Summary
Many acute and chronic anemias, including hemolysis, sepsis, and genetic bone marrow failure diseases such as Diamond-Blackfan Anemia (DBA), are not treatable with erythropoietin (Epo), because the colony-forming unit erythroid progenitors (CFU-Es) that respond to Epo are either too few in number or are not sensitive enough to Epo to maintain sufficient red blood cell production 1,2,3–5,6,7,8,9. Treatment of these anemias requires a drug that acts at an earlier stage of red cell formation and enhances the formation of Epo-sensitive CFU-E progenitors. Recently we showed that glucocorticoids specifically stimulate self-renewal of the early erythroid progenitor, the burst-forming unit erythroid (BFU-E), and increase the production of terminally differentiated erythroid cells 10,11. Here we demonstrate that activation of the peroxisome proliferator-activated receptor alpha (PPARα) by PPARα agonists, GW7647 and fenofibrate, synergizes with glucocorticoid receptor (GR) to promote BFU-E self-renewal. Over time these agonists greatly increase production of mature red blood cells in cultures both of mouse fetal liver BFU-Es and of mobilized human adult CD34+ peripheral blood progenitors, the latter employing a new and effective culture system that generates normal enucleated reticulocytes. While PPARα−/− mice show no hematological difference from wild-type mice in both normal and phenylhydrazine (PHZ)-induced stress erythropoiesis, PPARα agonists facilitate recovery of wild-type mice, but not PPARα−/− mice, from PHZ-induced acute hemolytic anemia. We also showed that PPARα alleviates anemia in a mouse model of chronic anemia. Finally, both in control and corticosteroid-treated BFU-E cells PPARα co-occupies many chromatin sites with GR; when activated by PPARα agonists, additional PPARα is recruited to GR-adjacent sites and presumably facilitates GR-dependent BFU-E self-renewal. Our discovery of the role of PPARα agonists in stimulating self-renewal of early erythroid progenitor cells suggests that the clinically tested PPARα agonists we used may improve the efficacy of corticosteroids in treating Epo resistant anemias.
The therapeutic effect of GCs in treating Epo-resistant anemias such as DBA is well documented, though steroid therapy is known for its severe side effects that limit its use 12,13. Given the physiological importance and attractive drug targets of nuclear receptors (NRs) 14–17, and in the hope of identifying other small molecule drugs that can either individually or in combination with GCs expand BFU-E progenitors, we analyzed the expression of 49 NR genes during mouse BFU-E self-renewal promoted by dexamethasone (DEX) 10. While expression of most NR genes was unchanged, seven were up-regulated and three down-regulated by more than 2-fold (Supplementary Table 1). Amongst these PPARα changed the most: a 5-fold induction (Extended Data Fig. 1a). While most NR agonists and antagonists had no significant effects on erythroid production in cultures of primary mouse BFU-Es, two PPARα agonists, GW7647 and fenofibrate, synergized with DEX to increase the output of erythroid cells (Supplementary Table 2 and Fig. 1a). In the absence of corticosteroids, neither GW7647 nor fenofibrate affected erythropoiesis. Fenofibrate is an FDA proved drug primarily used to treat hypercholesterolemia and hypertriglyceridaemia 18, and GW7647 is a PPARα agonist with higher receptor binding affinity and specificity (EC50 = 6 nM) 19.
Figure 1. PPARα signaling synergizes with glucocorticoid receptor to promote self-renewal of mouse BFU-E erythroid progenitors.
a, Wild-type (left) and PPARα−/− (right) BFU-E cells from E14.5 mouse fetal livers were isolated and cultured in SFELE medium with the indicated treatment. Cell numbers were counted every 3 days. b, Mouse BFU-E cells were cultured in DEX ± 10 μM GW7647 as indicated. Total erythroid cell numbers were counted at day 9. c, Colony forming assays were conducted to determine BFU-E colony numbers from 100 mouse BFU-E cells cultured under the indicated conditions. Colony forming assays were performed at 24-hour intervals. Error bars represent mean ± S.D. from three biological replicates; *p<0.05, **p<0.01, ***p<0.001, Student t test.
When added together with DEX, either GW7647 or fenofibrate led to a ~150-fold increase in erythroblast production, 3–5 fold greater than DEX alone (Fig. 1a). At the end of the culture, virtually all of the cells are erythroblast cells (Extended Data Fig. 1b). Importantly, the expression of PPARα declines markedly from the BFU-E stage to the CFU-E, consistent with the decline of DNase I hypersensitivity (DHS) of the PPARα promoter (Extended Data Fig. 1c, d).
Neither GW7647 nor fenofibrate was able to increase erythroblast production from isolated mouse CFU-E cells, suggesting a specific function of PPARα agonists on BFU-Es (Extended Data Fig. 1e). GW7647 also synergized with lower doses of DEX in promoting erythroid cell production (Fig. 1b). Importantly, the effects of GW7647 and fenofibrate require the PPARα receptor since BFU-E cells isolated from PPARα−/− mice 20 failed to respond to their treatment (Fig. 1a).
