Opposing actions of vesicle trafficking proteins on K+ channel gating implicate a ‘handoff’ in binding and channel control during vesicle fusion.
Abstract
SNARE (soluble N-ethylmaleimide-sensitive factor protein attachment protein receptor) proteins drive vesicle traffic, delivering membrane and cargo to target sites within the cell and at its surface. They contribute to cell homeostasis, morphogenesis, and pathogen defense. A subset of SNAREs, including the Arabidopsis thaliana SNARE SYP121, are known also to coordinate solute uptake via physical interactions with K+ channels and to moderate their gating at the plasma membrane. Here, we identify a second subset of SNAREs that interact to control these K+ channels, but with opposing actions on gating. We show that VAMPs (vesicle-associated membrane proteins), which target vesicles to the plasma membrane, also interact with and suppress the activities of the inward-rectifying K+ channels KAT1 and KC1. Interactions were evident in yeast split-ubiquitin assays, they were recovered in vivo by ratiometric bimolecular fluorescence complementation, and they were sensitive to mutation of a single residue, Tyr-57, within the longin domain of VAMP721. Interaction was also recovered on exchange of the residue at this site in the homolog VAMP723, which normally localizes to the endoplasmic reticulum and otherwise did not interact. Functional analysis showed reduced channel activity and alterations in voltage sensitivity that are best explained by a physical interaction with the channel gates. These actions complement those of SYP121, a cognate SNARE partner of VAMP721, and lead us to propose that the channel interactions reflect a “hand-off” in channel control between the two SNARE proteins that is woven together with vesicle fusion.
INTRODUCTION
SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins overcome the dehydration forces associated with lipid bilayer fusion in an aqueous environment, and they match vesicles with their destinations for targeting and delivery of membrane, protein, and soluble cargo. In animals, SNAREs contribute to neurotransmitter release; in yeast, they facilitate budding and growth; and in plants, they are important for the delivery of cell wall material to the apoplastic space and for plant cell expansion (Pratelli et al., 2004; Jahn and Scheller, 2006; Jürgens and Geldner, 2007; Lipka et al., 2007; Bassham and Blatt, 2008). SNAREs are small proteins of 100 to 300 amino acids in length. They contain an evolutionarily conserved SNARE motif comprising a linear sequence of roughly 60 residues. SNAREs have been classified as target and vesicle SNAREs, reflecting their functional localizations, and as Q (glutamine)- and R (arginine)-SNAREs based on the conserved amino acids contributing to the central layer of the core complex (Fasshauer et al., 1998; Bock et al., 2001). Q-SNAREs are further subdivided between Qa-, Qb-, Qc-, and Qbc-SNAREs based on homologies to synaptic SNAREs. Most SNAREs are anchored by a C-terminal transmembrane α-helix (Hong, 2005), and the N terminus of several SNAREs incorporates domains that may fold back on the SNARE motif to regulate its accessibility (Jahn and Scheller, 2006). In general, SNARE assembly brings together one copy each of the Qa-, Qb-, Qc-, and R-SNARE motifs, and it is the assembly of this SNARE core complex that drives vesicle fusion.
R-SNAREs, also known as vesicle-associated membrane proteins (VAMPs), are most commonly associated with trafficking vesicles. Barring a few notable exceptions (Rossi et al., 2004; Brunger, 2005), these proteins incorporate a C-terminal transmembrane anchor and a cytosolic domain to the N terminus of the protein that includes the SNARE motif. R-SNAREs of plants fall roughly into three groupings that have homologies to their counterparts in yeast and animals, namely, the Sec22-, YKT6-, and VAMP7-like R-SNAREs. Of the 12 VAMP-like R-SNAREs in the plant model Arabidopsis thaliana, the four VAMP71 proteins show the greatest similarity to mammalian VAMP7 and contribute primarily to endosomal trafficking; the remaining eight VAMP72 proteins appear specific to green plants and, with few exceptions, are thought to be responsible primarily for secretion at the plasma membrane (Uemura et al., 2004; Sanderfoot, 2007).
VAMPs have been associated with a number of physiological activities in plants. Along with other SNAREs, they have been identified with gravitropism (Kato et al., 2002; Yano et al., 2003), with nodule tissue formation (Ma et al., 2006), and with cytokinesis (Zhang et al., 2011; El Kasmi et al., 2013). VAMPs associate in SNARE complexes that contribute to defense responses (Collins et al., 2003; Kwon et al., 2008; Yun et al., 2013) and abiotic stress tolerance (Leshem et al., 2006, 2010). In general, these functions are linked directly with membrane vesicle traffic or can be understood as a direct consequence of its regulation.
By contrast, previous reports from this laboratory demonstrated that the Qa-SNARE SYP121 of Arabidopsis and tobacco (Nicotiana tabacum) mediates in the control of plasma membrane K+ channels (Leyman et al., 1999; Honsbein et al., 2009). In this case, a direct connection per se to vesicle traffic is not obvious. In Arabidopsis, SYP121 is one of three evolutionarily related Qa-SNAREs that are widely expressed throughout the vegetative plant and share overlapping functions in secretory vesicle traffic at the plasma membrane (Sanderfoot, 2007; Grefen and Blatt, 2008; Enami et al., 2009). Studies of Honsbein et al. (2009) and subsequent analyses of the channel binding site on the Qa-SNARE (Grefen et al., 2010a; Honsbein et al., 2011) have indicated that SYP121 control of the K+ channels is biochemically and functionally separable from its activity in vesicle traffic. Indeed, traffic and transport control by SYP121 can be uncoupled, leading to a hyperaccumulation of solute in the affected tissues (Sokolovski et al., 2008).
These findings are of interest because two R-SNAREs, VAMP721 and VAMP722, are known to assemble in SNARE core complexes with SYP121 and with its closest homolog SYP122 (Kwon et al., 2008; Karnik et al., 2013b). This overlap in cognate SNAREs at the Arabidopsis plasma membrane is significant for K+ channel control, if only because it indicates a degree of mechanistic interaction between the secretory pathways represented by the two Qa-SNAREs. It also raises the question of whether this interaction might extend to the R-SNAREs themselves. Here, we addressed this question to define the characteristics of R-SNARE interactions with the two K+ channels previously shown to bind with SYP121. We report that VAMP721 and VAMP722, but not VAMP723, interact with the channels, affecting channel gating and suppressing the K+ current within the physiological voltage range. Selective binding is associated with the VAMP longin domain, notably with Tyr-57, and substitutions with this residue alone are sufficient to engineer binding and K+ channel control in VAMP723. These and additional observations lead us to suggest that VAMP721 and VAMP722 binding with the channels contributes to a “hand-over” in channel control during SNARE complex assembly and vesicle fusion.
RESULTS
Specificities Associated with VAMP-K+ Channel Interactions in Vitro
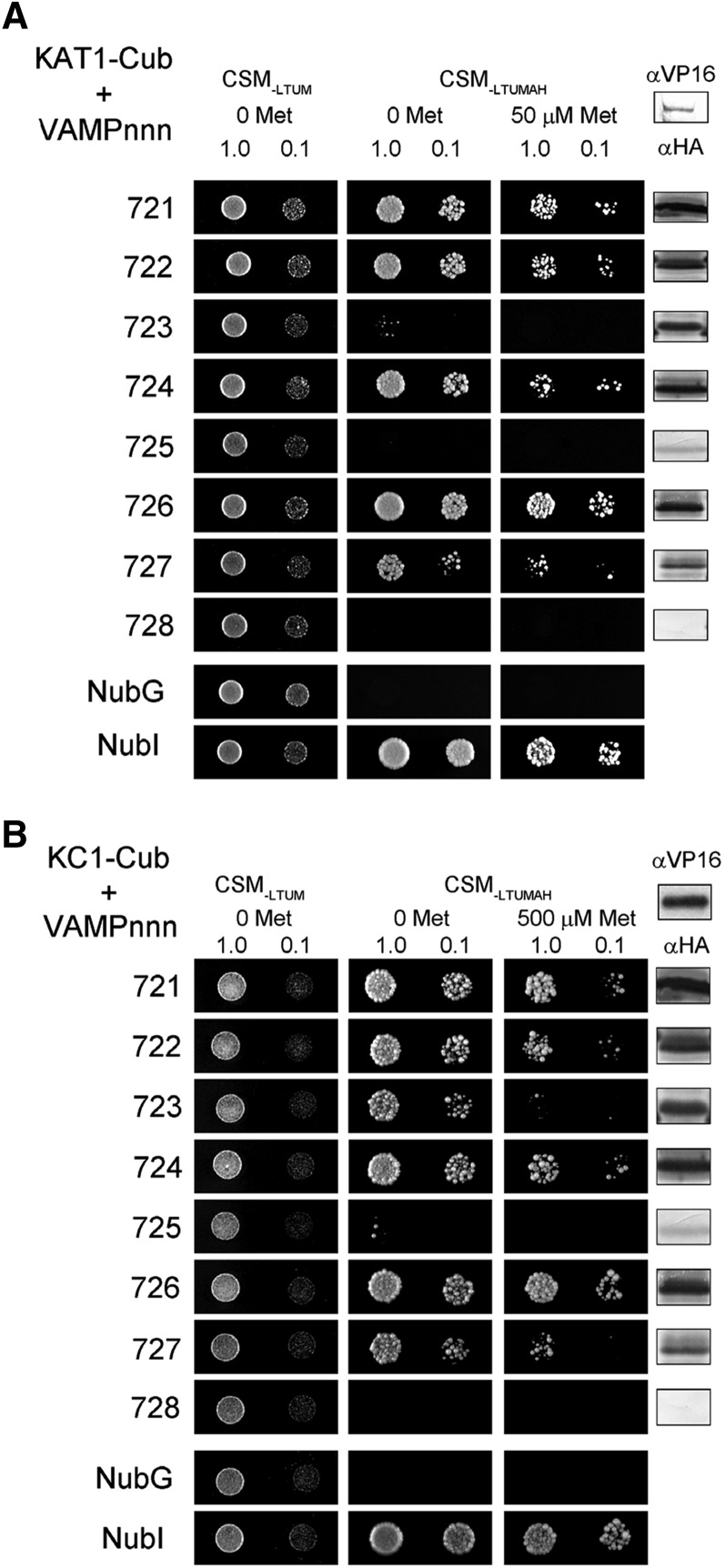
We used a mating-based split-ubiquitin system (SUS) (Grefen et al., 2009, 2010a; Honsbein et al., 2011) to explore interactions of the K+ channels KAT1 and KC1, previously shown to bind SYP121, with the VAMP72 subgroup that includes members localizing to the plasma membrane. SUS assays enable testing for interactions between full-length, integral membrane proteins without the need to generate constructs that isolate the soluble domains of the putative interactors. For SUS screens, K+ Channel-Cub fusions (where Cub comprises the C-terminal half of ubiquitin fused with PLV, a fusion of Protein A, the LexA DNA binding domain, and the VP16 activating domain) were placed under the expression control of the methionine-repressible promoter of yeast, Met-25. Nub-VAMP fusions were constructed with the N-terminal half of ubiquitin, NubG, incorporating a single Ile-to-Gly mutation that prevents spontaneous ubiquitin reassembly. Thus, only interaction of the protein fusions led to reassembly of the complete ubiquitin molecule, cleavage of the LexA-VP16 transactivator, and its migration to the nucleus to activate reporter gene expression (Grefen et al., 2009). Growth on interaction-selective media CSM-LTUMAH lacking Ade and His was used to test for interaction of bait (K+ channel) and prey (VAMP) proteins, and growth was further challenged with additions of Met to repress transcription of the K+ channel-Cub bait constructs as a test for binding specificity.
Figure 1 shows SUS assays for interaction of these VAMPs with the KAT1 (A) and KC1 (B) K+ channels and supporting immunoblot data from one of three independent experiments, each yielding similar results. We focus on comparing growth between preys (VAMPs) with each bait (K+ channel) construct. (A direct comparison between baits is hindered by differences in bait expression [compared with VP16 immunoblots] and, consequently, the different methionine concentrations needed for bait suppression.) The VAMP72 subgroup is largely associated with trafficking at the plasma membrane (VAMP721, VAMP722, VAMP724, VAMP725, and VAMP726; Uemura et al., 2004; Kwon et al., 2008), but also at the endoplasmic reticulum (VAMP723; Uemura et al., 2004) and, putatively, with traffic between the plasma membrane and early endosome (VAMP727; Uemura et al., 2004; Ebine et al., 2011). We found that the diploid yeast grew well on interaction-selective media when carrying the K+ channel-Cub fusions with Nub fusions of VAMP721, VAMP722, VAMP724, and VAMP726. Weaker growth was recovered with the Nub-VAMP727 fusion, notably in the presence of Met. Growth was not recovered with Nub-VAMP725 and Nub-VAMP728; however, expression of these prey constructs was weak or absent, precluding any meaningful conclusion. No growth was observed with either K+ channel-Cub fusion when mated with Nub-VAMP723, especially in the presence of Met, despite strong expression of the prey protein fusion. These results indicated consistent and selective interactions of the K+ channels, notably with the plasma membrane-associated VAMP721 and VAMP722, by contrast with VAMP723 that localizes to the endoplasmic reticulum. VAMP721 and VAMP722 are greater than 97% sequence identical (Supplemental Figure 1) and appear functionally redundant in vivo (Kwon et al., 2008; Zhang et al., 2011; El Kasmi et al., 2013). Therefore, we focused on the interaction with VAMP721 and used VAMP723 as a control to resolve the interacting domains and functionality of the channel-VAMP interactions.
Figure 1.
The KAT1 and KC1 K+ Channels Interact Strongly with the Plasma Membrane VAMPs of the VAMP72 Subgroup.
Yeast mating-based split-ubiquitin assay for interaction of VAMPs (VAMPnnn, where nnn is the numerical designator) with KAT1-Cub (A) and KC1-Cub (B) as baits. Yeast diploids were created with NubG fusion constructs of each of the VAMP proteins together with controls (NubG, negative; NubI [wild type], positive) spotted (left to right) on CSM medium without Leu, Trp, Ura, and Met (CSM-LTUM) to verify mating, CSM medium without Leu, Trp, Ura, Met, Ade, and His (CSM-LTUMAH) to verify adenine- and His-independent growth, and on CSM-LTUMAH with the addition of Met to verify interaction at lower K+ channel-Cub expression levels (50 μM for KAT1 and 500 μM for KC1). Diploid yeast was dropped at 1.0 and 0.1 OD600 in each case. Immunoblot analysis (5 μg total protein/lane) of the haploid yeast used for mating (right) using commercial HA antibody for the VAMP fusions and VP16 antibody for the K+ channel fusions.
VAMP721, but Not VAMP723, Interacts with KAT1 and KC1 K+ Channels in Vivo
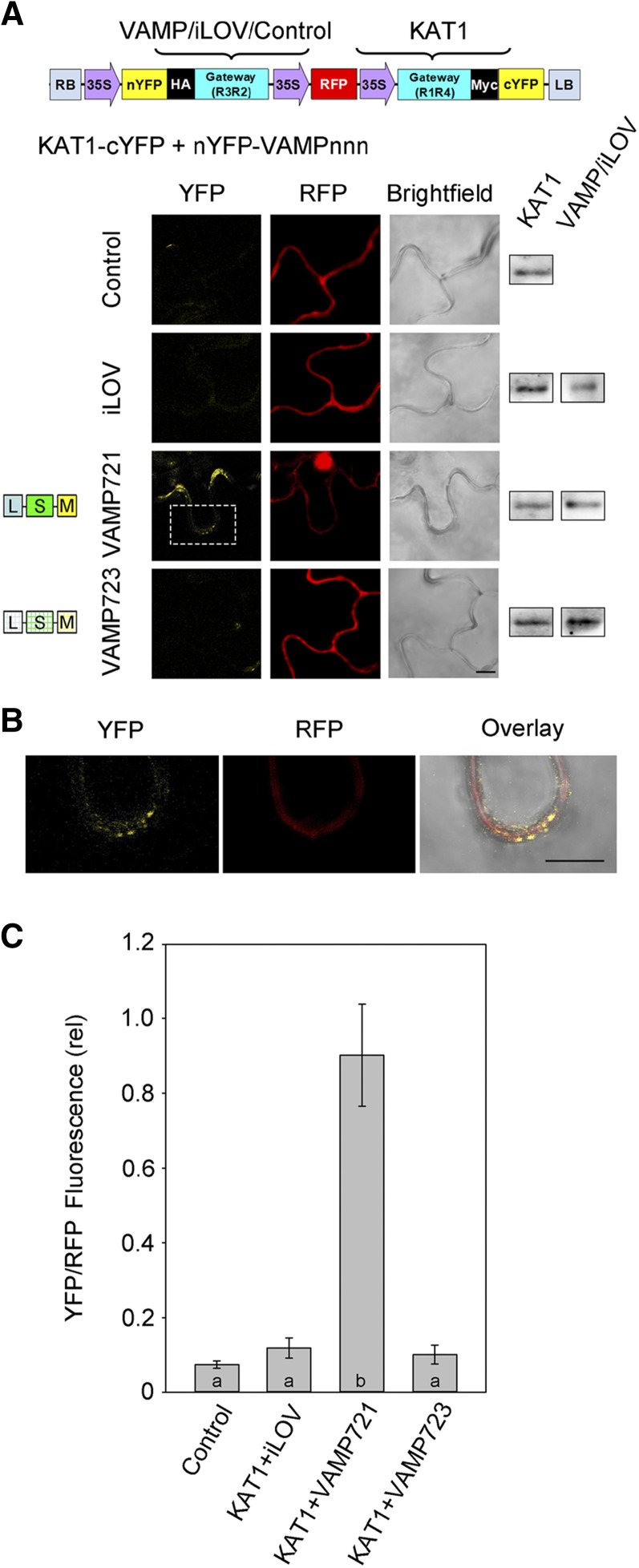
To assess VAMP-channel interactions in vivo, we cloned the coding sequences for VAMP721 and VAMP723 into the 2in1 vector system (Grefen and Blatt, 2012) for transient transformation, expression, and ratiometric bimolecular fluorescence complementation (rBiFC). The 2in1 rBiFC system incorporates a set of independent, Gateway-compatible cassettes, each with a 35S promoter and either the N- or C-terminal half of yellow fluorescent protein (YFP) (Figure 2A, top). The 2in1 system also includes a third 35S-driven cassette with the coding sequence for soluble red fluorescent protein (RFP), which provides a control for transformation and a reference for ratiometric quantification of BiFC. VAMP proteins incorporate a C-terminal membrane anchor; hence, when labeled at this end of the protein, any tag would be localized on the extracytosolic side of the membrane. Thus, we generated VAMPs tagged on the N terminus with the N-terminal half of YFP (nYFP-VAMPnnn) and we tagged the K+ channels at their C terminus with the C-terminal half of YFP (channel-cYFP).
