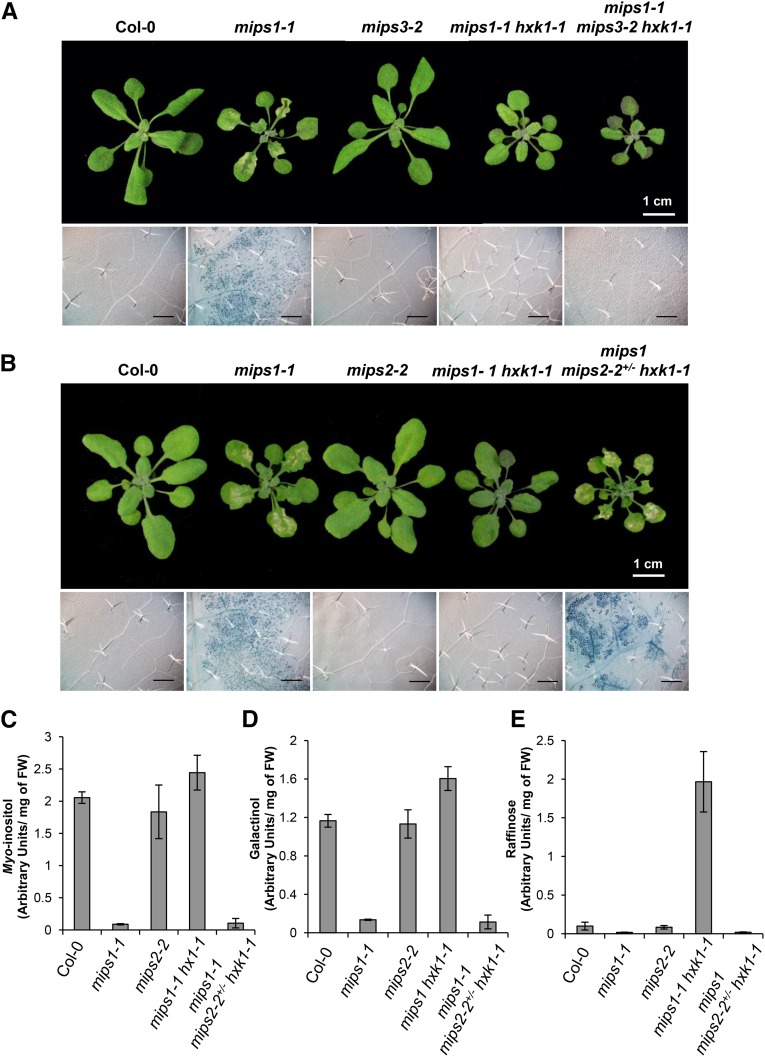
As shown Figure 7A, a mutation in MIPS3 was not able to rescue the mips1 cell death phenotype in the mips1-1 hxk1-1 double mutant background. As stated by Luo et al. (2011), the mips1 mips2 double mutant is embryo lethal. We thus generated a mips1-1 mips2-2+/− hxk1-1 line to avoid the embryo defective phenotype. As shown in Figure 7B, this triple mutant displayed lesions on leaves, indicating that in mips1-1 hxk1-1, cell death suppression is likely due to the action of MIPS2.
Figure 7.
Roles of MIPS2 and MIPS3 in Cell Death Suppression.
The suppressor mips1-1 hxk1-1 was crossed with mips2-2+/− and mips3-2 single mutants to obtain corresponding triple mutants.
(A) and (B) Rosette leaves pictures and Trypan blue staining after transfer in LD conditions of mips1-1 mips2-2+/− hxk1-1 and mips1-1 mips3-2 hxk1-1 mutants reveal cell death phenotypes. Photographs bar = 1 cm; Trypan blue bar = 500 µm.
(C) to (E) Metabolite levels, including myo-inositol (C), galactinol (D), and raffinose (E), were analyzed in the indicated lines by GC-MS. Means and standard deviations were calculated from four biological replicates. FW, fresh weight.
To confirm this result, we quantified MI, galactinol, and raffinose. While, as expected, in the mips2-2 null mutant the contents of these compounds were almost comparable to the wild-type control, MI, galactinol, and raffinose levels were similar in mips1-1 mips2-2+/− hxk1 to the levels observed in mips1-1, confirming that in this context, the action of MIPS2 is necessary for MI production (Figures 7C to 7E).
