Abstract
Metastatic dissemination drives the high mortality associated with melanoma. However, limitations in visualizing in vivo cell dynamics during metastatic invasion have limited our understanding of these cell behaviors. Recent evidence has revealed that melanoma cells exploit portions of their ancestral embryonic neural crest emigration program to facilitate invasion. What remains to be determined is how embryonic microenvironmental signals influence invasive melanoma cell behavior, and whether these signals are relevant to human disease. To address these questions, we interrogated the role of the neural crest microenvironment in dictating the spatio-temporal pattern of melanoma cell invasion in the chick embryo using 2-photon time-lapse microscopy. Results reveal that both permissive and inhibitory neural crest microenvironmental signals regulate the timing and direction of melanoma invasion to coincide with the neural crest migration pattern. These cues include bi-directional signaling mediated through the ephrin family of receptor tyrosine kinases. We demonstrate that EphB6 re-expression forces metastatic melanoma cells to deviate from the canonical migration pattern observed in the chick embryo transplant model. Furthermore, EphB6-expressing melanoma cells display significantly reduced metastatic potential in a chorioallantoic membrane (CAM) metastasis assay. These data on melanoma invasion in the embryonic neural crest and CAM microenvironments identify EphB6 as a metastasis suppressor in melanoma, likely acting at the stage of intravasation.
Keywords: melanoma, metastasis, neural crest, EphB6, cell migration, chick embryo, in vivo model
INTRODUCTION
The vast majority of all cancer-related deaths can be ascribed to metastasis. With tumor cell-microenvironment interactions at the forefront of metastatic disease progression, insufficient attention has been given to the role of the microenvironment in regulating cell migratory behaviors. This is primarily due to the inherent challenges associated with studying migratory behaviors throughout the metastatic cascade in vivo (1–3).
The embryonic neural crest offers a unique model system in which to study cell-microenvironment interactions in vivo. The neural crest is a highly invasive, multi-potent cell population that displays a regulated spatio-temporal migratory pattern in the vertebrate embryo (4). Neural crest cells emigrate from the dorsal neural tube in a rostral-to-caudal manner all along the vertebrate axis, including from rhombomere segments (r1–r7) of the hindbrain. During emigration, the neural crest follow stereotypical migratory pathways that are thought to be sculpted by both intrinsic and extrinsic guidance cues found within the neural tube and surrounding microenvironment (5, 6). Importantly, several tumor types, including melanoma and neuroblastoma, originate from neural crest-derived cells. This has led to the hypothesis that neural crest-related malignancies may be intrinsically predisposed to increased metastatic potential due to the inherent invasive abilities of the neural crest (7–9).
Our laboratory has developed a model system using the chick embryo to help overcome the significant roadblocks to studying the cellular and molecular dynamics of melanoma metastasis in vivo (10). This model system takes advantage of the accessibility of the embryonic microenvironment to in vivo imaging and molecular intervention, allowing us to directly investigate how melanoma cells respond to microenvironmental signals. We and others have shown that metastatic melanoma cells transplanted into the chick neural crest embryonic microenvironment migrate along stereotypic neural crest migratory pathways (7, 10–12). However, the mechanisms guiding their migration are not known. To address this, we recently performed a molecular analysis comparing transplanted melanoma cells and the neural crest, which revealed that metastatic melanoma cells revive portions of the embryonic neural crest emigration program (7). Thus, metastatic melanoma cells appear to hijack inherent neural crest-related developmental signaling pathways to enhance their metastatic potential. However, what remains unclear is how the embryonic microenvironment dictates melanoma cell migratory behavior. Specifically, what are the embryonic signals that guide melanoma migration, and can perturbation of those signals significantly alter migratory behavior?
Here, we asked to what extent the chick embryonic neural crest microenvironment regulates the timing and migratory patterning of transplanted melanoma cells. We also asked to what extent we could alter the migratory phenotype by perturbing cell-microenvironment interactions. We compared the invasion patterns of melanoma cells transplanted into the chick hindbrain at various developmental stages and axial positions. Single melanoma cell dynamics were observed in vivo using 2-photon microscopy. To perturb cell-microenvironment interactions, we examined how changes in Eph expression in transplanted melanoma cells affected cell invasion patterns. Lastly, to address the relevance of our studies to human disease, we assayed the tumorigenicity and metastatic potential of melanoma cells transplanted onto the highly vascularized chick chorioallantoic membrane (CAM). Our results highlight the importance of tumor cell-microenvironment interactions in promoting, inhibiting, and guiding tumor cell movements, and elucidate the anti-metastatic properties of EphB6 in vivo.
METHODS
Cell Culture
Cell lines were kindly provided by Dr. Mary Hendrix, Children’s Memorial Research Center, Chicago, IL. Cells were maintained in RPMI supplemented with 10% fetal bovine serum. For drop culture conditions, cells were resuspended in RPMI plus 10% fetal bovine serum at a concentration of 4e6 cells per ml. A 20μl drop was hung from the lid of a 35mm culture dish and incubated for 40 hours under standard culture conditions.
EphB6 Cloning, Lentiviral Infection and Western blot
The vector containing wildtype human EphB6 was kindly provided by Dr. Andrew Freywald, University of Saskatchewan, Saskatoon, SK, Canada. From this vector, EphB6 was PCR amplified and inserted into the Nhe1 and BamH1 sites of the lentiviral vector pCDH-CMV-MCS-EF1-RFP (System Biosciences) using the following PCR amplification primers: forward – 5′-AGTCGCTAGCATGGTGTGTAGCCTATGGGTGC-3′; reverse – 5′-GACTGGATCCTCAGACCTCCACTGAGCCCT-3′. Lentiviral particles were prepared using the ViraPower Lentiviral Packaging mix, Lipofectamine 2000, and the 293FT cell line according to the manufacturer’s directions (Invitrogen). Supernatant was collected at 48 and 72 hours post-transfection, and was concentrated using PEG precipitation. Briefly, the supernatant was collected, spun at 3,000 rpm for 5 minutes and then filtered using a 0.45um filter. For the precipitation, 3 ml of filter-sterilized 40% PEG-8,000 in PBS and 180 ul FBS was added to each 9 ml of supernatant. The virus was allowed to precipitate for at least 72 hours prior to pelleting at 1,500g for 10 minutes. The supernatant was removed and the tube was spun down again at 1,500g for 1 minute to remove residual PEG solution. The viral pellet was resuspended in culture media by gently pipetting up and down, before being stored at −80°C prior to use. Supernatant was concentrated 1:500 to 1:1,000, resulting in viral titers as high as 108 titerable units (TU)/ml. For infection, C8161 cells were seeded into 24-well plates at 1×104 cells/well, and allowed to adhere overnight. The next day, the cells were infected at various MOIs in culture media in the presence of 6 ug/ml polybrene (Sigma-Aldrich) overnight, before undergoing a media change. The cells were then expanded prior to transplant experiments. EphB6 protein expression was verified by Western blot. Equal amounts of cellular protein were subjected to SDS-PAGE and Western blot analysis using an antibody specific for EphB6 (EphB6 monoclonal antibody M03, clone 5D8, Abnova H00002051-M03).
Chick Embryo Transplants
Fertile White Leghorn chick eggs were acquired from Centurion Poultry (Lexington, GA). Eggs were incubated at 38°C for approximately 39 hours until the 9-somite stage of development. Eggs were windowed and embryos were visualized following the injection of a solution of 10% India ink in Howard Ringer’s solution below the blastodisc. Embryos were staged according to the criteria of Hamburger and Hamilton (Hamburger and Hamilton, 1951), denoted as stage 10 (HH10), for example. Transplantation of the embryos was performed as previously described (Kulesa et al., 2006). Briefly, C8161 hanging drop cultures were cut with a finely pulled glass needle to approximately 50um3 (corresponding roughly to 300 cells). Tumor cell transplants were grafted into the lumen of the chick embryo neural tube through a slit along the dorsal midline. Eggs were sealed with cellophane tape and incubated for 24hr.