Added together with DEX, 10 μM of either GW7647 or fenofibrate significantly increased BFU-E colony numbers and maintained high BFU-E numbers over one additional day relative to that achieved by DEX alone (Fig. 1c). Concentrations of GW7647 as low as 100 nM effectively synergized with 100 nM DEX in promoting BFU-E self-renewal (Extended Data Fig.1f).
Blood from PPARα−/− mice exhibits no hematological abnormalities, indicating that PPARα is not necessary for normal erythropoiesis (Supplementary Table 3). Consistent with this, the PPARα antagonist GW6471 does not interfere with DEX promoted BFU-E self-renewal, indicating that DEX does not promote BFU-E self-renewal through activating the transcriptional activity of PPARα (Extended Data Fig. 1g).
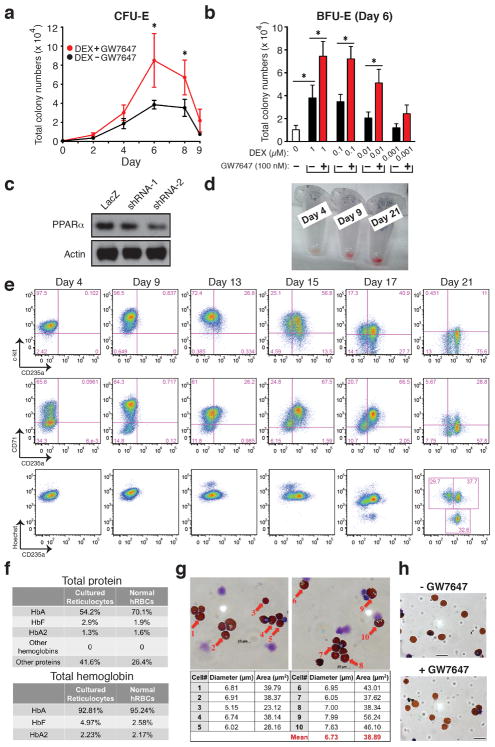
Adding GW7647 increases total cell number by an additional 4-fold, yielding a 120,000-fold expansion in our newly developed synchronized human CD34+ erythroid culture system (Supplementary Discussion and Fig. 2a). In these cultures the numbers of BFU-Es and CFU-Es are highest during days 5–8 (Fig. 2b, Extended Data Fig. 2a). Addition of GW7647 with DEX increased total BFU-E numbers, which in turn subsequently increased total CFU-E numbers in the culture. Similar to mouse BFU-E cultures, GW7647 synergized with very low concentrations of DEX in stimulating erythroid expansion of human CD34+ cells (Extended Data Fig. 2b). Addition of DEX and GW7647 affect expression of many erythroid-important genes (Supplementary Discussion).
Figure 2. Activation of PPARα signaling increased erythroid cell expansion in the ex vivo human CD34+ erythroid culture system.
a, Human CD34+ cells were cultured as described in Methods. Total cell numbers were quantified. b, BFU-E colony numbers were quantified by plating on methylcellulose 1000 cells at various time points during day 0–9 of the human CD34+ erythroid culture. c, (Left) BFU-E numbers or (Right) total cell number from control or PPARα knockdown human CD34+ cells cultured under the indicated condition were counted; d, Benzidine-Giemsa staining demonstrating cell morphology of the ex vivo human CD34+ erythroid differentiation system. (* p < 0.05; ** p < 0.01; Student t test. Error bars represent mean ± S.D. from three biological replicates.)
As expected, knocking down PPARα abrogated the ability of GW7647 to stimulate production of BFU-E cells or the total number of erythroid cells at the end of the culture (Fig. 2c, Extended Data Fig. 2c). Consistent with the absence of a role for PPARα in normal hematopoiesis, knocking down PPARα in human CD34+ cells did not affect the ability of DEX to stimulate BFU-E production or production of erythroid cells. Thus, in the human as well as in the mouse only ligand-activated, not unactivated, PPARα synergizes with DEX to promote BFU-E self-renewal during erythroid differentiation. We note that our cultures are highly synchronous based on cell surface marker expression (Extended Data Fig. 2e) as well as morphology (Fig. 2d). Neither DEX nor GW7647 had any effects on terminal differentiation or enucleation (Extended Data Fig. 2h).
GW7647 significantly increased the number of both BFU-E and CFU-Es in our CD34+ cell culture system following RPS19 knockdown, which recapitulates RPS19 haploinsufficiency in DBA patients 21,22 (Extended Data Fig. 3a). Addition of GW7647 substantially increased the fraction of CD71+ cells at day 9 of culture following RPS19 knockdown as well as total cell numbers and the fraction of CD235a+ cells at day 21 (Extended Data Figs. 3b, c). Taken together, our data suggest that GW7647 facilitates BFU-E self-renewal and production of immature erythroid progenitors in both wild-type and RPS19 knockdown human cells.