Although the metabolic events connecting a restoration of the MI content with changes in HXK1 activity are unknown, one simple explanation would be that in the mips1 background, the hxk1 mutation affects transcriptional regulation of MIPS2 by, for instance, modifying glucose homeostasis, leading to MIPS2 overexpression and subsequently to a restoration of MI production. RT-qPCR did not detect a real significant overaccumulation of the MIPS2 transcript in hxk1-1 and mips1-1 hxk1-1 (Supplemental Figure 5A), whereas its expression was slightly decreased in hxk1-2 and mips1-2 hxk1-2 compared with the corresponding wild type (Supplemental Figure 5B). Likewise, the expression of MIPS1 was not significantly affected in the hxk1-1 and hxk1-2 backgrounds. Nevertheless, in the single mips1-1 and mips1-2 mutants, MIPS2 transcript accumulation was ∼10 times lower than in the wild type in LD conditions and this downregulation was rescued in mips1-1 hxk1-1 and mips1-2 hxk1-2 double mutants (Supplemental Figures 5A and 5B). This suggests that PCD in mips1 could result from downregulation of MIPS2 in LD and that disruption of HXK1 activity could prevent the repression of MIPS2, thereby restoring normal MI accumulation and inhibiting lesion formation. This assumption is further substantiated by the fact that in mips1-1 hxk1-1 HXK1 complemented lines, MIPS2 expression was decreased, whereas in transgenic lines complemented with each HXK1 catalytically inactive form, MIPS2 expression increased (Supplemental Figure 5A), even though it did not fully reach levels observed in hxk1 mutants. Furthermore, loss of only one functional MIPS2 allele is sufficient to restore lesion formation in mips1 hxk1 mutants, indicating that fine-tuning of MIPS2 expression plays an important role in PCD control.
DISCUSSION
Due to their sessile nature, plants are confronted continuously with adverse conditions of abiotic and biotic nature. PCD is a vital component of plant immunity, although the molecular mechanisms that control it remain poorly understood. To identify novel regulators of plant PCD, we used a genetic approach and found that the hexokinase HXK1 is required for lesion formation in the mips1 mutant. Our work revealed that lesions triggered by a decrease in MI content could be genetically reverted in the mips1 hxk1 double mutant and reinduced by complementation with HXK1.
As demonstrated by cellular ion leakage and Trypan blue staining analysis, both of which measure cell death, hxk1 is epistatic over the mips1 mutation. The mechanisms linking carbohydrate metabolism with defense responses, including PCD, are beginning to be elucidated (Rojas et al., 2014). Analyses of PR gene induction in tobacco (Nicotiana tabacum) leaf disks and Arabidopsis liquid cultures in response to soluble sugars suggest a positive regulatory function of HXK1 (Herbers et al., 1996; Xiao et al., 2000). However, in Nicotiana benthamiana, downregulation of HXK1 by virus-induced gene silencing caused an increase in cell death (Kim et al., 2006), revealing that HXK1 can also function as a negative regulator of defense responses. Although the molecular events connecting the reduced MI content with changes in HXK1 activity remain to be elucidated, one simple explanation for the suppression of lesions in mips1 hxk1 mutants would be that MI levels affect the transcriptional regulation of HXK1, leading to its overexpression in the mips1 background and, subsequently, the constitutive activation of defense genes and PCD via unknown mechanisms. However, RT-qPCR did not detect an overaccumulation of HXK1 transcripts in either allele of mips1, which rules out this hypothesis.
Using catalytically inactive HXK1 mutants, we demonstrated that the glucose phosphorylation activity is necessary for the suppression of cell death. Although this result does not exclude the involvement of HXK1 signaling functions in the cell death suppression, it demonstrates that a metabolic deregulation is necessary. It is also consistent with the position of the hxk1-1 mutation within a highly conserved region involved in the fixation of the glucose, maybe leading to a decrease for its substrate affinity (Kuettner et al., 2010). The observation that MIPS2 expression in mips1 hxk1-1 mutants complemented with catalytically inactive forms of HXK1 is higher than in mips1 mutants but lower than in mips1 hxk1-1 may indicate that the signaling function of HXK1 also contributes, to a lesser extent, to the regulation of MIPS2. However, our results show that loss of HXK1 catalytic activity is sufficient to express MIPS2 above a threshold that prevents lesion formation.
Similarly, in the lesion mimic mutant cpr5, phenotypes related to glucose signaling and defense responses such as lesion formation and constitutive SA production were found to be independent of HXK1-mediated glucose signaling (Yoshida et al., 2002; Aki et al., 2007).
Metabolomics analysis further revealed a restoration of the MI content in mips1 hxk1 mutants. This result is consistent with our previous results showing that spraying plants with MI is enough to suppress lesion formation, indicating that inositol production, and not the MIPS1 protein itself, is required to prevent PCD in leaves. MI was even more overaccumulated in mutants deficient in HXK1 compared with the wild type, suggesting that normal HXK activity is required to downregulate the activity of enzymes involved in MI biosynthesis. Consistently, it has been shown that transgenic tomato (Solanum lycopersicum) plants overexpressing Arabidopsis HXK1 exhibit lower inositol accumulation in their fruits (Roessner-Tunali et al., 2003).
The overaccumulation of MI in the hxk1 mutant background may seem surprising since hexokinase enzymes catalyze the formation of G6P, the immediate substrate of MIPS enzymes. Transcriptional analysis revealed that this is not due to a major deregulation of MIPS1 or MIPS2 expression in this specific genetic background. However, hexokinase activity assays revealed only a slight decrease of G6P formation in the different hxk1 backgrounds, indicating that G6P levels are probably not drastically reduced in hxk1 mutants. Residual G6P formation is likely dependent on the activity of other endogenous hexokinases that may have important roles in glucose metabolism and may account for the HXK1 alteration being nonlethal as already noted by Moore et al. (2003).