Static Imaging
Individual embryos were removed from eggs with paper rings, rinsed with Ringer’s solution and placed dorsal side up within a thin ring of high-vacuum grease (Dow Corning) on 22×75mm microslides. Embryos were imaged using a Zeiss 710 multi-photon upright microscope. 2-photon z-stacks of dual labeled C8161 Gap43:CFP::H2B:YFP were acquired at a wavelength of 850nm. RFP-labeled EphB6+ C8161 cells were imaged with an excitation of 561nm. Images using a W Plan-Apochromat 20x/1.0 DIC objective (Zeiss).
Photoconversion
C8161 cells were dual labeled with H2B:mCherry and H2B:PSCFP2 (photo-switchable cyan fluorescent protein) by lentiviral infection. Cells were transplanted into the chick embryo neural tube as described above. A Teflon membrane window was installed in the egg as previously described (Kulesa et al., 2010a). H2B:PSCFP2-labeled cells were photoconverted within 1 hour following transplantation. Photoconversion was performed on a Zeiss 710 multi-photon upright microscope using 2-photon excitation at 780nm and a W Plan-Apochromat 20x/1.0 DIC objective (Zeiss).
Time-lapse Imaging
The time-lapse imaging platform, including sample preparation, is detailed in a previously published protocol (Kulesa et al., 2010a). Briefly, Teflon windows were were placed over the developing chick embryo and sealed with beeswax. The egg was maintained in an environmental box placed around a Zeiss 710 multi-photon upright microscope stage and heated to 38°C. The chamber was humidified with a sponge placed in a dish of water. Non-evaporating immersion liquid (Immersol W, Carl Zeiss) was used to bridge the W Plan-Apochromat 20x/1.0 DIC objective (Zeiss) with the Teflon membrane. Simultaneous 2-photon excitation of the dual labeled C8161 Gap43:CFP::H2B:YFP cells was achieved at a wavelength of 850nm. Z-stacks with a 10um z-slice were acquired every 7 minutes for 18 hours.
Chorioallantoic Membrane Metastasis Assay
Fertile White Leghorn chick eggs were incubated for 7hr, following which an artificial air sac was created in the egg by removing 4ml albumin from the egg with an 18-guage needle. The puncture hole was sealed with cellophane tape and the egg was further incubated for 7 days. At the time of transplantation, a small section (2×2mm) of the surface periderm of the CAM was removed with filter paper (Whatman) in a manner that did not compromise the integrity of the basal layer. Disruption of the periderm is required for tumor cells to gain access to the underlying mesoderm (Armstrong et al., 1982). Following periderm removal, eggs were allowed to recover for 2 hours prior to placing cells on the CAM. For transplantation, cells were suspended at a concentration of 1e5 cells/ul. 1e6 cells (10ul) were placed on the CAM. The egg was re-sealed with cellophane tape and incubated for 48hr. Following this incubation, the egg was bisected along the longitudinal axis, and the CAM from the lower half of the egg was placed in a 15ml conical tube and flash frozen in liquid nitrogen. The frozen tissue was then ground with a chilled mortar and pestle in the presence of liquid nitrogen. Genomic DNA was extracted from the ground tissue using the Wizard Genomic DNA Purification kit (Promega) according to the manufacturer’s protocol. Resulting DNA was quantified using a Nanodrop spectrophotometer.
qPCR
Human DNA was detected and quantified using Alu (YB8 subfamily) element-based polymerase chain reaction (Walker et al., 2003). qPCR was performed on an ABI7500 (Applied Biosystems Inc.) in optical 96-well reaction plates using the following 3-stage cycling protocol: 50°C for 2 minutes, 95°C for 10 minutes & 40 cycles of 95°C for 15 seconds and 68°C for 1 minute. The following Alu-specific primers and probe were utilized: forward – 5′-GTCAGGAGATCGAGACCATCCT-3′; reverse – 5′-AGTGGCGCAATCTCGGC-3′ and the following probe: 5′-6-FAM-AGCTACTCGGGAGGCTGAGGCAGGA-TAMRA-3′ as previously reported (van der Horst et al., 2004). Each 50μl qPCR reaction included 2ug genomic DNA, 25μl 2X Gene Expression Master Mix (Applied Biosystems Inc.), 25pmol forward primer, 25pmol reverse primer, and 12.5pmol probe. Resulting cycle threshold values (Ct) were compared to a serial dilution of human genomic DNA from C8161 cells, from which quantitative DNA amounts were extrapolated.
Data Analysis and Statistics, Image Processing
3D images acquired from 2-photon static and time-lapse imaging were analyzed with Imaris software (Bitplane Scientific Software). Analysis included spots detection cell tracking, Tracks translation, displacement vector analysis, and movie creation and editing. For the cylindrical coordinate conversion, Cartesian coordinates from Imaris were converted to cylindrical coordinates using standard mathematical methods. Densitometric scatterplots were created in MatLab. Histograms and curve-fitting analyses were created and performed using OriginPro software. Curve fitting was based on a Gaussian peak function, where Xc is the center of the peak and W equals 2 times the standard deviation of the Gaussian distribution (approximately 0.849 the width of the peak at half height). Statistical analysis was performed using Microsoft Excel and the Data Analysis Tools pack. For migratory distance comparisons, a single factor ANOVA was used to calculate the p-value. For the CAM metastasis assay, a 2-sample equal variance t-test with 2-tailed distribution was used. Statistically significant p-values are <0.05. Figure processing was performed with Adobe Photoshop CS3.
RESULTS
Melanoma cells transplanted into the chick embryonic neural crest microenvironment sense and respond to microenvironmental cues by following host cranial neural crest cell migratory pathways(7, 10). However, it remained unclear whether there was a temporal and/or spatial restriction to melanoma migration that corresponded to the developmental pattern of the host cranial neural crest.
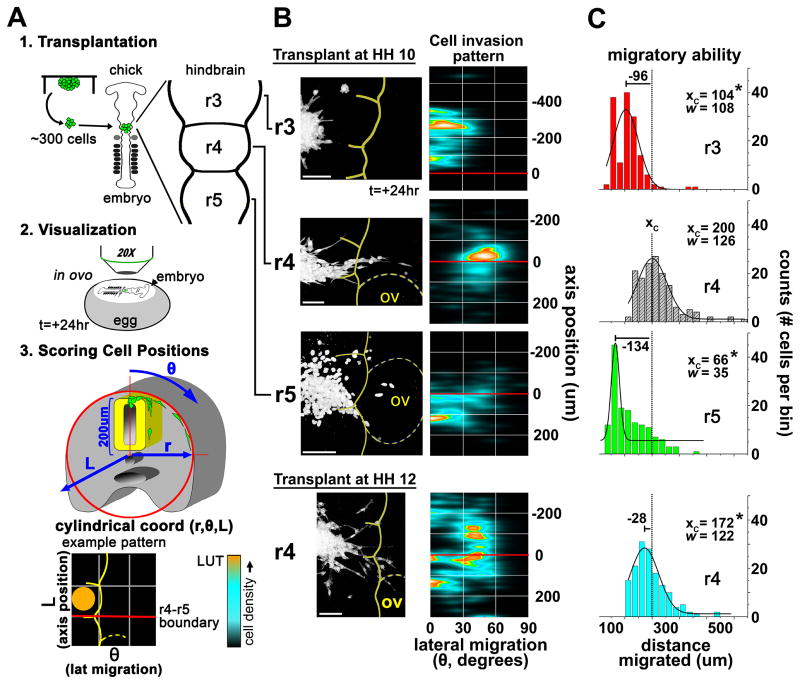
To examine this question, we transplanted human C8161 melanoma cells into different rhombomere segments at peak and off-peak times of cranial neural crest cell migration (Fig. 1). Cranial neural crest cells begin to migrate from r1 at Hamburger and Hamilton stage 8+ (HH8+), and migration typically ceases (from r7) by HH11+(13). We observed maximal invasion of C8161 cells when transplanted into r4 at HH10. Under these conditions, C8161 cells invaded the permissive area adjacent to r4, travelling as a discrete multi-cellular stream while respecting inhibitory cues present in and adjacent to r3 and r5 that are thought to help sculpt the neural crest migration pathway (Fig. 1B). To analyze migratory behaviors, positions of invasive cells from 9 different transplants were quantified using a cylindrical coordinate system (Fig. 1A, B). The embryonic dorsal midline and the r4–r5 rhombomere boundary were used as fiducial landmarks (Fig. 1A, B). The cell invasion pattern revealed a tight cluster of endpoint cell positions located along the r4 stream approximately 25um rostral to the r4–r5 boundary, demonstrating reproducible, guided directional migration (Fig. 1B). Following 24 hours, the average migratory distance was 200um, with some cells migrating nearly 600um (Fig. 1C).
