The Arabidopsis SWI/SNF chromatin-remodeling ATPase BRAHMA acts to maintain the stem cell niche in roots by modulating auxin distribution via regulation of the expression of PINs.
Abstract
BRAHMA (BRM), a SWI/SNF chromatin remodeling ATPase, is essential for the transcriptional reprogramming associated with development and cell differentiation in Arabidopsis thaliana. In this study, we show that loss-of-function mutations in BRM led to defective maintenance of the root stem cell niche, decreased meristematic activity, and stunted root growth. Mutations of BRM affected auxin distribution by reducing local expression of several PIN-FORMED (PIN) genes in the stem cells and impaired the expression of the stem cell transcription factor genes PLETHORA (PLT1) and PLT2. Chromatin immunoprecipitation assays showed that BRM could directly target to the chromatin of PIN1, PIN2, PIN3, PIN4, and PIN7. In addition, genetic interaction assays indicate that PLTs acted downstream of BRM, and overexpression of PLT2 partially rescued the stem cell niche defect of brm mutants. Taken together, these results support the idea that BRM acts in the PLT pathway to maintain the root stem cell niche by altering the expression of PINs.
INTRODUCTION
Plant roots form from a reservoir of undifferentiated cells, the root stem cells, in the root apical meristem. Within the root meristem, the quiescent center (QC), a small group of mitotically inactive cells, maintains the root stem cells. The QC generates unknown, non-cell-autonomous signals that prevent differentiation of the stem cells, through direct cell-cell contacts (van den Berg et al., 1997). This short-range signaling restrains the division of stem cells so that the stem cells do not become displaced from the growing root tip.
Two main pathways, the PLETHORA (PLT) pathway (Aida et al., 2004; Blilou et al., 2005) and the SHORT-ROOT (SHR)/SCARECROW (SCR)/RETINOBLASTOMA RELATED pathway (Helariutta et al., 2000; Nakajima et al., 2001; Sabatini et al., 2003; Wildwater et al., 2005), act to specify the stem cell niche. SHR and SCR also act upstream of the PLTs, as shr-2 and scr-4 mutants show decreased expression of PLT1 and PLT2 (Aida et al., 2004; Koizumi and Gallagher, 2013). SHR and SCR, which encode members of the plant-specific GRAS family of putative transcription factors, are required for stem cell maintenance. Genetic and molecular data indicate that SHR is expressed in the central stele tissue where the vasculature forms; SHR subsequently moves into the surrounding tissue layer to directly activate SCR expression by binding to the SCR promoter (Levesque et al., 2006; Cui et al., 2007). SCR forms a heterodimer with SHR to inhibit the binding of SHR at the SCR promoter. This feedback loop is thought to enable the rapid upregulation of SCR expression and limit the movement of SHR to a single cell layer adjacent to the stele.
The auxin-inducible PLT1 and PLT2 genes, which encode members of the AP2 class of transcription factors, are also essential for root stem cell niche maintenance (Aida et al., 2004; Galinha et al., 2007). PLTs function as dose-dependent regulators: High concentrations of PLTs maintain the QC and stem cell activity, intermediate concentrations regulate the division and differentiation of the transit-amplifying cells, and low concentrations allow differentiation to proceed (Galinha et al., 2007). Interestingly, the expression of PLT genes requires auxin response transcription factors and forms gradients that are thought to be a readout of an underlying auxin gradient (Aida et al., 2004; Galinha et al., 2007; Grieneisen et al., 2007). The PLT gradient is not a direct, proportionate readout of the auxin gradient. Rather, prolonged high auxin levels generate a narrow domain of PLT transcription, which generates a gradient of PLT protein through slow growth dilution and cell-to-cell movement (Mähönen et al., 2014). In addition, the gradient expression of PLTs is PIN dependent in controlling auxin-mediated root patterning. PIN proteins restrict PLT transcription in the basal embryo region to initiate root primordium formation. Conversely, the transcription of PINs that stabilizes the position of the stem cell niche is maintained by PLTs (Blilou et al., 2005; Grieneisen et al., 2007; Dinneny and Benfey, 2008).
The ATP-dependent SWI/SNF chromatin remodeling complexes affect gene expression by using the energy of ATP hydrolysis to alter the interactions between histones and DNA to create accessible DNA (Cairns, 2005). SWI and SNF were first identified from Saccharomyces cerevisiae by the examination of mating type switching (SWI) and sucrose nonfermenting (SNF) mutants (Neigeborn and Carlson, 1984; Stern et al., 1984). Biochemical analysis demonstrated that the yeast SWI/SNF complexes possess a catalytic subunit (ATPase) and 10 accessory core subunits (Cairns et al., 1994; Peterson et al., 1994) and can facilitate binding of transcription factors to nucleosomal DNA (Côté et al., 1994). Although the SWI/SNF complexes have been shown to play a central role in animal development and cell differentiation (Pedersen et al., 2001; Ohkawa et al., 2006), relatively little is known about their functions in plants. Nevertheless, genome analysis suggests that the Arabidopsis thaliana genome encodes more than 40 ATPases of the SNF2 family, four of which (BRM, SPLAYED [SYD], CHR12, and CHR23) belong to the SWI2/SNF2 subfamily based on phylogenetic analysis of the SNF2 ATPase catalytic domains (Knizewski et al., 2008). Several lines of evidence suggest that BRM is the ATPase of at least one of the putative SWI/SNF complexes in Arabidopsis. BRM has all the domains, including a C-terminal bromodomain that is a characteristic of ATPases of SWI/SNF complexes in yeast and Drosophila melanogaster (Knizewski et al., 2008).
Recent data suggest that BRM plays a crucial role in vegetative, embryonic, and reproductive plant development (Kwon et al., 2006; Jerzmanowski, 2007; Tang et al., 2008; Han et al., 2012; Wu et al., 2012; Efroni et al., 2013; Zhu et al., 2013; Vercruyssen et al., 2014). Indeed, Arabidopsis BRM is primarily expressed in meristems, organ primordia, and tissues with active cell division (Farrona et al., 2004). The brm mutant shows pleiotropic phenotypes, such as reduced plant size with short roots (Farrona et al., 2004; Hurtado et al., 2006), downward-curling leaves (Hurtado et al., 2006), hypersensitivity to abscisic acid (Han et al., 2012), and early flowering (Farrona et al., 2004, 2007). Recent work showed that the SYD and BRM ATPases interact with LEAFY and SEPALLATA3 proteins involved in controlling floral organ identity and act antagonistically with Polycomb repressors (Wu et al., 2012; Li et al., 2015). In addition, BRM associates with TCP4, ANGUSTIFOLIA3, and BREVIPEDICELLUS (BP) to regulate leaf development and inflorescence architecture (Efroni et al., 2013; Vercruyssen et al., 2014; Zhao et al., 2015).
In this study, we show that mutations of BRM led to defective root stem cell niche maintenance. BRM specifically bound to PIN loci and activated the expression of PIN genes. Overexpression of PLT2 partially rescued the stem cell niche defect of brm-3 mutants, indicating that BRM affects root stem cell niche maintenance mainly through the PLT pathway.