Using combinations of lines mutated in other MIPS genes, we demonstrated that restored MI biosynthesis in mips1 hxk1 required MIPS2, while MIPS3 was not involved. As a highly conserved enzyme, MIPS was found in all eukaryotes and a few prokaryotes. In the phylogenetic tree constructed with more than 100 eukaryotic MIPS protein sequences, MIPS from plants form an independent subgroup and multiple copies of the MIPS gene emerge only in land plant species (Luo et al., 2011). All three genes are functional and were able to rescue the yeast inositol auxotrophy mutant ino1 (Luo et al., 2011); moreover, the corresponding proteins share ∼90% identity at the amino acid level. Thus, the evolution of these small multigenic families likely represents diversification of temporal and spatial regulation rather than changes in amino acid sequences or catalytic capacity. Using RT-PCR analysis, we only detected a mild activation of MIPS2 expression in the mips1 hxk1 genetic background, and only after plants were transferred to LD conditions. However, while MIPS1 is expressed in cells throughout tissues such as leaves, MIPS2 expression is confined to vascular or vascular-associated cells (Donahue et al., 2010), and maybe subtle changes in its spatial expression could be more efficiently detected only by in situ analyses. This finding could also suggest that the rescue of MI content by MISP2 is due to a translational or posttranslational control mechanism. However, such regulation would be challenging to demonstrate, as it would require discriminating, in vivo, between enzymatic activities of the different MIPS isoforms. Moreover, it is still unclear why MIPS2 is able to rescue the mips1 cell death phenotype, by MI production, only in the context of the mips1 hxk1 background and not in the single mips1 mutant. According to our expression analysis, MIPS2 expression seems to be repressed when mips1 mutants are transferred to cell death-inducing conditions, and this downregulation appears to be partially alleviated by the hxk1 mutation. Although we were not able to show a significant increase of MIPS2 expression in the hxk1 single mutant in our culture conditions, Price et al. (2004) reported an induction of MIPS1 and MIPS2, but not of MIPS3 expression, in whole seedlings treated with glucose, indicating a possible transcriptional regulation of MIPS genes according to the carbohydrate metabolic status. Nevertheless, we were unable to show any reduction of lesion formation in mips1 via glucose treatment, suggesting that other signals contribute to the suppression of cell death in hxk1.
Interestingly, loss of a single MIPS2 allele was sufficient to restore lesion formation in mips1 hxk1 mutants, indicating that tight regulation of MIPS2 expression levels is crucial for the control of MI-dependent PCD. How much this regulation contributes to PCD onset remains to be fully established, but one can hypothesize that the control of MIPS2 activity by primary carbohydrate metabolism is a key component of MI-dependent PCD and that this pathway involves hexokinase enzymes, as energy provider or source of signaling molecules. Several studies have already suggested that plants have the ability to modulate their sugar pools to act as putative agents of immune reactions (Bolouri Moghaddam and Van den Ende, 2012).
Although our results provide insight into PCD control, how MI levels regulate this process is still an open question. Indeed, how MI accumulation modulates SA biosynthesis and this only in LD conditions remains unknown. An interesting research axis concerns the alteration of ceramides homeostasis since relative accumulation of ceramides and their derivatives appear as key elements for the control of plant PCD via SA-signaling pathway (Sánchez-Rangel et al., 2015). Donahue et al. (2010) reported elevated levels of ceramides and of hydroxyceramides in mips1, correlated with the decline of MI and of its derivative phosphatidylinositol. They have proposed that this could constitute a pro-death signal leading to SA production. However, this report does not explain why PCD appears in a light-dependent manner. In addition, since the suppression of cell death in mips1 hxk1 double mutants was due to a restoration of MI levels, it is very likely that the accumulation of ceramide and its derivatives was also restored in the mips1 hxk1 double mutant.
It is also worth noting that hxk1 suppresses the autoimmune response but not the root and cotyledon developmental defects of mips1, indicating that these pathways can be uncoupled. It confirms our previous work (Bruggeman et al., 2014), showing that although lesion formation was totally abolished in the mips1 oxt6 double mutant, defects in root elongation were still present and that rescue of the cotyledon phenotype was only partial, suggesting that these two developmental pathways are only partly dependent on the spontaneous cell death phenotype. This observation further supports the notion that HXK-dependent regulation of MIPS2 expression according to growth conditions is a key component of MI-dependent PCD regulation. To explain the non-rescue of root and cotyledon defects by the hxk1 mutation, one can hypothesize that the outcome of hxk1 deficiency differs between plantlets and mature plants.