Figure 1. Cytometric quantification of migratory behaviors of melanoma cells transplanted into varying regions and developmental stages of the chick embryonic hindbrain.
A) A cartoon depicting the experimental methods. Tumor cells are transplanted into various regions of the embryonic chick hindbrain and allowed to migrate for 24 hours. Cells are then imaged in high resolution and cell positions are quantified using a cylindrical coordinate system where length (L) represents the cell position along the rostral-caudal axis and theta (θ) represents the angle deviant from the vertical (dorsal-ventral) axis of the embryo and is a direct representation of lateral migratory distance. B) Cytometric quantification of transplanted cells in different regions of the hindbrain (r3, r4, r5) during active neural crest migration (HH10) and following the cessation of neural crest migration (HH12). The representative micrograph shows typical melanoma cell migration relative to the location and timing of the transplant. Meter bar=100um. The 30 most invasive cells from 9 transplants were used to populate the densitometric scatterplot that defines the Cell Invasion Pattern. The scatterplot uses embryonic landmarks as fiducal points representing the embryonic midline (y-axis) and the boundary between r4 and r5 (red line at axis position 0). C) A comparison of migratory ability (total cell displacement) is shown by the histogram and accompanying Gaussian fit. Xc is the center of the peak and represents the average X-value. W equals 2 times the standard deviation of the Gaussian distribution, or approximately 0.849 the width of the peak at half height. The dashed line in the histograms represents the Xc value of the HH10/r4 transplant. n=270 cells for each histogram. * p<0.05.
Importantly, guided directional migration was observed for other embryonic and neural crest-related cell types, including neuroblastoma, but not for the invasive breast cancer cell line MDA-MB-231 (Supplemental Figure 1). This suggests that embryonic neural crest signals are non-permissive to cells lacking the appropriate signaling mechanism.
Cranial neural crest streams are sculpted by both permissive and inhibitory signals present within the microenvironment, with inhibitory neural-crest-cell-free zones located adjacent to neural crest migratory streams (5, 14). As such, we investigated the extent to which inhibitory cues associated with r3 and r5 also regulate melanoma cell migration. For this, we transplanted C8161 cells into r3 and r5 and quantified their migratory behaviors. Cells transplanted into either r3 or r5 displayed a significant reduction in migration compared to cells transplanted into r4 (Fig. 1B, C). C8161 cells transplanted into r3 showed an average migratory distance of 104um, representing a 50% reduction in migration compared to cells transplanted into r4 (Fig. 1C). Cells transplanted into r5 displayed even greater inhibition, with an average migratory distance of only 66um. Those cells showing a propensity to migrate appeared as multi-cellular streams oriented rostrally or caudally toward adjacent neural crest migratory streams. This suggests that the microenvironments within and adjacent to r3 and r5 induce potent anti-migratory responses.
Cranial neural crest cells at the r4 axial level cease to emerge from the dorsal neural tube at approximately HH11+ (13). We asked whether age-related changes in the neural crest microenvironment would affect tumor cell behaviors in later-staged transplants. To test this, we transplanted C8161 cells into r4 at HH12 (Fig. 1B). We found that melanoma cells transplanted into r4 at later developmental stages spread radially from the transplant site, without respecting the neural crest cell-free zones (Fig. 1B). Furthermore, cells migrated independent of one another and did not appear to form discrete multi-cellular streams like those observed in cells transplanted at HH10 (Fig. 1B). Interestingly, with an average migratory distance of 172um, these cells also migrated less (based on total displacement) than cells transplanted at stage HH10 (Fig. 1C). From this, we conclude that guidance factors present in the embryonic microenvironment at HH10 do not appear to guide migration at later developmental stages.
The ability to migrate is cell-autonomous
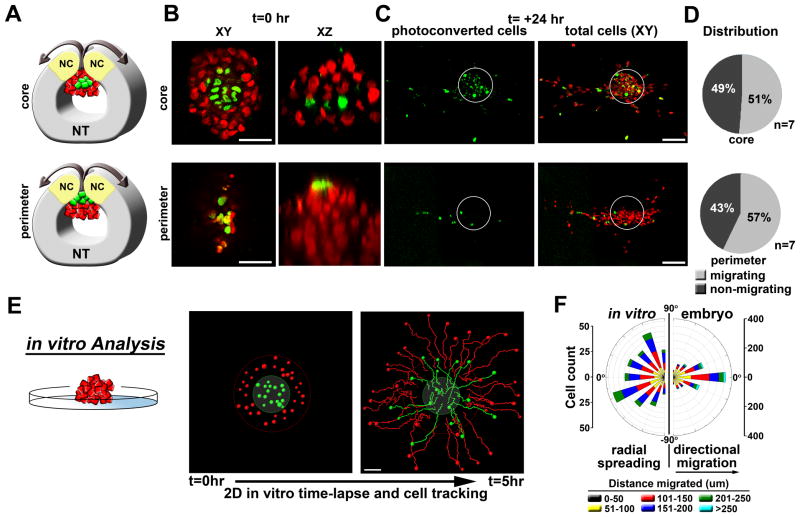
These results suggest that the embryonic neural crest microenvironment regulates melanoma migratory behaviors when transplantation is correlated with the developmental timing of cranial neural crest migration. We next asked whether the proximity of a melanoma cell to the microenvironment (neural tube lumen) influences melanoma cell invasion. To test this, we employed an in vivo selective cell labeling technique in which H2B-PSCFP2 (photoswitchable cyan fluorescent protein)-labeled C8161 cell clusters were transplanted into the embryo (r4, HH10). Using 2-photon microscopy, we photoconverted a small number of cells located either within the core or at the periphery of the transplant (Fig. 2A, B, Supplemental Video 1). Following 24 hours, photoconverted cells were scored as migratory or non-migratory. We found that migratory cells derived from either the periphery or core were similarly likely to be located in the leading population of migratory cells as those that started at the periphery (Fig. 2C, D). This contrasted with melanoma cell clusters placed on a basement membrane matrix in culture. In vitro, cells spread radially and without direction from the microenvironment, with the cells originating at the outer rim of the cluster remaining on the migratory front (Fig. 2E, F). Thus, the ability to sense and respond to migratory and directional cues within the embryonic neural crest microenvironment appears to be cell autonomous.
Figure 2. Migratory ability is cell-autonomous.
