TAP analysis to unravel protein complex composition in dividing and expanding tissue in a growing maize leaf reveals ANGUSTIFOLIA3 association with distinct GROWTH-REGULATING FACTORs to fine-tune growth.
Abstract
Most molecular processes during plant development occur with a particular spatio-temporal specificity. Thus far, it has remained technically challenging to capture dynamic protein-protein interactions within a growing organ, where the interplay between cell division and cell expansion is instrumental. Here, we combined high-resolution sampling of the growing maize (Zea mays) leaf with tandem affinity purification followed by mass spectrometry. Our results indicate that the growth-regulating SWI/SNF chromatin remodeling complex associated with ANGUSTIFOLIA3 (AN3) was conserved within growing organs and between dicots and monocots. Moreover, we were able to demonstrate the dynamics of the AN3-interacting proteins within the growing leaf, since copurified GROWTH-REGULATING FACTORs (GRFs) varied throughout the growing leaf. Indeed, GRF1, GRF6, GRF7, GRF12, GRF15, and GRF17 were significantly enriched in the division zone of the growing leaf, while GRF4 and GRF10 levels were comparable between division zone and expansion zone in the growing leaf. These dynamics were also reflected at the mRNA and protein levels, indicating tight developmental regulation of the AN3-associated chromatin remodeling complex. In addition, the phenotypes of maize plants overexpressing miRNA396a-resistant GRF1 support a model proposing that distinct associations of the chromatin remodeling complex with specific GRFs tightly regulate the transition between cell division and cell expansion. Together, our data demonstrate that advancing from static to dynamic protein-protein interaction analysis in a growing organ adds insights in how developmental switches are regulated.
INTRODUCTION
Recently, the mode of action of the Arabidopsis thaliana leaf growth-regulating protein ANGUSTIFOLIA3/GRF-INTERACTING FACTOR1 (AN3/GIF1), a transcriptional coactivator lacking direct DNA binding properties, was elucidated by the identification of its interacting partners through tandem affinity purification (TAP) followed by mass spectrometry (MS). AN3/GIF1 physically interacts with chromatin remodeling complexes containing the ATPases SPLAYED (SYD) and BRAHMA (BRM). TAP using AN3/GIF1 as bait identified novel players important for leaf growth, which was validated by the observation that some of these interacting proteins displayed growth-enhancing phenotypes when their expression was modified (Vercruyssen et al., 2014). In addition, using yeast two-hybrid and immunoprecipitation assays, AN3 was shown to interact with GROWTH-REGULATING FACTOR (GRF) proteins, a class of plant-specific transcriptional activators of which some members are posttranscriptionally regulated by microRNA396a (miR396a) (Kim and Kende, 2004; Liu et al., 2009; Debernardi et al., 2014). Ectopic expression of specific GRFs was shown to affect leaf size in Arabidopsis (Kim et al., 2003; Horiguchi et al., 2005; Liu et al., 2009; Debernardi et al., 2014), maize (Zea mays; Wu et al., 2014), Chinese cabbage (Brassica rapa ssp pekinensis; Wang et al., 2014), and rice (Oryza sativa; Kuijt et al., 2014), and modulation of the miR396a target site allowed the overexpression of miR396a-resistant GRFs, which resulted in altered growth phenotypes (Rodriguez et al., 2010; Wang et al., 2011; Debernardi et al., 2014; Liang et al., 2014).
Besides protein complexes involved in chromatin remodeling, TAP in Arabidopsis cell cultures revealed protein complexes involved in numerous biological processes, such as cell proliferation (Van Leene et al., 2007), protein degradation (Eloy et al., 2011, 2012), transcription elongation (Nelissen et al., 2010), endocytosis (Gadeyne et al., 2014), and plant hormone signaling (Pauwels et al., 2010). Furthermore, TAP allowed us to capture condition-dependent interactions specifically occurring in the presence of certain plant hormones, such as jasmonic acid (Pauwels et al., 2010) or abscisic acid (Antoni et al., 2013).
Understanding how protein complexes are regulated within growing organs, which undergo dynamic changes from cell division to cell expansion, would add a new layer of information to ultimately obtain a spatio-temporal understanding of growth-regulatory protein complexes. Most leaf growth processes in Arabidopsis occur when the leaf is <1 mm in length (Gonzalez et al., 2012), which makes this model plant unsuitable to study organ development with TAP. By contrast, the maize leaf offers a superior tool to study the dynamics of growth processes due to its larger size and the linear organization of its growth zone. The processes of cell division and cell expansion are spatially separated from the leaf base to the tip, resulting in a growth zone that encompasses several centimeters (Nelissen et al., 2013) and that can be easily sampled to obtain tissue enriched for dividing or expanding cellular domains. Combining this fine-sampling strategy with hormone measurements, transcriptomic studies, and genetic perturbations revealed a local accumulation of the plant hormone gibberellic acid (GA) at the transition between cell division and cell expansion, which is important for organ growth and final size (Nelissen et al., 2012). These data suggested that molecular differences occurring at this transition regulate the irreversible decision to exit cell division and enter cell expansion.
To reveal changes in composition of the AN3-associated SWI/SNF complex during the developmental transition between cell division and expansion in a growing organ, we developed in planta TAP in maize. TAP experiments on leaf and ear tissue using AN3/GIF1 as bait were performed and revealed that the AN3-associated chromatin remodeling complex was highly conserved between dicots and monocots. In addition, TAP purifications on tissue enriched for either dividing or expanding cells showed that the components of the core complex remained identical between the two zones but that the GRFs associated with the complex were zone dependent. This suggests that GRFs, known to have DNA binding activity, recruit the chromatin remodeling complex to specific target genes in a growth zone-dependent manner. The presence of the zone-enriched GRFs corresponded to their transcription profiles, which were shown to be tightly regulated by miR396a in the maize leaf growth zone (Candaele et al., 2014) and to their protein abundance along different positions of the maize leaf (Facette et al., 2013). Overexpression of an miRNA396a-resistant GRF1 that was significantly enriched in the division zone resulted in plants with larger leaves due to increased cell division. This is opposite to what was observed for the overexpression of GRF10, which was identified in both dividing and expanding cells, where cell expansion was favored over cell division (Wu et al., 2014). Here, we propose a model in which the balance between GRF1 and GRF10 in association with the AN3-associated chromatin remodeling complex determines the size of the division zone and, thus, the organization of the leaf growth zone.
RESULTS
TAP of the AN3-Associated SWI/SNF Protein Complex from Maize
PLAZA 3.0 (Proost et al., 2015) was used to determine the maize ortholog of AN3/GIF1 (GRMZM2G180246), and its coding sequence was fused at its C terminus with the GS-tag (Van Leene et al., 2008). This construct was expressed in B104 plants under the control of the weak constitutive maize UBI-L promoter (Coussens et al., 2012). Previous protein complex purifications from transgenic Arabidopsis seedlings had shown that in a wild-type background, weak overexpression of the bait protein is necessary to compete with the endogenous untagged protein for complex incorporation and successful protein complex purification (Van Leene et al., 2015). The transgenic AN3-GS1 line showing the highest accumulation of the fusion protein (Supplemental Figure 1) was selected for further analysis, and subsequent experiments were performed on segregating offspring of this line backcrossed to B104.
To delineate the sampling of dividing versus expanding tissue, we examined the growth phenotypes of the AN3-GS1 transgenic plants and determined the size of each zone (Nelissen et al., 2013). The AN3-GS1 line did not show an increase in final leaf size (P value = 0.63) or an altered leaf elongation rate (Supplemental Figures 2A and 2B), but the plants developed more slowly than the nontransgenic siblings. This observed growth delay was significant from leaf 4 onward (Supplemental Figure 2C). During steady state growth of the fourth leaf, the size of the division zone ranged from 1.11 to 1.35 cm in the AN3-GS1 transgenic plants and nontransgenic siblings, respectively (P value = 0.02), while the expansion zone typically exceeded 4 cm (Nelissen et al., 2012). For sampling, the most basal 4 cm (0 to 4 cm) of the leaf during steady state growth, corresponding to a mixture of both dividing and expanding cells, was considered as the entire “growth zone,” while collection of the most basal first centimeters (0 to 1 cm) and the fourth centimeter (3 to 4 cm) separately resulted in a clear enrichment of dividing cells and elongating cells, respectively. In addition to leaf samples, we sampled developing maize ears of ∼4 cm in size.
Affinity-enriched proteins were analyzed on an Orbitrap Velos or Q Exactive mass spectrometer and identified with the MaxQuant software. Nonspecific and sticky proteins were filtered out based on control TAP experiments performed on leaf and ear tissue from nontransgenic mock plants. Based on our experience in Arabidopsis, a small set of control experiments on wild-type tissue is a good start, but not optimal in filtering out nonspecific and sticky proteins. Therefore, we took advantage of our knowledge of the typical TAP background in Arabidopsis and filtered the identified proteins in a second step for proteins whose Arabidopsis orthologs were present in a list of nonspecific and sticky binders based on frequency of occurrence of the copurified proteins in 543 TAP experiments using 115 different baits performed in Arabidopsis with the same GS-based TAP procedure (Van Leene et al., 2015).
The resulting list of copurified proteins (Table 1; Supplemental Table 1) included many orthologs of SWI/SNF complex components as identified with AN3 as bait in Arabidopsis (Vercruyssen et al., 2014). Several additional proteins, for which no orthologs were found in the TAP experiments using the Arabidopsis AN3 as bait (Vercruyssen et al., 2014), seemed to be consistently associated with AN3 in the maize leaf and ear (Supplemental Table 1). As the previous TAP experiments were performed on a MALDI TOF/TOF proteomics analyzer, the AN3 TAP experiments in Arabidopsis were performed again in combination with more sensitive MS (LTQ Orbitrap Velos) (Supplemental Table 2), resulting in an increased overlap between the AN3 TAPs in both plant species (Table 1). These results show that our TAP/MS approach is a valid experimental procedure to isolate and identify functional protein complexes in maize.