In conclusion, our work defines an example of HXK1-mediated cell death in plants requiring catalytic activity of the enzyme. Genetic analysis demonstrated that it is not directly due to the function of HXK1 itself but to the cooperative action of two MIPS isoforms, with MIPS2 being responsible for the MI synthesis in the mips1 hxk1 background. The sessile nature of plants forces them to face an ever-changing environment instead of escaping from hostile conditions as animals do. To overcome this survival challenge, a fine monitoring and controlling of the status of general metabolism, including MI biosynthesis, is vital for these organisms. Frequently, evolutionary plant adaptation has involved the appearance of multigene families, comprising an array of enzymes, or sensing and signaling elements with highly conserved primary sequences but with differential spatio-temporal regulation. The demonstration of cooperative action of two MIPS enzymes under a particular metabolic status highlights a novel checkpoint of MI homeostasis in plants.
METHODS
Plant Materials and Growth Conditions
The T-DNA lines mips1-1 (SALK_023626), mips2-2 (SALK_031685), and mips3-2 (SALK_120131) were in the Col-0 background. The hxk1-2 (Flag346H03) and mips1-2 (Flag605F08) mutants were in the Ws background. All these T-DNA lines were obtained from public seeds banks (Samson et al., 2002; Alonso et al., 2003). All double mutants and triple mutants were obtained by crossing and genotyped with primers listed in Supplemental Table 1.
For primary root length and cotyledon analysis, seeds were sown on commercially available 0.5× MS medium (Basalt salt mixture M0221; Duchefa) solidified with 0.8% agar (Phyto-Agar HP696; Kalys) and grown in a LD (16 h light, 8 h night, 20°C) growth chamber. For glucose sensing assays, seeds were sown on solid 0.5× MS medium supplemented with 6% (w/v) of glucose or mannitol and seedlings were grown for 10 d.
For all other analyses, plants were grown for 1 week as described above and transferred to soil under SD conditions (8 h day, 16 h night, 200 µmol photons s−1 m−2, 21°C) for 2 weeks. Plants were subsequently transferred to LD conditions (16 h day, 8 h night, 200 µmol photons s−1 m−2, 21°C) for the indicated times.
Screen and Mapping of the somi1 Mutation
About 20,000 mips1-1 homozygous seeds were incubated for 4 h in 0.74% (w/v) ethyl methanesulfonate. The somi mutants were screened in the M2 generation in restrictive conditions.
To map the somi1 mutation, mips1-1 somi1 plants in the Col-0 background were crossed with an allelic mutant of mips1-1, the mips1-2 mutant, in the Ws background. First, a BSA was performed using F2 plants without (n = 22) or with lesions (n = 22) in LD. Primer sequences that flank polymorphism markers between Col-0 and Ws are described by Păcurar et al. (2012). Second, fine mapping of the mutation somi1 was performed by a high-throughput sequencing of nuclear DNA obtained from a bulk of 156 F2 plants without lesions in LD. Whole Arabidopsis thaliana genome resequencing was performed by the company Macrogen (10F, 254 Beotkkot-ro Geumcheon-gu, Seoul, Republic of Korea) using Illumina HiSeq2000 technology. The software CLC genomic workbench was used to analyze sequences and identify SNPs by comparison with the Col-0 genome as reference (TAIR10). After two backcrosses of mips1-1 somi1 with the mips1-1 single mutant to clean background mutations, CAPS markers were used on 30 mips1-1 somi1 plants to remove SNP mutations not linked to the somi1 locus. Primers are listed in Supplemental Table 1.
Cloning Methods
cDNA sequences of HXK1 were amplified from the clones G21654 and U16338, respectively, available in the public databank (http://www.arabidopsis.org). Primers listed in Supplemental Table 1 were used to amplify corresponding cDNA and to remove stop codons. cDNAs were cloned into the Gateway-compatible pEntr3C vector (Invitrogen) and transferred into the binary vector pGWB14 (Nakagawa et al., 2007) using the LR clonase (Invitrogen) according to manufacturer’s instructions. In this plasmid, the expression of cDNA is under the control of the 35S promoter and encoded proteins are fused with three HA tags.
To generate the two catalytically inactive HXK1 alleles, G104D and S177A, we performed a targeted mutagenesis of the HXK1 cDNA. Wild-type cDNA sequence in the plasmid pENTR3C was used as template for PCR amplification with the primers listed in Supplemental Table 1. These oligonucleotides create nucleotide substitutions to induce amino acid changes. The mutagenized cDNAs were then cloned into the pGWB14 vector as previously described.
Cell Death Assays: Electrolyte Loss and Trypan Blue Staining
For electrolyte loss, leaf disks were removed with a 5-mm punch and floated on distilled water for 10 min. Six leaf disks per genotype were transferred to a tube containing 2 mL of distilled water and agitated for 2 h. Conductivity (µs/cm2) was then measured with the CDM 210 conductivity meter (Radiometer Analytical). Means and sd were from four replicates per genotype per experiment.
For Trypan blue staining, rosette leaves were infiltrated three times under vacuum with lactophenol Trypan solution (2.5 mg/mL Trypan blue, 25% lactic acid, 23% water saturated phenol, 25% glycerol, and water). Samples were then washed with distilled water and heated over boiling water. After cooling, samples were cleared with a chloral hydrate solution (8 g of chloral hydrate were dissolved in 2 mL of glycerol [50%] and 1 mL of water); the solution was replaced several times. Samples were mounted in chloral hydrate solution and observed with a macroscope (Nikon AZ100 Multizoom).