We next tested the function of GW7647 in a PHZ-induced hemolytic anemia mouse model where the endogenous corticosteroid level becomes markedly increased 11. Wild-type mice were treated with either DMSO or GW7647 for 3 days followed by treatment with PHZ; they were then injected with DMSO or GW7647 for another 7 days (Supplementary Discussion and Extended Data Fig. 4a). Treatment of GW7647 resulted in significantly higher levels of hemoglobin (HGB), RBC numbers, and hematocrit (HCT) after PHZ injection compared to those in control mice (Fig. 3a). In contrast, white blood cell (WBC) counts were similar in the two groups (data not shown). Importantly, the function of GW7647 during stress erythropoiesis was dependent on PPARα, as PPARα−/− mice failed to respond to GW7647 treatment (Extended Data Fig. 4b).
Figure 3. PPARα agonist GW7647 is effective in vivo to alleviate anemic symptoms.
a, Wild-type mice were pretreated with DMSO or GW7647 (100 μg/kg) for 3 days (days −3 to −1) before PHZ injection on day 0. Mice were subject to daily DMSO or GW7647 injections during days 0 – 6. Hemoglobin (HGB), red blood cell numbers (RBC), and hematocrit (HCT) were measured in wild-type mice on the days indicated in panel. n = 6; b, Nan/+ mutant mice were injected either with DMSO or GW7647 (100 μg/kg) for 18 days. Each dot represents one mouse. (* p < 0.05; ** p < 0.01; *** p < 0.001; Student t test)
We also tested the ability of PPARα agonists to stimulate red cell production in a mouse model of chronic anemia, Nan (“neonatal anemia”) mice (Supplementary Discussion). While BFU-E numbers in spleens of Nan/+ mutant mice are similar to or slightly higher than that in wild-type mice, the average number of BFU-Es in the spleens of GW7647 injected Nan/+ mutant mice are higher than that in untreated mice (Extended Data Fig. 5b). Importantly, GW7647 injection increased hemoglobin level, hematocrits and red blood cell numbers in these anemic mice (Fig. 3b), suggesting that the PPARα agonist alleviates anemia in Nan/+ mutant mice by increasing BFU-E numbers in the spleen. GW7647 does not have any significant effects on either platelet or WBC numbers in peripheral blood from Nan/+ mice (Extended Data Fig. 5c).
To understand the molecular mechanism by which PPARα synergizes with GR to promote BFU-E self-renewal, we conducted ChIP-Seq analysis to interrogate the genome-wide chromatin occupancy of GR and PPARα in mouse BFU-E cells. Compared to untreated cells, GR and PPARα occupancy were both induced upon DEX treatment alone; addition of GW7647 further enhanced PPARα but not GR occupancy (Fig. 4a). These results indicate that GW7647 enhances the recruitment of PPARα to GR binding sites in BFU-Es. In BFU-Es co-treated with GW7647 and DEX, our ChIP-Seq detected 1058 GR peaks and 1623 PPARα peaks. GR and PPARα both predominantly occupied distal intergenic (> 63%) and intronic chromatin sites (> 25%) (Extended Data Fig. 6a). Importantly, in 719 peaks GR and PPARα localize in close proximity (Extended Data Fig. 6b); 67.9% of total GR peaks and 44.3% of PPARα peaks co-localize. The DNA sequences underlying these overlap peaks are enriched for DNA binding motifs for several transcription factors including PU.1, YY1, Smad3, Tal1, Klf4, Hif2α and Myb (Extended Data Fig. 6c); most of these transcription factors are known to play important roles in stem cell self-renewal 23. In addition, co-treatment of DEX and GW7647 also up-regulates many genes critical for stem cell self-renewal (Supplementary Discussion and Extended Data Figs. 7, 8).
Figure 4. PPARα and glucocorticoid signaling pathways regulate a common target gene ensemble.
a, Density maps of GR and PPARα ChIP-Seq signals in mouse BFU-E cells. GR-binding peaks were used as the reference to search for corresponding ChIP-Seq signals of PPARα; b, GR and PPARα occupancy across the PPARα locus in BFU-E cells treated as indicated. c, (Left) Protein level of GR, PPARα and β-actin in mouse BFU-E cells treated as indicated. (right) Densitometry quantification of GR and PPARα protein expression levels normalized to β-actin. Experiments were repeated three times (* p < 0.05; ** p < 0.01; Student t test). d, Co-immunoprecipitation demonstrating interaction between GR and PPARα in mouse BFU-E cells.