RESULTS
BRM Mutations Cause Stunted Root Growth and Reduced Root Meristem Size
To investigate the role of BRM in root development, we analyzed the phenotype of three brm mutant alleles, brm-1, brm-3, and brm-5. The brm-1 and brm-3 alleles carry T-DNA insertions in the first exon and 11th intron of BRM (Hurtado et al., 2006; Farrona et al., 2007), respectively; brm-5 is an ethyl methanesulfonate mutant (Tang et al., 2008). Previous studies indicated that disruption of BRM causes pleiotropic defects in shoots (Farrona et al., 2004) and short roots (Hurtado et al., 2006; Wu et al., 2012). Plants carrying the null mutant allele brm-1 are sterile, since brm-1 flowers fail to open at maturity and cannot set seeds (Kwon et al., 2006), whereas plants carrying the weak alleles brm-3 and brm-5 are fertile. We also observed the short root phenotype in brm-1, brm-3, and brm-5 mutant seedlings (Figure 1A). The primary root length of brm-1 mutant seedlings was significantly reduced compared with the weak alleles, brm-3 and brm-5, at 7 d after germination (DAG) (Figure 1A). The growth rate of the primary root was markedly reduced in brm-1, brm-3, and brm-5 mutants as early as 2 DAG (Figure 1B). At 10 DAG, the primary root lengths of brm-1, brm-3, and brm-5 seedlings were only ∼29, 54, and 60% of the wild type (Figure 1B). Compared with the wild type, brm-1, brm-3, and brm-5 seedlings showed significantly smaller meristem sizes at different DAG (Figure 1C). The differentiated epidermal cells of brm-1, brm-3, and brm-5 plants at 5 DAG were significantly smaller than those of the wild type (Figure 1D).
Figure 1.
BRM Deficiency Leads to Reduced Root Meristem Size and Stunted Root Growth.
(A) Phenotypes of wild-type (Columbia-0 [Col-0]), brm-3, brm-5, and brm-1 seedlings at 7 DAG. Bar = 1 cm.
(B) Primary root length of wild-type (Col-0), brm-3, brm-5, and brm-1 seedlings from 0 to 10 DAG. Data shown are means ± sd (n = 30).
(C) Root meristem size of the wild-type (Col-0), brm-3, brm-5, and brm-1 seedlings from 0 to 10 DAG. The root meristem size is expressed as the length from cortex cells in QC to the transition zone. Data shown are means ± sd (n = 25).
(D) The size of differentiated epidermis cells of the wild-type (Col-0), brm-3, brm-5, and brm-1 seedlings at 5 DAG. Data shown are means ± sd (for the wild type, brm-3, and brm-5, n = 40; for brm-1, n = 20).
Consistent with previous data (Jerzmanowski, 2007), more lateral roots were observed in brm-1, brm-3, and brm-5 mutant seedlings after 10 DAG (Supplemental Figure 1), suggesting that BRM may also play a role in lateral root development. The expression of CycB1;1 and CycB1;3, two markers for the G2/M phase of the cell cycle (Colón-Carmona et al., 1999), was reduced in brm-3 mutants (Supplemental Figure 2), indicating that the population of dividing cells is highly reduced in these mutants. Collectively, these results indicate that the short-root phenotype of brm mutants results from effects on both the cell division activity of root meristems and on cell elongation.
BRM Regulates Stem Cell Niche Maintenance
The observation that BRM is crucial for maintaining meristem sizes in roots prompted us to investigate its possible effects on the cellular organization of the QC and surrounding stem cells. The QC cells can be easily discerned by confocal microscopy in wild type (Figure 2A), and the pattern of cells in the root tips of brm-3 and brm-5 roots was not disrupted (Supplemental Figure 3A). However, the pattern of cells was disrupted in brm-1 root tips and the QC could not be identified morphologically (Figure 2A). In addition, the differentiated columella area next to columella stem cells (CSCs) in brm-1 mutants consists of irregularly shaped cells (Figure 2A; Supplemental Figure 3A). Furthermore, the presence of starch granules, which mark differentiated columella cells, were found in the CSC area in brm-1 roots (Figure 2B). These data indicate that the activity of CSC was decreased in brm-1 roots. Similar results were also observed in the brm-3 and brm-5 mutants (Figure 2B). Furthermore, the 4th differentiated columella cells in brm-3 and brm-5 were remarkably smaller compared with those of the wild type (Supplemental Figure 3B).
Figure 2.
BRM Mutants Show Defective Root Stem Cell Niche Maintenance.
(A) Cellular organization of wild-type and brm-1 root tips at 3 DAG using PI staining. The white arrow shows the QC cells. Bars = 50 μm.
(B) Lugol-stained (light blue) roots of the wild type (Col-0), brm-3, brm-5, and brm-1 at 3 DAG. Black and red arrows indicate QC and columella initials (CSC), respectively. Bars = 50 μm.
(C) The expression QC25 and QC46 GUS markers in the wild type (Col-0) and brm-3 at 3 DAG. Bars = 25 μm.
To further investigate whether the disruption of the stem cell niche is associated with misspecification of the QC, we monitored the expression of three independent QC-specific markers (QC25, QC46, and QC184) as indicated by the β-glucuronidase (GUS) reporter in brm mutants. We selected the hypomorphic brm-3 and brm-5 alleles for further analysis because they are fertile, which facilitated testing of homozygous mutant embryos. In wild-type plants, QC25 and QC46 were expressed in QC cells (Figure 2C). In a large proportion of brm-3 roots, however, QC25 and QC46 expression were either absent or highly reduced (Figure 2C, Table 1). Similarly, another QC marker QC184 was also aberrantly expressed in the brm-3 roots (Table 1). These results indicate that BRM is essential for proper QC identity and CSC activity.
Table 1. The Expression of QC Markers in the Wild Type and brm-3 Mutants.
| QC Markers | No Expression | Reduced Expression | Expanded Expression | Normal Expression | Total Numbers | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wild Type | brm-3 | Wild Type | brm-3 | Wild Type | brm-3 | Wild Type | brm-3 | Wild Type | brm-3 | |
| QC25 | 8.3% | 24.3%a | 16.4% | 32.4%a | 6.8% | 8.2% | 68.5% | 35.1% | 67 | 85 |
| QC46 | 10.5% | 31.2%a | 10.3% | 30.6%a | 11.3% | 13.7% | 67.9% | 24.5% | 52 | 62 |
| QC184 | 11.8% | 38.1%a | 13.2% | 29.4%a | 9.6% | 10.8% | 65.4% | 21.7% | 58 | 54 |
The GUS staining areas were measured using Digimizer image analysis software (http://www.digimizer.com). The staining areas between 50 and 90 μm2 were defined as “normal expression” (Supplemental Figure 10), <50 μm2 as “reduced expression,” and more than 90 μm2 as “expanded expression.”
One-way ANOVA (Student’s t test) analysis was performed, and statistically significant differences (P < 0.05) are indicated.