Table 1. TAP with Zm-AN3 as Bait.
| Leaf Entire Growth Zone | Leaf Division Zone | Leaf Expansion Zone | Ear | ||||
|---|---|---|---|---|---|---|---|
| Protein ID | Description | cm4 (2 exps) | cm1 (4 exps) | cm4 (4 exps) | Ear (6 exps) | At Ortholog | Arabidopsis TAP |
| Core subunits | |||||||
| GRMZM2G180246 | AN3/GIF1 | 2 | 4 | 4 | 6 | AT5G28640 | x |
| GRMZM2G163849 | BRM | 2 | 4 | 4 | 6 | AT2G46020 | x |
| GRMZM2G387890 | SYD | 2 | 4 | 4 | 6 | AT2G28290 | x |
| GRMZM2G467799 | SYD | 2 | 4 | 4 | 6 | AT2G28290 | x |
| GRMZM2G015384 | ARP4 | 2 | 4 | 4 | 6 | AT1G18450 | x |
| GRMZM2G015861 | ARP7 | 2 | 4 | 4 | 6 | AT3G60830 | x |
| GRMZM2G047038 | SWI3D (CHB3) | 2 | 4 | 4 | 6 | AT4G34430 | x |
| GRMZM2G119261 | SWI3D (CHB3) | 0 | 0 | 0 | 6 | AT4G34430 | x |
| GRMZM2G340756 | SWI3C (SWIRM) | 1 | 4 | 4 | 6 | AT1G21700 | x |
| GRMZM2G139760 | SWI3C (SWIRM) | 1 | 4 | 2 | 5 | AT1G21700 | x |
| GRMZM2G052416 | SWP73B (CHC1) | 2 | 4 | 4 | 6 | AT5G14170 | x |
| GRMZM2G033478 | G2484-1 | 2 | 4 | 4 | 6 | AT4G17330 | x |
| GRMZM2G473310 | G2484-1 | 2 | 4 | 4 | 6 | AT4G17330 | x |
| GRMZM2G103079 | Dentin sialophosphoprotein-related | 2 | 4 | 4 | 6 | AT5G07980 | x |
| GRMZM5G816791 | Dentin sialophosphoprotein-related | 2 | 4 | 4 | 4 | AT5G07980 | x |
| AC198518.3_FGP003 | GLTSCR | 2 | 4 | 4 | 6 | AT5G17510 | x |
| GRMZM2G018955 | BCL7 | 2 | 4 | 2 | 6 | AT4G22320 | x |
| GRMZM2G020548 | BRD1 | 1 | 4 | 2 | 4 | AT1G20670 | x |
| GRMZM2G312501 | BRD1 | 1 | 3 | 3 | 6 | AT1G20670 | x |
| GRMZM2G044044 | BRD1 | 0 | 2 | 1 | 0 | AT1G20670 | x |
| GRMZM2G336962 | BRD1 | 0 | 0 | 0 | 6 | AT1G20670 | x |
| GRMZM2G476652 | LFR | 1 | 3 | 3 | 6 | AT3G22990 | x |
| GRMZM2G079013 | LEUNIG HOMOLOG (LUH) | 0 | 0 | 0 | 5 | AT2G32700 | x |
| GRFs | |||||||
| GRMZM2G034876 | GRF1 | 2 | 4 | 2 | 6 | AT3G13960 | |
| GRMZM2G099862 | GRF2 | 0 | 0 | 0 | 6 | AT4G37740 | |
| GRMZM2G105335 | GRF3 | 0 | 2 | 0 | 6 | AT3G13960 | |
| GRMZM2G004619 | GRF4 | 0 | 3 | 4 | 0 | AT4G37740 | |
| GRMZM2G129147 | GRF5 | 1 | 2 | 0 | 6 | AT3G13960 | |
| GRMZM2G041223 | GRF6 | 2 | 3 | 0 | 6 | AT3G13960 | |
| GRMZM5G850129 | GRF7 | 2 | 4 | 0 | 2 | AT3G13960 | |
| GRMZM2G096709 | GRF10 | 2 | 4 | 3 | 6 | AT4G37740 | |
| GRMZM2G067743 | GRF11 | 0 | 0 | 0 | 6 | 0 | |
| GRMZM2G119359 | GRF12 | 0 | 0 | 0 | 6 | 0 | |
| GRMZM2G098594 | GRF14 | 0 | 0 | 0 | 4 | AT3G13960 | |
| GRMZM2G178261 | GRF15 | 0 | 4 | 0 | 6 | AT2G22840 | |
| GRMZM2G124566 | GRF17 | 2 | 4 | 1 | 6 | AT2G36400 | |
| GRMZM2G045977 | GRF18 | 0 | 0 | 0 | 6 | AT2G36400 | |
The data were compared with the data obtained in Arabidopsis (Vercruyssen et al., 2014). cm1 corresponds to dividing cells, while cm4 represents the enrichment in expanding cells. exps, experiments.
The AN3-Associated SWI/SNF Complex Is Conserved between Dicots and Monocots, in Leaves and Reproductive Organs
To gain insight in the maize AN3 TAP data, the copurified proteins were compared with the protein complexes identified using TAP with the Arabidopsis and the human ortholog of AN3 (SS18/SYT) (Middeljans et al., 2012) (Table 2). The human protein complex associated with SS18/SYT, known as BRG1-associated factor (BAF) complex, consists of SWI/SNF subunits and BAF-specific subunits. Orthologs of several human SWI/SNF subunits were identified with AN3 TAP in Arabidopsis and maize: the ATPases SYD and BRM, the actin-related proteins ARP4 and ARP7, SWIRM/SANT domain proteins SWI3C and SWI3D, and SWIB domain protein SWP73. Strikingly, the SNF5 component, strongly conserved between humans and yeast, was not retrieved in any plant AN3 TAP experiment, although there is a plant ortholog (BUSHY GROWTH). Also, the human BAF57 ortholog was not retrieved in plants.
Table 2. Conservation of the Identified Proteins Using TAP in Human, Arabidopsis, and Maize.
| Species |
||||
|---|---|---|---|---|
| Function or Domain | Homo sapiens | Arabidopsis | Maize | |
| SWI/SNF subunits | ATPase, helicase (SNF2) | BRM; BRG1 | BRM; SYD | BRM; SYD |
| Actin-like | BAF53A; BAF53B | ARP4; ARP7 | ARP4; ARP7 | |
| SWIRM SANT domain | BAF155 | SWI3C; SWI3D | SWI3C; SWI3D | |
| SWIRM SANT domain | BAF170 | SWI3B | SWI3C; SWI3D | |
| SWIB domain | BAF60A; BAF60B; BAF60C | SWP73 | SWP73 | |
| HMG | BAF57 | * | * | |
| SNF5 | BAF47 | * | * | |
| BAF-specific | N-terminal conserved region | SS18/SYT | AN3/GIF | AN3/GIF |
| ARID/BRIGHT domain | BAF250A; BAF250B | * | * | |
| Bromodomain | BRD9 | BRD | BRD | |
| N-terminal conserved region | BCL7A; BCL7B; BCL7C | BCL7B | BCL7B | |
| GLTSCR domain | GLTSCR1 | GLTSCR | GLTSCR | |
| PHD domain, Zn finger protein | BAF45B; BAF45C; BAF45D | * | * | |
| Zn finger protein | BCL11 | * | * | |
| p/BAF-specific | ARID/BRIGHT domain | BAF200 | * | * |
| Polybromo, HMG | BAF180 | * | * | |
| Bromodomain | BRD7 | BRD | BRD | |
| PHD domain, Zn finger protein | BAF45A | * | * | |
| Plant-specific | AT-rich interactive domain | * | LFR | LFR |
| Agenet domain | / | G2484-1 | G2484-1 | |
| LEUNIG HOMOLOG | * | LUH | LUH | |
| Dentin sialophosphoprotein-related protein | / | Dentin sialophospho | Dentin sialophospho | |
Homologs were identified by BLAST searches (NCBI). Asterisks indicate that homologs were present but not identified by TAP, and the slash means no clear homolog could be identified.
Plant orthologs to human BAF-specific BRD9, BCL7B, and GLTSCR1 were retrieved in both Arabidopsis and maize. In the human SS18 TAP, BCL7A was found instead of BCL7B, but no plant ortholog could be identified for BCL7A, using PLAZA3.0 dicots, containing 31 species (Proost et al., 2015) and PSI-BLAST (Altschul et al., 1997). Human BAF and P/BAF complexes were further characterized by the presence of ARID, polybromo/HMG, and PHD and Zn finger domain proteins (Middeljans et al., 2012), for which orthologs can be identified in each plant species, but none of them were identified in plant TAP experiments.
Conversely, the Agenet domain protein, G2484-1, and two dentin sialophosphoprotein-related proteins were consistently retrieved in plant TAP experiments using AN3 as bait, while no clear orthologs were found in the human genome. Additionally, LEAF and FLOWER RELATED (LFR) and LEUNIG HOMOLOG (LUH) were identified in plant TAP experiments, but the human ortholog was not retrieved as part of the human BAF complexes. Although the AN3-associated chromatin remodeling complex is conserved between Arabidopsis and maize, some differences were present mainly based on genome duplications, annotations, and gene copy number (Supplemental Table 3).
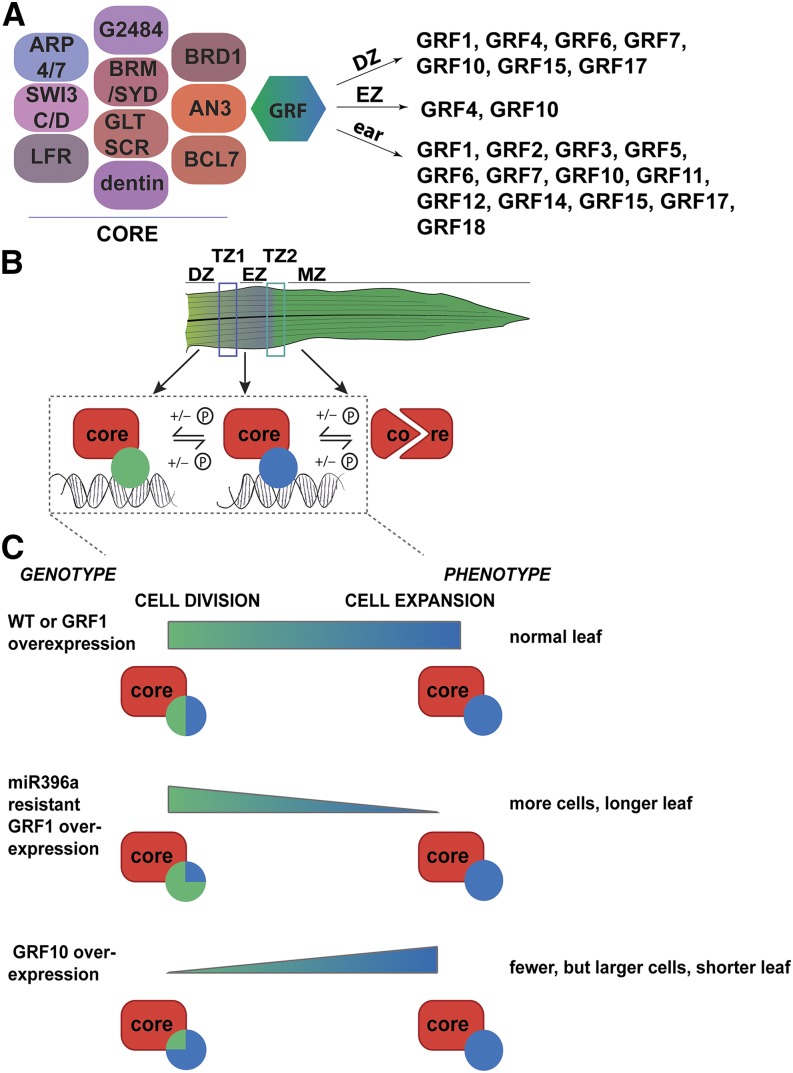
Taken together, these data show that the core SWI/SNF complex associated with AN3/GIF1 is conserved among eukaryotes, consisting of (homologs of) BRM, SYD, ARP4, ARP7, SWP73B, SWI3C, SWI3D, BRD1, BCL7, and GLTSCR. G2484-1, LFR, dentin sialophosphoprotein-related, and LUH (Table 1, Figure 1A) constitute the plant-specific core components.