Treatment of BFU-E cells with DEX leads to a slight increase in the level of the PPARα protein, and this is further increased to 1.6 times that of control cells by treatment with DEX and GW7647 (Fig. 4c). In contrast, the GR protein level is not altered in BFU-Es under these conditions. Consistent with these observations, addition of DEX to BFU-E cells induced the binding both of GR and PPARα to a chromatin site ~ 5 kb upstream of TSS of PPARα, presumably part of a PPARα gene enhancer, and the binding of PPARα to this chromatin site was further enhanced by addition of GW7647 (Fig. 4b). Our data suggest that there is a positive autoregulatory feedback loop of PPARα expression during BFU-E self-renewal promoted by DEX and GW7647.
Co-immunoprecipitation experiments demonstrated an interaction of GR and PPARα only in BFU-E cells treated with DEX together or not with GW7647 (Fig. 4d). While addition of DEX to cultures of BFU-E cells stimulated binding of PPARα to the GR, interactions between PPARα and GR were more pronounced following treatment with both DEX and GW7647. These results extend our ChIP-Seq data, indicating that there is a physical interaction between GR and PPARα and likely other proteins, that underlie DEX and GW7647 enhanced BFU-E self-renewal (Supplementary Discussion and Extended Data Fig. 9).
Given that little is known concerning endogenous PPARα ligands, the precise role of the PPARα in hematopoiesis, erythropoiesis in particular, remains elusive. Nonetheless, our finding that agonists of the PPARα lead to enhanced binding of PPARα to many chromatin sites and to an increase in corticosteroid-induced BFU-E self renewal and erythroid expansion, point to a previously unappreciated role for this nuclear receptor in hematopoiesis (Extended Data Fig. 10). The function of PPARα has been mainly studied in nutrient metabolism and energy homeostasis, and fenofibrate is already an FDA-approved drug for dyslipidemia treatment 18. Given that there is a very limited number of drugs that can be used to treat Epo-resistant anemias, the discovery of new drugs or repurposing current drugs to treat these diseases is challenging. Our surprising discovery suggests a novel function of PPARα in self-renewal of early committed erythroid progenitors, which potentially can lead to new therapeutics to treat Epo-resistant anemias such as DBA.
Methods
Reagents
Chemicals are obtained from Sigma. Human peripheral blood G-CSF mobilized hematopoietic stem/progenitor cells enriched for CD34+ were purchased from Fred Hutchinson Cancer Research Center (FHCRC), Seattle. StemSpan™ SFEM, CC100 cytokine cocktail, fetal bovine serum and bovine serum albumin (BSA) are purchased from STEMCELL Technologies. Holo human transferrin is from Sigma. Recombinant human and murine Stem cell factor (rhSCF, rmSCF) and Interleukin 3 (rhIL-3, rmIL-3), recombinant murine Interleukin 6 (rmIL-6) and recombinant murine insulin like growth factor-1 (rmIGF-1) are from Peprotech. Recombinant human erythropoietin (rhEpo) is from Amgen. Antibodies: Santa Cruz: GR: H300 (sc-8992), M-20 (sc-1004); PPARα: H-98 (sc-9000). β-actin: N-21(sc-130656); RPS19 antibody is from Abcam (ab57643). FACS antibodies are from eBioscience: anti-human c-kit PE (#12-1178), anti-human CD71 FITC (#11-0719), anti-human CD235a APC (#17-9987), anti-mouse CD71 PE (#12-0711), and anti-mouse Ter119 APC (#17-5921). Real time PCR Primers (mouse): Hbb-F: TTTAACGATGGCCTGAATCACTT; Hbb-R: CAGCACAATCACGATCATATTGC; Slc4a1-F: GGACAGATAGCATATAGAGACCTAACCA; Slc4a1-R: CGTAGTCTGTGGCTGTTTGCTC; Egln2-F: 5′ CTGGGCAACTACGTCATCAAT; Egln2-R: 5′ CCGCCATGCACCTTAACATC; Hmgcs2-F: 5′GAAGAGAGCGATGCAGGAAAC, Hmgcs2-R: 5′ GTCCACATATTGGGCTGGAAA; Kit-F: 5′ GTTCTGCTCCTACTGCTTCGC; Kit-R: 5′ TAACAGCCTAATCTCGTCGCC.
shRNA sequences: human PPARA shRNA-1 GGAGTTTATGAGGCCATATTC; human PPARA shRNA-2: GCTTTACGGAATACCAGTATT; mouse Pu.1 shRNA: CCATGTCCACAACAACGAGTT; ChIP-PCR primers: KIT ChIP-PCR 5′ F: GTCACAGCCACCAGAGAGAG; KIT ChIP-PCR 5′ R: TGGCAATGTTAAGAAGTGGTGG; PPARA ChIP-PCR 5′ F: CCAGGGCTACACAGAGAAAC; PPARA ChIP-PCR 3′ F: TAGCCTGAGAGTTGTGCCAA
RPS19 shRNA is described in a previously published study 21.