A) A cartoon depicting the 2-photon in ovo photoconversion of H2B:PSCFP2-labeled C8161 melanoma cells. Following transplantation into the chick embryo (r4,HH10), sub-populations of tumor cells in either the core or the perimeter of the graft were photoconverted with a 2-photon microscope using a wavelength of 780nm. B) Representative micrographs showing the photoconversion of tumor cells at t=0hr in either the core or the perimeter of the graft. XY and XZ views are provided. C) Representative micrographs showing the locations of migratory photoconverted tumor cells at t=24hr. The white circles outline the initial transplant graft location and show the non-migrating cells remaining in the neural tube. D) Pie charts depicting the percentage of photoconverted migratory verses non-migratory cells. Cells were scored as migrating or non-migrating based on their location within or without this circle. E) In vitro time-lapse analysis of a C8161 melanoma cell cluster placed on a basement membrane matrix. Cells were labeled based on position (red=periphery, green=core). Migratory tracks are shown at 5hr. F) A rose plot comparing cell directionality in vitro with directionality observed in the embryo. In the embryo, 90 degrees represents the anterior-posterior embryo axis. Cell positions were calculated for all migrating cells from 9 different transplants (r4,HH10, >1000 cells). Angles between cell trajectory and the horizontal r4 migratory stream were then determined. The size of each bar depicts the number of binned cells for a given angle. The colored segments depict the distance migrated by cells within the bin.
The embryonic neural crest microenvironment dictates migratory behaviors
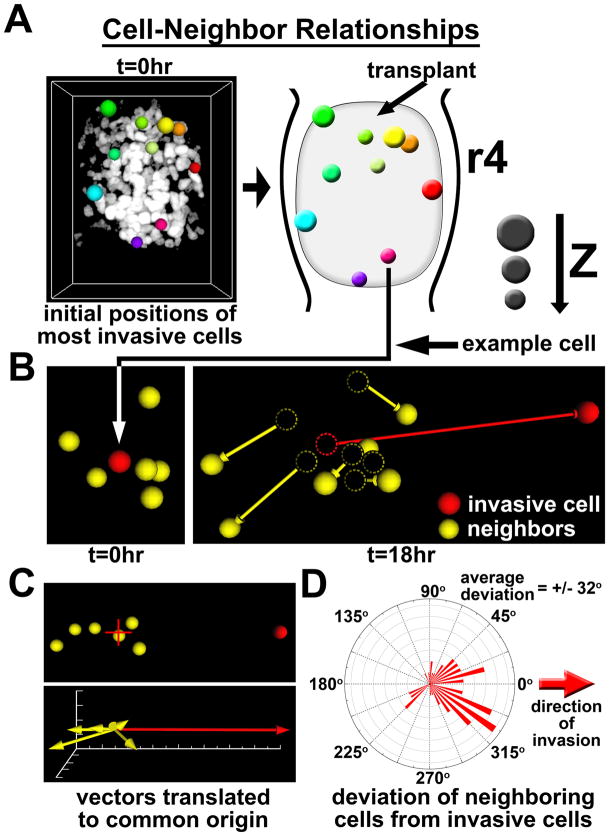
We next verified by in ovo 2-photon time-lapse microscopy that migratory cells unpredictably arose from anywhere within the transplant cluster, with no observed bias for any given region (Fig. 3A, Supplemental Video 2). Yet it remained unclear whether cell migrated independently or in association with neighboring cells. Thus, for each of the 10 most invasive cells identified in the transplant, we analyzed directional migration for the highly invasive cell’s 6 closest neighbors (Fig. 3B–D, Supplemental Video 3). This 3D analysis revealed that displacement vectors for neighboring cells rarely pointed in the same direction as the most invasive cell (Fig. 3C). We calculated the average deviation between the displacement vector of the invasive cell and those of its neighbors to be 32 degrees, indicating that neighboring cells do not tend to follow the direction of the highly migratory cells (Fig. 3D). We conclude that highly invasive tumor cells migrate independently of neighboring cells and do not adopt a follow-the-leader pattern of migration.
Figure 3. Invasive melanoma do not display follow-the-leader migratory behaviors.
A) Colored spheres represent the initial positions of the 10 most invasive cells (based on total displacement). z-depth can be gauged by the diameter of the sphere. Both the raw data image and a reference cartoon are provided. B) The migratory direction (displacement) of a representative highly invasive cell (red) was compared to the directions of its 6 closest neighboring cells (yellow). Initial positions are viewed at t=0hr. Final positions, including displacement vectors, are shown at t=18hr. The dashed circles represent the initial locations of each cell. C) Displacement vectors for all of the cells shown in section “D” were translated to a common origin and compared for changes in direction. The red + marks the origin. D) The deviation angles between the displacement vectors of the 10 most invasive cells and each of their six closest neighbors was calculated and graphed on a roseplot histogram. The red arrow indicates the direction of the highly invasive cells. The average deviation was +/− 32 degrees.
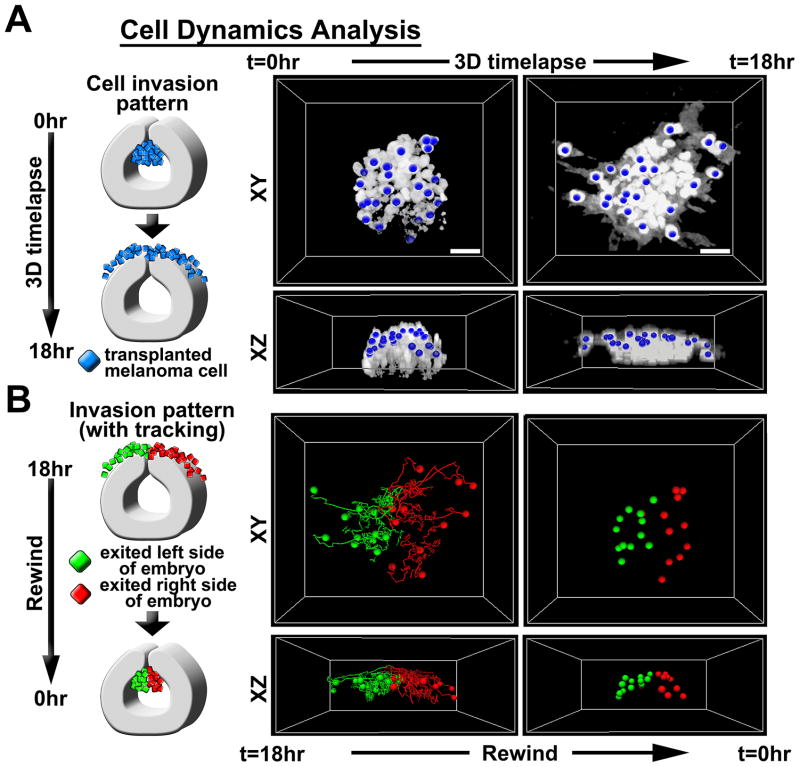
We then investigated whether the location of a cell within the cluster at the time of transplantation could predict the timing and trajectory of its invasion pattern (Fig. 4A–B, Supplemental Video 4). We pseudo-colored invasive melanoma cells at endpoint times based on whether the cell had migrated to the left or to the right of the embryo anterior-posterior axis along the dorsal midline. We then rewound the time-lapse to reveal the cell’s initial position (Fig. 4A–B). We observed that cells that had migrated to the right side of the embryo originated from the right half of the transplant cluster and vice versa, mimicking the host embryonic neural crest dispersion pattern. This suggested that transplanted melanoma cells were sorted by microenvironmental signals at the dorsal midline.
Figure 4. Invasive melanoma cells are guided by the embryonic neural crest microenvironment.