Figure 1.
Model of the Protein Complex Associated with AN3 as Identified in the TAP Experiments.
(A) Schematic representation of the core complex, based on data from Arabidopsis (Vercruyssen et al., 2014) and maize (leaf division zone [DZ], leaf elongation zone [EZ], and ear TAPs), and the maize tissue- or organ-specific GRFs as determined by quantitative analysis (leaf) and identification (ear).
(B) Schematic representation of the AN3/GIF1-associated chromatin remodeling core complex at the different positions in the leaf. The core complex is present at dividing and expanding tissue but is recruited to the target DNA by growth process-specific GRFs (blue is division zone specific; green is expansion zone specific). The components of the core complex, as well as the GRFs, are depleted from the mature leaf samples. For several components, differential phosphorylation throughout the leaf developmental gradient has been shown (Facette et al., 2013).
(C) Model of how the specific interaction of the GRFs with the AN3-associated chromatin remodeling complex regulates the transition from cell division to cell expansion, based on the phenotypes of the GRF10 (Wu et al., 2014) and the GRF1R overexpression lines.
Label-Free Quantification of the TAP Data Shows That GRFs Specifically Interact with the AN3/GIF1-Associated Chromatin Remodeling Complex in a Growth Process-Specific Manner
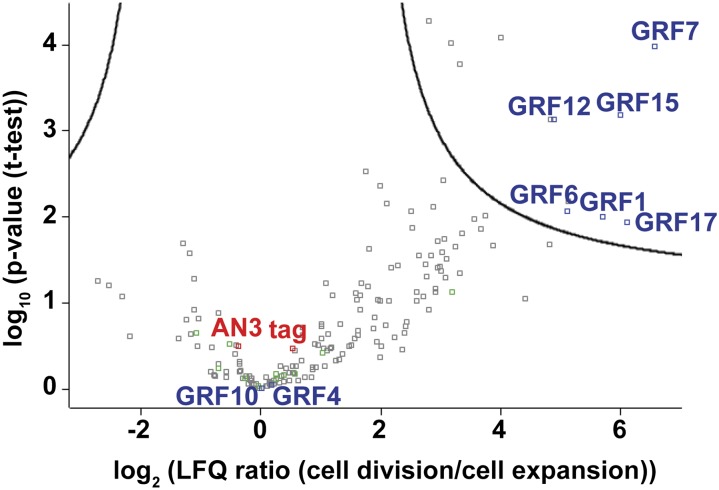
In contrast to the TAP experiments with AN3/GIF1 as bait in Arabidopsis cell cultures and whole seedlings (Vercruyssen et al., 2014), a considerable number of GRFs were copurified with AN3 in maize leaf and ear, highlighting the importance of applying this technology to appropriate developmental contexts. The majority of the GRFs appeared to be more predominantly present in the leaf division zone when compared with the leaf expansion zone (Table 1). To quantify growth-specific changes in the interacting proteins, relative intensity-based label-free quantification (MaxLFQ) with the MaxQuant software was used. Label-free quantification (LFQ) intensities were calculated, using unique peptides only, for all identified proteins in four independent TAP experiments on both dividing and expanding leaf tissue. Afterward, a t test analysis was performed on the LFQ intensities to retrieve significantly changing proteins between the division zone and the expansion zone. While bait, tag, and copurified core subunits did not change between the division and expansion zone, 12 proteins were significantly enriched in the dividing tissue (Figure 2; Supplemental Table 4). Among these proteins, we found GRF1, GRF6, GRF7, GRF12, GRF15, and GRF17, GRFs that all contain an miR396a target site (Candaele et al., 2014), while the miR396a-resistant GRFs, GRF4 and GRF10, did not show changes in their abundance between the two zones (Figure 2). The expression level of the GRF genes was evaluated along the leaf gradient using quantitative RT-PCR. Indeed, all GRF genes that contained the miR396a target site were highest expressed at the leaf base where cell division takes place and dropped to a basal level after the transition to cell expansion. Oppositely, GRF4 and GRF10 were expressed at high levels in both the division and expansion zone (Supplemental Figure 3).
Figure 2.
Relative Intensity-Based Label-Free Quantification in Maize Leaf: AN3/GIF1 TAP Copurified Proteins from Division Zone (1st cm) Compared with Expansion Zone (4th cm).
Significantly changing interactors of AN3/GIF1 are identified by a permutation-based FDR-corrected t test (threshold FDR = 0.01 and S0 = 0.9; Supplemental Table 4). The difference in average log2 LFQ intensities between division zone (group 1) and expansion zone (group 2) is plotted versus the significance (–log10 [P value]). The curve indicates the permutation-based FDR threshold. Positions of AN3/GIF1 bait, tag, GRFs, and core subunits (in green) are marked on the plot.
Overexpression of an miR396a-Resistant GRF1 (GRF1R) Increases the Number of Dividing Cells, Resulting in Larger Leaves
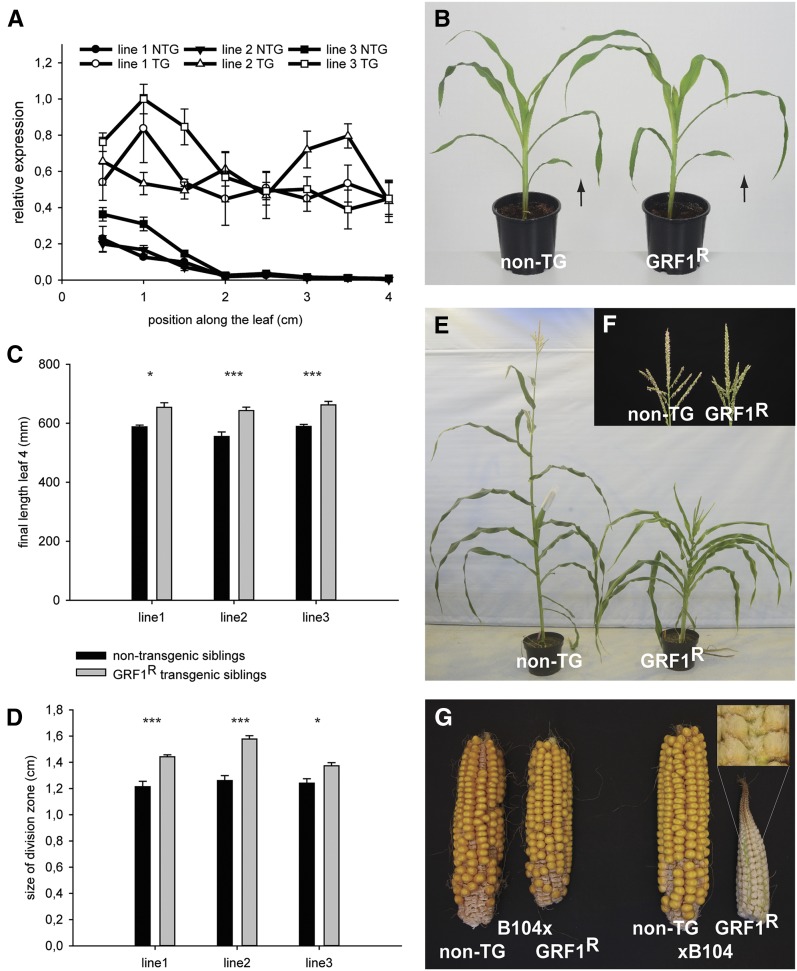
Of the six GRFs specifically associated with AN3 in the division zone in maize, only GRF1 is predominantly expressed in shoots and young leaves (Zhang et al., 2008). Recently, it was shown that overexpression of GRF1, which contains an miR396a target site, did not produce an altered phenotype in maize (Wu et al., 2014). This could be explained by the fact that the effect of overexpression is counteracted by the tight regulation of the transcript level by the action of miR396a. To test this hypothesis, we overexpressed an miR396a-resistant version of GRF1 (indicated as GRF1R) under the control of the strong constitutive EF1α promoter (Coussens et al., 2012). In three independent single locus lines, the relative levels of GRF1 were indeed higher in the transgenic siblings. This overexpression of GRF1 was seen, not only at the base of the leaf, but the levels of expression remained high in the expansion zone of the maize leaf (Figure 3A).
Figure 3.
Phenotypes of Nontransgenic and Transgenic GRF1R Segregating Plants of the Three Independent GRF1R Overexpression Lines.
(A) Expression levels of GRF1 in the GRF1R-overexpressing transgenic plants and the nontransgenic siblings over the maize leaf growth zone for three independent transgenic events. Quantitative RT-PCR was performed with primers specific for GRF1 and normalized relative to 18S rRNA. NTG, nontransgenic siblings; TG, transgenic siblings. Error bars represent se.
(B) Seedling phenotype of line 2. The arrows indicate leaf 3.
(C) Final length of leaf 4 of two independent transgenic events. Error bars represent se.
(D) Size of the division zone of two independent transgenic events. Three asterisks indicate P value < 0.01, and one asterisk indicates P value < 0.05. Error bars represent se.
(E) Overall plant height phenotype of line 1.
(F) Tassel phenotype of line 2.
(G) Ears of crosses between B104 and nontransgenic or transgenic plants and crosses between the nontransgenic and transgenic plants with B104. The inset panel shows an enlargement of the macrohairs formed on and between files of spikelets.