Figure 2.
The KAT1 K+ Channel Interacts in Vivo with VAMP721 but Not VAMP723.
(A) rBiFC analysis of YFP and RFP fluorescence collected from tobacco transformed using the pBiFCt-2in1-NC (Grefen and Blatt, 2012) 2in1 vector (schematic above). Images are (left to right) YFP (BiFC) fluorescence, RFP fluorescence, and bright field. Constructs (top to bottom) included coding sequences for KAT1-cYFP with either the empty cassette (control) or nYFP-X fusions with iLOV, VAMP721, and VAMP723. Immunoblot analysis using αHA and αmyc antibodies to verify fusion protein expression is shown (right). Bar = 10 μm.
(B) Expanded view of the image region denoted by the white box in (A) for nYFP-VAMP721 coexpressed with KAT1-cYFP. Images are (left to right) YFP fluorescence, RFP fluorescence, and bright field. Bar = 10 μm.
(C) rBiFC fluorescence signals from three independent experiments. Each bar represents the mean ± se of fluorescence intensity ratios from 10 images taken at random over the leaf surface. rBiFC signals were calculated as the mean fluorescence intensity ratio determined from each image set after subtracting the background fluorescence determined from an equivalent number of images taken from nontransformed tissues. Significance is indicated by letters at P < 0.01.
KAT1 expresses well and is correctly targeted to the plasma membrane when expressed in tobacco (Sutter et al., 2006, 2007). For reasons that are still unclear, KC1 is poorly mobilized from the endoplasmic reticulum in tobacco and becomes localized to the plasma membrane only when expressed in Arabidopsis (Duby et al., 2008; Honsbein et al., 2009). Therefore, we performed transient transformations with KAT1 and the VAMPs in tobacco leaves, and we transformed Arabidopsis to test interactions with KC1. YFP and RFP fluorescence was analyzed by confocal microscopy using standardized recording parameters, and rBiFC ratios were determined after subtraction of background fluorescence recorded from untransformed tissues in each case. Figure 2 summarizes representative images and supporting immunoblot data from one of three experiments in which KAT1 was coexpressed with the VAMPs, and it includes statistical analysis for all three experiments. As controls, we expressed KAT1-cYFP on its own and with nYFP-tagged iLOV in the 2in1 vector. iLOV is a soluble phototropin unrelated to the VAMPs and is expressed constitutively in Arabidopsis (Chapman et al., 2008). Consistent with the SUS assays, KAT1 coexpression with VAMP721 gave a strong rBiFC signal. YFP fluorescence was clearly evident in puncta distributed around the cell periphery (Figure 2B), consistent with the characteristic localization of the KAT1 channel at the plasma membrane (Sutter et al., 2006, 2007). Coexpression with VAMP723 yielded a low rBiFC ratio that was indistinguishable from the background signals observed on expressing KAT1-cYFP alone and together with nYFP-iLOV. Furthermore, a similar pattern in rBiFC ratios was observed using KC1 as the partner for interactions (Supplemental Figure 2). We concluded that VAMP721 normally interacts with the K+ channels, in vitro and in vivo, while VAMP723 does not.
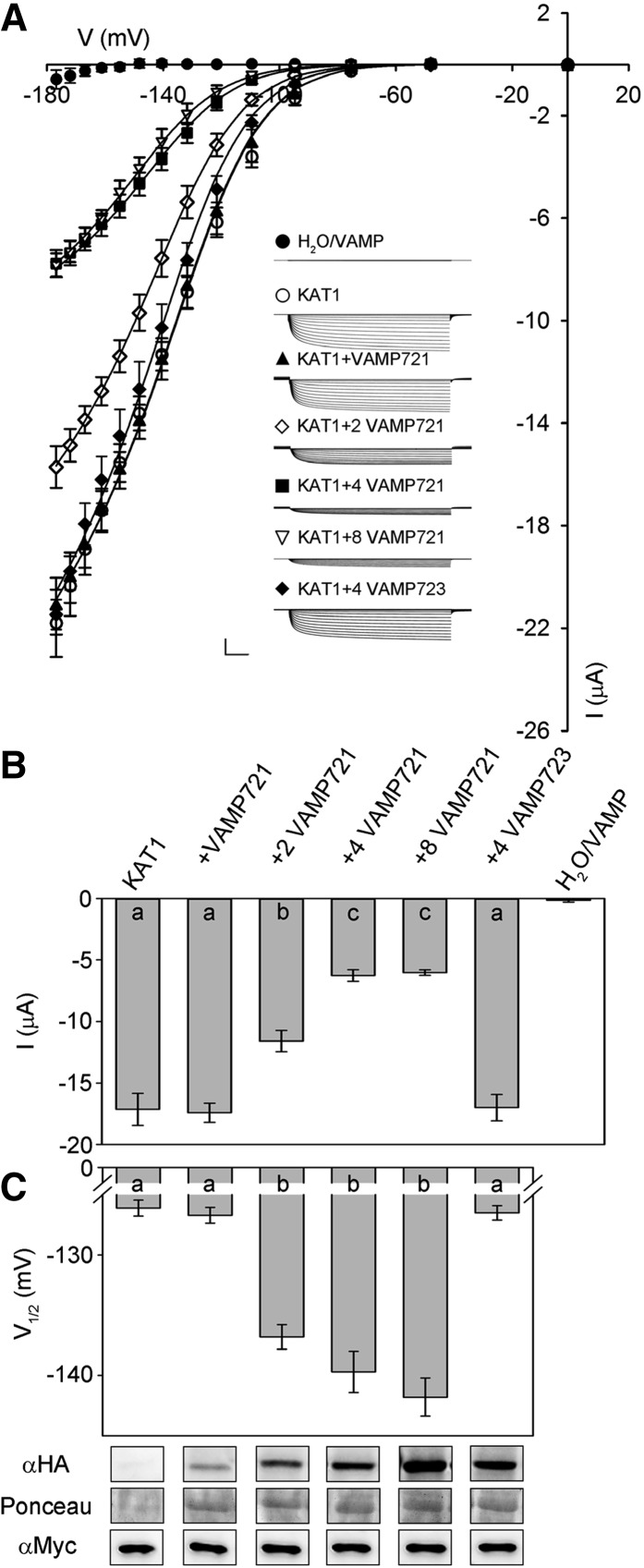
VAMP721 Affects KAT1 K+ Channel Current
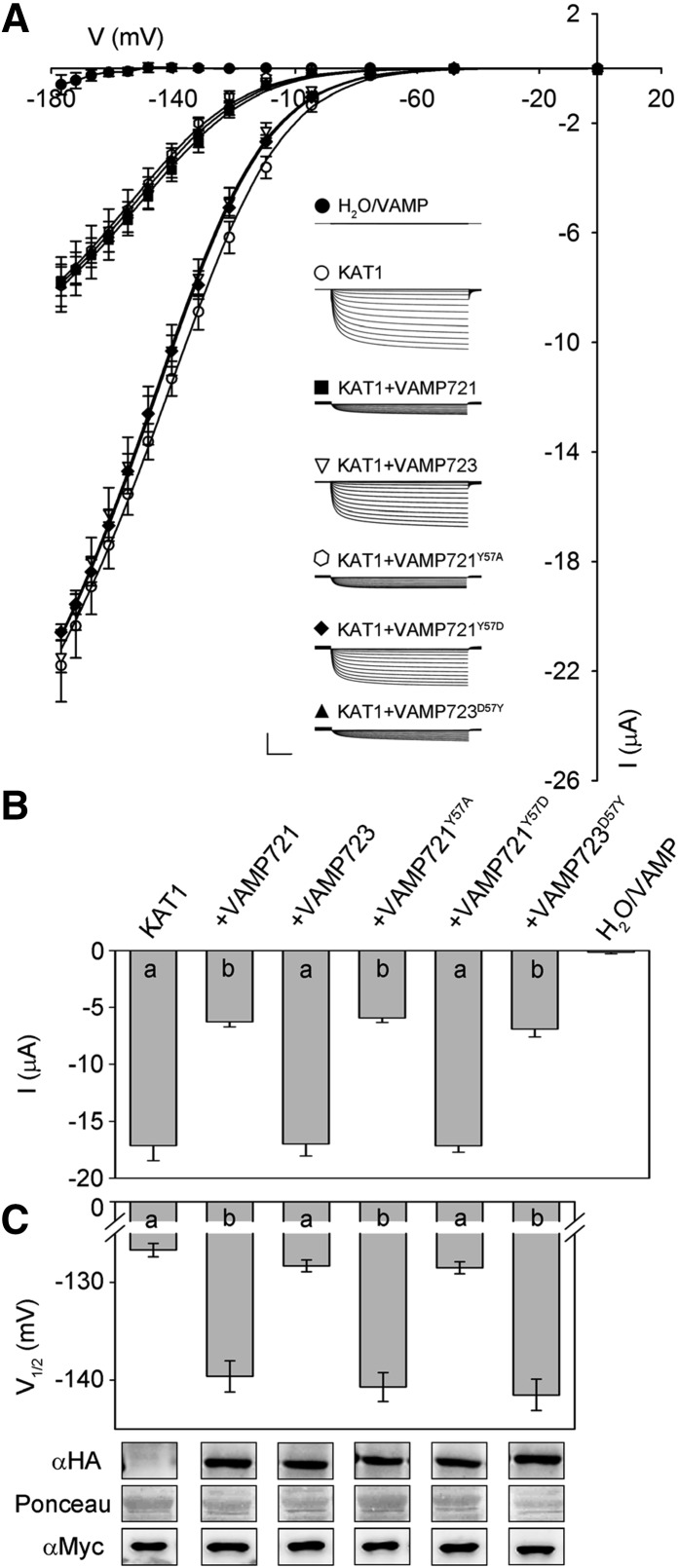
To explore the consequences of VAMP721 interaction on K+ channel activity, we recorded K+ currents under voltage clamp after heterologously expressing VAMP721 and VAMP723 with the KAT1 in Xenopus laevis oocytes. We also incorporated tags for immunoquantification. For VAMP721, measurements were performed on oocytes injected with KAT1 and VAMP cRNA in ratios of 1:1, 1:2, 1:4, and 1:8 (KAT1:VAMP721). Figure 3A presents the mean, steady state current-voltage relations for each set of injections along with representative current traces recorded under voltage clamp; Figures 3B and 3C and Table 1 summarize the current characteristics derived from these recordings and representative immunoblots for KAT1 and VAMP expression. Under voltage clamp, oocytes expressing VAMP721 and VAMP723 alone, and after water injection, showed only very small background currents. Oocytes injected with KAT1 cRNA showed a substantial current that activated with halftimes of around 300 ms at voltages near and negative of −100 mV, typical of KAT1 (Hoshi, 1995; Lefoulon et al., 2014). Coexpression with a 4-fold excess of VAMP723 had no apparent effect on the KAT1 current. However, coexpressing VAMP721 yielded a dose-dependent suppression of the KAT1 current that saturated at a 1:4 ratio of KAT1:VAMP721 (Figures 3B and 3C).
Figure 3.
Coexpressing VAMP721 Suppresses KAT1 K+ Current and Alters Channel Gating in X. laevis Oocytes.
(A) Representative current traces (insets) recorded under voltage clamp in 30 mM K+ and the mean steady state current-voltage curves derived from these recordings. Data are means ± se of seven or more experiments for each set of constructs with oocytes injected with water, expressing VAMP721 and VAMP723 alone (closed circles), KAT1 alone (open circles), KAT1 and VAMP721 injected with cRNA in ratios of 1:1 (filled triangles), 1:2 (open diamonds), 1:4 (filled squares), and 1:8 (open inverted triangles), and with KAT1 and VAMP723 injected with cRNA in a ratio of 1:4 (filled diamonds). Clamp cycles: holding voltage, −50 mV; voltage steps, 0 to −180 mV; tail voltage −50 mV. Scale: 10 μA (vertical), 1 s (horizontal). Solid curves are the results of joint, nonlinear least-squares fitting of the K+ currents (IK) to the Boltzmann function (Equation 1). Best and visually satisfactory fittings were obtained allowing V1/2 and gmax to vary between curves while holding the voltage sensitivity coefficient (apparent gating charge) δ in common between curves. Fitted parameter values are summarized in Table 1.
(B) Mean ± se for KAT1 K+ current amplitude at −160 mV, derived from the data in (A). Significance is indicated by letters at P < 0.01.
(C) Mean ± se for V1/2 derived from fittings to Equation 1 of the data in (A). Significance is indicated by letters at P < 0.01. Also shown (below) are immunoblots verifying VAMP (αHA antibody) and KAT1 (αmyc antibody) expression in oocytes collected after electrical recordings. Ponceau S stain was used to normalize VAMP expression levels for lanes with VAMP721 and yielded ratios of 1:2.06:3.77:6.92 after quantifying band densities using ImageJ.
Table 1. Coexpressing VAMP721 with the KAT1 K+ Channel in Xenopus Oocytes Suppresses the Current Amplitude and Alters Channel Gating.
| V1/2 (mV) | gmax (ms) | δ | |
|---|---|---|---|
| KAT1 | −126.1 ± 0.7 | 1.47 ± 0.01 | −1.70 ± 0.05 |
| KAT1+VAMP723 (1:4) | −128.5 ± 0.6 | ||
| KAT1+VAMP721 (1:1) | −126.7 ± 0.7 | ||
| KAT1+VAMP721 (1:2) | −136.8 ± 1.0* | 1.12 ± 0.02 | |
| KAT1+VAMP721 (1:4) | −139.7 ± 1.7* | 0.58 ± 0.01 | |
| KAT1+VAMP721 (1:8) | −141.5 ± 1.6* |
Parameter values are results of joint, nonlinear least-squares fitting of K+ currents in Figure 3. Fittings were carried out with the gating charge, δ, held in common, and values for the voltage giving half-maximal conductance V1/2 and conductance maximum gmax were allowed to vary between data sets. Data for KAT1 and KAT1+VAMP721 (1:1) were visually indistinguishable; therefore, gmax values were fitted jointly to simplify analysis. Similarly, gmax values for KAT1+VAMP721 (1:4) and KAT1+VAMP721 (1:8) were fitted jointly. Data are from seven or more separate experiments for each construct combination and are given as means ± se. Significance for V1/2 as the difference from KAT1 expressed alone at *P < 0.01.
To quantify the effects of VAMP721 expression on KAT1 gating, the mean, steady state current-voltage curves were fitted to a Boltzmann function of the form
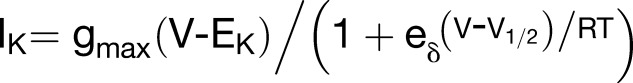 |
(1) |
where gmax is the conductance maximum, EK is the equilibrium voltage for K+, V1/2 is the voltage yielding half-maximal conductance, δ is the apparent gating charge or voltage sensitivity coefficient (Dreyer and Blatt, 2009), V is the membrane voltage, and R and T have their usual meanings. To avoid substantial indetermination, we applied standard methods for joint analysis with one or more selected parameters held in common between data sets (Press et al., 1992; Honsbein et al., 2009; Lefoulon et al., 2014). Statistically and visually best fittings (Figure 3A, solid lines) were obtained with δ held in common while allowing gmax and V1/2 to vary between data sets. These results yielded δ of −1.70 ± 0.05 and, with KAT1 expression alone, a V1/2 of −126 ± 1 mV and gmax of 1.47 ± 0.01 ms. With increasing VAMP721 dosage, gmax declined and V1/2 was displaced to more negative voltages (Figure 3C, Table 1). The analysis also showed that increasing the VAMP721 dosage beyond the 1:4 ratio had little influence on either parameter. Coexpression of KAT1 with VAMP721 at ratios of 1:4 and 1:8 yielded statistically equivalent values for V1/2 and current amplitudes. These results indicated that VAMP721 affects both the KAT1 current amplitude and its intrinsic gating properties in a stoichiometric fashion.
The Longin Domain of VAMP721 Is Essential for K+ Channel Interaction
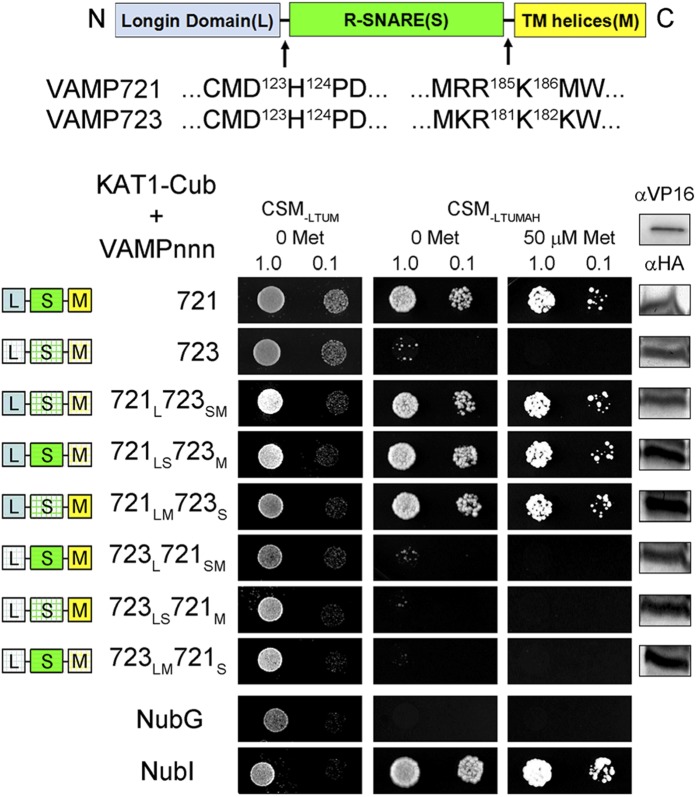
A comparison of the protein sequences for VAMP721, VAMP722, and VAMP723 shows a substantial degree of identity with few regions of divergence between the R-SNAREs (Supplemental Figure 1), and it offers few clues to any putative K+ channel binding site. Therefore, to isolate residues associated with channel binding, we undertook a series of domain-swapping experiments. Initially, VAMP721 and VAMP723 were divided into three regions (Figure 4) comprising the longin domain (L), the SNARE domain (S), and the transmembrane domain (M), with two breaks in the VAMP sequences at the D123H124 and the R185K186 (for VAMP723, R181K182 junctions; Supplemental Figure 1) that define the longin domain and membrane anchor boundaries. Constructs were prepared for all six nonredundant combinations of these domains from the two VAMPs (Figure 4, domain color-coding). Again, we used the SUS assay to test the interaction of each chimera with KAT1. Figure 4 summarizes one of three independent experiments, including both the positive and negative controls of the full-length VAMP721 and VAMP723 constructs, respectively. We found that diploid yeast growth was recovered provided that the VAMP chimera incorporated the longin domain of VAMP721. Interaction was retained, in each case, in the presence of Met, which suppresses the expression of the bait construct. However, in every case, chimeras incorporating the longin domain of VAMP723 failed to yield yeast growth.