A) 2-photon time-lapse cell tracking was used to determine the initial locations of migratory tumor cells. 3D time-lapse images were acquired in ovo using 2-photon microscopy. Images were analyzed using Imaris software to track cells over time. B) Cells were labeled at t=18hr based on whether they had migrated to the left or right of the embryonic dorsal midline. Cells on the left were labeled green and cells on the right were labeled red. Cell positions were then tracked backwards to identify the initial position at t=0hr. Both XY and XZ views are provided
EphB6 re-expression in melanoma cells alters migratory behavior
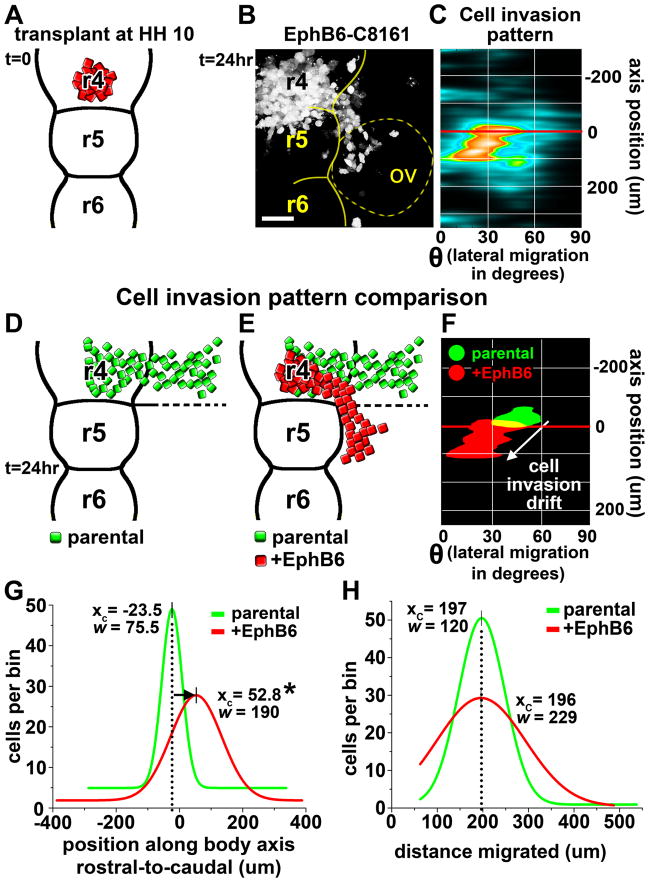
What embryonic neural crest microenvironmental factors are responsible for directing melanoma cell migratory behaviors? As Eph/ephrin signaling is known to direct embryonic neural crest migration (15–17), we sought to identify novel migratory phenotypes resulting from altered Eph expression. To accomplish this, we re-expressed EphB6, a kinase-defective Eph receptor, in C8161 melanoma cells. EphB6 protein expression was confirmed by Western blot (Supplemental Figure 2). We previously demonstrated that EphB6 gene expression was silenced in C8161 cells, but present in both primary melanocytes and poorly invasive C81–61 melanoma cells (7). Furthermore, EphB6 has been shown to dimerize with, and modulate the activities of, several other Eph receptors. These include EphB1, EphB2, EphB4, and EphA2, all of which are highly expressed in C8161 cells (7, 18–20). As such, we hypothesized that EphB6 re-expression would have significant effects on the interaction between transplanted C8161 cells and the chick microenvironment. To test this, we transplanted EphB6+ C8161 cells into the embryo (r4, HH10) and quantified the resulting cell invasion pattern (Fig. 5). EphB6+ C8161 cells initially exited the neural tube at the proper location rostral to the r4–r5 boundary. However, after encountering the microenvironment adjacent to r4, EphB6+ cells altered their trajectories away from the area adjacent to r4 and toward the area adjacent r5 (Fig. 5A–F). This resulted in a caudal shift in the mean position of migrating cells (approximately 75um) along the rostral-caudal axis (Fig. 5F, G). Importantly, the EphB6+ cells maintained the discrete multi-cellular streaming behavior observed in transplanted parental C8161 cells, suggesting that cell-cell interactions among the melanoma cells was not altered. Also, the overall distance that EphB6+ C8161 cells invaded into the embryonic neural crest microenvironment was unaffected (Fig. 5H).
Figure 5. Re-expression of EphB6 in C8161 melanoma cells results in aberrant directional migration without affecting migratory ability.
A) A cartoon depicting the transplant of EphB6+ C8161 melanoma cells into r4 at HH10. B) A representative micrograph showing migratory behaviors of transplanted EphB6+ C8161 cells. The neural tube boundary is outlined in yellow and the position of the otic vesicle (ov) is shown by the dotted yell line. Individual rhombomeres are also labeled. C) A densitometric scatterplot of the cell invasion pattern of the positions of the 30 most invasive cells from 10 transplant experiments. The y-axis represents the embryo midline. The x-axis represents lateral migration and is given in degrees (cylindrical coordinates). The r4/r5 boundary is depicted by the red line. D–E) Cartoons depicting the invasion patterns of parental C8161 cells (from Figure 1) and EphB6+ C8161 cells. F) An overlay of the cell invasion pattern from parental C8161 cells (green) and EphB6+ C8161 cells (red), highlighting the caudal shift in cell positions at t=24hr. G) The Gaussian fit to a histogram comparing cell positions of parental C8161 cells and EphB6+ C8161 cells along the rostral-caudal axis at t=24hr. H) The Gaussian fit to a histogram comparing total distance migrated (total displacement) between parental C8161 cells and EphB6+ C8161 cells. Xc is the center of the peak and represents the average X-value. W equals 2 times the standard deviation of the Gaussian distribution, or approximately 0.849 the width of the peak at half height. * p<0.05.
EphB6 is a metastasis suppressor in melanoma
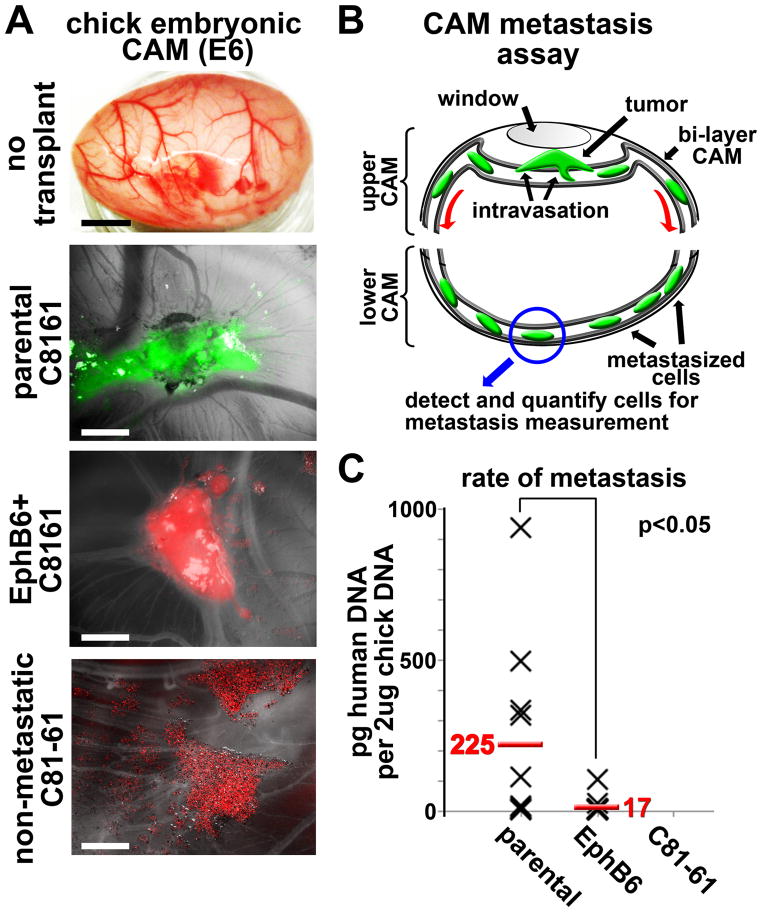
Following this observation, we investigated whether the altered cell directionality observed in the embryo reflects a change in tumorigenicity or metastatic potential (Fig. 6). Parental C8161 cells or EphB6+ C8161 cells were placed onto the highly vascularized CAM of an E10 chick embryo and incubated for 48 hours. We observed that EphB6+ C8161 cells maintained their ability to form tumors on the CAM in a similar manner to that observed with parental C8161 cells (Fig. 6A). The isogenic but poorly aggressive melanoma cell line C81–61 did not form tumors when placed on the CAM, but rather remained as a monolayer of dispersed cells (Fig. 6A).