Plants that overexpressed the GRF1R had longer leaves in three independent lines (line 1, 10.1%; line 2, 15.81%; line 3, 11%; P < 0.05), which was due to an increase in the size of the division zone (line 1, 15.8%; line 2, 24%; line 3, 9.6%; P < 0.05) (Figures 3B to 3D). As a similar phenotype was observed for three independent lines, kinematic analysis was performed on the line with the most pronounced phenotype (line 2). The size of the dividing cells was not significantly different; hence, the increase in the size of the division zone resulted in a significant increase of the number of dividing cells (23.65%, P < 0.05). The average duration of the cell cycle was prolonged in the GRF1R transgenic plants (29.4 h) compared with the nontransgenic siblings (24.9 h; 17.97%; P < 0.05). Given that there were more dividing cells of which the division rate was reduced in the GRF1R transgenic line, this might explain why the effect on final leaf length is less pronounced than that on the number of dividing cells. The mature cell size was not significantly affected in the GRF1R transgenic plants (5.73% increase, P = 0.08; Table 3).
Table 3. Effect of Overexpression of the miR396a-Resistant GRF1 (GRF1R) on Cellular Growth Parameters.
| Nontransgenic Siblings | GRF1R | P Value | % | |
|---|---|---|---|---|
| Size of division zone (cm) | 1.29 ± 0.02 | 1.60 ± 0.01 | 0.002 | 19.65 |
| Size of dividing cells (µm) | 27.39 ± 1.21 | 29.27 ± 2.46 | 0.543 | 6.42 |
| Number of dividing cells (cells) | 515.94 ± 13.80 | 637.99 ± 22.87 | 0.016 | 19.13 |
| LER (mm⋅h−1)a | 2079.84 ± 153.22 | 2305.43 ± 514.24 | 0.015 | 9.79 |
| Cell production (cell⋅h−1) | 14.38 ± 0.21 | 15.07 ± 0.27 | 0.119 | 4.63 |
| Cell division rate (cells cell−1 h−1) | 0.028 ± 0.0004 | 0.024 ± 0.0008 | 0.019 | −17.76 |
| Duration of cell cycle (h) | 24.87 ± 0.39 | 29.34 ± 1.04 | 0.037 | 15.25 |
| Mature cell size (µm) | 144.76 ± 2.10 | 153.06 ± 2.75 | 0.079 | 5.43 |
| Final leaf length (mm) | 555.2 ± 15.45 | 643 ± 11.68 | 0.004 | 13.65 |
LER, leaf elongation rate.
Two independent lines were monitored for additional phenotypes during plant development. Final plant height was reduced when GRF1R was overexpressed (Figure 3E). However, the growth-enhancing effect of GRF1R was still seen in subsequent leaves, since the blade of leaf 11 was increased by 8.1 and 7.6% (P < 0.05) in lines 1 and 2, respectively. GRF1R overexpression affected female fertility because both self- and cross-pollination with viable B104 pollen resulted in a reduced kernel set (Figure 3G). The overall arrangement of spikelets and florets appeared normal, especially at the ear base, but the glumes and the ear rachis were covered by large macrohairs (Figure 3G).
The GRF1R transgenic plants also displayed a tassel phenotype: The anthers did not dehisce well from within the glumes, although viable pollen was produced, since it could fertilize B104 ears (Figure 3F). To further investigate how GRF1R affected the morphology of the tassel, the organization of spikelets and florets was examined and the size of the branches and glumes was determined. Since all spikelets and florets appeared paired, we concluded that overexpression of GRF1R did not affect meristem determinacy. However, the growth of the tassel was positively affected in the GRF1R-overexpressing plants, since the length of the glumes was significantly increased. In addition, the lateral tassel branches seemed to be longer, although the number of spikelets formed per branch was not significantly different than that of nontransgenic siblings (Table 4).
Table 4. Effect of Overexpression of the miR396a-Resistant GRF1 (GRF1R) on Tassel Morphology (n = 4).
| GRF1R | Nontransgenic Plants | Difference (%) | P Value | |
|---|---|---|---|---|
| Average length spikelet (cm) | 1.09 | 0.75 | 31.38 | 0.00 |
| Length of branch (mm) | 230.25 | 192.00 | 16.61 | 0.02 |
| No. of spikelets/branch | 171.50 | 166.50 | 2.91 | 0.74 |
| No. of spikelets/cm | 0.74 | 0.87 | −17.30 | 0.24 |
DISCUSSION
High-Resolution Sampling Combined with TAP Followed by Label-Free Quantification Allowed for the Identification of Dynamic Changes in Protein Complex Composition in a Developmental Context
Most biological processes operate with a remarkable spatio-temporal specificity, and unraveling these processes at high resolution is often extremely technically challenging. Whereas it is now possible to microscopically focus on a subcellular process and to perform whole-transcriptome analysis with tissue or even cellular resolution, the analysis of protein complexes, using TAP, for example, still requires relatively large amounts of starting tissue. In Arabidopsis, a substantial number of protein complexes have been characterized using cell cultures as input material, but only a few protein complex studies in planta have been reported (Vercruyssen et al., 2014; Dedecker et al., 2015; Van Leene et al., 2015), and none address how these complexes change over time during development. To solve the requirement for more input material, plants with larger organs can be used, as was successfully illustrated in this report using maize to study the AN3/SWI/SNF complex that was previously shown to regulate Arabidopsis leaf growth (Vercruyssen et al., 2014). In previous studies in Arabidopsis, AN3 was shown to interact with GRFs by yeast two-hybrid and immunoprecipitation assays using cell cultures but not in planta. Here, using both zones of dividing and expanding cells from growing leaves, we demonstrated that the AN3/SWI/SNF complex associated with specific GRFs in a tissue-dependent manner. Indeed, our findings indicate that extending technologies such as the TAP technology (Van Leene et al., 2015) by transferring it from cell cultures to plants is often not sufficient to gain additional insights and may result in missing interactions if sampling is not performed at high resolution. For example, performing TAP on whole seedlings might dilute the sample, resulting in the inability to identify all relevant interactors. This is exemplified by the fact that using AN3 as bait in TAP experiments in Arabidopsis failed to identify any GRFs (Vercruyssen et al., 2014), even with more sensitive MS (Supplemental Table 2), while a more focused approach on subzones of the maize leaf resulted in the copurification of a number of GRFs showing developmental specificity. Although the TAP experiments in Arabidopsis cell cultures and seedlings provided only a static view of the protein complex, they were instrumental for identifying the interacting partners and for dissecting the function of AN3, identifying growth-enhancing genes (Vercruyssen et al., 2014).
Performing label-free quantification in different TAP experiments from the division and expansion zones made it possible to determine the changes in the dynamics in protein complex composition in a growing organ. Protein identification showed that the core components of the SWI/SNF complex associated with AN3 were present in both the division and the expansion zones, and in the developing ear, indicating that the core complex is conserved in several growing plant organs.
The Core AN3-Associated Chromatin Remodeling Complex Is Structurally and Functionally Conserved
Chromatin remodeling complexes have several roles in DNA replication, DNA repair, and gene regulation. Remodelers can affect transcription in different ways: They expose cis-elements in a nucleosome-free context so that they can be recognized by transcription factors or they assist the polymerase in advancing through the nucleosomes (Clapier and Cairns, 2009). Previously, the SWI/SNF complex was copurified in Arabidopsis (Vercruyssen et al., 2014), human (Middeljans et al., 2012), and yeast (Peterson et al., 1994). When comparing these data with the protein complex associated with the maize AN3 homolog, a remarkable conservation was noted. An SWI/SNF core complex can be defined consisting of ATPase components, actin-related proteins, and SANT and SWI proteins. The ATPase is necessary for nucleosome positioning (Gentry and Hennig, 2014), while SANT and SWI proteins are thought to be involved in histone interaction (Grüne et al., 2003; Sanchez and Zhou, 2009). The presence of actin-related proteins suggests a link with nuclear organization or signaling (Euskirchen et al., 2011).
The human BAF complexes are characterized by the presence of additional proteins, such as ARID, PHD, and Zn finger domain proteins. Orthologs of these transcription factors are present in plant genomes but were not retrieved in TAP experiments. Conversely, G2484-1, LUH, dentin sialophosphoprotein-related, and LFR proteins were identified in plant TAP experiments using AN3 as bait but were not identified using SS18/SYT as bait in human (Middeljans et al., 2012) (Table 2). The G2484-1 proteins contain Agenet or Tudor domains that are involved in binding to methylated histones (Lu and Wang, 2013). LFR is an Armadillo repeat-containing protein that most likely functions in protein-protein interactions. LFR is very broadly expressed in the different Arabidopsis organs but reaches its highest expression levels in young growing tissue. Mutation of LFR results in a delay in leaf appearance, narrow leaves, and even male sterility (Wang et al., 2009; Wang et al., 2012). LUH is a transcriptional corepressor belonging to the same class as LEUNIG (LUG) (Causier et al., 2012), which was also identified in all performed maize TAP experiments. LUH is functionally redundant to LUG in abaxial leaf identity and floral organ identity and binds histones to regulate chromatin structure (Shrestha et al., 2014). These observations suggest that the core components of the complex are rather conserved between humans and plants but that there are also kingdom-specific proteins that have diverged. Because the majority of the kingdom-specific proteins are involved in DNA (PHD and Zn finger, ARID domain proteins, and LUH) and chromatin binding (G2484-1), they seem to have a regulatory role in determining target specificity. It is remarkable that a transcriptional repressor is part of the AN3-associated SWI/SNF complex, as this complex is thought to have an activating role (Clapier and Cairns, 2009).
Results from the TAP experiments from two different plant species performed with state-of-the-art MS provide a baseline to begin to define which proteins associate with AN3 in plants (Tables 1 and 2). Allowing for minor differences due to genome duplications and the presence of large introns in SYD, the modular design of the core complex is very similar between Arabidopsis and maize, consisting of AN3, G2484-1, SYD, ARP4, ARP7, BRM, SWP73B, SWI3C, SWI3D, BRD1, GLTSCR, BCL7, LUH, LFR, G2484-1, and dentin sialophosphoprotein-related (Figure 1A). Moreover, independent of the starting tissues, cell cultures and seedlings in the case of Arabidopsis and developing leaves and ears in maize, the composition of the core complex remained the same. In conclusion, the data show that the core SWI/SNF complex associated with AN3/GIF1 is strongly conserved among eukaryotes and some plant-specific interacting proteins were identified.