Figure 4.
Interaction with the KAT1 K+ Channel Depends on the VAMP721 Longin Domain.
Yeast mating-based split-ubiquitin assay for interaction of NubG fusions of VAMP721 and VAMP723 chimeras with KAT1-Cub as the bait. Chimeras of VAMP721 and VAMP723 were constructed by exchange of three domains as indicated (above) corresponding to the longin domain (L), R-SNARE motif (S), and transmembrane (M) domain. Domain exchanges were made at the junction points indicated by arrows. Yeast diploids were created with NubG fusion constructs of each VAMP chimera and with controls (NubG, negative; NubI [wild type], positive) spotted (left to right) on CSM medium without Leu, Trp, Ura, and Met (CSM-LTUM) to verify mating, CSM medium without Leu, Trp, Ura, Met, Ade, and His (CSM-LTUMAH) to verify adenine- and His-independent growth, and on CSM-LTUMAH with the addition of 50 μM Met to verify interaction at lower K+ channel-Cub expression levels. Diploid yeast was dropped at 1.0 and 0.1 OD600 in each case. Immunoblot analysis (5 μg total protein/lane) of the haploid yeast used for mating (right) using commercial HA antibody for the VAMP fusions and VP16 antibody for the K+ channel fusion.
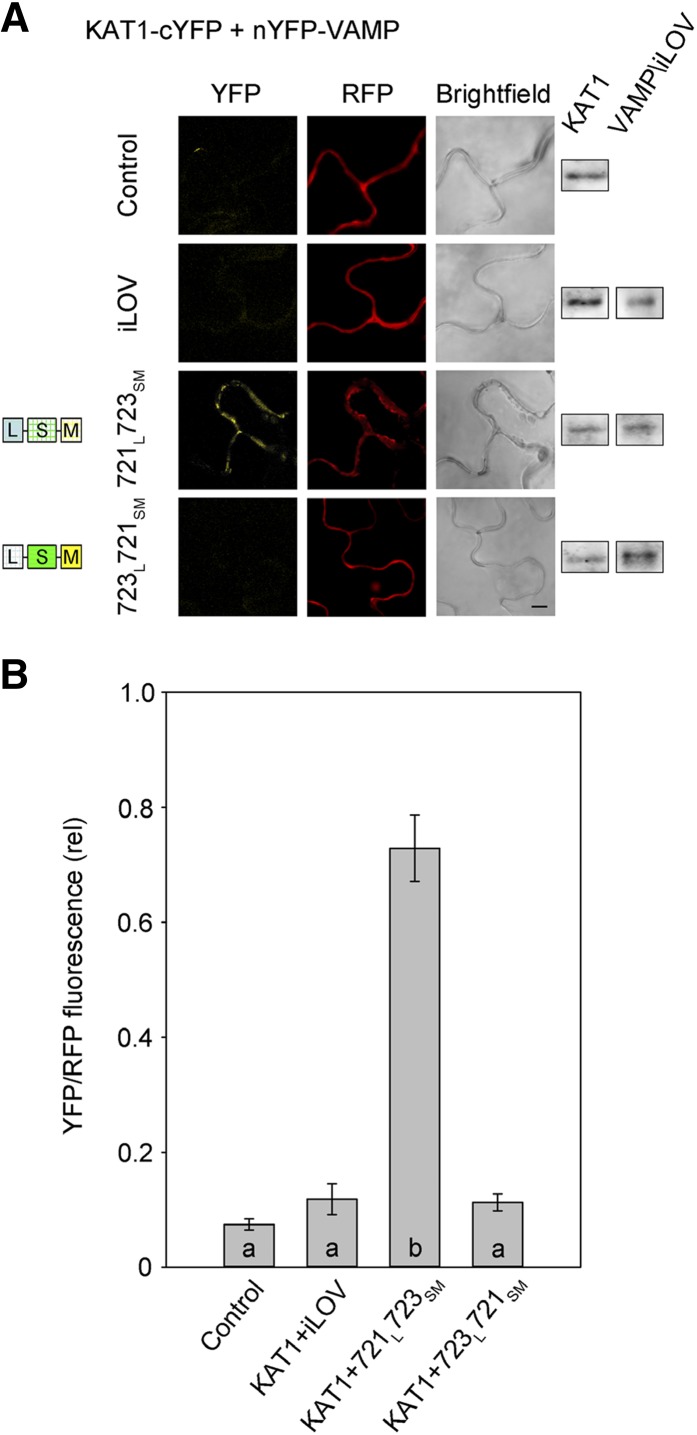
To validate the KAT1 interaction in vivo, we introduced chimeras incorporating the VAMP721 and VAMP723 longin domains, VAMP721L723SM and VAMP723L721SM, into the rBiFC 2in1 vector system (Figure 2) and used the constructs to transiently transform tobacco leaves. Figure 5 shows representative images and statistical analysis of rBiFC fluorescence ratios from three independent experiments together with measurements from tissues expressing the vector with KAT1 alone and with iLOV as the negative controls. The results demonstrate a highly significant rBiFC signal above the negative controls when KAT1 was coexpressed with VAMP721L723SM that includes the VAMP721 longin domain, but not with the complementary VAMP723L721SM chimera that includes the VAMP723 longin domain. Thus, we concluded that the longin domain of VAMP721 is a primary determinant of the physical interaction between VAMP721 and KAT1.
Figure 5.
Interaction of KAT1 K+ Channel Requires the VAMP721 Longin Domain.
(A) rBiFC analysis of YFP and RFP fluorescence collected from tobacco transformed using the pBiFCt-2in1-NC 2in1 vector (Figure 2). Images are (left to right) YFP (BiFC) fluorescence, RFP fluorescence, and bright field. Constructs (top to bottom) included coding sequences for KAT1-cYFP with either the empty cassette (control) or nYFP-X fusions of iLOV, VAMP721 longin domain with VAMP723 R-SNARE and TM helices (VAMP721L723SM), and VAMP723 longin domain with VAMP721 R-SNARE and TM helices (VAMP723L721SM). KAT1 and VAMP constructs were detected by αHA and αmyc antibodies to verify fusion protein expression (right). Bar = 10 μm.
(B) rBiFC fluorescence signals from three independent experiments. Each bar represents the mean ± se of fluorescence intensity ratios from 10 images taken at random over the leaf surface. rBiFC signals were calculated as the mean fluorescence intensity ratio determined from each image set after subtracting the background fluorescence determined from an equivalent number of images taken from nontransformed tissues. Significance is indicated by letters at P < 0.01.
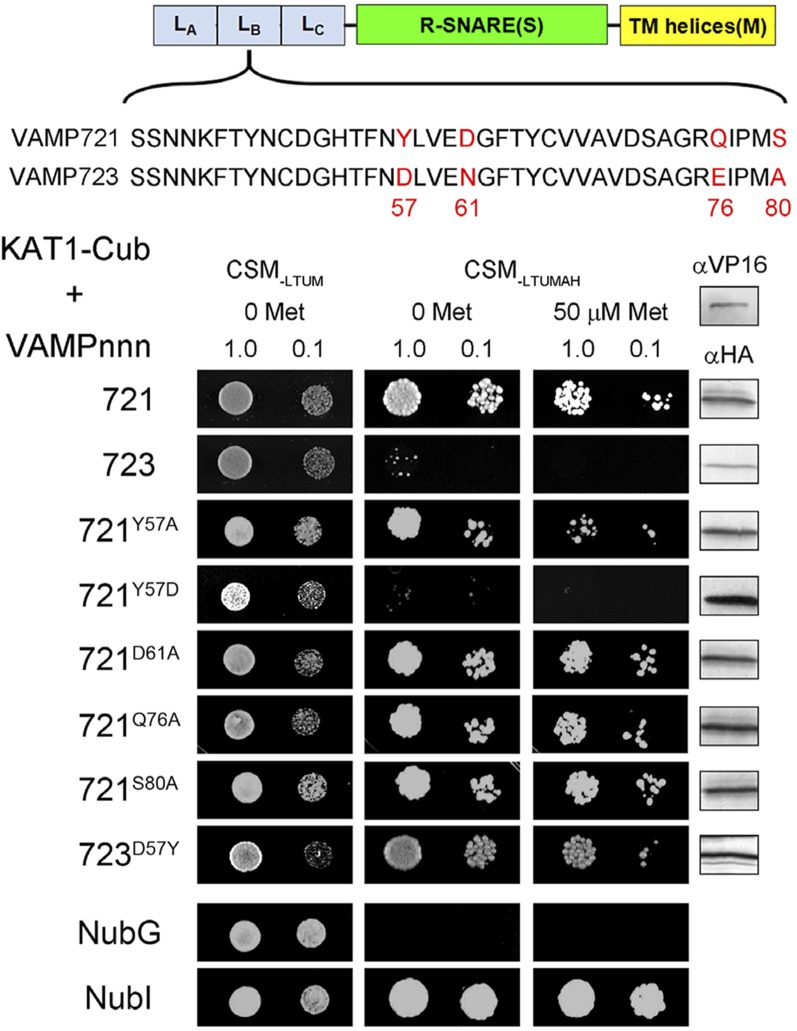
Tyr-57 of VAMP721 Is Essential for K+ Channel Interaction
We used a similar strategy of chimeric constructs to isolate the binding domain within the longin domain of VAMP721, dividing the longin domains of the two R-SNAREs between three roughly equal segments, LA (residues 1 to 40), LB (residues 41 to 80), and LC (residues 81 to 124). These segments were swapped using the VAMP721 sequence as the backbone for the remainder of the VAMP assembly. Again, six nonredundant chimeras were generated from the longin LALBLC segments of VAMP721 and VAMP723; each was tested in SUS assays for interaction with KAT1 and was validated with supporting immunoblot analysis. This second set of chimeras (Supplemental Figure 3) indicated that only residues within the central LB segment of the VAMP721 longin domain were essential for K+ channel interaction.
Within this segment there are four amino acids that differ between VAMP721 and VAMP723 (Figure 6; Supplemental Figure 1): Tyr-57, Asp-61, Gln-76, and Ser-80. Each of these residues in VAMP721 was substituted in turn with Ala and tested by SUS assay for interaction with KAT1. Figure 6 shows that yeast growth was recovered with the preys VAMP721D61A, VAMP721Q76A, and VAMP721S80A. However, growth of VAMP721Y57A was reduced in the presence of 50 μM Met, suggesting that this residue was important for interaction with the channel. The corresponding residue of VAMP723 is Asp. We therefore replaced Tyr-57 in VAMP721 with Asp and also undertook the reverse mutation in VAMP723 to generate VAMP721Y57D and VAMP723D57Y. The VAMP721Y57D substitution was sufficient to eliminate yeast growth, even in the absence of Met. Remarkably, the complementary mutation VAMP723D57Y yielded strong growth of the diploid yeast, indicating the interaction between VAMP723D57Y and KAT1. We also performed complementary residue exchanges, including VAMP721Y57F, VAMP721D61N, and VAMP721Q76E substitutions for analysis of interaction with KAT1 in SUS assays. The results (Supplemental Figure 4) indicated that these sites were not of primary importance for channel interaction.
Figure 6.
Interaction with the KAT1 K+ Channel Depends on Tyr-57 of the VAMP721 Longin Domain.
Yeast mating-based split-ubiquitin assay for interaction of NubG fusions of VAMP721, VAMP723, and selected single-site mutants as indicated with KAT1-Cub as the bait. Alignment of the longin domain within the central sequence of 20 residues is shown above with the divergent residues shown in red. Yeast diploids were created with NubG fusion constructs of each VAMP chimera and with controls (NubG, negative; NubI [wild type], positive) spotted (left to right) on CSM medium without Leu, Trp, Ura, and Met (CSM-LTUM) to verify mating, CSM medium without Leu, Trp, Ura, Met, Ade, and His (CSM-LTUMAH) to verify adenine- and His-independent growth, and on CSM-LTUMAH with the addition of 50 μM Met to verify interaction at lower K+ channel-Cub expression levels. Diploid yeast was dropped at 1.0 and 0.1 OD600 in each case. Immunoblot analysis (5 μg total protein/lane) of the haploid yeast used for mating (right) using commercial HA antibody for the VAMP fusions and VP16 antibody for the K+ channel fusion. Note the loss of growth with diploid yeast carrying the channel and VAMP721Y57D mutation.
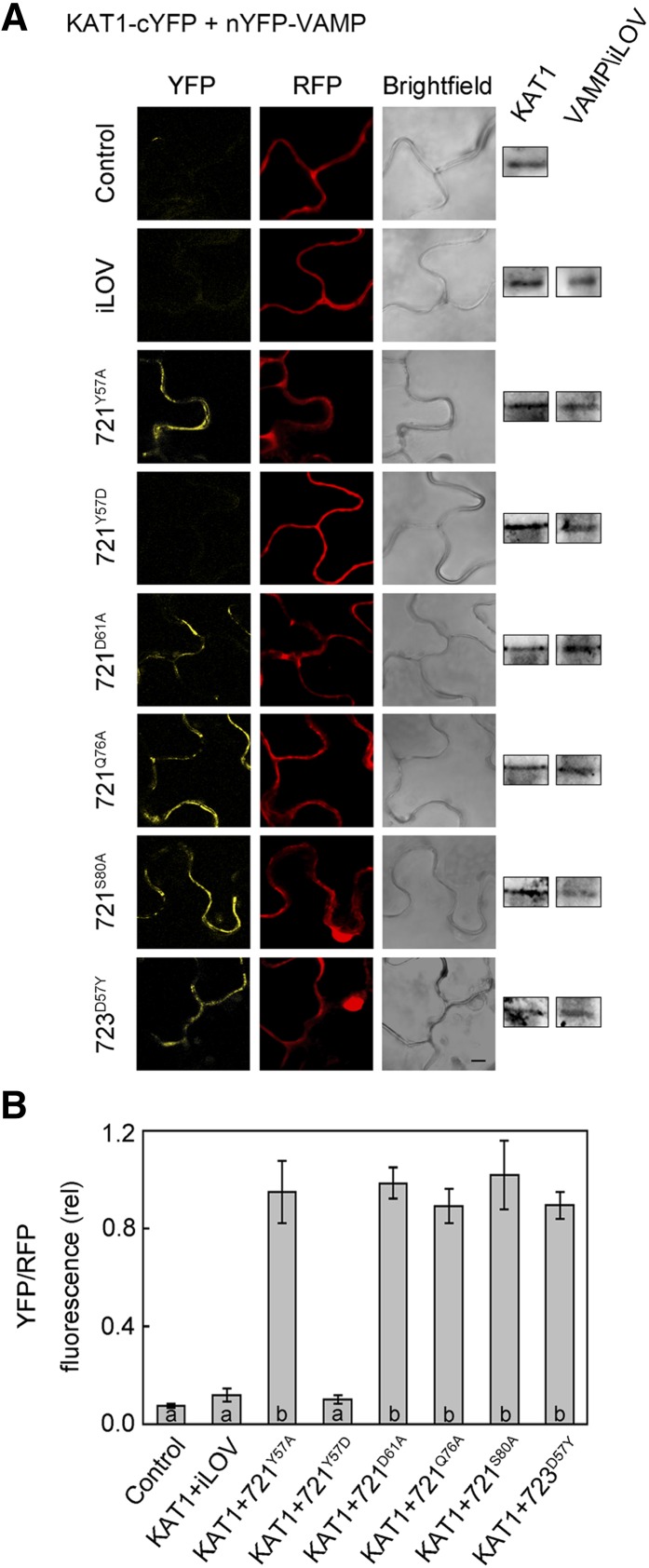
To validate these findings in vivo, we again incorporated the corresponding single-site substitutions in the 2in1 vector carrying KAT1-cYFP for transient transformation of tobacco and analysis by rBiFC. Figure 7A shows confocal fluorescence images collected for each of the VAMP mutants as nYFP fusions coexpressed with KAT1-cYFP from one of three experiments along with supporting immunoblot data for expression of the channel and VAMP constructs. A summary from all three independent experiments is shown in Figure 7B. In every case, we recovered strong rBiFC signals with VAMP721D61A, VAMP721Q76A, and VAMP721S80A, indicating that these single mutants had no effect on the interaction with KAT1. Although variable, a comparable rBiFC signal was also recovered with the VAMP721Y57A mutant. However, when the fusions of KAT1 and VAMP721Y57D were coexpressed, the rBiFC signal was statistically indistinguishable from the background of the negative controls expressing the KAT1 fusion on its own and with the complementary iLOV fusion. Coexpression of the rBiFC fusions of KAT1 with VAMP723D57Y also yielded a strong signal.
Figure 7.
Interaction of KAT1 in Vivo Depends on VAMP721 Residue Tyr-57.