Ex vivo human CD34+ erythroid culture
Our Human CD34+ cell erythroid differentiation method is composed of 4 phases over 21 days: Expansion (Day 0–4), Differentiation (Dif) I (Day 5–9), II (Day 10–13), and III (Day 14–21). Cells are thawed according to the FHCRC protocol, and cultured in Expansion medium (StemSpan™ SFEM, CC100 cytokine cocktail and 2% penicillin-streptomycin) at 105 cells/mL from Day 0–4. After expansion, cells are subsequently cultured in Iscove’s Modified Dulbecco’s Medium (IMDM)-based erythroid differentiation medium supplemented with different cytokines in Dif I, II and III. The medium base for all three differentiation phases comprises: IMDM, 15% FBS, 2mM glutamine, 1% BSA, 500 μg/mL holo human transferrin, 10 μg/mL recombinant human insulin and 2% penicillin-streptomycin. Day 5–9, cells are cultured in Dif I medium, which contains erythroid medium base, 1 μM Dexamethasone, 1μM β-estradiol, 5 ng/mL rhIL-3, 100 ng/mL rhSCF and 6U/mL rhEpo. Day 10–13, cells are grown in Dif II medium containing erythroid medium base, 50 ng/mL rhSCF and 6U/mL rhEpo. Day 14–21, cells are cultured in fibronectin-coated plates in Dif III medium, which is erythroid medium base supplemented with 2U/mL rhEpo. GW7647 was added in both Dif I (100 nM) and Dif II (10 nM) when indicated. Cell numbers reseeded in the beginning of Dif I, II and III are 105, 2*105, and 3*105/mL, respectively.
Mouse fetal liver BFU-E and CFU-E isolation, cell proliferation assay and virus transduction
Purification of BFU-E and CFU-E cells from murine fetal livers was performed as described before 10. Briefly, fetal liver erythroid cells were isolated between embryonic days 14.5 (E14.5) to E15.5. A pure erythroid progenitor population containing more than 90% of BFU-E and CFU-E cells were obtained through a negative magnetic bead selection for lineage markers including Ter119, B220, Mac-1, CD3, Gr-1, Sca-1, CD16/CD32, CD41, CD34 and positive selection for c-Kit. Afterwards c-Kit+ BFU-E (CD71 10% low) and CFU-E (CD71 20% high) cells were separated by flow cytometry. Purified BFU-E or CFU-E cells were seeded in serum-free erythroid liquid expansion medium (SFELE) (StemSpan™ SFEM with 100 ng/mL rmSCF, 40 ng/mL rmIGF-1, and 2 U/mL rhEpo) alone, or SFELE containing 100 nM DEX ± 10 μM GW7647. Erythroblasts produced from purified BFU-E and CFU-E were analyzed over time by FACS and benzidine-Giemsa staining as described 10.
For PU.1 knock down, primary BFU–Es purified from mouse E14.5 fetal liver were incubated with virus encoding a shRNA for LacZ or mouse Pu.1 at 37 °C overnight. After incubation, cells were cultured in SFELE medium with or without DEX and/or GW7647.
Colony forming assay
Murine BFU-E colony-forming assays were performed in MethoCult M3234 (StemCell Technologies) containing 10 U/mL rhEpo, 20 ng/mL rmIL-3, 20 ng/mL rmIL-6, and 50 ng/mL rmSCF, with or without 100 nM DEX ± 10 μM GW7647. For CFU-E colony-forming assays, cells were cultured in MethoCult M3234 containing 10 U/mL rhEpo, with or without 100 nM DEX ± 10 μM GW7647. The number of CFU-E or BFU-E colonies was scored after 3 days or 7 days in culture, respectively.
For human colony forming assays, cells were plated in MethoCult H4034 Optimum (StemCell Technologies) without additional cytokines. Cells were cultured for 12 – 14 days before BFU-E and CFU-E colonies were scored.
In vivo mice experiments to test PPARα agonists
129/SvImJ (wild-type), 129S4/SvJae (wild-type), or 129S4/SvJae-Pparα tm1Gonz/J female mice (The Jackson Laboratory) 6 – 8 weeks of age, randomized by weight, were pretreated with GW7647 (100 μg/kg) for 3 days (days −3 – −1). On day 0, the mice were injected with phenylhydrazine (PHZ) (60 mg/kg). GW7647 injection was continued during days 0 – 6. Whole blood samples were collected at each day of day 1 – 5, and day 7 and 9 for Complete Blood Count (CBC) analyses.
4 to 6 week old Nan/+ mutant mice (officially designated as Klf1Nan; http://www.informatics.jax.org/allele/MGI:1861107), obtained from the Jackson Laboratory, were injected with GW7647 (100 μg/kg) for 18 days. Mice were randomized by weight. Whole blood samples were collected every 3 days for CBC analyses.