In Arabidopsis, disrupting the composition of the complex resulted in numerous developmental abnormalities, indicating that this complex is active in numerous plant organs, ranging from leaves (Kim and Kende, 2004; Horiguchi et al., 2005; Vercruyssen et al., 2014) to reproductive organs (Lee et al., 2014), and during various developmental processes, ranging from embryogenesis (Kanei et al., 2012) to phase changes in leaves (Farrona et al., 2004). In humans, it was shown that BAF complexes are involved in many growth-related processes ranging from cancer initiation and progression (Euskirchen et al., 2012) to cardiovascular development (Han et al., 2011). While no clear leaf growth-related phenotypes were observed in plants overexpressing the maize homolog of AN3 fused to the TAP tag, perturbations of the GRFs resulted in obvious phenotypes, as was previously seen in Arabidopsis (Vercruyssen et al., 2014). This might be explained by the fact that a weaker constitutive promoter was used to drive expression of the tagged bait protein, while a strong constitutive promoter (Coussens et al., 2012) was used to increase the chances for phenotypes when overexpressing GRF1R.
More proteins were copurified with AN3 in a growing, developing ear compared with the leaf (Supplemental Table 1). Among these were DNA replication licensing factor MCM7 and several transcription factors, such as ARF6, ARF8, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE8, TOPLESS, and WRKY4. These results suggest that the AN3-associated SWI/SNF complex assists in transcriptional regulation through more transcription factors than only the GRFs. The copurification of a FRIGIDA-LIKE protein and several proteins involved in DNA methylation in the ear suggests that the AN3-associated chromatin remodeling complex is involved in the regulation of flowering genes, similar to what previously was reported for the FRIGIDA pathway (Choi et al., 2011; Yun et al., 2011). COACTIVATOR-ASSOCIATED ARGININE METHYLTRANSFERASE1/PROTEIN ARGININE METHYLTRANSFERASE4 also affects histone methylation in the FLOWER LOCUS C flowering pathway (Niu et al., 2007). Another copurified protein, trehalose-6-phosphate synthase, has previously been shown to be associated with inflorescence architecture (Satoh-Nagasawa et al., 2006).
The Copurification of GRF1 and GRF10 Corresponds to the Tight Developmental Regulation of the GRFs That Delimits mRNA and Protein Accumulation
Our results show that AN3 can associate with a wide range of GRFs but that the specificity of the interaction depends on the developmental context. In the division zone, a group of GRFs, consisting of GRF1, GRF6, GRF7, GRF12, GRF15, and GRF17, was significantly enriched compared with the expansion zone. MaxQuant intensities indicate that GRF1 is the most abundant GRF that associates with the AN3 chromatin remodeling complex in dividing tissue, while GRF1 levels drop below GRF4 and GRF10 levels in expanding tissue. Remarkably, some GRFs (GRF8, GRF9, GRF13, and GRF16) were never identified in the maize leaf or ear, which might indicate that they are either very low abundance or function in other organs, besides the maize leaf and ear. Much overlap was found between the GRFs identified in leaf and ear, indicating that some GRFs might be universally associated with growth processes such as division and expansion. Our experiments clarify which of the many GRFs that all show zone-specific expression profiles (Candaele et al., 2014; Supplemental Figure 3) are predominantly associated with AN3/GIF1, information that could not be deduced from the transcriptome data alone.
The dynamics of the transcript levels of the identified GRFs within the maize leaf growth zone (all GRF genes except GRF4 and GRF10 were more highly expressed at the division zone compared with the expansion zone) matched their dynamics in protein accumulation within the complex. GRF4 and GRF10 were shown to be insensitive to miR396a, resulting in a stable expression profile throughout the maize leaf growth zone (Candaele et al., 2014; Supplemental Figure 3). The GRFs associated with the AN3/GIF1 complex in the division zone (GRF1, GRF6, GRF7, GRF10, GRF15, and GRF17) showed high gene expression levels in the division zone, which gradually declined as miR396a transcript accumulation increased toward the transition between cell division and cell expansion (Candaele et al., 2014; Supplemental Figure 3). Taken together, these data show that the main differences in protein complex composition of the AN3/GIF1-associated SWI/SNF chromatin remodeling complex were reflected by the association of the complex to specific transcription factors, here from the GRF class, most likely determining the specificity of the complex functionality. A large-scale, quantitative analysis was performed on the maize leaf (phospho)proteome at four developmental zones that correspond to (1) dividing cells, (2) a mixture of dividing and expanding cells, (3) expanding cells of which some have undergone differentiation, and (4) mature leaf cells (Facette et al., 2013). Although the sampling was not done exactly the same way, the samples from zone 1 and 3 correspond to our samples for enriching dividing and expanding cells, respectively, allowing a comparison between two independent experiments. All proteins identified in the TAP experiments were identified in the proteome approach, except for GRF15 (which was present in the phosphoproteome) and GRF18.
In the “Facette” proteome study, the identified proteins were clustered in four major groups: (1) enriched in the mature tissue, (2) no change over the developmental gradient, (3) depleted in mature tissue and showing similar levels throughout the growth zone, and (4) depleted in mature tissue and showing enrichment at one of the three positions in the growth zone. Strikingly, all core components, but not the plant-specific proteins, and the GRFs belonged to clusters 3 and 4, which were depleted in the mature tissue, indicating that the core complex is associated with the growth zone, where both cell division and cell expansion occur. In addition, all GRFs were found to be enriched in one of the three positions within the growth zone, except for GRF4 and GRF10, which were insensitive to miRNA396a regulation and, thus, also had stable mRNA levels throughout the growth zone (Candaele et al., 2014).
Some of the components of the AN3-associated complex (GLTSCR-domain protein, BCL7B, SWI3C, SWI3D, both BRD1 proteins, and GRF4, GRF15, and GRF17) showed a phosphoprotein enrichment in zones 1 to 3 along the developmental gradient (Facette et al., 2013). Therefore, we hypothesize that this posttranslational modification can alter activity or even degradation (Figure 1B), as was observed for the BAF subunits in mammals (Clapier and Cairns, 2009). The protein abundance and differences in phosphorylation level of the majority of the components of the complex along the developmental gradient corresponded with the LFQ analysis of the TAP samples from division zone versus expansion zone. Our experiments show that in plants, the core complex is present in both dividing and expanding tissue but that the association with GRFs is dependent on the tissue context. In humans, it has been shown that the transition from neural progenitors to postmitotic neurons was characterized by a switch in isoforms of core-subunit composition (BAF45a was replaced by BAF45b and BAF45c, while BAF53a was replaced by BAF53b) (Lessard et al., 2007). This shows that methods capturing the dynamic changes in complex composition will be instrumental, since important developmental switches seem to be regulated by alterations in components or interacting proteins.
The Transition between Cell Division and Cell Expansion Is Highly Dependent on the Balance between the Cell Division-Enriched GRF1 and GRF10 That Is Present throughout the Growth Zone
From all the GRFs that were identified by TAP in the maize leaf, GRF1 and GRF10 have the highest transcript accumulation in immature leaves and shoot compared with other organs (Zhang et al., 2008). Since GRF1 was significantly enriched in the division zone and GRF10 was present throughout the entire growth zone, it would be interesting to examine what would happen upon perturbation of these proteins. Recently, it was shown that ectopic overexpression of GRF10, which is normally expressed in both dividing and expanding tissue and is not regulated by miRNA396a, resulted in altered growth phenotypes. The leaves of the GRF10 transgenic plants were decreased in length due to a reduction in cell number, which was partially compensated for by the enlarged cell sizes (Wu et al., 2014). This suggests that GRF10 affects the transition between cell division and cell expansion by stimulating cell expansion over cell division.
In the same study, the authors showed that overexpression of GRF1 did not result in phenotypes (Wu et al., 2014). However, it has previously been shown that mutation of the microRNA target site was required to accumulate GRF1 transcripts that result in altered growth phenotypes (Rodriguez et al., 2010; Wang et al., 2011; Debernardi et al., 2014; Liang et al., 2014). Here, we showed that the overexpression of an miR396a-resistant GRF1 (GRF1R) had the opposite phenotype to that of GRF10 overexpression, resulting in longer maize leaves: The size of the division zone and, hence, the number of dividing cells were significantly increased in three independent transgenic lines. Finally, no significant effect was observed for GRF1R overexpression on cell expansion. This indicates that GRF1 has a positive effect on the determination of the number of dividing cells and thus positions the transition zone more distally from the leaf base. Since fewer dividing cells and an increased cell expansion were observed in the GRF10 overexpression line (Wu et al., 2014), the transition between cell division and cell expansion was most likely shifted toward the base of the maize leaf. Therefore, we can hypothesize that there is an antagonistic effect of GRF1 and GRF10 to determine the position of the transition between cell division and cell expansion in the growth zone of the maize leaf.
As overexpression of GRF10 stimulates cell expansion and overexpression of GRF1R stimulates cell division, we propose a model that the balance between GRF1 and GRF10 determines the transition between cell division and cell expansion (Figure 1C). In wild-type leaf development, GRF1 mainly associates with the AN3 complex in the division zone, due to the strict developmental regulation of its expression. GRF10 is constitutively present throughout the growth zone and associates with AN3 in the expansion zone, while it may compete with GRF1 for occupancy in the complex in the division zone. Overexpression of GRF1R results in higher GRF1 levels that might outcompete GRF10 in the division zone, and since the expression of GRF1 is no longer strictly developmentally regulated, it can associate with AN3 for an extended position along the growth zone. Alternatively, GRF10 overexpression actively stimulates cell expansion by competing with GRF1 for binding with AN3 in the division zone and in that way favors the early onset of cell expansion, resulting in fewer but larger cells (Figure 1C). Taken together, we hypothesize that GRF1 and GRF10 compete with each other for binding with AN3 to regulate the position of the transition between cell division and cell expansion in the growing leaf.
Several phenotypes observed in the GRF1R transgenic lines were similar to those observed in plants with altered GA levels. Both the overexpression of GA20-Oxidase and GRF1R displayed longer leaves due to a distal shift in the transition zone. However, both transgenes had distinct effects on the final plant height: GA20-Oxidase overexpression increased internode elongation, while the GRF1R-overexpressing plants were semidwarfed. The tassel phenotype of GRF1R is reminiscent of other GA loss-of-function mutants, in which anthers fail to dehisce properly. Although anthers in GRF1R transgenic plants did not dehisce, fertile pollen was still produced. This was in contrast to ears in GRF1R transgenic plants, which showed reduced fertility. This more limited effect in male florets compared with female florets was also observed in GA-deficient mutants and might be explained by the fact that endogenous levels of GA are lower in the tassel stamen primordium (Wilson et al., 2011). In addition to GA, other phytohormones (e.g., jasmonic acid, auxin, and ethylene), enzymes involved in the breakdown, and programmed cell death of the septum and proteins involved in endothecium thickening and dehydration were shown to be involved in anther dehiscence (Wilson et al., 2011). The lack of anther dehiscence might be explained by GRF1R expression maintaining cells in the dedifferentiated state for a longer period. Alternatively, as GRF1R expression favored cell division over cell expansion in the maize leaf, it might have a similar role in delaying endothecium cell expansion, which is a prerequisite for anther dehiscence (Wilson et al., 2011). Since the pollen of GRF1R-overexpressing plants was fertile, we can exclude GRF1 playing a role in the development of functional pollen.