(A) rBiFC analysis of YFP and RFP fluorescence collected from tobacco transformed using the pBiFCt-2in1-NC 2in1 vector (Figure 2). Images are (left to right) YFP (BiFC) fluorescence, RFP fluorescence, and bright field. Constructs (top to bottom) included coding sequences for KAT1-cYFP alone and with nYFP-X fusions of iLOV, VAMP721Y57A, VAMP721Y57D, VAMP721D61A, VAMP721Q76A, VAMP721S80A, and VAMP723D57Y. KAT1 and VAMP constructs were detected by αHA and αmyc antibodies to verify fusion protein expression (right). Bar = 10 μm.
(B) rBiFC fluorescence signals from three independent experiments. Each bar represents the mean ± se of fluorescence intensity ratios from 10 images taken at random over the leaf surface. rBiFC signals were calculated as the mean fluorescence intensity ratio determined from each image set after subtracting the background fluorescence determined from an equivalent number of images taken from nontransformed tissues. Significance is indicated by letters at P < 0.01.
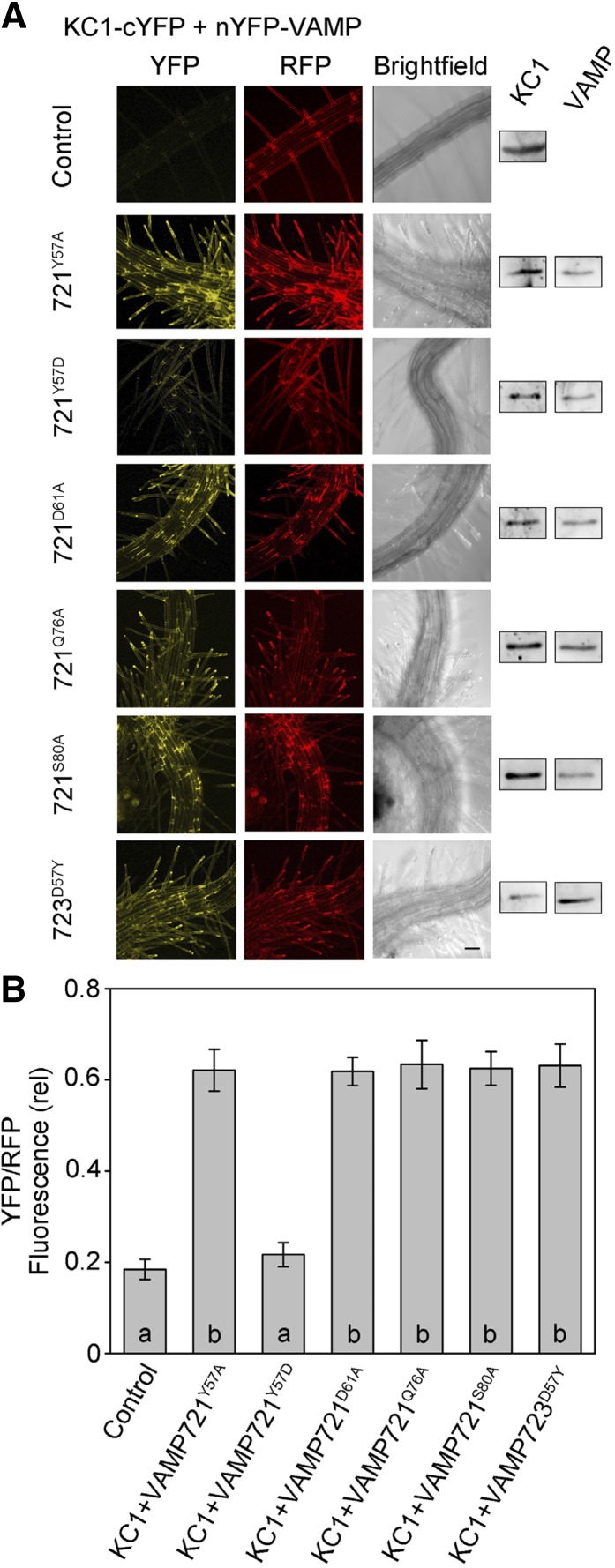
We repeated these experiments using the KC1 as the bait in SUS assays. In each of three independent experiments (Supplemental Figure 5), we found that VAMP721Y57D strongly suppressed diploid yeast growth when expression of the bait was reduced. Growth was not affected by any of the other mutants; furthermore, the complementary VAMP723D57Y mutant was sufficient to rescue growth, much as for KAT1. Similarly, we validated this specificity for interaction with KC1 in vivo by rBiFC fluorescence analysis. As before, rBiFC fusion constructs were generated in the 2in1 vector system to express nYFP-VAMP together with KC1-cYFP and RFP as a transformation marker. We transiently transformed Arabidopsis seedlings by cocultivation and analyzed rBiFC fluorescence in the root epidermis (Honsbein et al., 2009; Grefen et al., 2010a, 2010b; Blatt and Grefen, 2014). Figure 8 includes fluorescence image projections from one of four independent experiments along with a statistical summary of all four data sets determined from projections of YFP and RFP fluorescence intensities after background subtraction. As with KAT1, we found a substantial rBiFC signal for the wild-type VAMP721 but not VAMP721Y57D or VAMP723, while the complementary VAMP723D57Y mutant gave a strong rBiFC signal (Supplemental Figure 2).
Figure 8.
Interaction of KC1 in Vivo Depends on VAMP721 Residue Tyr-57.
(A) rBiFC analysis of YFP and RFP fluorescence collected from tobacco transformed using the pBiFCt-2in1-NC 2in1 vector (Figure 2). Images are (left to right) YFP (BiFC) fluorescence, RFP fluorescence, and bright field. Constructs (top to bottom) included coding sequences for KC1-cYFP alone and with nYFP-X fusions of iLOV, VAMP721Y57A, VAMP721Y57D, VAMP721D61A, VAMP721Q76A, VAMP721S80A, and VAMP723D57Y. KC1 and VAMP constructs were detected by αHA and αmyc antibodies to verify fusion protein expression (right). Bar = 10 μm.
(B) rBiFC fluorescence signals from three independent experiments. Each bar represents the mean ± se of fluorescence intensity ratios from 10 images taken at random over the leaf surface. rBiFC signals were calculated as the mean fluorescence intensity ratio determined from each image set after subtracting the background fluorescence determined from an equivalent number of images taken from nontransformed tissues. Significance is indicated by letters at P < 0.01.
One intriguing question arising from these data relates to the implicit relocalization of the mutant VAMPs, at least of VAMP723D57Y, and their distributions at the plasma membrane. To explore this issue, we expressed all four constructs as N-terminal fusions with green fluorescent protein (GFP) and analyzed their distributions in Arabidopsis following transient transformations. The results (Supplemental Figure 6) confirmed the localizations of VAMP721 and VAMP723 associated with the plasma membrane and endoplasmic reticulum, respectively. They also demonstrated that VAMP721Y57D relocated to an endomembrane network, while the VAMP723D57Y mutant relocated to the plasma membrane. In the latter case, we cannot rule out the possibility that channel binding facilitated a redistribution of VAMP723D57Y, but the results do argue against any effect of rBiFC fluorophore annealing in driving this relocation. Thus, we conclude that Tyr-57 is an essential and key determinant for R-SNARE binding with both of the K+ channels and is sufficient to engineer channel binding in the otherwise noninteracting VAMP723. It is also an important determinant for VAMP localization at the plasma membrane.
K+ Channel Activity Depends on Tyr-57 of VAMP721
To assess the functional consequences of the VAMP721 and VAMP723 mutations, we prepared constructs for heterologous expression and coinjected each of the cRNAs in turn with KAT1 cRNA in X. laevis oocytes. Again, we quantified the K+ currents under voltage clamp and analyzed the results for current relaxation, steady state current, as well as fitting the mean steady state current-voltage relations to Equation 1 to extract the gating parameters of V1/2 and δ. For comparative purposes, we injected cRNAs for each of the VAMP constructs in a 4-fold excess with KAT1 cRNA, and as before we confirmed expression by immunoblot analysis. Figure 9A includes representative current traces for each of the VAMP -K+ channel combinations and summarizes the mean steady state currents recorded from seven or more independent experiments in each case. Figure 9B presents the mean steady state currents recorded at −160 mV. Fittings to Equation 1 gave a common value for δ of −1.65 ± 0.04, and values for V1/2 derived from the analysis are shown in Figure 9C; best fittings are summarized in Table 2 and are shown as the solid lines in Figure 9A. When expressed with KAT1, VAMP721Y57A and the wild-type VAMP721 yielded K+ currents with similar characteristics, notably both a reduction in current amplitude at −160 mV, maximum conductance, and a negative displacement in V1/2. However, coexpression of KAT1 with the VAMP721Y57D gave K+ current characteristics similar to those for KAT1 alone and with its noninteracting partner VAMP723. KAT1 coexpression with VAMP723D57Y yielded K+ current characteristics that were statistically indistinguishable from those obtained on coexpression with wild-type VAMP721.
Figure 9.
VAMP721 Suppression of KAT1 K+ Current in X. laevis Oocytes Depends on Tyr-57.
(A) Representative current traces (insets) recorded under voltage clamp in 30 mM K+ and the mean steady state current-voltage curves derived from these recordings. Data are means ± se of seven or more experiments for each set of constructs with oocytes. VAMPs were injected in 4-fold excess with KAT1. Traces are cross-referenced to the current-voltage curves and are from oocytes expressing water, VAMP721, and VAMP723 alone (closed circles), KAT1 alone (open circles), KAT1 and VAMP721 (filled squares), KAT1 and VAMP723 (open inverted triangles), KAT1 and VAMP721Y57A (open hexagon), KAT1 and VAMP721Y57D (filled diamond), and KAT1 and VAMP723D57Y (filled triangle). Clamp cycles: holding voltage, −50 mV; voltage steps, 0 to −180 mV; tail voltage −50 mV. Scale: 10 μA (vertical), 1 s (horizontal). Solid curves are the results of joint, nonlinear least-squares fitting of the K+ currents (IK) to the Boltzmann function (Equation 1). Best and visually satisfactory fittings were obtained allowing V1/2 and gmax to vary between curves while holding the voltage sensitivity coefficient (apparent gating charge) δ in common between curves. Fitted parameter values are summarized in Table 2.
(B) Mean ± se for KAT1 K+ current amplitude at −160 mV, derived from the data in (A). Significance is indicated by letters at P < 0.01.
(C) Mean ± se for V1/2 derived from fittings to Equation 1 of the data in (A). Significance is indicated by letters at P < 0.01. Also shown (below) are immunoblots verifying VAMP (αHA antibody) and KAT1 (αmyc antibody) expression in oocytes collected after electrical recordings along with Ponceau S stain included as a loading control.
Table 2. Suppressing KAT1 K+ Current in Xenopus Oocytes by VAMP721 Depends on the Longin Domain Residue Tyr-57.
| V1/2 (mV) | gmax (ms) | δ | |
|---|---|---|---|
| KAT1 | −126.7 ± 0.7 | 1.47 ± 0.01 | −1.65 ± 0.04 |
| KAT1+VAMP721Y57D | −128.5 ± 0.6 | ||
| KAT1+VAMP723 | −128.3 ± 0.6 | ||
| KAT1+VAMP721 | −139.6 ± 1.5* | 0.57 ± 0.01 | |
| KAT1+VAMP721Y57A | −140.7 ± 1.4* | ||
| KAT1+VAMP723D57Y | −141.5 ± 1.6* |
Parameter values are results of joint, nonlinear least-squares fitting of K+ currents in Figure 9. Fittings were carried out with the gating charge, δ, held in common across all data sets, and values for gmax were held to common values between groups of three data sets as indicated. Values for the voltage giving half-maximal conductance V1/2 and conductance maximum gmax were allowed to vary between all of the data sets. Data are from seven or more separate experiments for each construct combination and are given as means ± se. Significance for V1/2 as the difference from KAT1 expressed alone *P < 0.01.
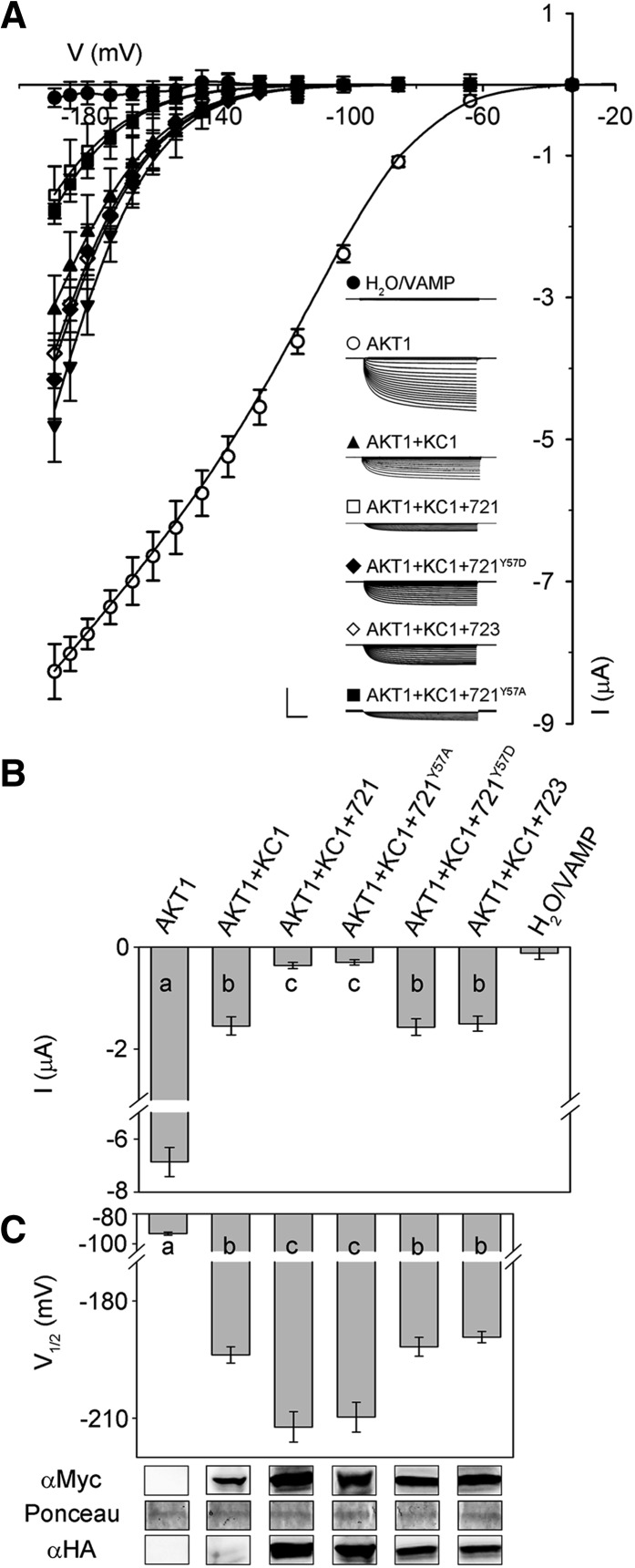
We repeated these experiments with KC1. KC1 is a “silent” K+ channel subunit when expressed on its own; it yields substantial inward-rectifying K+ currents only when coexpressed with other inward-rectifying K+ channels in X. laevis oocytes, and in vivo it is thought to function in a complex with the AKT1 K+ channel subunit to facilitate K+ uptake by the root epidermis (Duby et al., 2008; Geiger et al., 2009; Honsbein et al., 2009). AKT1 itself did not interact with VAMP721 or VAMP723 in SUS assays (Supplemental Figure 4). Therefore, we expressed KC1 in 1:1 ratio together with AKT1 as before (Honsbein et al., 2009; Grefen et al., 2010a), and to ensure activation of the channels in oocytes, we also coexpressed the protein kinase CIPK23 and calcineurin-like protein CBL1 (Xu et al., 2006). Additionally, we included cRNAs for each of the VAMP721 constructs in a 1:4 (KC1:VAMP) ratio. Figure 10A includes current traces from representative oocytes injected with each of the combinations of cRNAs along with the mean steady state current-voltage curves from all of the experiments, and Figure 10B summarizes the corresponding steady state currents at −160 mV. Again, the steady state current-voltage curves were fitted jointly to Equation 1, in this case holding both gmax and the voltage sensitivity coefficient, δ, in common between data sets (Honsbein et al., 2009; Grefen et al., 2010a). The solid lines in Figure 10A show the results of these fittings and values for V1/2 derived from the analysis are summarized in Table 3 and Figure 10C along with immunoblots validating the expression of KC1 and of the VAMP721 constructs.
Figure 10.
VAMP721 Suppression of KC1-AKT1 K+ Current in X. laevis Oocytes Depends on Tyr-57.
(A) Representative current traces (insets) recorded under voltage clamp in 30 mM K+ and the mean steady state current-voltage curves derived from these recordings. Data are means ± se of seven or more experiments for each set of constructs with oocytes. Oocytes were injected with water or VAMP721 or VAMP723 alone (closed circles), AKT1 alone (open circles), AKT1 with KC1 (filled triangles), AKT1 and KC1 with VAMP721 (open squares), AKT1 and KC1 with VAMP721Y57A (closed squares), AKT1 and KC1 with VAMP723 (open diamonds), and AKT1 and KC1 with VAMP721Y57D (closed diamonds). All oocytes were coinjected with CBL1 and CIPK23, essential for AKT1 function, in a 1:1:1 ratio with AKT1 (Xu et al., 2006; Honsbein et al., 2009). cRNAs were injected with AKT1:KC1:VAMP in a ratio of 1:1:4. Clamp cycles: holding voltage, −50 mV; voltage steps, 0 to −200 mV; tail voltage −50 mV. Scale: 5 μA (vertical), 1 s (horizontal). Solid curves are the results of joint, nonlinear least-squares fitting of the K+ currents (IK) to the Boltzmann function (Equation 1). Best and visually satisfactory fittings were obtained allowing V1/2 to vary between curves while holding the voltage sensitivity coefficient (apparent gating charge) δ and gmax in common between curves. Fitted parameter values are summarized in Table 3.
(B) Mean ± se of KC1-AKT1 K+ current amplitude at −160 mV, derived from (A). Significance is indicated by letters at P < 0.01.
(C) Mean ± se for V1/2 derived from fittings to Equation 1 of the data in (A). Significance is indicated by letters at P < 0.01. Also shown (below) are immunoblots verifying VAMP (αHA antibody) and KC1 (αmyc antibody) expression in oocytes collected after electrical recordings along with Ponceau S stain included as a loading control.