SA Quantification
Wild-type and mutant plants were grown under SD conditions for 3 weeks and subsequently transferred to LD conditions. About 100 mg of leaves was harvested just before transfer to LD (i.e., in SD conditions), and 4 d after transfer to LD conditions. Metabolites were extracted as described by Baillieul et al. (1995) with the following modifications. [C14]SA was added to each sample for extraction loss correction. Samples were dried in a Speed-Vac (Savant Instrument). One-half of dried samples was directly used to analyze free SA. The other dried samples were subjected to acidic hydrolysis to determine total SA (free plus conjugated forms). HPLC analyses were performed according to Simon et al. (2010) using the Waters system previously described. Peak purity determination and extraction loss correction were performed. Absolute SA quantification was obtained using a standard curve made with SA standards. Data were analyzed using Empower Pro Software (Waters). Corrections for losses were performed as described previously by Baillieul et al. (1995) using a LS 6500 Multi-Purpose Scintillation Counter (Beckman Coulter). Data presented here are the average of results obtained from four biological replicates.
RNA Extraction and RT-qPCR
Total RNA was extracted from rosette leaves 4 d after transfer to LD conditions, using the Nucleospin RNA kit (Macherey-Nagel) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 2 µg of total RNA using Improm-II Reverse Transcriptase (A3802; Promega) according to the manufacturer’s instructions. Then, 1/25th of the synthesized cDNA was mixed with 100 nM solution of each primer and LightCycler 480 Sybr Green I Master Mix (Roche Applied Science) for quantitative PCR analysis. Products were amplified and fluorescent signals were acquired with a LightCycler 480 detection system. The specificity of amplification products was determined by melting curves. Arabidopsis UBQ10 was used as an internal control for signal normalization. Exor4 relative quantification software (Roche Applied Science) automatically calculates the relative expression level of the selected genes with algorithms based on the ΔΔCt method. Data were from duplicates of at least two biological replicates. Primers used are described in Supplemental Table 1.
Metabolite Extraction and GC-MS Analysis
Analytical procedures were adapted from Roessner et al. (2000). About 100 mg of rosette leaves of the indicated lines were harvested 4 d after transfer to LD conditions. Samples were lyophilized and ground, and metabolites were extracted by adding 500 μL of cold methanol, containing 250 µM of ribitol as the internal standard. Extracts were then warmed to 70°C for 15 min and vortexed regularly. Next, 500 μL of chloroform was added and samples were vortexed and incubated at 37°C for 5 min. Then, 450 μL of ultrapure water, containing 250 µM of ribitol, was added to the extracts and the samples were centrifuged for 5 min at 13,200g, at 4°C, to separate phases. The upper phase was collected (600 µL) and 50 μL of the supernatant was evaporated overnight under vacuum.
Sample Derivatization
Next, 50 μL of a pyridine solution was added to the evaporated supernatant, containing 20 mg/mL of methoxyamine hydrochloride and 5 µL/mL of a 2 mg/mL n-alkane mixture (retention index standards: decane, dodecane, pentadecane, octadecane, nonadecane, docosane, octacosane, dotriacontane, and hexatriacontane). The tubes were sealed and heated to 40°C for 90 min. Then, 80 μL of N,O-bis(trimethylsilyl)trifluoroacetamide was silylated for 30 min at 40°C.
GC-MS Analysis
Analyses were performed by gas chromatography (Agilent 6890) and quadrupole mass spectrometry with electron impact ionization (Agilent 5973 Network). A volume of 1 μL was injected on an Agilent J&W HP-5ms column (diameter, 0.25 mm; film thickness, 0.25 µm; length, 30 m) with a 0.6-mL/min helium flow. The injection parameters were as follows: splitless injection at 230°C with a 1-min purge at 20 mL/min. The temperature gradient was 1 min at 70°C; 9 min until 320°C; and then 10 min at 320°C, and the solvent delay was 5.4 min. The source was set to 250°C and 70 eV, scanning at 20 scans/min, from 70 to 600 m/z. Acquisition was performed with Chemstation software (Agilent).
Data Treatment
Semiautomated integration of chromatograms was performed with Quanlynx software (Waters) after conversion of the raw data into NetCDF format. A model ion was chosen manually to perform a correct integration for 138 peaks, with the following criteria: Gaussian shape, high relative abundance in the mass spectrum, and absence of saturation in any sample. Three abundant compounds, i.e., sucrose, citrate, and malate, gave a saturated signal for any ion; the samples were thus reinjected in split mode (1:30) to integrate these compounds. Metabolites were annotated according to the Chemical Analysis Working Group (Sumner et al., 2007): 41 metabolites were identified using pure reference standards, 34 putatively annotated compounds on the basis of both spectral and retention index matches with public database (The Golm Metabolome Database; Kopka et al., 2005), 41 compounds related to known compounds or chemical classes on the unique basis of spectral match, and 10 unknown analytes. Peak surfaces were then normalized according to the internal standard peak surface (ribitol) and sample fresh weight. Data are the average of the results obtained from four biological replicates.