Figure 6. Re-expression of EphB6 in C8161 melanoma cells causes a significant loss of metastatic potential but does not affect tumorigenicity in a chorioallantoic membrane (CAM) metastasis assay.
A) Images of a native chick embryo CAM and representative tumors from parental C8161 cells (green) and EphB6+ C8161 cells (red) grown on the CAM. 1e6 cells in suspension (10ul volume) were dropped onto the CAM at day E10. Tumor formation occurred over 48 hours. Non-metastatic C81–61 cells did not form tumors. The black meter bar represents 1cm and the white meter bars represent 2.5mm. B) A cartoon depicting the CAM metastasis assay. Tumor cells are placed onto the upper CAM through a window in the egg shell. The egg is resealed for 48 hours. The lower CAM is removed and genomic DNA is harvested from the tissue. Metastatic human cells are detected by qPCR using primers that amplify a human-specific Alu element. C) A scatterplot showing detected amounts of human DNA per 2ug chick CAM DNA. Each X represents one biological replicate. A t-test was used to calculate statistical significance.
To test whether EphB6 re-expression directly affects metastasis, we tested the ability of tumor cells to invade into the host vasculature. To assay intravasation, the bottom half of the CAM was removed 48 hours post-transplantation and assayed for the presence of human DNA. Due to physical and temporal constraints, tumor cells must intravasate into the blood or lymph circulation in order to populate the bottom half of the CAM within 48 hours (21, 22). PCR amplification of a human-specific Alu repeat sequence allowed us to quantify the presence of metastatic human cells (23, 24). We evaluated 16 replicates from both parental C8161 cells and EphB6+ C8161 cells. We observed that 50% of the parental C8161 samples showed high metastatic ability, with maximal rates observed at more than 900pg human DNA per 2ug chick DNA (Fig. 6C). Parental C8161 cells showed an average of 225pg human DNA per sample over 48 hours (corresponding roughly to 45 cells per sample, based on 1 cell = 5pg DNA, Fig. 6C). In contrast, EphB6+ C8161 cells displayed a marked reduction in metastatic ability (17pg per sample, or 3.4 cells). These results support the hypothesis that EphB6 acts as a metastasis suppressor in melanoma cells and suggest intravasation as the step likely blocked by EphB6.
DISCUSSION
In the present study, we sought to employ an embryonic model system to examine the role of the microenvironment in promoting and directing metastatic invasion in vivo. Using a dynamic in vivo imaging platform to assess single cell behaviors, we discovered 3 main results. First, embryonic neural crest microenvironmental signals regulate the timing, trajectory, and order of metastatic melanoma dissemination after transplantation into the chick hindbrain. Second, cues within the chick neural tube and surrounding microenvironment regulated the migratory behaviors of invasive melanoma cells, but we could not predict, based on location, which cells would respond in an invasive manner to those cues. Third, re-expression of a kinase-defective Eph receptor, EphB6, in melanoma cells induced aberrant directional tumor cell migration from the hindbrain and loss of metastatic potential within the chick CAM vascular bed. Our results are the first comprehensive details of in vivo human melanoma cell dynamics at the single cell level within the embryonic neural crest and vascularized CAM microenvironments.
Our work approaches discerning just how susceptible tumor cells are to microenvironmental migration cues and whether an understanding of those cues might help predict metastatic behaviors. The early embryonic hindbrain utilizes a complex set of signaling mechanisms to control and guide the invasion of the neural crest (5). These same signals appear to be sufficient to regulate melanoma invasion microenvironment in both time and space, including directional and non-permissive cues. Melanoma cells transplanted at a developmental stage after neural crest cells have emigrated from the hindbrain, or into odd-numbered rhombomere hindbrain segments (that typically support fewer migrating neural crest cells) fail to replicate the behaviors of cells transplanted into r4 at stage HH10 (Fig. 1). These observations demonstrate that confined microenvironments can directly regulate tumor cell migratory behaviors such as migratory capacity and directionality, and suggest that an understanding of these signals may provide insight into the metastatic abilities of tumor cells.
The establishment of non-permissive tissue boundaries is critical for maintaining discrete neural crest migratory routes. What are these cues and can we use them to obstruct the metastatic process? The neural crest provides a superb model system in which to study cell-microenvironment exchanges during cell invasion.
One key question was whether the microenvironment dictated which cells would migrate, as the majority of transplanted tumor cells remain in the neural tube 24 hours post-transplantation. If the microenvironment was responsible for inducing the migratory phenotype, we would expect to observe a reduction in migratory efficiency in cells at the core of the transplant that are insulated from direct contact with the embryo. Likewise, if all cells responded to the microenvironment equally, we would expect cells at the perimeter to migrate farther simply because they could more rapidly delaminate from the tumor graft (as was observed in vitro). However, we were able to show that migratory cells originated from the core of the transplant at nearly the same rate as cells located on the periphery, and that these core cells could be found at the leading front of invasion (Fig. 2). This suggests that heterogeneities among the transplanted cells enabled some cells to recognize the proper migratory cues present in the microenvironment, independent of cell position.
Dynamic in vivo 2-photon time-lapse microscopy provided further insights into how the embryonic microenvironment regulates tumor cell migration. First, we observed that transplanted cells are compliant to signals establishing a dorsal midline boundary (Fig. 4). This same midline segregation is observed during neural crest migration and is thought to be induced by direct cell guidance mechanisms, including Slit-Robo signaling (25). We postulate that a similar guidance mechanism segregates migrating tumor cells. Also, cells do not maintain strict neighbor relationships, but act independently in responding to microenvironmental signaling cues (Fig. 3). We conclude that the ability to migrate is cell-autonomous while specific migratory behaviors are dictated by the microenvironment.
Several guidance mechanisms are used by the embryo to sculpt discrete neural crest migratory streams and direct cells to precise destinations (26). Among these mechanisms, Eph/ephrin signaling has been well described in guiding neural crest migration in both the hindbrain and the trunk (15–17, 27). In the hindbrain, distinct combinations of Ephs and ephrins are thought to help establish boundaries that sculpt neural crest streams en route to their respective branchial arches.
Aberrant Eph/ephrin signaling has also been reported to mediate cancer metastasis (28). In the current study, we show that EphB6 re-expression altered tumor cell invasion directionality and significantly reduced metastatic potential (Fig 6). EphB6 has been identified as reduced or absent in several metastatic solid tumor types, including breast cancer, lung cancer, and melanoma (29–31). As Eph/ephrin signaling is commonly associated with cell-cell repulsion, it is tempting to speculate that the ephrin signature expressed by the host neural crest or the underlying mesenchymal cells induces an inhibitory response in EphB6+ C8161 cells. This would act to divert the melanoma cells away from the r4 migratory route. This is supported by work from Matsuoka and colleagues which demonstrates that EphB6 is repelled by high concentrations of the ligand ephrin-B2, which is thought to be expressed by migrating cranial neural crest cells (17, 32). However, it has also been suggested that varying concentrations of EphB6 may induce a switch from repulsion to attraction (32). Indeed, ephrin-B1 has been shown to be expressed by non-neural-crest cells lining neural crest migratory boundaries in the chick hindbrain, including areas around the otic vesicle (17). Thus, it is possible that typically-repulsive ephrin-B1 signals have become attractive to EphB6+ C8161 cells.