Table 3. Suppressing KC1-AKT1 K+ Current in X. laevis Oocytes by VAMP721 Depends on the Longin Domain Residue Tyr-57.
| V1/2 (mV) | gmax (ms) | δ | |
|---|---|---|---|
| AKT1 | −93.1 ± 1.0 | 0.52 ± 0.01 | −1.45 ± 0.03 |
| AKT1+KC1 | −186.8 ± 2.1 | ||
| AKT1+KC1+VAMP721 | −212.2 ± 3.2* | ||
| AKT1+KC1+VAMP723 | −189.2 ± 1.4 | ||
| AKT1+KC1+VAMP721Y57A | −209.7 ± 3.1* | ||
| AKT1+KC1+VAMP721Y57D | −187.1 ± 2.4 |
Parameter values are results of joint, nonlinear least-squares fitting of K+ currents in Figure 10. Fittings were carried out with the gating charge, δ, and gmax held in common, and values for the voltage giving half-maximal conductance V1/2 were allowed to vary between data sets. Data are from seven or more separate experiments for each construct combination and are given as means ± se. Significance for V1/2 as the difference from KC1-AKT1 expressed alone at *P < 0.01.
As observed previously (Duby et al., 2008; Honsbein et al., 2009), AKT1 expression on its own yielded an inward-rectifying K+ current with V1/2 near −100 mV; expressing AKT1 together with KC1 gave a substantive K+ current only at voltages negative of −140 mV. We found that coexpression with VAMP721 and VAMP721Y57A reduced the current at any one voltage, apparently displacing the steady state current-voltage curve to the right, but coexpression with VAMP723 and VAMP721Y57D was without effect. To avoid indetermination in our analysis, we extended fittings with Equation 1 to these currents asking the question whether satisfactory fittings could be obtained on holding gmax and δ in common so that only V1/2 varied between data sets (Honsbein et al., 2009; Grefen et al., 2010a). The analysis yielded statistically and visually satisfactory fittings, as shown in Figure 10A with V1/2 varying from −186 ± 1 mV in the control to −212 ± 3 mV on coexpressing VAMP721. Coexpression with VAMP723 did not affect the V1/2 for the K+ current, nor did coexpressing VAMP721Y57D. These results underline the pattern in regulation of the KC1-AKT1 K+ current by the VAMP constructs as we observed for the KAT1 current. Notably, they incorporated the same constraints in fitting that were previously applied in studies of SYP121-mediated control of KC1-AKT1 gating (Honsbein et al., 2009; Grefen et al., 2010a), giving us confidence to compare the effects of VAMP721 expression with that of SYP121 (see Discussion).
VAMP721 and K+ Channel Interaction Inhibits the K+ Current in the Plant
Current carried by K+ channels formed of KC1 and AKT1 subunits are normally observed in Arabidopsis root epidermal cells (Ivashikina et al., 2001; Honsbein et al., 2009). However, analysis of the current in vivo does not lend itself to work with one or more VAMP null mutants, if only because VAMP721 and VAMP722 are functionally redundant and the vamp721/vamp722 double mutant is embryo lethal (Kwon et al., 2008). Therefore, to test the effects of VAMP721, we overexpressed VAMP721, VAMP723, and VAMP721Y57D in the root epidermis using the bicistronic vector pUB-Bic-Dest (Chen et al., 2011) after tagging each construct N-terminally with HA. pUB-Bic-Dest incorporates a second, independent cassette to express soluble GFP and thereby allows selection during experiments of cells carrying the vector and expressing the constructs after transient transformations by cocultivation with Agrobacterium tumefaciens. VAMP expression was further verified by immunoblot analysis.
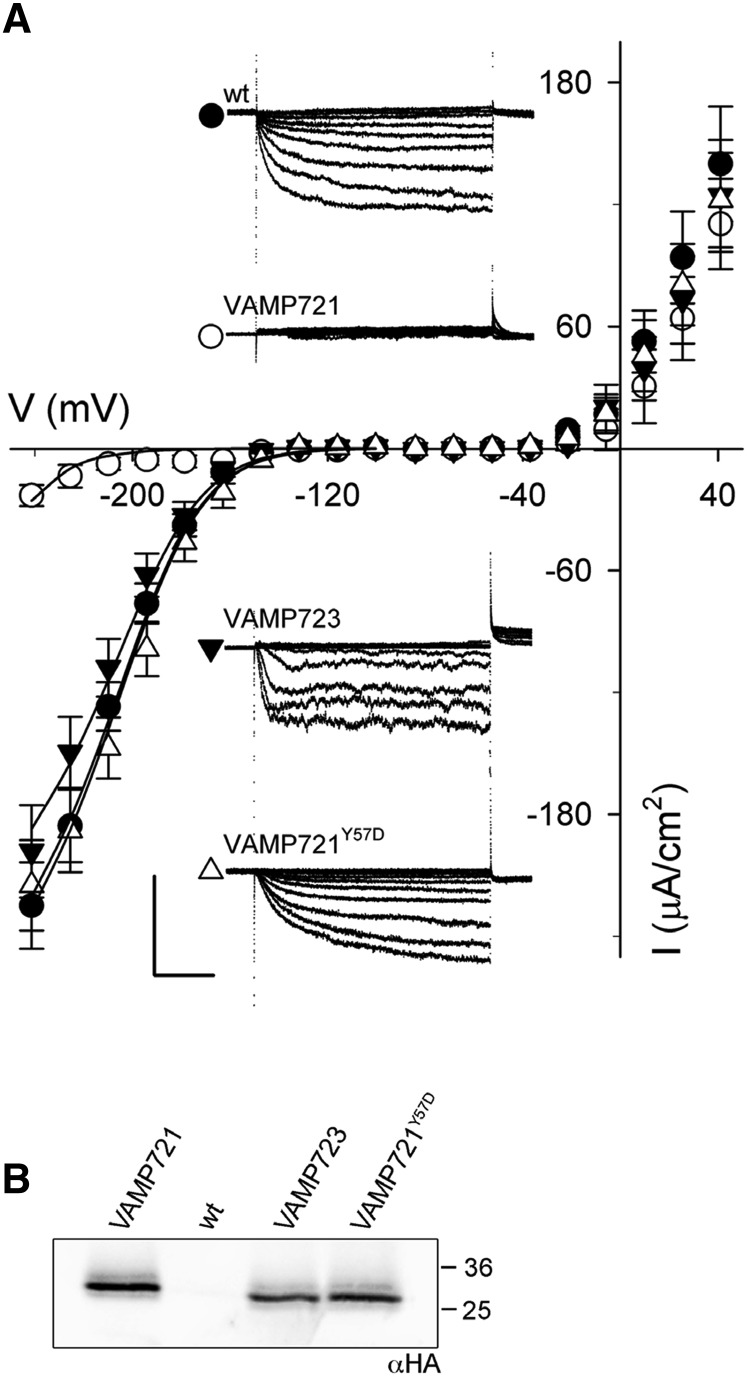
Again, we recorded the K+ current under voltage clamp and compared the currents in each case. Figure 11A includes representative current traces for each of the VAMP constructs and summarizes the mean steady state currents recorded from five or more independent experiments in each case. As before, we extracted the gating characteristics of the K+ current by joint fittings to Equation 1, holding values for δ in common between data sets. A brief inspection of the raw current traces (Figure 11A, insets) and steady state currents shows that the K+ current was strongly suppressed when VAMP721 was expressed, yielding only a small current at all but the most negative voltages; by contrast, currents similar to the wild-type (untransformed) plants were recorded when VAMP723 or the VAMP721Y57D mutant was expressed (Figure 11B). Fittings to Equation 1 as in Figure 10, gave statistically and visually satisfactory results with a common value for δ, a single value for V1/2, and with gmax varying only marginally between data sets for the wild type, VAMP723, or the VAMP721Y57D mutant. Incorporating the data set for VAMP721-expressing plants was best achieved by allowing only V1/2 to differ from the wild type. These results are summarized in Table 4, and the fittings are shown as the solid lines in Figure 11A. They concur with the analysis of the KC1-AKT1 current in oocytes and lead us to conclude that VAMP721 acts directly on channel gating to suppress the open channel.
Figure 11.
VAMP721, but Not VAMP723 or VAMP721Y57D, Suppresses the Inward-Rectifying K+ Current Selectively in Arabidopsis Root Epidermis.
(A) Representative current traces (insets) recorded under voltage clamp in 5 mM Ca2+-MES, pH 6.1, with 10 mM K+ and the mean steady state current-voltage curves derived from these recordings. Data are means ± se of five or more experiments for each set of constructs, VAMP721 (open circles), VAMP723 (filled inverted triangles), and VAMP721Y57D (open triangle), expressed in Arabidopsis seedling root epidermis. Included are measurements from wild-type (untransformed) seedlings (closed circles). Clamp cycles: holding voltage, −50 mV; voltage steps, +40 to −240 mV; tail voltage −50 mV. Scale: 200 μA cm−2 (vertical), 1 s (horizontal). Solid curves are the results of joint, nonlinear least-squares fitting of the K+ currents (IK) to the Boltzmann function (Equation 1). Best and visually satisfactory fittings were obtained allowing V1/2 to vary only for the VAMP721 data set and for gmax to vary between wild-type, VAMP723, and VAMP721Y57D data sets while holding the voltage sensitivity coefficient (apparent gating charge) δ in common between curves. Fitted parameter values are summarized in Table 3.
(B) Immunoblot analysis verifying VAMP transgene (αHA antibody) expression in seedlings pooled for each construct.
Table 4. Overexpressing VAMP721, but Not VAMP723, Suppresses the Inward-Rectifying K+ Current in Arabidopsis Root Epidermal Cells.
| V1/2 (mV) | gmax (ms) | δ | |
|---|---|---|---|
| Wild type (nontransformed) | −195.7 ± 1.4 | 1.38 ± 0.04 | −1.43 ± 0.06 |
| +VAMP723 | 1.27 ± 0.04 | ||
| +VAMP721Y57D | 1.41 ± 0.03 | ||
| +VAMP721 | −277.2 ± 4.9* |
Parameter values are results of joint, nonlinear least-squares fitting of K+ currents in Figure 11. Fittings were carried out with the gating charge, δ, held in common. Values for gmax were allowed to vary between data sets recorded on overexpressing VAMP721, VAMP723, and VAMP721Y57D. gmax was held in common between the wild-type (nontransformed) and VAMP721-overexpressing roots. Voltages giving half-maximal conductance V1/2 were allowed only between data sets for the wild-type and VAMP721-overexpressing plants. Data are from five or more separate experiments for each construct combination and are given as means ± se. Significance for V1/2 as the difference from the wild-type (nontransformed) plants at *P < 0.01.
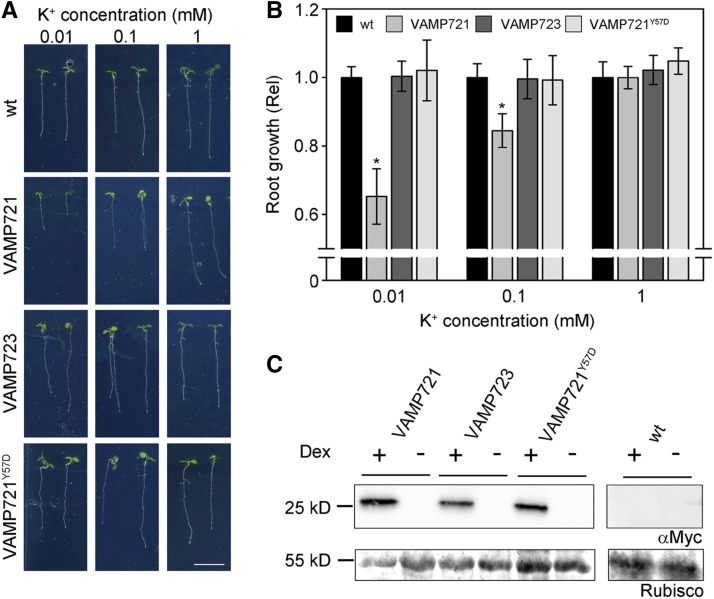
Finally, we asked whether the effects on the K+ current might translate to changes in K+-dependent growth. When germinated in the presence of NH4+, which suppresses K+ uptake via high-affinity transport, growth in Arabidopsis becomes dependent on the activity of the channels assembled from KC1 and AKT1 subunits and sensitive to the K+ concentration outside (Hirsch et al., 1998). Under these conditions, alterations in K+ channel gating can have substantial effects on growth, especially at submillimolar K+ concentrations when suppressing K+ channel activity can lead to greatly reduced seedling growth (Honsbein et al., 2009). We generated independent, stable lines of Arabidopsis to overexpress myc-tagged constructs of VAMP721, VAMP723, and VAMP721Y57D under dexamethasone (Dex)-inducible control (Aoyama and Chua, 1997) using the pDXIsY-Dest vector, which gives strong and sustained expression at concentrations above 1 μM and within 12 h of treatment (Karnik et al., 2015). Plants of three independent transformed lines for each construct were analyzed for root growth at submillimolar to millimolar [K+] in the presence and absence of 2 mM NH4+. In the absence of Dex induction, we found no significant differences in growth at 0.01, 0.1, and 1 mM [K+] between the untransformed and transgenic lines carrying any of the VAMP constructs, either in the absence or presence of NH4+, and only a marginal, but statistically insignificant, reduction in growth at the lowest [K+] (Supplemental Figure 7). Similarly, growth in the presence of Dex was unaffected in lines expressing the VAMP723 and VAMP721Y57D constructs; only plants expressing the VAMP721 construct showed substantial reductions in root growth in the presence of NH4+ with 0.01 and 0.1 mM [K+] (Figure 12). Thus, the effects on the K+ channels of VAMP721 and its Tyr-57 mutant appear to translate directly to growth when the channels are the dominant pathway for K+ uptake.
Figure 12.
Overexpressing VAMP721, but Not VAMP723 or VAMP721Y57D, Suppresses Root Growth Dependent on Channel-Mediated K+ Uptake at Submillimolar K+.
(A) Arabidopsis seedlings grown for 10 d on vertical 0.7% agar plates with defined minimal salts medium containing 0.01, 0.1, and 1 mM K+ with 2 mM NH4+ to block high-affinity K+ uptake and 20 μM Dex to induce VAMP721, VAMP723, and VAMP721Y57D expression. Shown are seedlings of wild-type (wt) and representative Arabidopsis transgenic lines. Bar = 1 cm.
(B) Root length analysis of Arabidopsis seedlings for wild-type and pooled lines expressing VAMP721, VAMP723, and VAMP721Y57D, including the seedlings in (A). Data are means ± se from three independent experiments with >20 seedlings for each line. Root lengths for lines of each transgene have been pooled and are normalized to the wild type at the same K+ concentration. Asterisks indicate significance at P < 0.01.
(C) VAMP721, VAMP723, and VAMP721Y57D overexpression with Dex and their absence without Dex induction, verified by gel electrophoresis of total protein (5 μg/lane) and immunoblot with αmyc antibody. Ponceau S stain of the gel showing Rubisco bands is included as a loading control.
DISCUSSION
There is now a substantial body of information for Qa-SNAREs in Arabidopsis and other plants of their roles in cell division and polarization, and in endomembrane and secretory trafficking (Lipka et al., 2007; Bassham and Blatt, 2008). These proteins contribute to a number of related physiological activities, notably in osmotic homeostasis (Zhu et al., 2002; Drakakaki et al., 2012), tropic growth (Kato et al., 2002; Morita et al., 2002), and pathogen defense (Collins et al., 2003; Kalde et al., 2007; Zhang et al., 2007; Kwon et al., 2008), each of which relies on the mechanics of vesicle traffic. A subset of Qa-SNAREs has proven important also in controlling K+ and other solute uptake through mechanisms that are unrelated to vesicle traffic per se. We originally identified the Qa-SNARE SYP121 (=SYR1/PEN1) of tobacco and Arabidopsis through its activity in hormonal regulation of K+ and Cl− channels that facilitate solute flux for stomatal movements and cellular expansion (Leyman et al., 1999, 2000; Geelen et al., 2002). Subsequent studies demonstrated that SYP121 binds directly with two inward-rectifying K+ channels, KC1 and KAT1, thereby altering channel gating, channel-mediated K+ uptake, and growth (Honsbein et al., 2009; Grefen et al., 2010a; Honsbein et al., 2011). These characteristics extend to stomatal function in which the syp121 mutant exhibits a strong phenotype in gas exchange, carbon assimilation, and growth under moderate relative humidity (Eisenach et al., 2012). Recent work has extended these findings to interactions of SYP121 with selected aquaporins at the plasma membranes of Arabidopsis and maize (Zea mays) (Besserer et al., 2012; Hachez et al., 2014), implying a role in coordinating both solute and water uptake for growth.
Much less is known of the physiology and functional associations that pertain to the cognate SNARE partners in plants, notably the VAMPs. VAMPs have been implicated in endosomal biogenesis (Ueda et al., 2004), gravitropism (Kato et al., 2002; Yano et al., 2003), nodulation (Ma et al., 2006), pathogen defense (Kwon et al., 2008; Yun et al., 2013), salt and drought tolerance (Zhu et al., 2002; Leshem et al., 2006), and cytokinesis (Zhang et al., 2011; El Kasmi et al., 2013), again with the expectation that they act through SNARE-driven vesicle fusion to affect these processes. The near-identical homologs VAMP721 and VAMP722 are known to assemble SNARE core complexes with SYP121 to drive vesicle fusion at the Arabidopsis plasma membrane (Kwon et al., 2008; Karnik et al., 2013b). Thus, the juxtaposition with the noncanonical functions of SYP121 raises the possibility that these VAMPs interact with the same K+ channels that bind the Qa-SNARE. We have now explored this possibility to define the characteristics of VAMP interactions with the K+ channels, KAT1 and KC1. We report that (1) VAMP721 and VAMP722, but not VAMP723, interact with the channels, and we show that VAMP721 affects channel gating and the K+ current within the physiological voltage range, and it suppresses seedling growth when channel-mediated K+ uptake predominates; (2) selective binding is associated with the longin domain of the VAMP, specifically with Tyr-57; and (3) substitutions of this residue alone are sufficient to engineer binding and K+ channel control in the otherwise noninteracting VAMP723. We conclude that interaction of these VAMPs with the K+ channels, like that of SYP121, is complementary to their roles in vesicle traffic. We suggest, too, that VAMP721 and VAMP722 binding with the channels may contribute to a hand-over in channel control during SNARE complex assembly and vesicle fusion, thus coordinating K+ uptake with secretory membrane traffic.