Mutation of BRM Affects the Expression of PINs in the Root Tip
In Arabidopsis root development, auxin regulates pattern formation as well as the orientation and extent of cell division (Sabatini et al., 1999). Polar auxin transport is a major factor in organ formation, such as the initiation of lateral roots and leaf primordia (Benková et al., 2003; Friml et al., 2003; Reinhardt et al., 2003). To investigate whether the brm-3 short root phenotype was related to auxin, we first monitored auxin accumulation using the DR5:GFP (green fluorescent protein) reporter (Ulmasov et al., 1997). We found that even though the expression of DR5:GFP in the brm-3 root tip showed reduced expansion compared with the wild type, the expression maxima of DR5:GFP localized in the center of the root meristem (Figures 3A to 3B). Next, we examined the auxin efflux transporter genes, PINs, which play an important role in stem cell niche maintenance (Friml et al., 2003; Blilou et al., 2005). The expression levels of PIN1 and PIN2, as shown by the PIN1:GFP and PIN2:GFP promoter fusion reporters (Figures 3C to 3D; Supplemental Figure 4A), were obviously reduced in the brm-3 root tip as marked by fluorescence (Figure 3E). Meanwhile, the transcript levels of PIN1 and PIN2 were also reduced in the brm-3 root tips and brm-1 seedlings (Figure 3F; Supplemental Figure 4B). In addition, similar to the previous microarray data using 18-d-old brm-1 seedlings (Archacki et al., 2013), the levels of PIN3, PIN4, and PIN7 were also reduced in brm-3 root tips (Figure 3F). Collectively, our data suggest that the short-root phenotype of brm was likely caused by reduced expression of the auxin transport-related genes and the concomitant alteration of local auxin distribution in the root tip.
Figure 3.
Mutations of BRM Affects the Contents of Auxin and the Expression of PIN Genes
(A) Expression pattern of the DR5pro:GFP reporters in the wild type (Col-0) and brm-3 at 3 DAG. Bars = 50 μm.
(B) Quantification of DR5pro:GFP fluorescence in the wild type (Col-0) and brm-3. Data shown are means ± sd (n = 25). Asterisks denote Student’s t test significant difference between wild-type and mutant plants (P ≤ 0.05).
(C) and (D) PIN1pro:PIN1:GFP and PIN2pro:PIN2:GFP expression in the wild type (Col-0) and brm-3 at 3 DAG. Bars = 50 μm
(E) Quantification of PIN1pro:PIN1:GFP and PIN2pro:PIN2:GFP fluorescence in the wild type (Col-0) and brm-3. Data shown are means ± sd. Asterisks denote Student’s t test significant difference between wild-type and mutant plants (P < 0.05).
(F) Real-time RT-PCR analysis of the expression of PIN genes in wild-type (Col-0), brm-3, and brm-5 roots. The total RNAs were isolated from roots of the wild type (Col-0), brm-3, and brm-5 (3 DAG), and UBQ10 was used as a control. Data are means ± sd of three biological repeats. Asterisks indicate significant differences compared with wild type (P < 0 .05; Student’s t test).
BRM Affects PLT1 and PLT2 Expression
As noted above, in Arabidopsis, two main pathways are involved in root stem cell niche maintenance (Aida et al., 2004). The SHR/SCR pathway provides positional information along the radial axis, whereas the PLT1/PLT2 pathway provides longitudinal information. To determine whether the disturbed stem cell niche in the brm mutants was caused by misregulation of these stem cell niche-defining transcription factors, we examined the expression pattern of these genes in brm-3 at 3 DAG. We first monitored the expression of SHR/SCR using SHRpro:GFP/SCRpro:GFP in the brm-3 mutants. The expression and localization of SHR (Supplemental Figure 5A) and SCR (Supplemental Figure 5C) were not affected in brm-3 mutants compared with the wild type. In addition, the root lengths of brm-3 shr-1 and brm-3 scr-1 double mutants were shorter than those of brm-3, shr-1, and scr-1 single mutants (Supplemental Figures 5B and 5D). Similar results were also observed in roots of plants carrying the null allele brm-1 (Supplemental Figures 5E to 5F). These data support that BRM acts in parallel with the SHR/SCR pathway.
To test whether mutations of BRM affect the expression of PLT genes, we quantified the PLT1 and PLT2 transcripts in the roots of wild-type, brm-3, brm-5, and brm-1 seedlings using reverse transcription-quantitative PCR (RT-qPCR) assays. The data showed that PLT1 and PLT2 transcripts were markedly reduced in brm mutants (Figure 4A; Supplemental Figure 4B). Similarly, yellow fluorescent protein (YFP) levels of the translational fusions, PLT1:YFP and PLT2:YFP, were also reduced in brm-3 compared with the wild type (Figures 4B to 4C), suggesting that loss of BRM activity also affects PLT1 and PLT2 expression at the protein level. Taken together, our results reveal that the defective stem cell niche maintenance in the brm mutants is correlated with a dramatic misregulation of PLT1 and PLT2 expression.
Figure 4.
Mutations of BRM Affect the Expression of PLT1 and PLT2.
(A) Real-time RT-PCR analysis of the expression of PLT1 and PLT2 in wild-type (Col-0), brm-3, and brm-5 roots. Total RNAs were isolated from 3 DAG roots of the wild type (Col-0), brm-3, and brm-5, and UBQ10 was used as a control. Data presented are means ± sd of three biological repeats. Asterisks indicate significant differences compared with the wild type (P < 0 .05; Student’s t test).
(B) PLT1:YFP and PLT2:YFP expression in wild-type (Col-0) and brm-3 root tips at 3 DAG. Bars = 50 μm.
(C) Quantification of PLT1:YFP and PLT2:YFP fluorescence in the wild type (Col-0) and brm-3. Data shown are means ± sd (n = 20). Asterisks denote Student’s t test significant difference between wild-type and mutant plants (P < 0.05).
(D) Root tips of the wild type (Col-0), brm-3, plt1-4 plt2-2, and brm-3 plt1-4 plt2-2 single, double, and triple mutants at 3 DAG. Bars = 50 μm.
(E) Statistics of meristem cell number of the indicated genotypes at 3 DAG. Data shown are means ± sd (n = 20). Different letters are used to indicate means that are significantly different (P < 0.05, Student’s t test).
To assess whether BRM acts in the PLT pathway, brm-3 plants were crossed to plt1 plt2 double mutant plants (Aida et al., 2004). The root lengths and meristem sizes of brm-3 plt1-4 plt2-2 and brm-1 plt1-4 plt2-2 triple mutants were similar to those of plt1-4 plt2-2 double mutants (Figures 4D and 4E; Supplemental Figure 6), confirming that BRM acts in the PLT pathway.