The phenotypes in the tassel and the ear could be caused by prolonged cell division, such that full differentiation needed for fertilization was not yet achieved in the transgenic plants. If GRF1R indeed stimulates cell division in leaf, tassel, and ear, it could indicate that the mechanisms of regulation of cell division are conserved among the different plant organs and that fine-tuned modification of cell division, which is often distinct from constitutive overexpression, is instrumental for timely completion of the developmental program. Indeed, it has previously been demonstrated that hyperproliferation, as exemplified by the overexpression of CYCLIND3, could result in inhibition of differentiation (Dewitte et al., 2003).
Interestingly, both GRF1 and GRF10 were identified in the TAP experiments that were performed on developing ears, while only for GRF1R overexpression, ear phenotypes were observed. This could suggest that growth in the ear is regulated by different GRFs than in the leaf, despite the copurification of GRF10 in both organs. This is in accordance to data in Arabidopsis where it was shown that all nine GRF genes are expressed in both leaves and flowers, while only mutations in some GRF genes cause leaf phenotypes (Kim et al., 2003).
Remarkably, despite the different effect on the processes of cell division and cell expansion during leaf growth, and on final leaf length, overexpression of both GRF1R and GRF10 (Wu et al., 2014) resulted in a similar effect of reduced final plant height. This indicates that precise sampling is needed to reveal the role of genes/proteins in a particular part of the plant but that this cannot be extrapolated to other developmental processes. Also, when overexpressing GA20-Oxidase in maize, it was observed that elevated levels of bioactive GA had opposing roles in stimulating cell division in the leaf and cell expansion in the internodes (Nelissen et al., 2012). It has already been shown that both excessive and reduced cell division can result in smaller organs (De Veylder et al., 2002; Dewitte et al., 2003; Horiguchi et al., 2005; Borghi et al., 2010; Gonzalez et al., 2012; Kalve et al., 2014).
Toward Dynamics in Interactomics
In this work, we have implemented TAP in maize and thus generated a tool to identify protein complexes in a crop species. Beyond the implementation of this technology, combining discrete regions of the maize leaf growth zone with the technical advances of MS allowed for the analysis of the dynamics of the protein-protein interactions in a growing organ. Transitioning from gathering information on static protein complexes to gaining insights into the dynamics of the complex composition is of great benefit for all developmental biologists, whether they are studying Drosophila melanogaster wing development, worm growth, bone cartilage growth, or plant organ size regulation. As a next step, the information extracted from querying interactome dynamics could be used to devise inventive strategies using chimeric constructs to improve growth.
METHODS
Transgenic Lines
To render GRF1 resistant for miR396a, the target site (5′-AACCGTTCAAGAAAGCCTGTGGAA-3′) was altered by changing nucleotides at the wobble position (5′-AATCGCTCCAGGAAACCAGTCGAG-3′), without affecting the protein sequence encoded. The ZmUBI-L:AN3-GSTEV and BdEF1α:GRF1R constructs were introduced into the B104 inbred background using Agrobacterium tumefaciens-mediated transformation of immature embryos (Coussens et al., 2012). For AN3-GS, we selected the lines in which the T-DNA was present in a single locus and for which a good protein expression level (at 23.6 kD + tag) was observed in the transgenic plants from a segregating population. Leaf samples were always taken during steady state growth of the leaf (2 d after the appearance of leaf 4), and ear samples were harvested when the growing ear was between 4 and 5 cm in size. As we wanted to zoom into the dynamics of the complex composition along the growth zone, we first determined the protein levels that could be extracted from the leaf tissue from the maize (Zea mays) leaf growth zone sampled with centimeter resolution. To obtain sufficient input for the TAP experiments, we pooled 50 samples of one centimeter, both for the division zone (first centimeter) and the expansion zone (fourth centimeter). On average, 10.8 mg (±0.75 mg) total protein could be extracted from a pool of 50 basal centimeter samples representing the dividing tissue, while 5.8 mg (±0.072 mg) was extracted from elongating tissue. Subsequently we determined the protein levels in these samples and found that the growth zone leaf tissue contained between 1.86% protein/fresh weight for the division zone and 0.69% protein/fresh weight for the expanding tissue. Ears of maximal 4 cm in size were sampled for TAP.
Two transgenic lines were selected corresponding to an 18- and 2-fold increase in AN3 mRNA level in AN3-GS1 and AN3-GS2, respectively. AN3-GS TAP fusions were detected with peroxidase antiperoxidase soluble complex antibody (Sigma-Aldrich P1291; 1/2500 dilution). For the loading control, rabbit antibody against Histone H3 was used (Merck Millipore 17-10046; 1/1000 dilution). Of these two lines, AN3-GS1 showed the highest accumulation of the fusion protein correlating with mRNA levels (Supplemental Figure 1), and subsequent experiments were performed on segregating offspring of this line. Growth analyses were performed according to Nelissen et al. (2013). For EF1α:GRF1R, three independent single-locus events were used for growth analysis (Nelissen et al., 2013) and phenotyping. The expression level of the different GRFs along the leaf gradient of B104 (during steady state growth, 2 d after leaf four appearance) was evaluated using primers specific for each GRF, and 18S rRNA was used as a housekeeping gene. The primers are listed in Supplemental Table 5. For each sample, three biological and three technical repeats were assessed. The expression profile of the different GRF genes is presented relative to the most basal sample, while the GRF1 expression profile to show the difference in expression level between the transgenic GRF1R plants and nontransgenic siblings was relative to the sample with the maximal expression value.
TAP
For one TAP purification, 50 1-cm samples of the fourth leaf centimeter 1 or 4, or five 1-cm samples of ear centimeter 1 or 4 were collected and pooled. Leaf or ear material was ground in liquid nitrogen with mortar and pestle. The fine powder was transferred to a precooled 12-mL tube and 1:2 (w:v) ice-cold extraction buffer (25 mM Tris-HCl, pH 7.6, 15 mM MgCl2, 150 mM NaCl, 15 mM p-nitrophenyl phosphate, 60 mM β-glycerophosphate, 0.1% Nonidet P-40, 0.1 mM Na3VO4, 1 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 1 μM E64, EDTA-free Ultra Complete tablet [Roche; 1/10 mL], 5% ethylene glycol, and 0.1% benzonase) was added. Extracts were mixed three times for 30 s with an Ultra-Turrax for homogenization using a precooled dispersing probe (10G). The mixture was incubated for 30 min on a tube rotator at 4°C. Extracts were centrifuged two times for 20 min at 20,800g in a precooled centrifuge at 4°C. The supernatant was collected and filtered through a GF-prefilter combined with a 0.45-μm syringe filter. The protein concentration was determined via a standard Bradford method. For leaf samples, a typical total protein input of 5 to 7 mg was obtained, while for ear samples, this was ∼15 mg. The protein extract was incubated for 1 h at 4°C on a tube rotator with 25 μL IgG Sepharose beads (GE Healthcare) equilibrated in extraction buffer. The mixture was transferred to a Mobicol column with a 35-µm bottom filter (ImTec) assembled on a vacuum manifold, and the unbound fraction was carefully removed by applying vacuum. The beads were washed five times on the Mobicol column with 750 μL wash buffer (10 mM Tris-HCl, pH 7.6, 150 mM NaCl, 0.1% Nonidet P-40, 0.5 mM EDTA, 1 μM E64, 1 mM phenylmethylsulfonyl fluoride, and 5% ethylene glycol). The Mobicol column was removed from the vacuum manifold and closed at the bottom with a plug. Wash buffer (100 μL) and 10 units of TEV protease were added to the beads, and the Mobicol column was closed with the Luer-lock cap. The beads were incubated with TEV protease for 1 h at 16°C on a tube rotator. After 30 min, a boost of 10 units of TEV protease was added. The plug at the bottom was removed and the Mobicol column was transferred to a 1.5-mL Protein LoBind tube. The eluate was collected by a 1-min centrifugation step at 450g at 4°C. The IgG beads were washed with 100 μL wash buffer, and the wash/eluate was collected by repeating the 1-min centrifugation step at 450 g at 4°C. The IgG eluate was added to 25 μL Streptavidin beads (GE Healthcare), equilibrated in wash buffer. The mixture was incubated for 1 h at 4°C on a tube rotator. Afterward, the mixture was transferred to a Mobicol column, and the unbound fraction was carefully removed by applying vacuum. The beads were washed three times with 833 μL wash buffer (2.5 mL in total) by gravity. The Mobicol column was closed and the complexes were eluted from the Streptavidin beads by adding 30 μL 1× NuPAGE sample buffer (Invitrogen) supplemented with 1× NuPAGE reducing agent and 20 mM desthiobiotin (Sigma-Aldrich) and incubated for 10 min at room temperature with regular mixing. The plug at the bottom was removed and the Mobicol column was transferred to a 1.5-mL Protein LoBind tube. The TAP eluate was collected by a 1-min centrifugation step at 450g and 4°C.
In-Gel Protein Digestion
The TAP was eluated for 10 min at 70°C and loaded on a precast 4 to 12% gradient NuPAGE Bis-Tris gel (Invitrogen). The gel was run at 200 V for 7 min, and the proteins were visualized by colloidal Coomassie Brilliant Blue G 250 staining. The gel was destained twice for 1 h in 25 mL HPLC grade water. The polypeptide disulfide bridges were reduced by incubating the gel for 40 min in 25 mL of 6.66 mM DTT in 50 mM NH4HCO3 and sequentially the thiol groups were alkylated by incubating the gel for 30 min in 25 mL 55 mM IAM in 50 mM NH4HCO3 in the dark. The gel was washed for 30 min in 25 mL HPLC-grade water and transferred to a glass plate. The broad protein zone containing all of the eluted proteins was cut out per lane. The gel fragment was sliced into ∼18 gel plugs and transferred to a 1.5-mL Protein LoBind tube. The gel plugs were washed with 600 μL HPLC grade water and dehydrated in 600 μL 95% acetonitrile (v/v) for 10 min. The gel plugs were rehydrated in 600 μL HPLC-grade water twice for 10 min. The gel particles were rehydrated in 90 μL digest buffer containing 1.125 μg trypsin (MS Gold; Promega), 50 mM NH4HCO3, and 10% acetonitrile (v/v) for 30 min at 4°C. Afterward, proteins were digested for 3.5 h at 37°C. The trypsin solution was transferred to a fresh 1.5-mL Protein LoBind tube and was placed in the sonication bath for 5 min. The gel plugs were dehydrated with 300 μL 95% acetonitrile (v/v) for 10 min. The acetonitrile solution was removed from the gel plugs and added to the removed trypsin solution. The trypsin digest was dried in a cooled SpeedVac (∼2 to 3 h).