The Kv K+ Channels KAT1 and KC1 Associate with a Subset of Plasma Membrane R-SNAREs
Of the eight proteins in the VAMP72 subgroup, both K+ channels interacted strongly with VAMP721, VAMP722, VAMP724, and VAMP726 (Figure 1), all of which associate with secretory traffic at the plasma membrane. By contrast, no substantive interaction was observed with VAMP723, which has been localized to the endoplasmic reticulum, and SUS assays indicated only a weak interaction with VAMP727, which has been associated with traffic between early endosomes and the plasma membrane (Uemura et al., 2004). Little is known of the cognate SNAREs that interact with VAMP724 and VAMP726. VAMP727 colocalizes with SYP22 on early endosome and prevacuolar compartment membranes (Ebine et al., 2008), and it is known to relocate to the plasma membrane and bind with SYP121 (Ebine et al., 2011). Thus, while at first glance VAMP specificity for the channels appears weaker than among the Qa-SNAREs (Honsbein et al., 2009, 2011), it is nonetheless consistent with current knowledge of the cognate SNARE distributions and almost certainly reflects functional overlaps among the VAMPs that have yet to be explored. Indeed, SYP121 was recently identified with traffic at the trans-Golgi network, possibly indicating a cycling of the Qa-SNARE itself or suggesting additional roles for the Qa-SNARE in membrane vesicle cycling between the plasma membrane, early endosomes, and the Golgi (Drakakaki et al., 2012).
Most telling is our finding that the specificity of K+ channel interactions, at least among the VAMP72 subgroup, depends on the longin domain of the R-SNAREs (Figures 4 to 8). Several SNAREs have been identified to interact directly with K+ and Ca2+ channels in animals, interactions that have been proposed to underpin potentiation in neuronal signaling (Bezprozvanny et al., 1995, 2000; Ji et al., 2002; Cui et al., 2004). Notably the principal Qa-SNARE Syn1A, which mediates in traffic at the neuronal plasma membrane, will bind with Kv2.1 K+ channels in vitro and has been shown to subtly modulate channel activity when expressed in X. laevis oocytes (Tsuk et al., 2005). Mammalian VAMP2 is a cognate partner of Syn1A. It also binds with the Kv2.1 channel in vitro and enhances channel inactivation when expressed in oocytes (Lvov et al., 2008). However, channel binding by Syn1A is associated with the H3 (SNARE motif) domain of the Qa-SNARE (Bezprozvanny et al., 2000; Cui et al., 2004; Tsuk et al., 2005). This domain assembles with the cognate SNAREs to form the core complex during vesicle fusion. Furthermore, Kv2.1 binding to VAMP2 is lost on assembly of the SNARE core complex in vitro (Tsuk et al., 2008), suggesting that the channel also binds with the SNARE motif of VAMP2. Therefore, only by postulating an elaborate exchange of partners have these observations be integrated with the physiology of neurotransmission (Dai et al., 2012). An added concern is that the SNARE motif of Qa-SNAREs, and most likely of other SNAREs, is notoriously promiscuous in protein binding in vitro (Fletcher et al., 2003).
By contrast, selective binding of VAMP721 with the KAT1 and KC1 K+ channels in Arabidopsis is clearly isolated to the longin domain, well removed from the SNARE motif that assembles with SYP121 in the SNARE core complex (Lipka et al., 2007; Bassham and Blatt, 2008). This finding parallels our earlier discovery that SYP121 binds the channels through its N-terminal FxRF motif, again well-removed from the SNARE motif of this Qa-SNARE (Grefen et al., 2010a; Honsbein et al., 2011). We found that VAMP721 binding was associated with the single amino acid residue Tyr-57, located centrally within the longin domain; furthermore, exchange of residues between VAMP721 and VAMP723, generating the VAMP723D57Y mutant, was sufficient to engineer channel interaction in this otherwise noninteracting R-SNARE (Figures 6 to 8; Supplemental Figures 4 and 5). These findings do not rule out contributions from other residues within the longin domain or elsewhere in the VAMP sequence, but they underline the central importance of this residue to the interaction and its functional consequences for channel activity (Figures 3 and 9 to 12).
It is of interest that Tyr-45 of the neuronal Tl-VAMP/VAMP7 (Vivona et al., 2010) plays a crucial role in maintaining the VAMP in a closed conformation and unavailable for binding in a SNARE core complex with Syn1A and SNAP25. The residue is phosphorylated by the c-Src kinase in vitro and in vivo the phosphomimetic Tl-VAMPY45E mutant activates exocytosis and increases the VAMP binding affinity with the cognate SNAREs (Burgo et al., 2013). Our results point to an analogous role for Tyr-57 of VAMP721 and VAMP722, suggesting that an alternation of charge at this site is critical for VAMP-channel interaction. The VAMP721Y57A and VAMP721Y57F mutations had little effect on channel association in yeast, in vivo, and on its functional consequences for channel activity. Furthermore, the VAMP723D57Y exchange mutant engineered channel binding and its control with this otherwise noninteracting VAMP (Figures 6 to 10; Supplemental Figures 4 and 5). The findings suggest that the negative charge introduced by Asp at this location, rather than the residue conjugation or physical size, is an important determinant for the VAMP-channel association, and the findings are therefore consistent with the idea that this residue might be a target for regulation via protein phosphorylation.
The longin domain of Tl-VAMP/VAMP7 in animals plays a dual role in regulating availability for binding in the SNARE core complex, and it interacts with the AP3 adaptor complex of clathrin-coated vesicles, directing the VAMP for endocytosis to late endosomes (Martinez-Arca et al., 2003). An analogous function in targeting has been proposed for the VAMP7 R-SNAREs in Arabidopsis. Uemura et al. (2005) reported that truncation of the longin domain of VAMP711 led to a redirection of the protein to the plasma membrane; however, swapping longin domains between plasma membrane and vacuolar VAMP7 proteins gave mixed results, indicating that the longin domain alone was not sufficient to define the targeting of these R-SNAREs.
Our results are only understandable if mutation of Asp-57 to Tyr-57 in VAMP723 leads to its relocation to the plasma membrane where it interacts with the K+ channels (Figures 6 to 8). These data do not speak to the question of whether channel interaction per se contributes to this relocation, but it seems unlikely, if only because cycling of the K+ channels is very slow by comparison with that expected for the VAMPs at the plasma membrane (Steegmaier et al., 2000; Ueda et al., 2004; Lipka et al., 2007). We note, too, that distributions for both rBiFC complexes with VAMP721 were consistent with the corresponding K+ channel distributions (Sutter et al., 2006, 2007; Honsbein et al., 2009), thus precluding any substantial alterations in the localizations of the channels that might have arisen through rBiFC annealing. Consistent with this interpretation we found that, as a GFP fusion, the single-site mutation of VAMP721Y57D was sufficient to lead to its redistribution within the endomembrane network independent of any rBiFC fusion constructs; conversely, mutation of VAMP723 to give the GFP-VAMP723D57Y fusion led to its relocation to the cell periphery (Supplemental Figure 6). Regardless of the mechanism, it is clear that the single site mutations in VAMP721 and VAMP723 are sufficient to dictate their interactions with the channels at the plasma membrane.
Functional Consequences of R-SNARE Interaction with the K+ Channels
Substantial overlaps exist between the several plasma membrane VAMPs, making a genetic dissection of their functions difficult. Indeed, the amino acid sequences of VAMP721 and VAMP722 show greater than 97% identity (Supplemental Figure 1). Single null mutants of these genes are apparently without phenotype, but the vamp721 vamp722 double mutant is embryo lethal, consistent with the (near) complete functional redundancy of VAMP721 and VAMP722 (Kwon et al., 2008; Zhang et al., 2011). A key to understanding the functional relationship between VAMP721 and the K+ channels therefore must be drawn at present from our findings of K+ current modulation, both on heterologous expression in X. laevis oocytes and on overexpression in vivo. In the oocytes, we found that expressing VAMP721 suppressed the K+ currents carried by KAT1 (Figures 3 and 9) and by channels assembled from KC1 and AKT1 subunits (Figures 10 and 11). For KAT1, the effect was distributed between the ensemble channel conductance, gmax, and its voltage sensitivity, as evidenced by a significant shift in V1/2. Furthermore, VAMP721 action was dose dependent, saturating near a ratio of 1:4 (Figure 3), indicating a stoichiometric relationship between the channel subunit and the VAMP. For KC1, we assumed that gmax was not affected, in line with previous analysis of the channel and its association with AKT1 and SYP121 (Duby et al., 2008; Honsbein et al., 2009; Grefen et al., 2010a). In this case, too, the currents were well-fitted with a shift in V1/2 on VAMP721 coexpression. Recordings from Arabidopsis root epidermal cells yielded complementary results. The dominant inward K+ current in these cells is normally carried by channels assembled from KC1 and AKT1 subunits (Duby et al., 2008; Dreyer and Blatt, 2009; Honsbein et al., 2009). We found that overexpressing VAMP721, but not VAMP723 and VAMP721Y57D, strongly suppressed the inward-rectifying K+ current, consistent with a substantial shift in V1/2 (Figure 11). These effects in suppressing the K+ current translated directly to reduced seedling growth at low [K+] when channel-mediated uptake was the primary pathway for assimilation of the cation (Figure 12). Furthermore, the effects of VAMP721 overexpression were limited to the inward-rectifying K+ current and were without significant effect on the outward K+ current that is carried by the GORK channel. Therefore, we can discount a general effect of VAMP overexpression on transporter activity, delivery to, or cycling from, the plasma membrane.
For any one channel, changes in current amplitude, and hence in gmax, may be understood as the consequence either of a change in the population of channels at the membrane or of a change in channel gating. However, the mid-point voltage, V1/2, for gating is intimately connected with the biophysical properties of the K+ channel voltage sensor (Hille, 2001; Dreyer and Blatt, 2009; Lefoulon et al., 2014). Therefore, it is difficult to reconcile the effects of VAMP721 (and VAMP723D57Y) with membrane traffic or changes in the number of channels at the plasma membrane. For the same reason, the observations cannot be understood as a result of binding and titration of the channels by the R-SNARE. Again, simply altering the number of channels available for activation at negative voltages could be expected to affect current amplitude, but not its intrinsic gating characteristics. Instead, the shifts in V1/2 are a forceful argument for a direct, physical interaction with the mechanism of voltage sensing by these channels, possibly with the voltage sensor domain itself (Dreyer and Blatt, 2009; Lefoulon et al., 2014).
Implications for the Mechanics of Membrane Fusion
The effect of VAMP721 expression on the K+ currents does stand out in contrast to that of SYP121. We previously reported that SYP121 binding with the KC1-AKT1 channel complex strongly promoted the K+ current by shifting V1/2 toward positive voltages and enhancing the voltage sensitivity of gating, both on heterologous expression and in vivo (Honsbein et al., 2009; Grefen et al., 2010a). Additionally, Eisenach et al. (2012) found that basal KAT1 currents were reduced by 50% or more in guard cells of the Arabidopsis syp121 mutant when compared with the currents in the guard cells of wild-type plants. While the latter observations may reflect, in part, a suppression in channel traffic to the plasma membrane, this is clearly not the case for SYP121 binding with KC1. Indeed, the effects of SYP121 in both examples are diametrically opposite to the actions of VAMP721 on the K+ currents and growth (Figures 3 and 9 to 12). This functional juxtaposition between the two cognate SNAREs begs a question whether the counterpoint in control of K+ channel gating might be woven together with the roles of VAMP721, VAMP722, and SYP121 in vesicle fusion.
Vesicle tethering and docking are generally recognized to precede SNARE complex assembly, positioning the vesicle close to the site of fusion (Zárský et al., 2009); only later do the cognate SNAREs engage to drive the target and vesicle membrane surfaces together. Vesicles arriving by actin-mediated transport are tethered within 50 to 100 nm of the fusion site by factors that include proteins of the exocyst complex (Hála et al., 2008; Fendrych et al., 2010). Docking is poorly defined in plants, but in yeast it facilitates exocyst binding with Sec1 (Novick et al., 2006), a member of the Sec1/Munc18 (SM) protein family that initially clamps many Qa-SNAREs in a so-called closed or inactive conformation. Docking probably recruits additional components, such as Munc13 (Ma et al., 2013) of nerve, to displace SM binding and initiate SNARE assembly. Significantly, the channel binding site on SYP121 in Arabidopsis overlaps with that for the SM protein SEC11 (=KEULE; Grefen et al., 2010a; Karnik et al., 2013b). A recent study highlights the possible role for the K+ channels to initiate a handover in SEC11 binding at this site and release SYP121 for SNARE assembly (Karnik et al., 2013b).
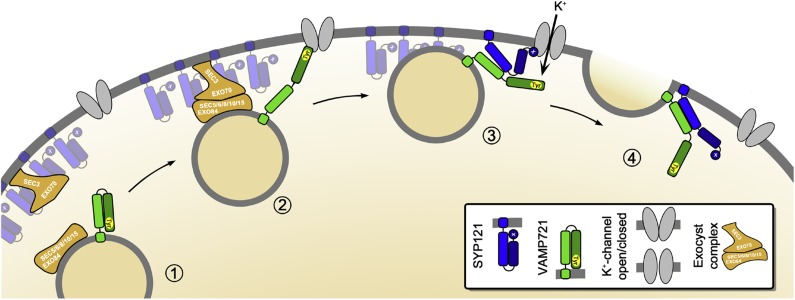
Some time ago we concluded that SYP121 interaction with the K+ channels could be understood as a mechanism for their mutual control, coordinating the rates of solute uptake and of surface area expansion as the cell grows. By analogy with James Watt’s application of the mechanical governor (Maxwell, 1868), this Governor Hypothesis (Grefen and Blatt, 2008) satisfies a number of experimental observations, including the uncoupling of solute uptake from cell expansion and runaway solute accumulation in cells expressing a soluble fragment of SYP121 (Geelen et al., 2002; Sokolovski et al., 2008; Honsbein et al., 2009). Necessarily, the hypothesis has been short of temporal detail. Our findings now anticipate a sequence in which channel binding, alternately with one SNARE and then the other, periodically activates the channel to enhance K+ flux while engaging the SNAREs for assembly. We illustrated one of several alternative sequences in Figure 13. Here, vesicle tethering leads to docking when VAMP721 binds with the K+ channel to bring the vesicle within 8 to 10 nm of the plasma membrane surface, and it recruits SYP121. Interaction of the exocyst complex with the SYP121-bound SEC11 then facilitates SEC11 exchange with the K+ channel, concurrently promoting K+ influx and activating both SNAREs for their assembly in the core complex to drive the final stages of membrane fusion.
Figure 13.
Vesicle Tethering, Docking, and Fusion Associated with K+ Channel Binding and Control by SYP121 and Its Cognate R-SNAREs VAMP721 and VAMP722.
The hypothetical sequence brings together our understanding of the exocyst in tethering in Arabidopsis (Fendrych et al., 2010), of the channels as focal points for vesicle traffic (Sutter et al., 2006, 2007), and of SYP121 recruitment by the K+ channels from larger “icebergs” of the Qa-SNARE at the plasma membrane (Sieber et al., 2007; Murray and Tamm, 2009; Honsbein et al., 2011). Here vesicle approach (1) and tethering by the exocyst complex leads to docking when VAMP721 binds with the K+ channel (2) to bring the vesicle within 8 to 10 nm of the plasma membrane surface. Once docked via the VAMP longing domain, the K+ channel-VAMP complex recruits SYP121 leading to an exchange of binding (3). Not shown is the anticipated exchange in SYP121 binding between SEC11 and the K+ channel (Karnik et al., 2013b). These events concurrently promote K+ channel activation (3) and bring the SNAREs together for their assembly in the core complex to drive the final stages of membrane fusion (4).
This is an attractive, if hypothetical, sequence. It brings together our understanding of the exocyst in tethering in Arabidopsis (Fendrych et al., 2010), of the channels as focal points for vesicle traffic (Sutter et al., 2006, 2007), and of SYP121 recruitment by the K+ channels, a variation of the so-called icebreaker model (Sieber et al., 2007; Murray and Tamm, 2009; Honsbein et al., 2011), leading up to vesicle fusion. It also anticipates an exchange in SEC11 and K+ channel binding at the SYP121 N terminus to drive SNARE complex assembly (Karnik et al., 2013b). However, there are other alternatives, including the possibility that K+ channel binding proceeds in reverse order from SYP121 to VAMP721 during vesicle fusion. How might these alternatives be distinguished? We need now to identify the R-SNARE binding site(s) on the channel proteins. This knowledge will provide tools essential to resolving the sequence and scales of SNARE-channel interactions in relation to cell expansion.
METHODS
Molecular Biology
The pGT-nHA-Dest vector was created by amplifying the vector backbone of pGT-Dest (Chen et al., 2011) with primers CG-S-pGT-Hpal and CG-A-pGT-SnaBI, thereby adding an HpaI site at the 3′ and a SnaBI site at the 5′ termini of the expression cassette. The Gateway-myc sequence of vector pNX35-Dest (Grefen and Blatt, 2012) was amplified using primers BZ-S-PmeI-2xHA and BZ-A-HpaI-GW. T4 DNA ligase (NEB) was used to carry out blunt-end ligation and subsequent transformation and selection was in ccdB-survival Escherichia coli cells (Life Technologies). The vector sequence was verified by restriction endonuclease digestion and sequencing (GATC).