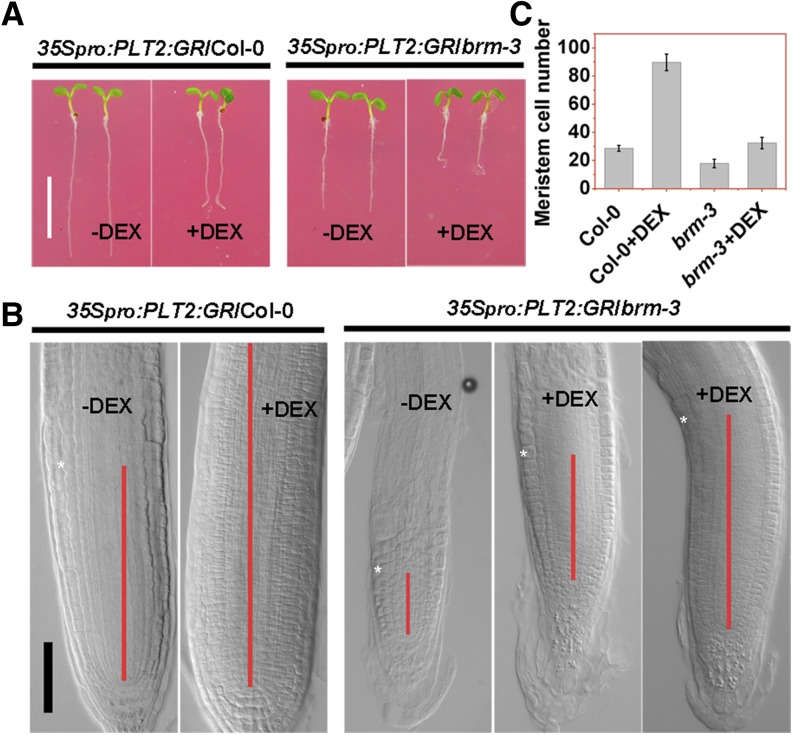
We further determined whether overexpression of PLT genes can rescue the brm-3 mutant phenotype. The inducible expression construct 35Spro:PLT2:GR (Galinha et al., 2007) was introduced into brm-3 through genetic crossing. A short-term induction (2 d) of PLT2 expression by adding dexamethasone (DEX) did not severely affect the growth of wild-type and mutant seedlings (Figure 5A). Consistent with previous reports (Galinha et al., 2007; Kornet and Scheres, 2009), a short-term induction of PLT2 expression by DEX led to a substantial increase of the meristem cell number in wild-type roots (Figures 5B and 5C). In the brm-3 background, the cell number of the meristem also significantly increased after DEX induction (Figures 5B and 5C), similar to the wild type with DEX induction (Figure 5C). About 48.1% of brm-3 seedlings (n = 56) possess meristem sizes similar to the wild type without DEX induction (Figure 5B). In addition, DEX induction substantially improved the length of columella cells in the 35Spro:PLT2:GR/brm-3 seedlings (Figure 5B). Together, these data indicate that overexpression of PLT2 can, at least partially, rescue the root meristem defects of brm-3, supporting the idea that BRM acts in the PLT pathway.
Figure 5.
Overexpression of PLT2 Partially Rescues the Root Meristem Defects of brm-3.
(A) Phenotypes of 5 DAG 35Spro:PLT2:GR/wild type (Col-0) and 35Spro:PLT2:GR/brm-3 seedlings without or with 2 μM DEX (+DEX) treatment for 2 d. Bar = 0.5 cm.
(B) The root tips of 5 DAG 35Spro:PLT2:GR/Col-0 and 35Spro:PLT2:GR/brm-3 seedlings without or with 2 μM DEX (+DEX) treatment for 2 d. Pink bars represent the root meristem of different plants extending from the QC to the transition zone. The white asterisks marked the meristem boundary where cortical cells rapidly expand. Bars = 50 μm.
(C) Quantification of the number of cortical cells in the meristem at 5 DAG, induced by 35Spro:PLT2:GR in the wild type (Col-0) and brm-3 2 d after 2 μM DEX treatment at 3 DAG. Data shown are average and sd (n = 20).
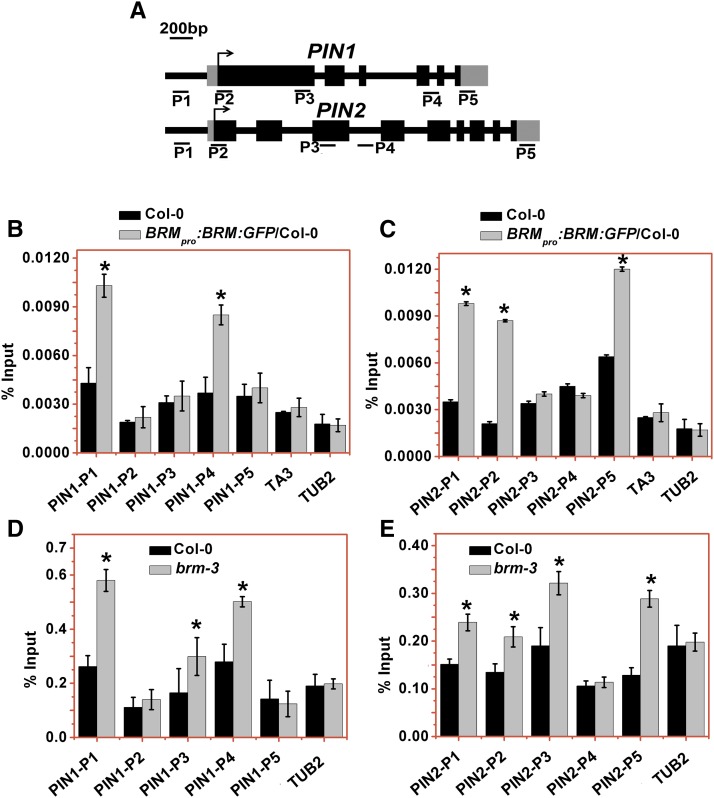
BRM Directly Targets to PIN Loci in Roots
As described above, we observed markedly reduced transcript levels of PINs and PLTs in brm-3 roots. Next, we investigated whether the effect of BRM on the mRNA accumulation of PINs and PLTs is direct or indirect. To test for binding of BRM to PIN and PLT loci, a GFP-tagged BRM (BRMpro:BRM:GFP) (Smaczniak et al., 2012; Li et al., 2015) that fully rescued the root meristem defects of the brm-1 null mutant (Supplemental Figure 7) was used in chromatin immunoprecipitation (ChIP) assays. Roots of 3 DAG seedlings were selected to investigate the enrichment of BRM in the different regions of the PINs and PLTs in the BRMpro:BRM:GFP plants. ChIP-qPCR was used to determine the regions enriched by ChIP with an anti-GFP antibody, and the length of the amplicon is shown in Supplemental Table 2. As shown in Figure 6, BRM bound to the promoter and the fourth to fifth exon regions of PIN1 (Figure 6B) as well as the promoter, transcriptional start region, and 5′-untranslated region (UTR) of PIN2 (Figure 6C). Similarly, BRM also bound to PIN3, PIN4, and PIN7 (Supplemental Figure 8A). These data suggest that these genes are the direct target genes regulated by BRM. In contrast, BRM did not bind to PLT1 and PLT2 (Supplemental Figure 9).
Figure 6.
Binding of BRM to PIN1 and PIN2 Loci and the Prevalent H3K27Me3 Levels in brm-3 Roots.
(A) Schematic diagram of PIN1 and PIN2. Black boxes, gray boxes, and black lines indicate the exon, UTR, and promoter and intron of indicated genes, respectively. The regions analyzed by ChIP-qPCR are indicated by P1-P5, and the length of amplicon is shown in Supplemental Table 2.