Liquid Chromatography-Tandem MS Analysis
The obtained peptide mixtures were introduced into a liquid chromatography-tandem MS system through an Ultimate 3000 RSLCnano LC (Thermo Fisher Scientific) in-line connected to either an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) or a Q Exactive mass spectrometer (Thermo Fisher Scientific). First, the sample mixture was bound on a trapping column (made in-house, 100-μm i.d. × 20 mm, 5 μm beads, C18 Reprosil-HD; Dr. Maisch). After flushing from the trapping column, the sample was loaded on an analytical column (made in-house, 75 μm i.d. × 150 mm, 5-μm beads, C18 Reprosil-HD; Dr. Maisch) packed in the needle (PicoFrit SELF/P PicoTip emitter, PF360-75-15-N-5; New Objective). Peptides were loaded with loading solvent (0.1% trifluoroacetic acid in water) and separated with a linear gradient from 98% solvent A′ (0.1% formic acid in water) to 40% solvent B′ (0.1% formic acid in water/acetonitrile, 20/80 [v/v]) in 30 min at a flow rate of 300 nL/min. This was followed by a 5-min wash reaching 99% solvent B′. The mass spectrometer was operated in data-dependent, positive ionization mode, automatically switching between MS and tandem MS acquisition for the 10 most abundant peaks in a given MS spectrum. In the LTQ-Orbitrap Velos, full-scan MS spectra were acquired in the Orbitrap at a target value of 1E6 with a resolution of 60,000. The 10 most intense ions were then isolated for fragmentation in the linear ion trap, with a dynamic exclusion of 20 s. Peptides were fragmented after filling the ion trap at a target value of 1E4 ion counts. In the Q Exactive, the source voltage was 3.4 kV, and the capillary temperature 275°C. One MS1 scan (m/z 400 to 2000, AGC target 3 × 106 ions, maximum ion injection time 80 ms) acquired at a resolution of 70,000 (at 200 m/z) was followed by up to 10 tandem MS scans (resolution 17,500 at 200 m/z) of the most intense ions fulfilling predefined selection criteria (AGC target 5 × 104 ions, maximum ion injection time 60 ms, isolation window 2 D, fixed first mass 140 m/z, spectrum data type: centroid, underfill ratio 2%, intensity threshold 1.7 × E4, exclusion of unassigned, 1, 5 to 8, >8 charged precursors, peptide match preferred, exclude isotopes on, dynamic exclusion time 20 s). The HCD collision energy was set to 25% normalized collision energy, and the polydimethylcyclosiloxane background ion at 445.120025 D was used for internal calibration (lock mass).
Data Analysis and Background Filtering
The raw files were processed in three sets (leaf, ear, and controls) using MaxQuant software (version 1.4.1.2; Cox and Mann, 2008). Data were searched with the built-in Andromeda search engine against the Zea mays database ZmB73_5a_WGS_translationsplus. This database contains all entries from the ZmB73_5a_WGS_translations database (downloaded from ftp://ftp.gramene.org/pub/gramene/maizesequence.org) concatenated with sequences of all types of possible contaminants in TAP or proteomics experiments in general. These include the common repository of adventitious protein sequences, a list of proteins commonly found in proteomics experiments, present either by accident or by unavoidable contamination of protein samples (The Global Proteome Machine, www.thegpm.org/crap/). Additionally, commonly used tag sequences and typical TAP contaminants, such as sequences derived from the resins or the proteases used, were added. The ZmB73_5a_WGS_translationsplus database contains in total 136,770 sequence entries and is accessible at www.psb.ugent.be/tapdata.
Variable modifications were set to oxidation of methionines and N-acetylation of protein N termini. Fixed modifications were set to carbamidomethylation of cysteines. Mass tolerance on precursor (peptide) ions was set to 4.5 ppm and on fragment ions to 0.5 D or to 20 ppm for LTQ Orbitrap Velos or Q Exactive data, respectively. Enzyme was set to Trypsin/P, allowing for two missed cleavages, and cleavage was allowed when arginine or lysine was followed by proline. The complete list with the search parameters in MaxQuant can be found in Supplemental Table 6. PSM and protein identifications were filtered using a target-decoy approach at false discovery rate (FDR) of 1%. The protein group output files from MaxQuant are available as Supplemental Data Sets 1A and 1B, for leaf and ear, respectively. Proteins identified with at least two peptides, of which at least one was unique, were retained (Table 1).
Correcting for False Positives and Nonspecific Interactors
Lists of copurified proteins from TAP experiments contain besides specific, bona fide interactors, also nonspecific interactors that copurify with many unrelated bait proteins and false positives that are copurified through binding on the resin instead of on the bait protein. The best way to discriminate between specific and nonspecific or false positives is to perform a large series of TAP experiments with many different baits in order to determine the bait-independent recurring copurified proteins (Keilhauer et al., 2015; Van Leene et al., 2015). However, upon startup of the technology in a new species, where only a small data set was available and such a background list had to be built from scratch, performing mock purifications was the only way to obtain a basic background list. Accordingly, we performed a set of eight mock TAPs on wild-type B104 protein extracts obtained from leaf and ear. All proteins identified with at least one high confidence peptide from the mock GS purifications on wild-type B104 extracts (from leaf and ear) identified with Q Exactive were used to build the maize background list (Supplemental Data Set 1C). Proteins that were present in this background list were filtered out from TAP experiments in maize. Additionally, relying on the fact that the type of background proteins is largely determined by two factors, namely, their abundance in the protein sample and the type of purification used, we took advantage of the plethora of GS purifications performed in Arabidopsis thaliana cell cultures and seedlings to further filter GS background in maize. Of all proteins identified in maize TAP experiments, the orthologous proteins in Arabidopsis were determined by PLAZA 3.0 dicots (Proost et al., 2015), and if the orthologous Arabidopsis protein was known to be GS background in Arabidopsis, the maize protein was also considered GS background and filtered out from maize TAP experiments. Proteins filtered out in this way are listed in Supplemental Data Set 1D. In the future, when TAP data from more baits will become available for maize, a filtering based on frequency of occurrence of the copurified proteins based on a large TAP data set in maize can be applied (Van Leene et al., 2015).
Label-Free Quantification (MaxLFQ) of Leaf AN3 TAP Samples
Relative LFQ of proteins from leaf AN3 TAP samples was done using the MaxLFQ algorithm (Cox et al., 2014) integrated in the MaxQuant software. For this purpose, at least one unique peptide was required per protein identification, and only unique peptides were used to establish MaxLFQ values. The minimum LFQ ratio count was set to 1. Data analysis was performed using Perseus software (version 1.5.1.6). The protein groups file from MaxQuant was loaded into Perseus, and LFQ intensity values were logged. Proteins only identified by site and reverse database hits were removed. AN3 leaf TAP samples were grouped into division zone (four TAPs) and expansion zone (four TAPs). Proteins that did not display three valid values in at least one group were filtered out to improve confidence levels. Missing values were imputed following a normal distribution around the detection limit of the mass spectrometer. Next, a t test was performed to determine statistical outliers between division zone and expansion zone groups. A permutation-based FDR correction was applied to correct for the multiple hypothesis testing problem. Protein groups and t test results with all significantly enriched proteins in the division zone are available in Supplemental Data Set 1A and Supplemental Table 4, respectively.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the accession numbers listed in Table 1 and Supplemental Tables 1, 2, 4, and 5.
Supplemental Data
Supplemental Figure 1. Expression Analysis of the AN3-GS1 Maize Line That Was Used for TAP.
Supplemental Figure 2. Phenotype of the AN3-GS1 Plants Compared with Their Nontransgenic Siblings.
Supplemental Figure 3. Expression Levels of the GRF Genes over the Maize Leaf Growth Zone in B104 at Steady State Growth of Leaf 4.
Supplemental Table 1. Tandem Affinity Purification with Zm-AN3 as Bait.
Supplemental Table 4. Significantly Changing Proteins between the Division Zone and the Expansion Zone.
Supplemental Table 5. Sequence of the qPCR Primers.
Supplemental Table 6. MaxQuant Search Parameters.
Supplemental Data Set 1A. Protein Groups Output from MaxQuant Search on 10 AN3 LEAF TAP Experiments (2x 4 cm 4 [Entire Growth Zone]; 4x cm1 [Division Zone]; 4x cm4 [Expansion Zone]) Analyzed on Orbitrap Velos.
Supplemental Data Set 1B. Protein Groups Output from MaxQuant Search on 6 AN3 EAR TAP Experiments Analyzed on Q Exactive.
Supplemental Data Set 1C. ZM Background Proteins, Identified from Four Leaf and Four Ear Mock TAP Experiments.
Supplemental Data Set 1D. Additional Proteins That Are Considered as Background because the Arabidopsis Ortholog Is Present in the Arabidopsis TAP Background List.
Supplementary Material
Acknowledgments
We thank Karel Spruyt for help with the pictures, Annick Bleys for help in preparing with the article, and Nathalie Gonzalez and Liesbeth Vercuyssen for critical reading of the article. This work was supported by the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013; ERC Grant Agreement [339341-AMAIZE]11), Ghent University (“Bijzonder Onderzoeksfonds Methusalem project” BOF08/01M00408 and Multidisciplinary Research Partnership “Biotechnology for a Sustainable Economy” Grant 01MRB510W), the Interuniversity Attraction Poles Programme (IUAP P7/29 “MARS”) initiated by the Belgian Science Policy Office, and the Research Foundation-Flanders (postdoctoral fellowship to J.V.L.).
AUTHOR CONTRIBUTIONS
H.N., D.I., and G.D.J. designed the research. H.N., D.E., K.D., G.P., J.D.B., M.V., and J.C. performed research. H.N., D.E., J.V.L., M.M., K.G., D.I., and G.D.J. analyzed data. M.v.B. and K.V. provided bioinformatics support. D.E., A.W., S.G., and K.G. provided statistical support. J.D.B., S.A., and M.V.L. did transformations. H.N., D.E., D.I., and G.D.J. wrote the article.
Glossary
- TAP
tandem affinity purification
- MS
mass spectrometry
- GA
gibberellic acid
- LFQ
label-free quantification
- FDR
false discovery rate
Footnotes
Articles can be viewed online without a subscription.