All mouse procedures were approved by the Animal Care and Use Committees of the Massachusetts Institute of Technology (Cambridge, MA).
Loss-of-function assay in human CD34+ erythroid culture system via lentiviral transduction
The lentiviral backbone vector pLKO.1 and packaging plasmids were transfected into 293T cells. Supernatants containing viral particles were harvested at 48 and 72 hrs. Primary human CD34+ hematopoietic cells were transduced with lentivirus one day after thawing with the presence of 2 μg/mL polybrene (Sigma). 24 hrs after viral transduction, cells were selected by growing in culture medium containing 1 μg/mL puromycin (Sigma) for 2 days.
For RPS19 knock down, 5x105 CD34 cells at day 1 of culture were infected with lentivirus encoding GFP and an shRNA targeting human RPS19 21 or a scrambled shRNA. Cells were then treated with DMSO or 0.01 μM, 0.1 μM or 1 μM GW7647. After 48 hrs, GFP positive cells were isolated by flow cytometry and returned to culture. Colony forming assay were conducted at day 6. CD71 expression was analyzed at day 9 and CD235 expression was analyzed at day 21 to determine the percentage of erythroid cells. Total cell numbers were also counted at the end of each differentiation stage.
ChIP-Seq and de novo motif discovery
Mouse ChIP-Seq experiments in BFU-Es were conducted as described before 11. 107 mouse BFU-E cells purified from E14.5 fetal liver with or without treatment were used per IP. Besides specific antibodies, we included species-matched IgG as control, and species-matched IgG yielded little to no signals. Purified DNA was prepared for sequencing according to a modified version of the Solexa genomic DNA protocol. ChIP-Seq fragments as well as inputs were barcoded and sequenced on an Illumina HiSeq sequencer. Approximately 30 million 40 nt long single reads per sample were obtained. Adapters were removed and reads shorter than 20 nt were discarded. Reads were mapped with Bowtie1 24. Peaks were called with MACS 1.4 using the corresponding input for each sample and “mfold” set to 5,30 25. We selected peaks with fold enrichment ≥10. Plots showing density of reads around the peak summits were done with ngsplots. For de novo motif discovery, Homer was used with default setting to find motifs in GR and PPARα overlapping peaks 26.
RNA-Seq
BFU-E cells from embryonic day 14.5 (E14.5) mouse fetal livers were isolated as described 3. Total RNA was purified from the mouse BFU-Es. Samples for Paired-End mRNA-seq were prepared using the Solexa kit according to the manufacturer’s instructions. RNA samples from two replicas of cells untreated, or treated with DEX ± GW7647 for 12 hrs were sequenced on an Illumina HiSeq sequencer. Around 50 million 100 nucleotide (nt) paired-end reads were obtained from each sample. Reads were trimmed to remove low quality reads using FASTQ quality trimmer with quality threshold of 20 (−t 20) and minimum length to keep 25 nt (−l 25). Reads that still had both pairs after the trimming step were mapped with TopHat 27 using gene models from ENSEMBL Genes 67, Mus musculus genes NCBIM37 (Mus_musculus.NCBIM37.67). The number of reads mapped to each gene was obtained with HTseq-count. Differential expression was assayed using DE-seq 28.
For each of the 719 peaks bound by both GR and PPARα in the presence of GW7647 and DEX, we found the closest gene using the closestBed tool 29. We filtered out any gene at a distance higher than 10 Kb from a peak.
Immunoprecipitation
Mouse BFU-Es were isolated from E14.5 mouse fetal livers. Cells were cultured in SFELE medium alone or treated with 100 nM Dex with or without 10 μM GW7647 for 12 hrs. Whole cell lysates were prepared from 4 x 107 cells using RIPA lysis buffer (sc-24948, Santa Cruz Biotechnology) with protease inhibitor cocktail. The lysates were pre-cleared by incubating with protein A-Sepharose for 1 h at 4°C and centrifugation. The supernatant was immunoprecipitated with 1 μg rabbit IgG or anti-GR antibody overnight at 4°C. Immune complexes were collected by incubation with protein A-Sepharose for 4 hrs at 4°C and washed for 5 times at 4°C with lysis buffer. The immune complexes adsorbed to the beads were centrifuged and the supernatant was removed. 50 μL of 1x loading buffer was added to the samples and boiled at 95°C for 5 minutes. Proteins were resolved by SDS-PAGE and immunoblotted by GR and PPARα antibodies.
Quantitative Real-Time RT-PCR
Total RNA from mouse BFU-Es was purified with TRIzol (Invitrogen). cDNA was prepared from 1 μg RNA. Reaction mixtures (15 μL) contained 2.0 μL of cDNA, 7.5 μL of SYBR green master mix (Applied Biosystems) and appropriate primers. Product was monitored by SYBR green fluorescence. Control reactions lacking RT yielded little to no signal. Relative expression levels were determined from a delta-delta CT method and were normalized to 18S rRNA expression.