(B) and (C) ChIP-qPCR analysis of enrichment of BRMpro:BRM:GFP to the different regions of PIN1 (B) and PIN2 (C) in Col-0 and BRMpro:BRM:GFP/Col-0 roots. An anti-GFP antibody was used for the immunoprecipitation. BRMpro:BRM:GFP/Col-0 is a transgenic line expressing GFP-tagged BRM under the control of the BRM native promoter. TA3 and TUB2 were used as negative controls. Data are mean values ± sd of three replicates. Similar results were obtained for at least two additional independent experiments. Asterisks denote Student’s t test significant difference between Col-0 and BRMpro:BRM:GFP/Col-0 roots (P < 0.05).
(D) and (E) ChIP-qPCR analysis of H3K27me3 levels at the regions of PIN1 (D) and PIN2 (E) in Col-0 and brm-3 mutant roots. TUB2 was used as negative control. Data are mean values ± sd of three replicates. Similar results were obtained for at least two additional independent experiments. Asterisks denote Student’s t test significant difference between Col-0 and brm-3 roots.
Previous data showed that BRM interacts with LEAFY and SEPALLATA3 proteins to alter floral organ identity by acting antagonistically with Polycomb repressors (Wu et al., 2012). BRM also antagonizes the function of Polycomb group (PcG) proteins during plant development (Li et al., 2015). PcG proteins are involved in the establishment and maintenance of the repressed chromatin state, by introducing the H3K27me3 mark. Increased levels of H3K27me3 in the brm-3 roots were observed in the promoter, the first exon, and the fourth to fifth exon regions of PIN1 (Figure 6D). Furthermore, increased levels of H3K27me3 in the brm-3 roots were also observed in the promoter, the transcriptional start region, the third exon, and the 5′-UTR of PIN2 (Figure 6E), supporting that BRM antagonizes PcG function in regulation of PIN1 and PIN2. In contrast, levels of H3K27me3 in the brm-3 roots were not changed in PIN3, PIN4, and PIN7 (Supplemental Figure 8B). Previous studies indicated that PIN3, PIN4, and PIN7 are not associated with H3K27me3 in 10- to 14-d-old seedlings (Zhang et al., 2007). Similarly, we also found that the levels of H3K27me3 in PIN3, PIN4, and PIN7 were very low in both wild-type and brm-3 roots (Supplemental Figure 8B).
DISCUSSION
BRM Acts in the PLT Pathway to Regulate Root Stem Cell Niche Maintenance
BRM, the putative enzymatic motor subunit of the SWI/SNF chromatin-remodeling complex in plants, plays an essential role in cell patterning and differentiation (Farrona et al., 2004). In this study, we demonstrated that BRM regulates stem cell niche maintenance during root development in Arabidopsis. BRM mutations led to markedly reduced growth of the primary roots as well as smaller meristem sizes, indicating that BRM is involved in root development (Figures 1A to 1C). The cellular organization of the QC and surrounding stem cells was also disrupted in brm-1 root tips (Figures 2A and 2B). Although the QC acts as an organizer of root meristematic cells (Dolan et al., 1993; van den Berg et al., 1997; Aida et al., 2004), a low proliferation rate is observed, indicating that it can be a source for new stem cells (Dolan et al., 1993; van den Berg et al., 1995). Therefore, the fate of stem cells surrounding the QC can be used as a readout of the QC’s organizing activity. By measuring the expression of QC-specific markers and the differentiation of stem cells, we found the requirement for BRM in the maintenance of root stem cell niche. The aberrant expression of QC-specific markers such as QC25, QC46, and QC184 indicates that BRM is essential for maintaining proper identity and activity of the QC (Figure 2C, Table 1).
In Arabidopsis roots, two main pathways specify and maintain the identity and function of QC and the associated stem cells: the SHR/SCR pathway and the auxin/PLT pathway. The SHR/SCR pathway provides positional signal along the radial axis, whereas the PLT pathway provides the longitudinal signal (Scheres, 2007; Benjamins and Scheres, 2008; Petricka and Benfey, 2008). Our study suggests that BRM acts in the auxin/PLT pathway (Supplemental Figures 5 and 6). The BRM mutation leads to alteration in auxin contents (Figures 3A and 3B) and local expression levels of several PIN genes (Figures 3C to 3F; Supplemental Figure 4). In addition, the BRM mutation significantly impairs PLT1/2 expression at both the transcriptional and protein levels (Figures 4A to 4C), indicating that BRM plays an important role in mediating auxin-induced expression of PLT1/2. Furthermore, the brm-3 plt1-1 plt2-2 and brm-1 plt1-1 plt2-2 triple mutants have similar phenotypes to the plt1-1 plt2-2 double mutants, supporting that BRM acts in the PLT1/2 pathway (Figures 4D and 4E; Supplemental Figure 6). Overexpression of PLT2 can partially bypass the root meristem defects of brm-3 (Figure 5). Taken together, these observations demonstrated that BRM regulates PLT-mediated root stem cell niche maintenance.
PINs Are the Direct Target Genes Regulated by BRM
Auxins are involved in a wide range of developmental responses in plants. Graded concentrations of auxins established and maintained by auxin transport proteins are essential for embryonic, root, and shoot organogenesis (Friml et al., 2002; Benková et al., 2003; Reinhardt et al., 2003). The gradient of auxin along the roots is due to the collective activities and topology of the PIN proteins, the AUX1/LAX family proteins (Blilou et al., 2005; Grieneisen et al., 2007; Ugartechea-Chirino et al., 2010), and the multidrug-resistant/P-glycoprotein family proteins (Blakeslee et al., 2007). Auxin regulates the maintenance of the QC and the activity of the root meristem through PLTs (Galinha et al., 2007). Expression of PLTs is induced by PIN-driven auxin gradients; conversely, PIN transcription is maintained by PLT proteins to stabilize the position of the stem cell niche (Blilou et al., 2005; Grieneisen et al., 2007). Our data show that the expression of PINs and PLTs is reduced in brm mutant roots (Figures 3C to 3F and 4A to 4C). ChIP analysis showed that BRM bound to different regions of PIN1, PIN2, PIN3, PIN4, and PIN7 chromatin (Figures 6B and 6C; Supplemental Figure 8A). Since no DNA binding domain was found in BRM protein, BRM may bind to PINs by interacting with transcription factors and other DNA binding proteins. By using yeast two-hybrid screening assays, it was demonstrated that BRM could interact with a number of transcription factors (Wu et al., 2012). More recently, it was demonstrated that BRM interacts with BP to regulate the expression of KNAT2 and KNAT6 in control of inflorescence architecture (Zhao et al., 2015). Further research is required to investigate the molecular mechanism how BRM is targeted to other genomic loci such as PINs.