Open reading frames for KAT1, KC1, and the VAMPs (VAMP711, VAMP712, VAMP713, VAMP714, VAMP721, VAMP722, VAMP723, VAMP724, VAMP725, VAMP726, VAMP727, and VAMP728) were amplified with gene-specific primers that included Gateway attachment sites (attB1/attB2, attB3/B2, or attB1/B4). Subsequent BP reactions in pDONR207, pDONR221-P3P2, and pDONR221-P1P4 (Life Technologies) yielded Entry clones that were verified via sequencing. KAT1 and KC1 sequences were obtained without stop codon to allow C-terminal fusions, whereas VAMPs were amplified to include their native stop codon. Gateway Destination clones were generated using LR Clonase II (Life Technologies) by LR reaction according to the manufacturer’s instructions.
Chimeric clones of VAMPs were constructed using overlap extension PCR (Chavez et al., 1999). Point mutants were generated by site-directed mutagenesis (Karnik et al., 2013a). Primers of different point mutants were designed by SDM-Assist software to include unique “silent” restriction sites along with the desired mutation for downstream identification via restriction endonuclease digestion. All primers used in this article are listed in Supplemental Table 1.
For SUS assays, KAT1 and KC1 were recombined in pMetYC-Dest (Grefen et al., 2009). VAMPs, chimeras, and point mutants were recombined in pNX35-Dest. For electrophysiological analysis in oocytes, KAT1 was recombined in pGT-Dest (Grefen et al., 2010a) and KC1 in pGTmyc-Dest, fusing a C-terminal myc tag, while VAMPs were recombined in pGT-nHA-Dest. AKT1, CBL1, and CIPK23 constructs were used as described previously (Honsbein et al., 2009). For electrophysiological analysis in plants, these constructs were linked with an N-terminal HA tag via PCR reaction and introduced into the pUB-Bic-Dest vector (Chen et al., 2011). For VAMP localization studies, each construct was introduced via LR reaction into pUBN-GFP-DEST (Grefen et al., 2010b). For rBiFC analysis, all constructs were introduced via 2in1-LR reaction in pBiFCt-2in1-NC as described previously (Grefen and Blatt, 2012). The VAMPs were tagged N-terminally with nYFP, and KAT1 and KC1 were tagged C-terminally with cYFP. Gateway Entry and Destination clones were amplified using Top10 cells (Life Technologies) with the appropriate antibiotic (gentamycin at 20 mg/L for Entry clones and spectinomycin at 100 mg/L for Destination clones).
Plant Growth and Transformation
Wild-type tobacco (Nicotiana tabacum) plants were grown in soil at 26°C and 70% relative humidity on a 16-h-day/8-h-night cycle. Plants with three to four fully expanded leaves after 4 to 6 weeks of growth were selected for infiltration. Tobacco leaves were infiltrated with Agrobacterium tumefaciens GV3101 carrying the desired construct as described previously (Tyrrell et al., 2007). Seeds of Arabidopsis thaliana wild-type (Columbia-0) were sterilized in 10% (v/v) NaHClO and 0.1% Triton X-100 and stratified at 4°C for 48 h. Thereafter the seed was sown in sterile 0.05% Murashige and Skoog medium minus organics, and 3-d-old seedlings were transformed by cocultivation using methods developed from those previously described for Agrobacterium rhizogenes (Campanoni et al., 2007; Li et al., 2009) as detailed recently for Agrobacterium GV3101 (Grefen et al., 2010b; Blatt and Grefen, 2014).
Stable Arabidopsis lines for Dex-inducible expression were generated by floral dip (Clough and Bent, 1998) with Agrobacterium GV3101 carrying VAMP721, VAMP723, and VAMP721Y57D in the pDXIsY-Dest vector (Karnik et al., 2015), and transformed seed were selected by growth on 50 mg/L phosphinothricin (Basta; Bayer Cropscience). Homozygous T4 lines were verified for expression of the transgene by immunoblot analysis, and three independently transformed lines were chosen for experiments. For phenotypic analysis, plants were grown on vertical 0.7% (w/v) agar plates and in liquid culture with defined minimal salts medium including 0.01, 0.1, and 1 mM K+ and either with or without 2 mM NH4+ (Honsbein et al., 2009). Plants were germinated and grown at 22°C and 100 μmol m−2 s−1 PAR for 10 d. As required, 20 μM Dex was included in the medium. Root lengths were determined in three independent experiments from >20 seedlings per line for each treatment and were quantified using EZ-Rhizo (Armengaud et al., 2009). In the absence of statistically significant differences between lines with any one construct, we pooled these data.
Mating-Based SUS Assays
The haploid yeast strains THY.AP4 and THY.AP5 (Obrdlik et al., 2004) were transformed as described previously (Grefen et al., 2009). Yeast mating-based split-ubiquitin assays were performed as follows: Pools of 10 to 15 yeast colonies were selected and inoculated in selective media (CSM-LM for THY.AP4 and CSM-MTU for THY.AP5) for overnight growth at 180 rpm and 28°C. Liquid cultures were harvested and resuspended in YPD medium. Yeast mating was performed in sterile PCR tubes by mixing equal aliquots of cultures containing KAT1-Cub or KC1-Cub in THY.AP4 with the appropriate NubG-VAMPs in THY.AP5. Aliquots of 5 μL from each mixture were dropped on YPD plates and incubated at 28°C overnight. Colonies were transferred from YPD onto CSM-LMTU plates and incubated at 28°C for 2 to 3 d. Diploid colonies were selected and inoculated in liquid CSM-LMTU media and grown at 180 rpm and 28°C overnight. Thereafter, yeast was harvested and resuspended in sterile water. Serial dilutions at OD600 1.0 and 0.1 in water were dropped, 6 μL per spot, on CSM-AHLMTU plates with added methionine in increasing concentrations. Plates were incubated at 28°C and images were taken after 3 d. Yeast was also dropped on a CSM-LMTU control plate to confirm mating efficiency, and cell density and growth were monitored after 24 h at 28°C. To verify expression, yeast cells were harvested in aliquots equal to those used for the dilution series and extracted for protein gel blot analysis using commercial HA antibody (Sigma-Aldrich) for NubG and commercial VP16 antibody (Abcam) for the Cub-fusions.
Electrophysiology
For electrical recordings using Xenopus laevis oocytes, plasmids were linearized and capped cRNA was synthesized in vitro using T7 mMessage Machine (Ambion). cRNA quality was confirmed by denaturing gel electrophoresis. cRNA was mixed to ensure equimolar ratios unless otherwise noted, and mixtures were adjusted to standard volume using RNase-free water.
Stage VI oocytes were isolated from mature X. laevis, and the follicular cell layer was digested with 2 mg/mL collagenase (type 1A; Sigma-Aldrich) for 20 min before injection. Injected oocytes were incubated in ND96 (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1 mM CaCl2, and 10 mM HEPES-NaOH, pH 7.4) supplemented with gentamycin (5 mg/L) at 18°C for 3 d before electrophysiological recordings. Whole-cell currents were recorded under voltage clamp using an Axoclamp 2B two-electrode clamp circuit (Axon Instruments) as described previously (Leyman et al., 1999; Sutter et al., 2006). Measurements were performed under continuous perfusion with 30 mM KCl and 66 mM NaCl with additions of 1.8 mM CaCl2 and 10 mM HEPES-NaOH, pH 7.2. A standard voltage clamp cycle was used with a holding voltage of −50 mV and 14 test voltage steps from 0 to −180 mV or −200 mV. Oocytes yielding currents were collected and total membrane protein isolated according to Sottocornola et al. (2006) using 20 μL of extraction buffer per oocyte. To verify expression, commercial antibodies (Abcam) for myc and HA were used in immunoblots for KC1 and the VAMPs, respectively.
Recordings from Arabidopsis root epidermal cells were performed on 6- to 8-d-old seedlings 3 to 5 d after transforming by cocultivation with Agrobacterium. Transformations were with indicated VAMP constructs in pUB-Bic-Dest. Seedlings were bathed in solutions of 10 mM KCl with 5 mM Ca2+-MES, pH 6.1 [adjusted with Ca(OH)2, free [Ca2+] = 1 mM] and voltage clamp recordings made use of standard two-electrode methods (Meharg et al., 1994; Honsbein et al., 2009; Chen et al., 2011). Measurements were performed on mature epidermal cells in cell files lacking root hairs to avoid electrical coupling and clamp-current dissipation by root hairs and between cells as described previously (Chen et al., 2011). All recordings were analyzed and leak currents subtracted using Henry III software (Leyman et al., 1999; Sutter et al., 2006; Y-Science, University of Glasgow). Expression was verified on a cell-by-cell basis using the coexpressed GFP marker (Chen et al., 2011).
Confocal Microscopy
For GFP localization and rBiFC assays, confocal images were collected using a Zeiss LSM510-META confocal microscope with 20×/0.75-NA and 40×/1.3-NA objectives. Excitation intensities, filter settings, and photomultiplier gains were standardized. GFP was excited with the 488-nm line of an Argon2 laser and fluorescence collected after passage over a 545-nm dichroic and through a 505- to 530-nm band-pass filter. YFP and RFP were excited by the 514-nm line of an argon laser and the 543-nm line of a HeNe laser, respectively. YFP fluorescence was collected after passage through a 515-nm dichroic mirror and collected by the META head between 521 and 565 nm. RFP fluorescence was collected after passing through a 545-nm dichroic mirror and 560- to 615-nm band-pass filter. Chlorophyll fluorescence from tobacco leaves was identified after excitation by the 514-nm line of the argon laser by collection after passing through a 545-nm dichroic mirror and 560-nm long-pass filter. rBiFC fluorescence ratios were calculated as described previously (Grefen and Blatt, 2012; Karnik et al., 2013b) after subtraction of background fluorescence recorded from nontransformed tissues prepared in parallel.
Plant Extraction and Immunoblot Analysis
Arabidopsis seedlings and tobacco leaf sections were harvested, weighed, flash-frozen, and ground in liquid N2. Equal weights of tissue were suspended by sonication 1:1 (w/v) in homogenization buffer containing 500 mM sucrose, 10% glycerol, 20 mM EDTA, 20 mM EGTA, 1× Protease Inhibitor (Roche), 10 mM ascorbic acid, 5 mM DTT, and 50 mM Tris-HCl, pH 7.4. Suspended samples were centrifuged at 13,000g for 10 min at 4°C, supernatants were diluted 1:1 in loading buffer (Karnik et al., 2013b), and proteins were separated by SDS-PAGE on 10% acrylamide gels. Immunoblot analysis was performed, after transfer to nitrocellulose filters and blocking, using primary commercial antibodies αGFP (1:5000), αHA (1:2000), αmyc (1:5000), and αVP16 (1:10,000), and secondary goat anti-rabbit antibodies (Abcam). Detection was by ECL Advance Detection Kit (GE Healthcare), and all blots were recorded digitally for subsequent analysis using ImageJ v. 1.48 (http://rsb.info.nih.gov/ij/).
Statistics
Statistical analysis of independent experiments is reported as means ± se as appropriate with significance determined by Student’s t test or ANOVA. Joint, nonlinear least-squares fittings were performed using a Marquardt-Levenberg algorithm (Marquardt, 1963) as implemented in SigmaPlot v.11.2 (SPSS Software).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: KAT1 (At5g46240), KC1 (At4g32650), VAMP711 (At4g32150), VAMP712 (At2g25340), VAMP713 (At5g11150), VAMP714 (At5g22360), VAMP721 (At1g04750), VAMP722 (At2g33120), VAMP723 (At2g33110), VAMP724 (At4g15780), VAMP725 (At2g32670), VAMP726 (At1g04760), VAMP727 (At3g54300), and VAMP728 (At3g24890).
Supplemental Data
Supplemental Figure 1. Alignment of the amino acid sequences for the Arabidopsis VAMP proteins.
Supplemental Figure 2. VAMP721, but not VAMP723, interacts with the KC1 K+ channel in vivo.
Supplemental Figure 3. The central section of the VAMP721 longin domain is essential for interaction with KAT1.
Supplemental Figure 4. Single-site mutants VAMP721D61N, VAMP721Y57F, and VAMP721Q76E have no substantive effect on interaction with KAT1 or KC1, and AKT1 shows no interaction with VAMP721 or VAMP723.
Supplemental Figure 5. The VAMP721Y57D mutant suppresses interaction with the KC1 K+ channel.
Supplemental Figure 6. The single-site mutants VAMP721Y57D and VAMP723D57Y affect the localization of the R-SNAREs.
Supplemental Figure 7. Arabidopsis lines carrying VAMP721, VAMP723, and VAMP721Y57D under the Dex-inducible promoter show no substantive effects on root growth dependent on channel-mediated K+ uptake at submillimolar K+ in the absence of dexamethasone.
Supplementary Material
Acknowledgments
We thank Kristin Aderhold, Amparo Ruiz-Pardo, and George Boswell for support in immunoblot analysis and plant and X. laevis maintenance. Cecile Lefoulon and Annegret Honsbein provided guidance early on for experiments with oocytes. This work was supported by a Chinese Scholarship Council studentship to B.Z. and by grants BB/H0024867/1, BB/I024496/1, BB/K015893/1, BB/L001276/1, and BB/M01133X/1 to M.R.B. from the Biotechnology and Biological Sciences Research Council of the UK. C.G. was supported by an Emmy Noether Fellowship of the Deutsche Forschungsgemeinschaft GR 4251/1-1.
AUTHOR CONTRIBUTIONS
B.Z. carried out the SUS assays with C.G. and the confocal and oocyte studies with M.R.B. B.Z., C.G., and R.K. designed the constructs and vectors. R.K. and B.Z. undertook immunoblot analyses. Y.W. carried out transformations and analysis of currents in Arabidopsis root epidermis. B.Z. generated and analyzed the Dex-inducible lines. N.W. and B.Z. carried out localization studies. B.Z. and M.R.B. wrote the article with R.K. and C.G.
Glossary
- SUS
split-ubiquitin system
- rBiFC
ratiometric bimolecular fluorescence complementation
- Dex
dexamethasone
Footnotes
Articles can be viewed online without a subscription.