Extended Data
Extended Figure 1. PPARα agonist GW7647 does not have adverse effects on erythroid differentiation and has no effects on CFU-E cells.
a, Gene expression changes of nuclear receptors in BFU-E cells from RNA-Seq results published before 10. b, Flow cytometry analyses of CD71 and Ter119 markers to demonstrate erythroid differentiation of mouse BFU-E cells after 9 days of culture with the indicated additions. c, PPARα gene expression in BFU-E, CFU-E and Ter119+ erythroblasts. BFU-E, CFU-E and Ter119+ erythroid cells were isolated from E14.5 mouse fetal livers as described 24. Total RNA was purified for quantitative PCR analysis. PPARα gene expression was normalized to mouse 18S rRNA in different stages. (Error bars represent mean ± S.D. from three independent experiments.) d, DNase I hypersensitivity (HS) analysis at PPARα promoter region in different mouse cells from Encode. e, Production of mouse erythroblasts from isolated CFU-E cells. Wild-type mouse CFU-E cells from E14.5 fetal livers were untreated (black line) or treated with DEX (blue line), GW7647 (red line) or fenofibrate (green line). (Error bars represent mean ± S.D. from three independent experiments.) f, Colony forming assays were conducted at 48 hrs after compound treatment to determine BFU-E colony numbers from 100 mouse BFU-E cells cultured under the indicated conditions. (* p < 0.05; Student t test. Error bars represent mean ± S.D. from three independent experiments.) g, At day 3, BFU-E colony numbers from 100 purified mouse BFU-E cells were quantified by colony forming assays. 100 purified mouse BFU-E cells were untreated or treated with DEX alone or DEX in combination with agonists or antagonists targeting PPAR receptors (α, γ or β). BFU-E colonies were quantified after 8 days in culture. (* p < 0.05; ** p < 0.01; Student t test. Error bars represent mean ± S.D. from three independent experiments.) h, Real-time PCR analysis of gene expression in DEX treated and DEX+GW7647 treated wild-type or PPARα−/− mouse BFU-E cells. (* p < 0.05; *** p < 0.001; Student t test. Error bars represent mean ± S.D. from three independent experiments.)
Extended Figure 2. Human CD34+ Erythroid Differentiation System.
a, Total CFU-E colonies formed during day 0–9. CFU-E colony numbers were quantified by plating 1000 cells from various time points during day 0–9 of the human CD34+ erythroid culture on methylcellulose. CFU-E colonies were quantified after 12–14 days. Total CFU-E colony numbers in culture under conditions without GW7647 (black line) or with GW7647 (red line) were calculated using the total cell numbers at corresponding time points in Figure 2a. b, Human CD34+ cells were treated at day 1 with 100 nM GW7647 with or without DEX at the concentration indicated in the figure. At day 6, total cell numbers were counted and cells were collected for BFU-E colony assays. c, Protein expression of PPARα demonstrating shRNA knock-down efficiency via lentiviral transduction. LacZ shRNA is used as a control. shRNA-1 and -2 are both specific for PPARα. shRNA-2 has higher knock-down efficiency. d, Cell pellets of 1 million cells demonstrating hemoglobin accumulation during the differentiation process. e, Flow cytometry analyses of erythroid markers during the 21-day human CD34+ erythroid culture. (top row) c-kit vs. CD235a; (middle row) CD71 vs. CD235a. Note the sequential induction of c-kit, CD71 and CD235a, as well as the sequential down-regulation of c-kit and CD71, (bottom row) Enucleated reticulocytes are CD235a+Hoechst−, nuclei are CD235a−Hoechst+, and nucleated erythroblasts are CD235a+Hoechst+. Enucleation rate is 32.6/(32.6+37.7)× 100%= 46.4%. f, Summary of high-performance liquid chromatography (HPLC) results using hemolysates of cultured reticulocytes and normal human RBCs (control). (top) total protein composition of hemolysates. (bottom) hemoglobin composition of hemolysates. Cultured reticulocytes contain more than 90% of adult globins. g, Size measurement of enucleated reticulocytes by both diameter and area. (Scale bar=10μm) h, Benzidine-Giemsa staining of human reticulocytes cultured with or without GW7647. (Scale bar=12μm) (* p < 0.05; Student t test. Error bars represent mean ± S.D. from three independent experiments.)