BRM Acts Antagonistically with PcG in the Regulation of PIN1 and PIN2
ATP-dependent chromatin-remodeling complexes control DNA accessibility by positioning, moving, or exchanging nucleosomes via ATP-dependent alterations in histone-DNA contacts (Jiang and Pugh, 2009; Hargreaves and Crabtree, 2011). Based on the sequence similarity of their conserved ATPase subunits, they are classified into four distinct families: ISWI (ISW1a, ISW1b, and ISW2), INO80/SWR1, CHD, and SWI/SNF (including RSC) (Hota and Bartholomew, 2011). In yeast, SWI/SNF binds almost exclusively to promoters and activates its direct targets concomitant with nucleosome displacement. However, human BAF complexes are most often found in intergenic regions where they both activate and repress genes (Hargreaves and Crabtree, 2011). Similarly, the plant BRM also plays a dual role in gene transcription, since 1090 genes were downregulated, while 1115 genes were upregulated in brm-1 mutants (Archacki et al., 2013). In yeast, the SWI/SNF complex was considered as a general activator of transcription, working in coordination with sequence-specific transactivators and the histone acetyltransferase GCN5 (Biggar and Crabtree, 1999). Indeed, stable promoter occupancy by the SWI/SNF complex requires the acetylation of the chromatin template by the histone acetyltransferase and the acetylated-lysine binding activity of the bromodomain of SWI2/SNF2 is required in this process (Hassan et al., 2002; Yamada et al., 2004; Ogiwara et al., 2011; Watanabe et al., 2013). These data indicate that histones around some loci are hyperacetylated by histone acetyltransferases, and acetylated histones are preferential targets of ATP-dependent chromatin remodeling proteins. Similar to our results, the plant histone acetyltransferases GCN5 and ADA2b regulate the expression of PLTs to modulate root development (Kornet and Scheres, 2009), indicating that the Arabidopsis SWI/SNF complex containing BRM may act collaboratively with histone acetyltransferases in regulating gene expression in root development.
In Drosophila, BRM was initially classified as a Trithorax group (TrxG) protein since it activates the transcription of homeotic genes and thus antagonizes the function of PcG during fly development (Tamkun et al., 1992; Hargreaves and Crabtree, 2011). Recent studies in plants indicated that SWI2/SNF2 ATPases SYD and BRM counteract PcG function in gene expression (Wu et al., 2012). These data indicate that the PcG-TrxG antagonistic regulation of gene expression is conserved between plants and metazoans, although the structures of these genes are not conserved. Nevertheless, SWI/SNF complexes also appear to cooperate together with Polycomb complexes to repress transcription at some loci (Farrona et al., 2011; Li et al., 2015). Our results show that the H3K27me3 levels of BRM binding regions were increased in PIN1 and PIN2 loci (Figure 6), supporting that BRM acts antagonistically with PcG functions in regulating gene expression during root development. However, the H3K27me3 levels of PIN3, PIN4, and PIN7 loci were not changed in brm-3 mutants (Supplemental Figure 8B), suggesting that H3K27me3 is not associated with increased PIN3, PIN4, and PIN7 expression in the mutant. Further research is required to investigate the molecular mechanism of BRM and PcG interaction in gene regulation.
METHODS
Plant Materials and Growth Conditions
The following marker lines and mutants were used: brm-1 (SALK_030046), brm-3 (SALK_088462), and brm-5 (Hurtado et al., 2006; Farrona et al., 2007; Tang et al., 2008); QC25, QC46, and QC18 (Sabatini et al., 2003); SHRpro:GFP (Helariutta et al., 2000); SCRpro:GFP (Wysocka-Diller et al., 2000); PLT1pro:PLT1:YFP and PLT2pro:PLT2:YFP (Galinha et al., 2007); DR5pro:GFP (Benková et al., 2003); PIN1pro:PIN1:GFP (Benková et al., 2003); PIN2pro:PIN2:GFP (Blilou et al., 2005); shr-1 (Benfey et al., 1993); scr-1 (Di Laurenzio et al., 1996); and plt1-4 plt2-2 (Aida et al., 2004).
Seeds were surface-sterilized for 2 min in 75% ethanol, followed by 5 min in 1% NaClO solution and rinsed five times with sterile water, plated on Murashige and Skoog (MS) medium with 1.5% sucrose and 0.8% agar, and then stratified at 4°C in the dark for 2 d. Plants were growth under long-day conditions (16 h light/8 h dark) at 22°C in a Phytotron.
Root Meristem Size Analysis
Seeds were germinated and grown on vertically oriented plates from 1 to 14 d. Roots were examined at different days after germination depending on the experiment. Approximately 30 to 50 seedlings were examined in at least three independent experiments. Roots were mounted in chloral hydrate and then root meristem sizes were determined by counting the number of cortex cells in a file extending from the QC to the first elongated cell (Perilli and Sabatini, 2010).
Histology and Microscopy
Roots were cleared in HCG solution (chloroacetaldehyde:water:glycerol = 8:3:1) for several minutes before microscopy analysis. For Lugol staining, roots were incubated in Lugol solution (Sigma-Aldrich) for 3 to 5 s, then washed by water and mounted in HCG solution for microscopy. Histochemical staining for GUS activity in homozygous transgenic plants was performed according to the described method (Jefferson et al., 1987). Whole seedlings were immersed in the GUS staining solution (1 mM X-glucuronide in 100 mM sodium phosphate, pH 7.2, 0.5 mM ferricyanide, 0.5 mM ferrocyanide, and 0.1%Triton X-100) and incubated at 37°C in the dark from 2 to 8 h depending on the experimental requirement. To determine the expression pattern of QC markers, we measured the GUS staining area using Digimizer image analysis software (http://www.digimizer.com). The staining areas between 50 and 90 μm2 were defined as “normal expression,” those lower than 50 μm2 as “reduced expression,” and those bigger than 90 μm2 as “expanded expression.” Plants on MS medium were photographed using the Leica DFC 490 stereomicroscope and Leica DM5000B microscope. Images were processed with Adobe Photoshop CS 8.0 software.
Homozygous transgenic plants were used for confocal imaging. Cell walls were labeled with propidium iodide (PI) as described (Truernit and Haseloff, 2008). Roots were counterstained with 10 μg mL−1 PI (Sigma-Aldrich) for 5 min, washed once in distilled water, and mounted in water for confocal microscopy. Confocal images were taken using a Zeiss LSM 710 laser scanning microscope with the following excitation (Ex) and emission (Em) wavelengths (Ex/Em): 561 nm/591 to 635 nm for PI, 488 nm/505 to 530 nm for GFP, and 514 nm/530 to 600 nm for YFP. The objective lenses 20× and 40× were used. Fluorescence was quantified with the LAS AF Lite program on confocal sections acquired with the same microscope settings. Approximately 10 to 15 images were examined, and at least two independent experiments were performed. The statistical significance was evaluated by Student’s t test analysis.
Gene Expression Analyses
Total RNA from 3 DAG roots was extracted with Trizol reagent (Invitrogen) according to the manufacturer’s protocol and used to synthesize cDNA. The gene-specific primers used for real-time PCR are listed in Supplemental Table 1. Each sample was quantified at least in triplicate and normalized using Ubiquitin10 (UBQ10) as an internal control.