Hexokinase Activity Assay
Wild-type, mutant, and transgenic plants were grown under SD conditions for 3 weeks and subsequently transferred to LD conditions. At least 200 mg of rosette leaves was harvested 2 d after transfer to LD. Frozen tissues were ground in liquid nitrogen and resuspended in 2 mL of extraction buffer (50 mM Bicine-NaOH buffer, pH 8.5, 5 mM MgCl2, 0.1% [v/v] Triton X-100, and a few microliters of protease inhibitor cocktail). The extracts were centrifuged for 5 min at 10,000 rpm at 4°C, and the supernatants were desalted with PD10 columns (GE Healthcare). The extracts were eluted from the column with 3.5 mL of elution buffer (same as extraction buffer, but lacking Triton). The desalted extracts were directly used for protein quantification or activity assays. For glucokinase activity measurement, desalted extracts were mixed with the glucose reaction buffer (50 mM Bicine-NaOH buffer, pH 8.5, 5 mM MgCl2, 1 mM NADP, 2.5 mM ATP, 2 mM glucose, and 1 unit/mL of G6PDH enzyme [Roche]) in a 1-mL cuvette, and absorbance at 340 nm was measured continuously for 30 min with a spectrophotometer. For the fructokinase activity assay, desalted extracts were mixed with the fructose reaction buffer (50 mM Bicine-NaOH buffer, pH 8.5, 5 mM MgCl2, 1 mM NADP, 2.5 mM ATP, 2 mM fructose, 1 unit/mL of G6PDH, and 2 units/mL of PGI enzyme [Roche]), and measurements were performed as for the glucokinase activity. Data presented here are the average of the results obtained from four biological replicates.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: MIPS1 (AT4G39800), MIPS2 (AT2G22240), MIPS3 (AT5G10170), and HXK1(At4g29130).
Supplemental Data
Supplemental Figure 1. Phenotypic analyses of the mips1-1 hxk1-1 suppressor mutant.
Supplemental Figure 2. Position of threonine 231 in HXK1 primary protein sequences.
Supplemental Figure 3. Relative expression of PR1 and PR5.
Supplemental Figure 4. HXK1 metabolic and signaling functions in the mutants hxk1-2 and mips1-2 hxk1-2.
Supplemental Figure 5. Effect of mips1, mips2, and hxk1 mutations and of complementation on MIPS2 expression.
Supplementary Material
Acknowledgments
We thank S. Merlot (ISV, CNRS, Gif/Yvette, France) for assistance with bioinformatic and sequence analyses and S. Nessler (IBBMC, Université Paris-Sud, Gif/Yvette, France) for assistance with HXK1 structural analysis. We thank T. Messier and Q. Hocheux for assistance during phenotypical characterization.
AUTHOR CONTRIBUTIONS
Q.B. performed experiments, analyzed data, and wrote the article. F.P., L.d.B., C.M., M.G., R.L., M.B., C.B., and C.R. performed experiments and analyzed data. M.D. designed research and wrote the article.
Glossary
- MI
myo-inositol
- HXK
hexokinase
- G6P
glucose-6-P
- MIPS
myo-inositol 1-phosphate synthase
- PCD
programmed cell death
- SA
salicylic acid
- LD
long-day
- SD
short-day
- Col-0
Columbia-0
- Ws
Wassilewskija
- BSA
bulk segregant analysis
- SNP
single-nucleotide polymorphism
- CAPS
cleaved amplified polymorphic sequence
- RT-qPCR
quantitative RT-PCR
- MS
Murashige and Skoog
- GC-MS
gas chromatography-mass spectrometry
References
- Aki T., Konishi M., Kikuchi T., Fujimori T., Yoneyama T., Yanagisawa S. (2007). Distinct modulations of the hexokinase1-mediated glucose response and hexokinase1-independent processes by HYS1/CPR5 in Arabidopsis. J. Exp. Bot. 58: 3239–3248. [DOI] [PubMed] [Google Scholar]
- Alcázar-Román A.R., Wente S.R. (2008). Inositol polyphosphates: a new frontier for regulating gene expression. Chromosoma 117: 1–13. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Baillieul F., Genetet I., Kopp M., Saindrenan P., Fritig B., Kauffmann S. (1995). A new elicitor of the hypersensitive response in tobacco: a fungal glycoprotein elicits cell death, expression of defence genes, production of salicylic acid, and induction of systemic acquired resistance. Plant J. 8: 551–560. [DOI] [PubMed] [Google Scholar]
- Berkey R., Bendigeri D., Xiao S. (2012). Sphingolipids and plant defense/disease: the “death” connection and beyond. Front. Plant Sci. 3: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolouri Moghaddam M.R., Van den Ende W. (2012). Sugars and plant innate immunity. J. Exp. Bot. 63: 3989–3998. [DOI] [PubMed] [Google Scholar]
- Bruggeman Q., Garmier M., de Bont L., Soubigou-Taconnat L., Mazubert C., Benhamed M., Raynaud C., Bergounioux C., Delarue M. (2014). The polyadenylation factor subunit CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR30: a key factor of programmed cell death and a regulator of immunity in Arabidopsis. Plant Physiol. 165: 732–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouch S., Noctor G. (2010). Myo-inositol abolishes salicylic acid-dependent cell death and pathogen defence responses triggered by peroxisomal hydrogen peroxide. New Phytol. 188: 711–718. [DOI] [PubMed] [Google Scholar]