References
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni R., Gonzalez-Guzman M., Rodriguez L., Peirats-Llobet M., Pizzio G.A., Fernandez M.A., De Winne N., De Jaeger G., Dietrich D., Bennett M.J., Rodriguez P.L. (2013). PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 161: 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi L., Gutzat R., Fütterer J., Laizet Y., Hennig L., Gruissem W. (2010). Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell 22: 1792–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candaele J., Demuynck K., Mosoti D., Beemster G.T.S., Inzé D., Nelissen H. (2014). Differential methylation during maize leaf growth targets developmentally regulated genes. Plant Physiol. 164: 1350–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B., Ashworth M., Guo W., Davies B. (2012). The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Kim J., Hwang H.-J., Kim S., Park C., Kim S.Y., Lee I. (2011). The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23: 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C.R., Cairns B.R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78: 273–304. [DOI] [PubMed] [Google Scholar]
- Coussens G., Aesaert S., Verelst W., Demeulenaere M., De Buck S., Njuguna E., Inzé D., Van Lijsebettens M. (2012). Brachypodium distachyon promoters as efficient building blocks for transgenic research in maize. J. Exp. Bot. 63: 4263–4273. [DOI] [PubMed] [Google Scholar]
- Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13: 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26: 1367–1372. [DOI] [PubMed] [Google Scholar]
- Debernardi J.M., Mecchia M.A., Vercruyssen L., Smaczniak C., Kaufmann K., Inzé D., Rodriguez R.E., Palatnik J.F. (2014). Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 79: 413–426. [DOI] [PubMed] [Google Scholar]
- Dedecker M., Van Leene J., De Jaeger G. (2015). Unravelling plant molecular machineries through affinity purification coupled to mass spectrometry. Curr. Opin. Plant Biol. 24: 1–9. [DOI] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Beemster G.T.S., de Almeida Engler J., Ormenese S., Maes S., Naudts M., Van Der Schueren E., Jacqmard A., Engler G., Inzé D. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21: 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W., Riou-Khamlichi C., Scofield S., Healy J.M.S., Jacqmard A., Kilby N.J., Murray J.A.H. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloy N.B., de Freitas Lima M., Van Damme D., Vanhaeren H., Gonzalez N., De Milde L., Hemerly A.S., Beemster G.T.S., Inzé D., Ferreira P.C.G. (2011). The APC/C subunit 10 plays an essential role in cell proliferation during leaf development. Plant J. 68: 351–363. [DOI] [PubMed] [Google Scholar]
- Eloy N.B., et al. (2012). SAMBA, a plant-specific anaphase-promoting complex/cyclosome regulator is involved in early development and A-type cyclin stabilization. Proc. Natl. Acad. Sci. USA 109: 13853–13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen G., Auerbach R.K., Snyder M. (2012). SWI/SNF chromatin-remodeling factors: multiscale analyses and diverse functions. J. Biol. Chem. 287: 30897–30905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen G.M., Auerbach R.K., Davidov E., Gianoulis T.A., Zhong G., Rozowsky J., Bhardwaj N., Gerstein M.B., Snyder M. (2011). Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 7: e1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facette M.R., Shen Z., Björnsdóttir F.R., Briggs S.P., Smith L.G. (2013). Parallel proteomic and phosphoproteomic analyses of successive stages of maize leaf development. Plant Cell 25: 2798–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona S., Hurtado L., Bowman J.L., Reyes J.C. (2004). The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131: 4965–4975. [DOI] [PubMed] [Google Scholar]
- Gadeyne A., et al. (2014). The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156: 691–704. [DOI] [PubMed] [Google Scholar]
- Gentry M., Hennig L. (2014). Remodelling chromatin to shape development of plants. Exp. Cell Res. 321: 40–46. [DOI] [PubMed] [Google Scholar]
- Gonzalez N., Vanhaeren H., Inzé D. (2012). Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci. 17: 332–340. [DOI] [PubMed] [Google Scholar]
- Grüne T., Brzeski J., Eberharter A., Clapier C.R., Corona D.F.V., Becker P.B., Müller C.W. (2003). Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol. Cell 12: 449–460. [DOI] [PubMed] [Google Scholar]
- Han P., Hang C.T., Yang J., Chang C.-P. (2011). Chromatin remodeling in cardiovascular development and physiology. Circ. Res. 108: 378–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G., Kim G.-T., Tsukaya H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43: 68–78. [DOI] [PubMed] [Google Scholar]
- Kalve S., De Vos D., Beemster G.T.S. (2014). Leaf development: a cellular perspective. Front. Plant Sci. 5: 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanei M., Horiguchi G., Tsukaya H. (2012). Stable establishment of cotyledon identity during embryogenesis in Arabidopsis by ANGUSTIFOLIA3 and HANABA TARANU. Development 139: 2436–2446. [DOI] [PubMed] [Google Scholar]
- Keilhauer E.C., Hein M.Y., Mann M. (2015). Accurate protein complex retrieval by affinity enrichment mass spectrometry (AE-MS) rather than affinity purification mass spectrometry (AP-MS). Mol. Cell. Proteomics 14: 120–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Kende H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 13374–13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Choi D., Kende H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36: 94–104. [DOI] [PubMed] [Google Scholar]
- Kuijt S.J.H., et al. (2014). Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant Physiol. 164: 1952–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.H., Wynn A.N., Franks R.G., Hwang Y.S., Lim J., Kim J.H. (2014). The Arabidopsis thaliana GRF-INTERACTING FACTOR gene family plays an essential role in control of male and female reproductive development. Dev. Biol. 386: 12–24. [DOI] [PubMed] [Google Scholar]
- Lessard J., Wu J.I., Ranish J.A., Wan M., Winslow M.M., Staahl B.T., Wu H., Aebersold R., Graef I.A., Crabtree G.R. (2007). An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55: 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., He H., Li Y., Wang F., Yu D. (2014). Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol. 164: 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Song Y., Chen Z., Yu D. (2009). Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant. 136: 223–236. [DOI] [PubMed] [Google Scholar]
- Lu R., Wang G.G. (2013). Tudor: a versatile family of histone methylation ‘readers’. Trends Biochem. Sci. 38: 546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middeljans E., Wan X., Jansen P.W., Sharma V., Stunnenberg H.G., Logie C. (2012). SS18 together with animal-specific factors defines human BAF-type SWI/SNF complexes. PLoS ONE 7: e33834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H., Rymen B., Coppens F., Dhondt S., Fiorani F., Beemster G.T.S. (2013). Kinematic analysis of cell division in leaves of mono- and dicotyledonous species: a basis for understanding growth and developing refined molecular sampling strategies. Methods Mol. Biol. 959: 247–264. [DOI] [PubMed] [Google Scholar]
- Nelissen H., Rymen B., Jikumaru Y., Demuynck K., Van Lijsebettens M., Kamiya Y., Inzé D., Beemster G.T.S. (2012). A local maximum in gibberellin levels regulates maize leaf growth by spatial control of cell division. Curr. Biol. 22: 1183–1187. [DOI] [PubMed] [Google Scholar]
- Nelissen H., et al. (2010). Plant Elongator regulates auxin-related genes during RNA polymerase II transcription elongation. Proc. Natl. Acad. Sci. USA 107: 1678–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L., Lu F., Pei Y., Liu C., Cao X. (2007). Regulation of flowering time by the protein arginine methyltransferase AtPRMT10. EMBO Rep. 8: 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C.L., Dingwall A., Scott M.P. (1994). Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc. Natl. Acad. Sci. USA 91: 2905–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S., Van Bel M., Vaneechoutte D., Van de Peer Y., Inzé D., Mueller-Roeber B., Vandepoele K. (2015). PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. 43: D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R.E., Mecchia M.A., Debernardi J.M., Schommer C., Weigel D., Palatnik J.F. (2010). Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137: 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R., Zhou M.-M. (2009). The role of human bromodomains in chromatin biology and gene transcription. Curr. Opin. Drug Discov. Devel. 12: 659–665. [PMC free article] [PubMed] [Google Scholar]
- Satoh-Nagasawa N., Nagasawa N., Malcomber S., Sakai H., Jackson D. (2006). A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441: 227–230. [DOI] [PubMed] [Google Scholar]
- Shrestha B., Guragain B., Sridhar V.V. (2014). Involvement of co-repressor LUH and the adapter proteins SLK1 and SLK2 in the regulation of abiotic stress response genes in Arabidopsis. BMC Plant Biol. 14: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., Witters E., Inzé D., De Jaeger G. (2008). Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci. 13: 517–520. [DOI] [PubMed] [Google Scholar]
- Van Leene J., et al. (2015). An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nat. Protoc. 10: 169–187. [DOI] [PubMed] [Google Scholar]
- Van Leene J., et al. (2007). A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol. Cell. Proteomics 6: 1226–1238. [DOI] [PubMed] [Google Scholar]
- Vercruyssen L., et al. (2014). ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26: 210–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Qiu N., Ding Q., Li J., Zhang Y., Li H., Gao J. (2014). Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genomics 15: 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Gu X., Xu D., Wang W., Wang H., Zeng M., Chang Z., Huang H., Cui X. (2011). miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J. Exp. Bot. 62: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-T., Yuan C., Yuan T.-T., Cui S.-J. (2012). The Arabidopsis LFR gene is required for the formation of anther cell layers and normal expression of key regulatory genes. Mol. Plant 5: 993–1000. [DOI] [PubMed] [Google Scholar]
- Wang Z., Yuan T., Yuan C., Niu Y., Sun D., Cui S. (2009). LFR, which encodes a novel nuclear-localized Armadillo-repeat protein, affects multiple developmental processes in the aerial organs in Arabidopsis. Plant Mol. Biol. 69: 121–131. [DOI] [PubMed] [Google Scholar]
- Wilson Z.A., Song J., Taylor B., Yang C. (2011). The final split: the regulation of anther dehiscence. J. Exp. Bot. 62: 1633–1649. [DOI] [PubMed] [Google Scholar]
- Wu L., Zhang D., Xue M., Qian J., He Y., Wang S. (2014). Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height. J. Integr. Plant Biol. 56: 1053–1063. [DOI] [PubMed] [Google Scholar]
- Yun H., Hyun Y., Kang M.-J., Noh Y.-S., Noh B., Choi Y. (2011). Identification of regulators required for the reactivation of FLOWERING LOCUS C during Arabidopsis reproduction. Planta 234: 1237–1250. [DOI] [PubMed] [Google Scholar]
- Zhang D.-F., Li B., Jia G.-Q., Zhang T.-F., Dai J.-R., Li J.-S., Wang S.-C. (2008). Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in maize (Zea mays L.). Plant Sci. 175: 809–817. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.