ChIP Assays
Roots (∼0.3 g) from 3 DAG seedlings grown on vertically oriented plates with MS medium were collected for ChIP assays (Gendrel et al., 2005; Liu et al., 2012). After fixation with formaldehyde, the chromatin was sheared to an average length of 500 bp by sonication and then immunoprecipitated with GFP-Trap_A agarose beads (ChromoTek) or H3K27me3 antibody (Millipore 07-449). After cross-linking was reversed, the amount of precipitated DNA fragments and input DNA was detected by quantitative real-time PCR using specific primers listed in Supplemental Table 2. The percentage of input was calculated by determining 2−ΔCt (=2−[Ct(ChIP)−Ct(Input)]). The exon region of retrotransposon TA3 (Han et al., 2012) and TUB2 was used as negative control.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome initiative or GenBank/EMBL databases under the following accession numbers: BRM (AT2G46020), SCR (AT3G54220), SHR (AT4G37650), PLT1 (AT3G20840), PLT2 (AT1G51190), PIN1 (AT1G73590), PIN2 (AT5G57090), PIN3 (AT1G70940), PIN4 (AT2G01420), PIN7 (AT1G23080), CycB1;1 (AT4G37490), and CycB1;3 (AT3G11520).
Supplemental Data
Supplemental Figure 1. The lateral root number in wild-type, brm-1, brm-3, and brm-5 roots 10 DAG.
Supplemental Figure 2. The expression of CycB1;1 and CycB1;3 in 3 DAG wild-type, brm-3, and brm-5 roots.
Supplemental Figure 3. Cellular organization of wild-type (Col-0), brm-3, brm-5, and brm-1 root tips at 3 DAG.
Supplemental Figure 4. The expression of PIN2 in wild-type (Col-0) and brm-3 root tips at 3 DAG and the expression of root development related genes in wild-type (Col-0) and brm-1 5-d-old seedlings.
Supplemental Figure 5. The expression pattern of SHR and SCR in the wild type and brm-3 and the phenotype of shr-1 brm-3, scr-1 brm-3, shr-1 brm-1, and scr-1 brm-1 double mutants.
Supplemental Figure 6. The phenotype of brm-1 plt1-4 plt2-2 and brm-3 plt1-4 plt2-2 triple mutants at 7 DAG.
Supplemental Figure 7. The phenotype of Col-0 and BRMpro:BRM:GFP/brm-1 seedlings at 10 DAG.
Supplemental Figure 8. ChIP-qPCR analysis of BRM targeting to PIN3, PIN4, and PIN7 and H3K27me3 levels of PIN3, PIN4, and PIN7 loci in brm-3 mutant roots.
Supplemental Figure 9. BRM does not target to PLT1 and PLT2 directly.
Supplemental Figure 10. The normal expression of QC25 and QC46 in wild-type (Col-0) and brm-3 root tips at 3 DAG.
Supplemental Table 1. Primers used for RT-qPCR.
Supplemental Table 2. Primers used for ChIP-qPCR.
Supplementary Material
Acknowledgments
We thank Philip Benfey (Duke University) for providing 35Spro:PLT2:GR seeds, Ben Scheres (Utrecht University) for shr-1 and scr-1 seeds, Lizhen Tao (South China Agricultural University) for plt1-1/plt2-1 seeds, and the ABRC for kindly providing seeds used in this study. This work was supported by grants from the National Basic Research Program of China (973 Program No. 2012CB910900), the National Natural Science Foundation of China (No. 31201106 and No. 31371308), and the Ministry of Science and Technology of Taiwan (101-2311-B-002-012-MY3 and 103-2321-B-002-039).
AUTHOR CONTRIBUTIONS
S.Y. and K.W. conceived this project and designed all research. S.Y., C.L., L.Z., S.G., J.L., M.Z., C.-Y.C., X.L., and M.L. performed the research. S.Y.,Y.C., C.L., C.Y., and K.W. analyzed data. S.Y. and K.W. wrote the article.
Glossary
- QC
quiescent center
- DAG
days after germination
- CSC
columella stem cell
- RT-qPCR
reverse transcription-quantitative PCR
- DEX
dexamethasone
- ChIP
chromatin immunoprecipitation
- UTR
untranslated region
- MS
Murashige and Skoog
- PI
propidium iodide
- Col-0
Columbia-0
References
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.S., Amasino R., Scheres B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. [DOI] [PubMed] [Google Scholar]
- Archacki R., et al. (2013). BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin-mediated responses in Arabidopsis. PLoS ONE 8: e58588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey P.N., Linstead P.J., Roberts K., Schiefelbein J.W., Hauser M.T., Aeschbacher R.A. (1993). Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119: 57–70. [DOI] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. (2008). Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 59: 443–465. [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Biggar S.R., Crabtree G.R. (1999). Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J. 18: 2254–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee J.J., et al. (2007). Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19: 131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. [DOI] [PubMed] [Google Scholar]
- Cairns B.R. (2005). Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr. Opin. Genet. Dev. 15: 185–190. [DOI] [PubMed] [Google Scholar]
- Cairns B.R., Kim Y.J., Sayre M.H., Laurent B.C., Kornberg R.D. (1994). A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc. Natl. Acad. Sci. USA 91: 1950–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A., You R., Haimovitch-Gal T., Doerner P. (1999). Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20: 503–508. [DOI] [PubMed] [Google Scholar]
- Côté J., Quinn J., Workman J.L., Peterson C.L. (1994). Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265: 53–60. [DOI] [PubMed] [Google Scholar]
- Cui H., Levesque M.P., Vernoux T., Jung J.W., Paquette A.J., Gallagher K.L., Wang J.Y., Blilou I., Scheres B., Benfey P.N. (2007). An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L., Wysocka-Diller J., Malamy J.E., Pysh L., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433. [DOI] [PubMed] [Google Scholar]
- Dinneny J.R., Benfey P.N. (2008). Plant stem cell niches: standing the test of time. Cell 132: 553–557. [DOI] [PubMed] [Google Scholar]
- Dolan L., Janmaat K., Willemsen V., Linstead P., Poethig S., Roberts K., Scheres B. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84. [DOI] [PubMed] [Google Scholar]
- Efroni I., Han S.K., Kim H.J., Wu M.F., Steiner E., Birnbaum K.D., Hong J.C., Eshed Y., Wagner D. (2013). Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24: 438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona S., Hurtado L., Reyes J.C. (2007). A nucleosome interaction module is required for normal function of Arabidopsis thaliana BRAHMA. J. Mol. Biol. 373: 240–250. [DOI] [PubMed] [Google Scholar]
- Farrona S., Hurtado L., Bowman J.L., Reyes J.C. (2004). The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131: 4965–4975. [DOI] [PubMed] [Google Scholar]
- Farrona S., Hurtado L., March-Díaz R., Schmitz R.J., Florencio F.J., Turck F., Amasino R.M., Reyes J.C. (2011). Brahma is required for proper expression of the floral repressor FLC in Arabidopsis. PLoS ONE 6: e17997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153. [DOI] [PubMed] [Google Scholar]
- Friml J., Benková E., Blilou I., Wisniewska J., Hamann T., Ljung K., Woody S., Sandberg G., Scheres B., Jürgens G., Palme K. (2002). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673. [DOI] [PubMed] [Google Scholar]
- Galinha C., Hofhuis H., Luijten M., Willemsen V., Blilou I., Heidstra R., Scheres B. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057. [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218. [DOI] [PubMed] [Google Scholar]
- Grieneisen V.A., Xu J., Marée A.F.M., Hogeweg P., Scheres B. (2007). Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013. [DOI] [PubMed] [Google Scholar]
- Han S.K., Sang Y., Rodrigues A., Wu M.F., Rodriguez P.L., Wagner D., BIOL425 F2010 (2012). The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 24: 4892–4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves D.C., Crabtree G.R. (2011). ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 21: 396–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.H., Prochasson P., Neely K.E., Galasinski S.C., Chandy M., Carrozza M.J., Workman J.L. (2002). Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111: 369–379. [DOI] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., Hauser M.T., Benfey P.N. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567. [DOI] [PubMed] [Google Scholar]