- Chen H., Xiong L. (2010). myo-Inositol-1-phosphate synthase is required for polar auxin transport and organ development. J. Biol. Chem. 285: 24238–24247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin P.L., Gatzek S., Wheeler G.L., Dowdle J., Raymond M.J., Rolinski S., Isupov M., Littlechild J.A., Smirnoff N. (2006). Arabidopsis thaliana VTC4 encodes L-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme. J. Biol. Chem. 281: 15662–15670. [DOI] [PubMed] [Google Scholar]
- Donahue J.L., Alford S.R., Torabinejad J., Kerwin R.E., Nourbakhsh A., Ray W.K., Hernick M., Huang X., Lyons B.M., Hein P.P., Gillaspy G.E. (2010). The Arabidopsis thaliana Myo-inositol 1-phosphate synthase1 gene is required for Myo-inositol synthesis and suppression of cell death. Plant Cell 22: 888–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg F., Bolden A.H., Loewus F.A. (1964). Inositol formation by cyclization of glucose chain in rat testis. Biochem. Biophys. Res. Commun. 14: 419–424. [DOI] [PubMed] [Google Scholar]
- GhoshDastidar K., Chatterjee A., Chatterjee A., Majumder A.L. (2006). Evolutionary divergence of L-myo-inositol 1-phosphate synthase: significance of a “core catalytic structure”. Subcell. Biochem. 39: 315–340. [PubMed] [Google Scholar]
- Gillaspy G.E. (2011). The cellular language of myo-inositol signaling. New Phytol. 192: 823–839. [DOI] [PubMed] [Google Scholar]
- Gillaspy G.E. (2013). The role of phosphoinositides and inositol phosphates in plant cell signaling. Adv. Exp. Med. Biol. 991: 141–157. [DOI] [PubMed] [Google Scholar]
- Granot D., Kelly G., Stein O., David-Schwartz R. (2014). Substantial roles of hexokinase and fructokinase in the effects of sugars on plant physiology and development. J. Exp. Bot. 65: 809–819. [DOI] [PubMed] [Google Scholar]
- Herbers K., Meuwly P., Métraux J.P., Sonnewald U. (1996). Salicylic acid-independent induction of pathogenesis-related protein transcripts by sugars is dependent on leaf developmental stage. FEBS Lett. 397: 239–244. [DOI] [PubMed] [Google Scholar]
- Jang J.C., León P., Zhou L., Sheen J. (1997). Hexokinase as a sugar sensor in higher plants. Plant Cell 9: 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.D., Sussex I.M. (1995). 1 L-myo-inositol 1-phosphate synthase from Arabidopsis thaliana. Plant Physiol. 107: 613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Lim J.H., Ahn C.S., Park K., Kim G.T., Kim W.T., Pai H.S. (2006). Mitochondria-associated hexokinases play a role in the control of programmed cell death in Nicotiana benthamiana. Plant Cell 18: 2341–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J., et al. (2005). GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21: 1635–1638. [DOI] [PubMed] [Google Scholar]
- Kraakman L.S., Winderickx J., Thevelein J.M., De Winde J.H. (1999). Structure-function analysis of yeast hexokinase: structural requirements for triggering cAMP signalling and catabolite repression. Biochem. J. 343: 159–168. [PMC free article] [PubMed] [Google Scholar]
- Kuettner E.B., Kettner K., Keim A., Svergun D.I., Volke D., Singer D., Hoffmann R., Müller E.C., Otto A., Kriegel T.M., Sträter N. (2010). Crystal structure of hexokinase KlHxk1 of Kluyveromyces lactis: a molecular basis for understanding the control of yeast hexokinase functions via covalent modification and oligomerization. J. Biol. Chem. 285: 41019–41033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrasse D., Jégu T., Meng P.H., Mazubert C., Hudik E., Delarue M., Charon C., Crespi M., Hirt H., Raynaud C., Bergounioux C., Benhamed M. (2013). Dual function of MIPS1 as a metabolic enzyme and transcriptional regulator. Nucleic Acids Res. 41: 2907–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Yao N., Song J.T., Luo S., Lu H., Greenberg J.T. (2003). Ceramides modulate programmed cell death in plants. Genes Dev. 17: 2636–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewus F.A., Murthy P.P.N. (2000). myo-Inositol metabolism in plants. Plant Sci. 150: 1–19. [Google Scholar]
- Lorence A., Chevone B.I., Mendes P., Nessler C.L. (2004). myo-inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol. 134: 1200–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Qin G., Zhang J., Liang Y., Song Y., Zhao M., Tsuge T., Aoyama T., Liu J., Gu H., Qu L.J. (2011). D-myo-inositol-3-phosphate affects phosphatidylinositol-mediated endomembrane function in Arabidopsis and is essential for auxin-regulated embryogenesis. Plant Cell 23: 1352–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Bloom L.M., Zhu Z.M., Walsh C.T., Botstein D. (1989). Isolation and characterization of mutations in the HXK2 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 9: 5630–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng P.H., Raynaud C., Tcherkez G., Blanchet S., Massoud K., Domenichini S., Henry Y., Soubigou-Taconnat L., Lelarge-Trouverie C., Saindrenan P., Renou J.P., Bergounioux C. (2009). Crosstalks between myo-inositol metabolism, programmed cell death and basal immunity in Arabidopsis. PLoS ONE 4: e7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michell R.H. (2008). Inositol derivatives: evolution and functions. Nat. Rev. Mol. Cell Biol. 9: 151–161. [DOI] [PubMed] [Google Scholar]