Importantly, the reduction in metastatic potential observed in EphB6+ cells did not likely result from a migratory phenotype since EphB6+ C8161 cells traveled a similar distance as parental cells in our transplant model (Fig. 5). As such, we hypothesize that EphB6 may function to perturb the process of intravasation, since our CAM assay uniquely assesses intravasation potential by specifically identifying and quantifying tumor cells that were able to intravasate into the CAM vasculature (Fig. 6 and Fig. 7-model). Specifically, we postulate that EphB6 may function to regulate the tumor cell – endothelial cell interaction, possibly receiving a repulsive signal following engagement with ephrins expressed on endothelial cells. Endothelial cells have been reported to express multiple Ephs and ephrins, including ephrin-B2, which has been shown to have a repulsive effect on EphB6 (32, 33). Eph/ephrin signaling has also recently been reported to play a role in the transendothelial migration of some immune cells (33, 34). Thus, we hypothesize that the endothelium becomes an impenetrable barrier to cells expressing a specific Eph/ephrin signature that includes EphB6, while the absence of EphB6 may facilitate passage across the endothelium (Fig. 7-model).
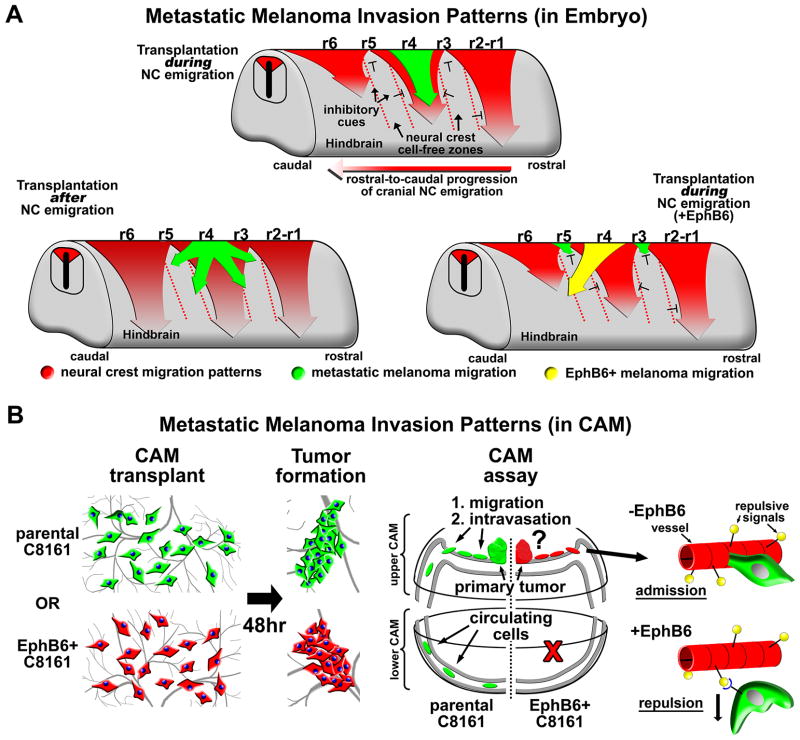
Figure 7. A model comparison of metastatic melanoma invasion patterns in the embryonic hindbrain and on the vascularized embryonic chorioallantoic membrane (CAM).
A) Three different cartoons comparing metastatic melanoma invasion patterns from different rhombomeres in the hindbrain both during and after neural crest migration. The embryonic hindbrain directs the formation of discrete neural crest migratory pathways emerging from specific hindbrain segments called rhombomeres, listed as r1 through r6. Neural crest migration proceeds in a rostral-to-caudal progression. Neural crest invasion patterns are depicted by red arrows. Hashed red lines highlight neural crest cell-free zones that are thought to be established by the presence of inhibitory cues within the microenvironment. When metastatic melanoma cells are transplanted into the dorsal neural tube during active neural crest migration, melanoma cells invade along neural crest migratory pathways. Melanoma invasion patterns are regulated spatially and temporally by the developing embryo. This pattern is disrupted in melanoma cells expressing EphB6. Melanoma invasion patterns are shown for both parental C8161 cells (green) and EphB6+ C8161 cells (yellow). B) C8161 melanoma cells in suspension are dropped onto the CAM of an E10 chick embryo. Cells aggregate over 48 hours to form a tumor. At this time, cells that are capable of intravasation can be detected in the lower CAM tissue. The re-expression of EphB6 on C8161 cells abrogates metastasis, possibly by inhibiting C8161 cells from entering the CAM vasculature.
In summary, the focus of this study was to understand how metastatic tumor cells respond to and interact with their microenvironment at the level of a single cell. We show that the embryonic neural crest microenvironment exerts a powerful, controlling influence on the migratory behaviors of an aggressive metastatic melanoma (C8161). This regulation is enacted both temporally and spatially by the embryo, and was most evident by the induction of a spatially controlled symmetric exit pattern of melanoma cells from the neural tube. These data highlight the idea that successful invasion (and metastasis) of tumor cells requires that a cell be able to interpret complex microenvironmental cues encountered along the route of invasion and either be guided or adapt to overcome those cues that are meant to disrupt or block migration. As an example, we show that EphB6 acts specifically as a metastasis suppressor. Because EphB6+ cells maintain their invasiveness and tumorigenicity, we postulate that metastasis suppression occurs as EphB6 disrupts intravasation by forcing the tumor cell to recognize a non-permissive endothelial boundary. These findings were made possible by studying melanoma metastasis within the accessible embryonic neural crest microenvironment.
Supplementary Material
IMPLICATIONS.
This article links cellular metastasis to behaviors observed in the ancestrally-related embryonic neural crest and demonstrates the powerful influence of the embryonic microenvironment in regulating cell migratory behavior.
Acknowledgments
CMB thanks the National Institutes of Health Ruth L. Kirschtein Postoctoral Fellowship Program (award 5F32CA144297) for funding, and partial support from the Stowers Institute for Medical Research. PMK thanks the kind generosity of the Stowers Institute for Medical Research for support. We gratefully acknowledge the laboratory of Dr. Mary Hendrix for providing the cell lines used in these experiments and for thoughtful and constructive discussion. We also gratefully acknowledge the laboratory of Dr. Andrew Freywald for providing the EphB6 expression vector used for these studies.