References
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612. [DOI] [PubMed] [Google Scholar]
- Armengaud P., Zambaux K., Hills A., Sulpice R., Pattison R.J., Blatt M.R., Amtmann A. (2009). EZ-Rhizo: integrated software for the fast and accurate measurement of root system architecture. Plant J. 57: 945–956. [DOI] [PubMed] [Google Scholar]
- Bassham D.C., Blatt M.R. (2008). SNAREs: cogs and coordinators in signaling and development. Plant Physiol. 147: 1504–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer A., Burnotte E., Bienert G.P., Chevalier A.S., Errachid A., Grefen C., Blatt M.R., Chaumont F. (2012). Selective regulation of maize plasma membrane aquaporin trafficking and activity by the SNARE SYP121. Plant Cell 24: 3463–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezprozvanny I., Scheller R.H., Tsien R.W. (1995). Functional impact of syntaxin on gating of N-type and Q-type calcium channels. Nature 378: 623–626. [DOI] [PubMed] [Google Scholar]
- Bezprozvanny I., Zhong P., Scheller R.H., Tsien R.W. (2000). Molecular determinants of the functional interaction between syntaxin and N-type Ca2+ channel gating. Proc. Natl. Acad. Sci. USA 97: 13943–13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatt, M.R., and Grefen, C. (2014). Applications of fluorescent marker proteins in plant cell biology. In Arabidopsis Protocols, J.J. Sanchez-Serrano J. Salinas, eds (New York: Humana Press, Springer), pp. 487–507. [DOI] [PubMed] [Google Scholar]
- Bock J.B., Matern H.T., Peden A.A., Scheller R.H. (2001). A genomic perspective on membrane compartment organization. Nature 409: 839–841. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. (2005). Structure and function of SNARE and SNARE-interacting proteins. Q. Rev. Biophys. 38: 1–47. [DOI] [PubMed] [Google Scholar]
- Burgo A., Casano A.M., Kuster A., Arold S.T., Wang G., Nola S., Verraes A., Dingli F., Loew D., Galli T. (2013). Increased activity of the vesicular soluble N-ethylmaleimide-sensitive factor attachment protein receptor TI-VAMP/VAMP7 by tyrosine phosphorylation in the Longin domain. J. Biol. Chem. 288: 11960–11972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanoni P., Sutter J.U., Davis C.S., Littlejohn G.R., Blatt M.R. (2007). A generalized method for transfecting root epidermis uncovers endosomal dynamics in Arabidopsis root hairs. Plant J. 51: 322–330. [DOI] [PubMed] [Google Scholar]
- Chapman S., Faulkner C., Kaiserli E., Garcia-Mata C., Savenkov E.I., Roberts A.G., Oparka K.J., Christie J.M. (2008). The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc. Natl. Acad. Sci. USA 105: 20038–20043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez R.A., Gray A.T., Zhao B.B., Kindler C.H., Mazurek M.J., Mehta Y., Forsayeth J.R., Yost C.S. (1999). TWIK-2, a new weak inward rectifying member of the tandem pore domain potassium channel family. J. Biol. Chem. 274: 7887–7892. [DOI] [PubMed] [Google Scholar]
- Chen Z., Grefen C., Donald N., Hills A., Blatt M.R. (2011). A bicistronic, Ubiquitin-10 promoter-based vector cassette for transient transformation and functional analysis of membrane transport demonstrates the utility of quantitative voltage clamp studies on intact Arabidopsis root epidermis. Plant Cell Environ. 34: 554–564. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Collins N.C., Thordal-Christensen H., Lipka V., Bau S., Kombrink E., Qiu J.L., Hückelhoven R., Stein M., Freialdenhoven A., Somerville S.C., Schulze-Lefert P. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977. [DOI] [PubMed] [Google Scholar]
- Cui N., Kang Y., He Y., Leung Y.M., Xie H., Pasyk E.A., Gao X., Sheu L., Hansen J.B., Wahl P., Tsushima R.G., Gaisano H.Y. (2004). H3 domain of syntaxin 1A inhibits KATP channels by its actions on the sulfonylurea receptor 1 nucleotide-binding folds-1 and -2. J. Biol. Chem. 279: 53259–53265. [DOI] [PubMed] [Google Scholar]
- Dai X.Q., et al. (2012). The voltage-dependent potassium channel subunit Kv2.1 regulates insulin secretion from rodent and human islets independently of its electrical function. Diabetologia 55: 1709–1720. [DOI] [PubMed] [Google Scholar]
- Drakakaki G., van de Ven W., Pan S., Miao Y., Wang J., Keinath N.F., Weatherly B., Jiang L., Schumacher K., Hicks G., Raikhel N. (2012). Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res. 22: 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer I., Blatt M.R. (2009). What makes a gate? The ins and outs of Kv-like K+ channels in plants. Trends Plant Sci. 14: 383–390. [DOI] [PubMed] [Google Scholar]
- Duby G., Hosy E., Fizames C., Alcon C., Costa A., Sentenac H., Thibaud J.B. (2008). AtKC1, a conditionally targeted Shaker-type subunit, regulates the activity of plant K+ channels. Plant J. 53: 115–123. [DOI] [PubMed] [Google Scholar]
- Ebine K., Okatani Y., Uemura T., Goh T., Shoda K., Niihama M., Morita M.T., Spitzer C., Otegui M.S., Nakano A., Ueda T. (2008). A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 20: 3006–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K., et al. (2011). A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 13: 853–859. [DOI] [PubMed] [Google Scholar]
- Eisenach C., Chen Z.H., Grefen C., Blatt M.R. (2012). The trafficking protein SYP121 of Arabidopsis connects programmed stomatal closure and K⁺ channel activity with vegetative growth. Plant J. 69: 241–251. [DOI] [PubMed] [Google Scholar]
- El Kasmi F., Krause C., Hiller U., Stierhof Y.-D., Mayer U., Conner L., Kong L., Reichardt I., Sanderfoot A.A., Jürgens G. (2013). SNARE complexes of different composition jointly mediate membrane fusion in Arabidopsis cytokinesis. Mol. Biol. Cell 24: 1593–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enami K., Ichikawa M., Uemura T., Kutsuna N., Hasezawa S., Nakagawa T., Nakano A., Sato M.H. (2009). Differential expression control and polarized distribution of plasma membrane-resident SYP1 SNAREs in Arabidopsis thaliana. Plant Cell Physiol. 50: 280–289. [DOI] [PubMed] [Google Scholar]
- Fasshauer D., Sutton R.B., Brunger A.T., Jahn R. (1998). Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA 95: 15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M., Synek L., Pecenková T., Toupalová H., Cole R., Drdová E., Nebesárová J., Sedinová M., Hála M., Fowler J.E., Zársky V. (2010). The Arabidopsis exocyst complex is involved in cytokinesis and cell plate maturation. Plant Cell 22: 3053–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher S., Bowden S.E.H., Marrion N.V. (2003). False interaction of syntaxin 1A with a Ca2+-activated K+ channel revealed by co-immunoprecipitation and pull-down assays: implications for identification of protein-protein interactions. Neuropharmacology 44: 817–827. [DOI] [PubMed] [Google Scholar]
- Geelen D., Leyman B., Batoko H., Di Sansebastiano G.P., Moore I., Blatt M.R. (2002). The abscisic acid-related SNARE homolog NtSyr1 contributes to secretion and growth: evidence from competition with its cytosolic domain. Plant Cell 14: 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D., Becker D., Vosloh D., Gambale F., Palme K., Rehers M., Anschuetz U., Dreyer I., Kudla J., Hedrich R. (2009). Heteromeric AtKC1⋅AKT1 channels in Arabidopsis roots facilitate growth under K+-limiting conditions. J. Biol. Chem. 284: 21288–21295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C., Blatt M.R. (2008). SNAREs—molecular governors in signalling and development. Curr. Opin. Plant Biol. 11: 600–609. [DOI] [PubMed] [Google Scholar]
- Grefen C., Blatt M.R. (2012). A 2in1 cloning system enables ratiometric bimolecular fluorescence complementation (rBiFC). Biotechniques 53: 311–314. [DOI] [PubMed] [Google Scholar]
- Grefen C., Obrdlik P., Harter K. (2009). The determination of protein-protein interactions byt the mating-based split-ubiquitin system (mbSUS). Methods Mol. Biol. 479: 1–17. [DOI] [PubMed] [Google Scholar]
- Grefen, C., Chen, Z.H., Honsbein, A., Donald, N., Hills, A., and Blatt, M.R. (2010a). A novel motif essential for SNARE interaction with the K+ channel KC1 and channel gating in Arabidopsis. Plant Cell 22: 3076–3092. [DOI] [PMC free article] [PubMed]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M.R. (2010b). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64: 355–365. [DOI] [PubMed] [Google Scholar]
- Hachez C., Laloux T., Reinhardt H., Cavez D., Degand H., Grefen C., De Rycke R., Inzé D., Blatt M.R., Russinova E., Chaumont F. (2014). Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2;7 to modulate the cell membrane water permeability. Plant Cell 26: 3132–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hála M., Cole R., Synek L., Drdová E., Pecenková T., Nordheim A., Lamkemeyer T., Madlung J., Hochholdinger F., Fowler J.E., Zárský V. (2008). An exocyst complex functions in plant cell growth in Arabidopsis and tobacco. Plant Cell 20: 1330–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. (2001). Ionic Channels of Excitable Membranes. (Sunderland, MA: Sinauer Press; ). [Google Scholar]
- Hirsch R.E., Lewis B.D., Spalding E.P., Sussman M.R. (1998). A role for the AKT1 potassium channel in plant nutrition. Science 280: 918–921. [DOI] [PubMed] [Google Scholar]
- Hong W. (2005). SNAREs and traffic. Biochim. Biophys. Acta 1744: 120–144. [DOI] [PubMed] [Google Scholar]
- Honsbein A., Blatt M.R., Grefen C. (2011). A molecular framework for coupling cellular volume and osmotic solute transport control. J. Exp. Bot. 62: 2363–2370. [DOI] [PubMed] [Google Scholar]
- Honsbein, A., Sokolovski, S., Grefen, C., Campanoni, P., Pratelli, R., Paneque, M., Chen, Z.H., Johansson, I., and Blatt, M.R. (2009). A tripartite SNARE-K+ channel complex mediates in channel-dependent K+ nutrition in Arabidopsis. Plant Cell 21: 2859–2877. [DOI] [PMC free article] [PubMed]
- Hoshi T. (1995). Regulation of voltage dependence of the KAT1 channel by intracellular factors. J. Gen. Physiol. 105: 309–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashikina N., Becker D., Ache P., Meyerhoff O., Felle H.H., Hedrich R. (2001). K+ channel profile and electrical properties of Arabidopsis root hairs. FEBS Lett. 508: 463–469. [DOI] [PubMed] [Google Scholar]
- Jahn R., Scheller R.H. (2006). SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7: 631–643. [DOI] [PubMed] [Google Scholar]
- Ji J., et al. (2002). The 25-kDa synaptosome-associated protein (SNAP-25) binds and inhibits delayed rectifier potassium channels in secretory cells. J. Biol. Chem. 277: 20195–20204. [DOI] [PubMed] [Google Scholar]
- Jürgens G., Geldner N. (2007). The high road and the low road: trafficking choices in plants. Cell 130: 977–979. [DOI] [PubMed] [Google Scholar]
- Kalde M., Nühse T.S., Findlay K., Peck S.C. (2007). The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc. Natl. Acad. Sci. USA 104: 11850–11855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik A., Karnik R., Grefen C. (2013b). SDM-Assist software to design site-directed mutagenesis primers introducing “silent” restriction sites. BMC Bioinformatics 14: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik R., Grefen C., Bayne R., Honsbein A., Köhler T., Kioumourtzoglou D., Williams M., Bryant N.J., Blatt M.R. (2013a). Arabidopsis Sec1/Munc18 protein SEC11 is a competitive and dynamic modulator of SNARE binding and SYP121-dependent vesicle traffic. Plant Cell 25: 1368–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik R., Zhang B., Waghmare S., Aderhold C., Grefen C., Blatt M.R. (2015). Binding of SEC11 indicates its role in SNARE recycling after vesicle fusion and identifies two pathways for vesicular traffic to the plasma membrane. Plant Cell 27: 675–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, T., Morita, M.T., Fukaki, H., Yamauchi, Y., Uehara, M., Niihama, M., and Tasaka, M. (2002). SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell 14: 33–46. [DOI] [PMC free article] [PubMed]
- Kwon C., et al. (2008). Co-option of a default secretory pathway for plant immune responses. Nature 451: 835–840. [DOI] [PubMed] [Google Scholar]
- Lefoulon C., Gutla P.V., Honsbein A., Wang Y., Grefen C., Riedelsberger J., Karnik R., Gonzalez W., Blatt M.R. (2014). Voltage-sensor transitions of the inward-rectifying K+ channel KAT1 indicate a latching mechanism biased by hydration within the voltage sensor. Plant Physiol. 166: 960–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshem Y., Golani Y., Kaye Y., Levine A. (2010). Reduced expression of the v-SNAREs AtVAMP71/AtVAMP7C gene family in Arabidopsis reduces drought tolerance by suppression of abscisic acid-dependent stomatal closure. J. Exp. Bot. 61: 2615–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshem Y., Melamed-Book N., Cagnac O., Ronen G., Nishri Y., Solomon M., Cohen G., Levine A. (2006). Suppression of Arabidopsis vesicle-SNARE expression inhibited fusion of H2O2-containing vesicles with tonoplast and increased salt tolerance. Proc. Natl. Acad. Sci. USA 103: 18008–18013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyman B., Geelen D., Blatt M.R. (2000). Localization and control of expression of Nt-Syr1, a tobacco SNARE protein. Plant J. 24: 369–381. [DOI] [PubMed] [Google Scholar]
- Leyman B., Geelen D., Quintero F.J., Blatt M.R. (1999). A tobacco syntaxin with a role in hormonal control of guard cell ion channels. Science 283: 537–540. [DOI] [PubMed] [Google Scholar]
- Li J.F., Park E., von Arnim A.G., Nebenführ A. (2009). The FAST technique: a simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka V., Kwon C., Panstruga R. (2007). SNARE-ware: the role of SNARE-domain proteins in plant biology. Annu. Rev. Cell Dev. Biol. 23: 147–174. [DOI] [PubMed] [Google Scholar]
- Lvov A., Chikvashvili D., Michaelevski I., Lotan I. (2008). VAMP2 interacts directly with the N terminus of Kv2.1 to enhance channel inactivation. Pflugers Arch. 456: 1121–1136. [DOI] [PubMed] [Google Scholar]
- Ma C., Su L., Seven A.B., Xu Y., Rizo J. (2013). Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science 339: 421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.F., Tamai K., Yamaji N., Mitani N., Konishi S., Katsuhara M., Ishiguro M., Murata Y., Yano M. (2006). A silicon transporter in rice. Nature 440: 688–691. [DOI] [PubMed] [Google Scholar]
- Marquardt D. (1963). An algorithm for least-squares estimation of nonlinear parameters. J. Soc. Ind. Appl. Math. 11: 431–441. [Google Scholar]
- Martinez-Arca S., Rudge R., Vacca M., Raposo G., Camonis J., Proux-Gillardeaux V., Daviet L., Formstecher E., Hamburger A., Filippini F., D’Esposito M., Galli T. (2003). A dual mechanism controlling the localization and function of exocytic v-SNAREs. Proc. Natl. Acad. Sci. USA 100: 9011–9016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell J.C. (1868). On governors. Proc. R. Soc. Lond. 16: 270–283. [Google Scholar]
- Meharg A.A., Maurousset L., Blatt M.R. (1994). Cable correction of membrane currents recorded from root hairs of Arabidopsis thaliana L. J. Exp. Bot. 45: 1–6. [Google Scholar]
- Morita, M.T., Kato, T., Nagafusa, K., Saito, C., Ueda, T., Nakano, A., and Tasaka, M. (2002). Involvement of the vacuoles of the endodermis in the early process of shoot gravitropism in Arabidopsis. Plant Cell 14: 47–56. [DOI] [PMC free article] [PubMed]
- Murray D.H., Tamm L.K. (2009). Clustering of syntaxin-1A in model membranes is modulated by phosphatidylinositol 4,5-bisphosphate and cholesterol. Biochemistry 48: 4617–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Medkova M., Dong G., Hutagalung A., Reinisch K., Grosshans B. (2006). Interactions between Rabs, tethers, SNAREs and their regulators in exocytosis. Biochem. Soc. Trans. 34: 683–686. [DOI] [PubMed] [Google Scholar]
- Obrdlik P., et al. (2004). K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl. Acad. Sci. USA 101: 12242–12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli R., Sutter J.U., Blatt M.R. (2004). A new catch in the SNARE. Trends Plant Sci. 9: 187–195. [DOI] [PubMed] [Google Scholar]
- Press W., Flannerly B., Teukolsky S., Vetterling W. (1992). Numerical Recipes: The Art of Scientific Computing. (Cambridge, UK: Cambridge University Press; ). [Google Scholar]
- Rossi V., Picco R., Vacca M., D’Esposito M., D’Urso M., Galli T., Filippini F. (2004). VAMP subfamilies identified by specific R-SNARE motifs. Biol. Cell 96: 251–256. [DOI] [PubMed] [Google Scholar]
- Sanderfoot A. (2007). Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants. Plant Physiol. 144: 6–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber J.J., Willig K.I., Kutzner C., Gerding-Reimers C., Harke B., Donnert G., Rammner B., Eggeling C., Hell S.W., Grubmüller H., Lang T. (2007). Anatomy and dynamics of a supramolecular membrane protein cluster. Science 317: 1072–1076. [DOI] [PubMed] [Google Scholar]
- Sokolovski S., Hills A., Gay R.A., Blatt M.R. (2008). Functional interaction of the SNARE protein NtSyp121 in Ca2+ channel gating, Ca2+ transients and ABA signalling of stomatal guard cells. Mol. Plant 1: 347–358. [DOI] [PubMed] [Google Scholar]
- Sottocornola B., Visconti S., Orsi S., Gazzarrini S., Giacometti S., Olivari C., Camoni L., Aducci P., Marra M., Abenavoli A., Thiel G., Moroni A. (2006). The potassium channel KAT1 is activated by plant and animal 14-3-3 proteins. J. Biol. Chem. 281: 35735–35741. [DOI] [PubMed] [Google Scholar]
- Steegmaier M., Lee K.C., Prekeris R., Scheller R.H. (2000). SNARE protein trafficking in polarized MDCK cells. Traffic 1: 553–560. [DOI] [PubMed] [Google Scholar]
- Sutter J.U., Campanoni P., Tyrrell M., Blatt M.R. (2006). Selective mobility and sensitivity to SNAREs is exhibited by the Arabidopsis KAT1 K+ channel at the plasma membrane. Plant Cell 18: 935–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter J.U., Sieben C., Hartel A., Eisenach C., Thiel G., Blatt M.R. (2007). Abscisic acid triggers the endocytosis of the arabidopsis KAT1 K+ channel and its recycling to the plasma membrane. Curr. Biol. 17: 1396–1402. [DOI] [PubMed] [Google Scholar]
- Tsuk S., Lvov A., Michaelevski I., Chikvashvili D., Lotan I. (2008). Formation of the full SNARE complex eliminates interactions of its individual protein components with the Kv2.1 channel. Biochemistry 47: 8342–8349. [DOI] [PubMed] [Google Scholar]
- Tsuk S., Michaelevski I., Bentley G.N., Joho R.H., Chikvashvili D., Lotan I. (2005). Kv2.1 channel activation and inactivation is influenced by physical interactions of both syntaxin 1A and the syntaxin 1A/soluble N-ethylmaleimide-sensitive factor-25 (t-SNARE) complex with the C terminus of the channel. Mol. Pharmacol. 67: 480–488. [DOI] [PubMed] [Google Scholar]
- Tyrrell M., Campanoni P., Sutter J.U., Pratelli R., Paneque M., Sokolovski S., Blatt M.R. (2007). Selective targeting of plasma membrane and tonoplast traffic by inhibitory (dominant-negative) SNARE fragments. Plant J. 51: 1099–1115. [DOI] [PubMed] [Google Scholar]
- Ueda T., Uemura T., Sato M.H., Nakano A. (2004). Functional differentiation of endosomes in Arabidopsis cells. Plant J. 40: 783–789. [DOI] [PubMed] [Google Scholar]
- Uemura T., Sato M.H., Takeyasu K. (2005). The longin domain regulates subcellular targeting of VAMP7 in Arabidopsis thaliana. FEBS Lett. 579: 2842–2846. [DOI] [PubMed] [Google Scholar]
- Uemura T., Ueda T., Ohniwa R.L., Nakano A., Takeyasu K., Sato M.H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29: 49–65. [DOI] [PubMed] [Google Scholar]
- Vivona S., Liu C.W., Strop P., Rossi V., Filippini F., Brunger A.T. (2010). The longin SNARE VAMP7/TI-VAMP adopts a closed conformation. J. Biol. Chem. 285: 17965–17973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., Li, H.D., Chen, L.Q., Wang, Y., Liu, L.L., He, L., and Wu, W.H. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360. [DOI] [PubMed]
- Yano D., Sato M., Saito C., Sato M.H., Morita M.T., Tasaka M. (2003). A SNARE complex containing SGR3/AtVAM3 and ZIG/VTI11 in gravity-sensing cells is important for Arabidopsis shoot gravitropism. Proc. Natl. Acad. Sci. USA 100: 8589–8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun H.S., Kwaaitaal M., Kato N., Yi C., Park S., Sato M.H., Schulze-Lefert P., Kwon C. (2013). Requirement of vesicle-associated membrane protein 721 and 722 for sustained growth during immune responses in Arabidopsis. Mol. Cells 35: 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zárský V., Cvrcková F., Potocký M., Hála M. (2009). Exocytosis and cell polarity in plants - exocyst and recycling domains. New Phytol. 183: 255–272. [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhang H., Liu P., Hao H., Jin J.B., Lin J. (2011). Arabidopsis R-SNARE proteins VAMP721 and VAMP722 are required for cell plate formation. PLoS ONE 6: e26129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Feechan A., Pedersen C., Newman M.A., Qiu J.L., Olesen K.L., Thordal-Christensen H. (2007). A SNARE-protein has opposing functions in penetration resistance and defence signalling pathways. Plant J. 49: 302–312. [DOI] [PubMed] [Google Scholar]
- Zhu J., Gong Z., Zhang C., Song C.P., Damsz B., Inan G., Koiwa H., Zhu J.K., Hasegawa P.M., Bressan R.A. (2002). OSM1/SYP61: a syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. Plant Cell 14: 3009–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.