- Hota S.K., Bartholomew B. (2011). Diversity of operation in ATP-dependent chromatin remodelers. Biochim. Biophys. Acta 1809: 476–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado L., Farrona S., Reyes J.C. (2006). The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol. Biol. 62: 291–304. [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerzmanowski A. (2007). SWI/SNF chromatin remodeling and linker histones in plants. Biochim. Biophys. Acta 1769: 330–345. [DOI] [PubMed] [Google Scholar]
- Jiang C., Pugh B.F. (2009). Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10: 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knizewski L., Ginalski K., Jerzmanowski A. (2008). Snf2 proteins in plants: gene silencing and beyond. Trends Plant Sci. 13: 557–565. [DOI] [PubMed] [Google Scholar]
- Koizumi K., Gallagher K.L. (2013). Identification of SHRUBBY, a SHORT-ROOT and SCARECROW interacting protein that controls root growth and radial patterning. Development 140: 1292–1300. [DOI] [PubMed] [Google Scholar]
- Kornet N., Scheres B. (2009). Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 21: 1070–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C.S., Hibara K., Pfluger J., Bezhani S., Metha H., Aida M., Tasaka M., Wagner D. (2006). A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development 133: 3223–3230. [DOI] [PubMed] [Google Scholar]
- Levesque M.P., Vernoux T., Busch W., Cui H., Wang J.Y., Blilou I., Hassan H., Nakajima K., Matsumoto N., Lohmann J.U., Scheres B., Benfey P.N. (2006). Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Chen C., Gao L., Yang S., Nguyen V., Shi X., Siminovitch K., Kohalmi S.E., Huang S., Wu K., Chen X., Cui Y. (2015). The Arabidopsis SWI2/SNF2 chromatin Remodeler BRAHMA regulates polycomb function during vegetative development and directly activates the flowering repressor gene SVP. PLoS Genet. 11: e1004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yu C.W., Duan J., Luo M., Wang K., Tian G., Cui Y., Wu K. (2012). HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 158: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen A.P., ten Tusscher K., Siligato R., Smetana O., Díaz-Triviño S., Salojärvi J., Wachsman G., Prasad K., Heidstra R., Scheres B. (2014). PLETHORA gradient formation mechanism separates auxin responses. Nature 515: 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Sena G., Nawy T., Benfey P.N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311. [DOI] [PubMed] [Google Scholar]
- Neigeborn L., Carlson M. (1984). Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara H., Ui A., Otsuka A., Satoh H., Yokomi I., Nakajima S., Yasui A., Yokota J., Kohno T. (2011). Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene 30: 2135–2146. [DOI] [PubMed] [Google Scholar]
- Ohkawa Y., Marfella C.G.A., Imbalzano A.N. (2006). Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. EMBO J. 25: 490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen T.A., Kowenz-Leutz E., Leutz A., Nerlov C. (2001). Cooperation between C/EBPalpha TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 15: 3208–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli S., Sabatini S. (2010). Analysis of root meristem size development. Methods Mol. Biol. 655: 177–187. [DOI] [PubMed] [Google Scholar]
- Peterson C.L., Dingwall A., Scott M.P. (1994). Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc. Natl. Acad. Sci. USA 91: 2905–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka J.J., Benfey P.N. (2008). Root layers: complex regulation of developmental patterning. Curr. Opin. Genet. Dev. 18: 354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260. [DOI] [PubMed] [Google Scholar]
- Sabatini S., Heidstra R., Wildwater M., Scheres B. (2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17: 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472. [DOI] [PubMed] [Google Scholar]
- Scheres B. (2007). Stem-cell niches: nursery rhymes across kingdoms. Nat. Rev. Mol. Cell Biol. 8: 345–354. [DOI] [PubMed] [Google Scholar]
- Smaczniak C., et al. (2012). Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 109: 1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M., Jensen R., Herskowitz I. (1984). Five SWI genes are required for expression of the HO gene in yeast. J. Mol. Biol. 178: 853–868. [DOI] [PubMed] [Google Scholar]
- Tamkun J.W., Deuring R., Scott M.P., Kissinger M., Pattatucci A.M., Kaufman T.C., Kennison J.A. (1992). brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68: 561–572. [DOI] [PubMed] [Google Scholar]
- Tang X., Hou A., Babu M., Nguyen V., Hurtado L., Lu Q., Reyes J.C., Wang A., Keller W.A., Harada J.J., Tsang E.W.T., Cui Y. (2008). The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol. 147: 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Haseloff J. (2008). A simple way to identify non-viable cells within living plant tissue using confocal microscopy. Plant Methods 4: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugartechea-Chirino Y., Swarup R., Swarup K., Péret B., Whitworth M., Bennett M., Bougourd S. (2010). The AUX1 LAX family of auxin influx carriers is required for the establishment of embryonic root cell organization in Arabidopsis thaliana. Ann. Bot. (Lond.) 105: 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C., Willemsen V., Hage W., Weisbeek P., Scheres B. (1995). Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 378: 62–65. [DOI] [PubMed] [Google Scholar]
- van den Berg C., Willemsen V., Hendriks G., Weisbeek P., Scheres B. (1997). Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390: 287–289. [DOI] [PubMed] [Google Scholar]
- Vercruyssen L., et al. (2014). ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26: 210–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Radman-Livaja M., Rando O.J., Peterson C.L. (2013). A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science 340: 195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildwater M., Campilho A., Perez-Perez J.M., Heidstra R., Blilou I., Korthout H., Chatterjee J., Mariconti L., Gruissem W., Scheres B. (2005). The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349. [DOI] [PubMed] [Google Scholar]
- Wu M.F., Sang Y., Bezhani S., Yamaguchi N., Han S.K., Li Z., Su Y., Slewinski T.L., Wagner D. (2012). SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc. Natl. Acad. Sci. USA 109: 3576–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka-Diller J.W., Helariutta Y., Fukaki H., Malamy J.E., Benfey P.N. (2000). Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127: 595–603. [DOI] [PubMed] [Google Scholar]
- Yamada T., Mizuno K., Hirota K., Kon N., Wahls W.P., Hartsuiker E., Murofushi H., Shibata T., Ohta K. (2004). Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 23: 1792–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Clarenz O., Cokus S., Bernatavichute Y.V., Pellegrini M., Goodrich J., Jacobsen S.E. (2007). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Yang S., Chen C.Y., Li C., Shan W., Lu W., Cui Y., Liu X., Wu K. (2015). Arabidopsis BREVIPEDICELLUS interacts with the SWI2/SNF2 chromatin remodeling ATPase BRAHMA to regulate KNAT2 and KNAT6 expression in control of inflorescence architecture. PLoS Genet. 11: e1005125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Rowley M.J., Böhmdorfer G., Wierzbicki A.T. (2013). A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol. Cell 49: 298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.