Footnotes
Conflict of Interest: The authors report no conflict of interest
References
- 1.Spano D, Heck C, De Antonellis P, Christofori G, Zollo M. Molecular networks that regulate cancer metastasis. Semin Cancer Biol. 2012;22:234–49. doi: 10.1016/j.semcancer.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Condeelis J, Weissleder R. In vivo imaging in cancer. Cold Spring Harb Perspect Biol. 2010;2:a003848. doi: 10.1101/cshperspect.a003848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polacheck WJ, Zervantonakis IK, Kamm RD. Tumor cell migration in complex microenvironments. Cell Mol Life Sci. 2013;70:1335–56. doi: 10.1007/s00018-012-1115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Douarin NM, Kalcheim C. The Neural Crest. 2. Cambridge: Cambridge University Press; 1999. [Google Scholar]
- 5.Trainor PA, Krumlauf R. Patterning the cranial neural crest: hindbrain segmentation and Hox gene plasticity. Nat Rev Neurosci. 2000;1:116–24. doi: 10.1038/35039056. [DOI] [PubMed] [Google Scholar]
- 6.Trainor PA, Sobieszczuk D, Wilkinson D, Krumlauf R. Signalling between the hindbrain and paraxial tissues dictates neural crest migration pathways. Development (Cambridge, England) 2002;129:433–42. doi: 10.1242/dev.129.2.433. [DOI] [PubMed] [Google Scholar]
- 7.Bailey CM, Morrison JA, Kulesa PM. Melanoma revives an embryonic migration program to promote plasticity and invasion. Pigment Cell Melanoma Res. 2012;25:573–83. doi: 10.1111/j.1755-148X.2012.01025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nature genetics. 2005;37:1047–54. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nesbit M, Setaluri V, Herlyn M. Biology of Melanocytes and Melanoma. In: Balch CM, editor. Cutaneous melanoma. 3. St. Louis, Mo: Quality Medical Publishing; 1998. p. xxii.p. 596. [Google Scholar]
- 10.Kulesa PM, Kasemeier-Kulesa JC, Teddy JM, Margaryan NV, Seftor EA, Seftor RE, et al. Reprogramming metastatic melanoma cells to assume a neural crest cell-like phenotype in an embryonic microenvironment. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3752–7. doi: 10.1073/pnas.0506977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendrix MJ, Seftor EA, Seftor RE, Kasemeier-Kulesa J, Kulesa PM, Postovit LM. Reprogramming metastatic tumour cells with embryonic microenvironments. Nature reviews. 2007;7:246–55. doi: 10.1038/nrc2108. [DOI] [PubMed] [Google Scholar]
- 12.Busch C, Krochmann J, Drews U. The chick embryo as an experimental system for melanoma cell invasion. PloS one. 2013;8:e53970. doi: 10.1371/journal.pone.0053970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lumsden A, Sprawson N, Graham A. Segmental origin and migration of neural crest cells in the hindbrain region of the chick embryo. Development (Cambridge, England) 1991;113:1281–91. doi: 10.1242/dev.113.4.1281. [DOI] [PubMed] [Google Scholar]
- 14.Kulesa P, Ellies DL, Trainor PA. Comparative analysis of neural crest cell death, migration, and function during vertebrate embryogenesis. Dev Dyn. 2004;229:14–29. doi: 10.1002/dvdy.10485. [DOI] [PubMed] [Google Scholar]
- 15.Krull CE, Lansford R, Gale NW, Collazo A, Marcelle C, Yancopoulos GD, et al. Interactions of Eph-related receptors and ligands confer rostrocaudal pattern to trunk neural crest migration. Current biology: CB. 1997;7:571–80. doi: 10.1016/s0960-9822(06)00256-9. [DOI] [PubMed] [Google Scholar]
- 16.Kasemeier-Kulesa JC, Bradley R, Pasquale EB, Lefcort F, Kulesa PM. Eph/ephrins and N-cadherin coordinate to control the pattern of sympathetic ganglia. Development (Cambridge, England) 2006;133:4839–47. doi: 10.1242/dev.02662. [DOI] [PubMed] [Google Scholar]
- 17.Mellott DO, Burke RD. Divergent roles for Eph and ephrin in avian cranial neural crest. BMC developmental biology. 2008;8:56. doi: 10.1186/1471-213X-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freywald A, Sharfe N, Roifman CM. The kinase-null EphB6 receptor undergoes transphosphorylation in a complex with EphB1. J Biol Chem. 2002;277:3823–8. doi: 10.1074/jbc.M108011200. [DOI] [PubMed] [Google Scholar]
- 19.Fox BP, Kandpal RP. A paradigm shift in EPH receptor interaction: biological relevance of EPHB6 interaction with EPHA2 and EPHB2 in breast carcinoma cell lines. Cancer genomics & proteomics. 2011;8:185–93. [PubMed] [Google Scholar]
- 20.Truitt L, Freywald T, DeCoteau J, Sharfe N, Freywald A. The EphB6 receptor cooperates with c-Cbl to regulate the behavior of breast cancer cells. Cancer research. 2010;70:1141–53. doi: 10.1158/0008-5472.CAN-09-1710. [DOI] [PubMed] [Google Scholar]
- 21.Kim J, Yu W, Kovalski K, Ossowski L. Requirement for specific proteases in cancer cell intravasation as revealed by a novel semiquantitative PCR-based assay. Cell. 1998;94:353–62. doi: 10.1016/s0092-8674(00)81478-6. [DOI] [PubMed] [Google Scholar]
- 22.Deryugina EI, Quigley JP. Chick embryo chorioallantoic membrane model systems to study and visualize human tumor cell metastasis. Histochem Cell Biol. 2008;130:1119–30. doi: 10.1007/s00418-008-0536-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker JA, Kilroy GE, Xing J, Shewale J, Sinha SK, Batzer MA. Human DNA quantitation using Alu element-based polymerase chain reaction. Anal Biochem. 2003;315:122–8. doi: 10.1016/s0003-2697(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 24.Zijlstra A, Mellor R, Panzarella G, Aimes RT, Hooper JD, Marchenko ND, et al. A quantitative analysis of rate-limiting steps in the metastatic cascade using human-specific real-time polymerase chain reaction. Cancer research. 2002;62:7083–92. [PubMed] [Google Scholar]
- 25.Ypsilanti AR, Zagar Y, Chedotal A. Moving away from the midline: new developments for Slit and Robo. Development (Cambridge, England) 2010;137:1939–52. doi: 10.1242/dev.044511. [DOI] [PubMed] [Google Scholar]
- 26.Kulesa PM, Bailey CM, Kasemeier-Kulesa JC, McLennan R. Cranial neural crest migration: new rules for an old road. Developmental biology. 2010;344:543–54. doi: 10.1016/j.ydbio.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith A, Robinson V, Patel K, Wilkinson DG. The EphA4 and EphB1 receptor tyrosine kinases and ephrin-B2 ligand regulate targeted migration of branchial neural crest cells. Current biology: CB. 1997;7:561–70. doi: 10.1016/s0960-9822(06)00255-7. [DOI] [PubMed] [Google Scholar]
- 28.Astin JW, Batson J, Kadir S, Charlet J, Persad RA, Gillatt D, et al. Competition amongst Eph receptors regulates contact inhibition of locomotion and invasiveness in prostate cancer cells. Nature cell biology. 2010;12:1194–204. doi: 10.1038/ncb2122. [DOI] [PubMed] [Google Scholar]
- 29.Hafner C, Bataille F, Meyer S, Becker B, Roesch A, Landthaler M, et al. Loss of EphB6 expression in metastatic melanoma. International journal of oncology. 2003;23:1553–9. [PubMed] [Google Scholar]
- 30.Fox BP, Kandpal RP. Transcriptional silencing of EphB6 receptor tyrosine kinase in invasive breast carcinoma cells and detection of methylated promoter by methylation specific PCR. Biochemical and biophysical research communications. 2006;340:268–76. doi: 10.1016/j.bbrc.2005.11.174. [DOI] [PubMed] [Google Scholar]
- 31.Yu J, Bulk E, Ji P, Hascher A, Tang M, Metzger R, et al. The EPHB6 receptor tyrosine kinase is a metastasis suppressor that is frequently silenced by promoter DNA hypermethylation in non-small cell lung cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:2275–83. doi: 10.1158/1078-0432.CCR-09-2000. [DOI] [PubMed] [Google Scholar]
- 32.Matsuoka H, Obama H, Kelly ML, Matsui T, Nakamoto M. Biphasic functions of the kinase-defective Ephb6 receptor in cell adhesion and migration. J Biol Chem. 2005;280:29355–63. doi: 10.1074/jbc.M500010200. [DOI] [PubMed] [Google Scholar]
- 33.Ivanov AI, Romanovsky AA. Putative dual role of ephrin-Eph receptor interactions in inflammation. IUBMB Life. 2006;58:389–94. doi: 10.1080/15216540600756004. [DOI] [PubMed] [Google Scholar]
- 34.Trinidad EM, Ballesteros M, Zuloaga J, Zapata A, Alonso-Colmenar LM. An impaired transendothelial migration potential of chronic lymphocytic leukemia (CLL) cells can be linked to ephrin-A4 expression. Blood. 2009;114:5081–90. doi: 10.1182/blood-2009-03-210617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.