- Mitsuhashi N., Kondo M., Nakaune S., Ohnishi M., Hayashi M., Hara-Nishimura I., Richardson A., Fukaki H., Nishimura M., Mimura T. (2008). Localization of myo-inositol-1-phosphate synthase to the endosperm in developing seeds of Arabidopsis. J. Exp. Bot. 59: 3069–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B., Zhou L., Rolland F., Hall Q., Cheng W.H., Liu Y.X., Hwang I., Jones T., Sheen J. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Păcurar D.I., Păcurar M.L., Street N., Bussell J.D., Pop T.I., Gutierrez L., Bellini C. (2012). A collection of INDEL markers for map-based cloning in seven Arabidopsis accessions. J. Exp. Bot. 63: 2491–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J., Laxmi A., St Martin S.K., Jang J.C. (2004). Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16: 2128–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzio J.A., Lorence A., Chevone B.I., Nessler C.L. (2003). L-Gulono-1,4-lactone oxidase expression rescues vitamin C-deficient Arabidopsis (vtc) mutants. Plant Mol. Biol. 53: 837–844. [DOI] [PubMed] [Google Scholar]
- Roessner U., Wagner C., Kopka J., Trethewey R.N., Willmitzer L. (2000). Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 23: 131–142. [DOI] [PubMed] [Google Scholar]
- Roessner-Tunali U., Hegemann B., Lytovchenko A., Carrari F., Bruedigam C., Granot D., Fernie A.R. (2003). Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol. 133: 84–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas C.M., Senthil-Kumar M., Tzin V., Mysore K.S. (2014). Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F., Baena-Gonzalez E., Sheen J. (2006). Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 57: 675–709. [DOI] [PubMed] [Google Scholar]
- Samson F., Brunaud V., Balzergue S., Dubreucq B., Lepiniec L., Pelletier G., Caboche M., Lecharny A. (2002). FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30: 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rangel D., Rivas-San Vicente M., de la Torre-Hernández M.E., Nájera-Martínez M., Plasencia J. (2015). Deciphering the link between salicylic acid signaling and sphingolipid metabolism. Front. Plant Sci. 6: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C., Langlois-Meurinne M., Bellvert F., Garmier M., Didierlaurent L., Massoud K., Chaouch S., Marie A., Bodo B., Kauffmann S., Noctor G., Saindrenan P. (2010). The differential spatial distribution of secondary metabolites in Arabidopsis leaves reacting hypersensitively to Pseudomonas syringae pv. tomato is dependent on the oxidative burst. J. Exp. Bot. 61: 3355–3370. [DOI] [PubMed] [Google Scholar]
- Sumner L.W., et al. (2007). Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3: 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T., Takahashi S., Shinozaki K. (2006). Inositols and their metabolites in abiotic and biotic stress responses. Subcell. Biochem. 39: 239–264. [DOI] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L.I., Sharon M., Zheng C., Robinson C.V., Estelle M., Zheng N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645. [DOI] [PubMed] [Google Scholar]
- Torabinejad J., Gillaspy G.E. (2006). Functional genomics of inositol metabolism. Subcell. Biochem. 39: 47–70. [DOI] [PubMed] [Google Scholar]
- Torabinejad J., Donahue J.L., Gunesekera B.N., Allen-Daniels M.J., Gillaspy G.E. (2009). VTC4 is a bifunctional enzyme that affects myoinositol and ascorbate biosynthesis in plants. Plant Physiol. 150: 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Ende W. (2013). Multifunctional fructans and raffinose family oligosaccharides. Front. Plant Sci. 4: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., et al. (2008). An inositolphosphorylceramide synthase is involved in regulation of plant programmed cell death associated with defense in Arabidopsis. Plant Cell 20: 3163–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Sheen J., Jang J.C. (2000). The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol. Biol. 44: 451–461. [DOI] [PubMed] [Google Scholar]
- Yoshida K.T., Fujiwara T., Naito S. (2002). The synergistic effects of sugar and abscisic acid on myo-inositol-1-phosphate synthase expression. Physiol. Plant. 114: 581–587. [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.