Extended Figure 3. GW7647 increases erythroid progenitors and CD235a+ cells in RPS19 knock down human progenitor cells.
a, Human CD34+ hematopoietic progenitors were transduced with lentivirus encoding GFP and either a scrambled shRNA or an shRNA targeting RPS19. Then transduced cells were treated with or without 100 nM GW7647. After 48 hrs, GFP+ cells were sorted by FACS and plated for BFU-E and CFU-E colony forming assays. RPS19 knocking down efficiency is shown at the bottom. (* p < 0.05; Student t test. Error bars represent mean ± S.D. from three independent experiments.) b. Sorted GFP+ cells were returned to culture with the indicated concentration of GW7647. (Top) Percentage of CD71+ cells at day 9 in was determined by FACS; (Bottom) Percentage of CD235a+ cells at day 21 was determined by FACS. (* p < 0.05; Student t test. Error bars represent mean ± S.D. from three independent experiments.) c. Total cell numbers generated from one GFP positive cell at the indicated times of culture. (* p < 0.05; Student t test. Error bars represent mean ± S.D. from three independent experiments.)
Extended Figure 4. GW7647 improves the anemia in two mouse models of anemia.
a, Experimental scheme for phenylhydrazine-induced hemolytic anemia, used also in Figure 3a. Wild-type or PPARα−/− mice were pretreated with DMSO (control) or GW7647 (100 μg/kg) for 3 days (days −3 to −1) before phenylhydrazine (PHZ) injection on day 0. Mice were subject to daily DMSO or GW7647 injections during days 0 – 6. Red arrows indicate days of blood sample collection. b, PPARα−/− mice were treated with DMSO or GW7647 and then injected with PHZ. Hemoglobin (HGB), red blood cell numbers (RBC), and hematocrit (HCT) were measured on the days indicated in panel; (Error bars represent mean ± S.D. from six mice.) c, BFU-E and CFU-E colony forming assays of spleen or bone marrow cells. Wild-type or PPARα−/− mice were treated with PHZ and DMSO (control) or GW7647 (100 μg/kg) as described in the legend to panel a. (* p < 0.05; Student t test. Error bars represent mean ± S.D. from three independent experiments.) d, Spleen and bone marrow cells were harvested. Representative flow cytometry analysis of spleen erythroblasts isolated from GW7647- or DMSO-treated WT mice at day 3 following PHZ injection. FSC-A, forward scatter area.
Extended Figure 5. GW7647 increases BFU-E numbers in Nan/+ mutant mice.
a, Corticosteroid levels in serum were measured in WT and Nan/+ mutant mice. Each dot represents one mouse. (* p < 0.05; Student t test. Error bars represent mean ± S.D. from all mice.) b, Increase of BFU-E numbers in spleens from GW7647 treated WT and Nan/+ mutant mice at day 18. (* p < 0.05; Student t test.) c, Total numbers of white blood cells (WBC) and platelets from peripheral blood samples were measured at day 0 and day 18. Each dot represents one mouse. (Error bars represent mean ± S.D. from all mice.)
Extended Figure 6. Bioinformatic analyses of mouse BFU-E cells.
a, Genome-wide distribution of GR and PPARα chromatin occupancy sites in BFU-E cells. ChIP-Seq analyses of GR and PPARα occupancy in mouse BFU-E cells isolated from DEX and GW7647 treated wild-type E14.5 fetal livers. TSS, transcription start site. TTS, transcription termination site. UTR, untranslated region. Distal intergenic, regions greater than 3kb from TSS. b, Venn diagram showing overlap between GR and PPARα chromatin occupancy sites. c, De novo motif searching of the overlapped chromatin sites occupied by GR and PPARα. The table depicts transcription factor binding motifs enriched at GR and PPARα overlapping sites relative to genomic background and associated p values. d, Real-time PCR analysis of Pu.1 gene expression in mouse BFU-E cells transduced with virus encoding either LacZ shRNA or Pu.1 shRNA. (* p < 0.05; Student t test. Error bars represent mean ± S.D. from three independent experiments.) e, Colony forming assays were conducted to determine BFU-E colony numbers from 100 mouse BFU-E cells infected with virus encoding either LacZ shRNA or Pu.1 shRNA. Cells were cultured in SFELE medium with or without DEX ± GW7647. Colony forming assays were performed at 48 hrs. (* p < 0.05; Student t test. Error bars represent mean ± S.D. from three independent experiments.)
Extended Figure 7.
a, Real-time RT-PCR analysis of Kit gene expression in wild-type and PPARα−/− mouse BFU-E cells untreated or treated with DEX with or without addition of GW7647 (* p < 0.05; Student t test. Error bars represent mean ± S.D. from three independent experiments.); b, Human CD34+ cells were treated with or without GW7647 as described in the legend to Figure 2. (top) At day 9 of culture, cell surface KIT and CD71 expression were analyzed by flow cytometry. (bottom) A representative histogram of KIT expression in cells treated or untreated with GW7647; c, ChIP-Seq occupancy signal map of GR and PPARα across the Kit locus in BFU-E cells.
Extended Figure 8.

Pathway analysis of RNA-Seq data of genes that are up- or down-regulated by more than 50%, comparing cultures treated with DEX alone or DEX+GW7647.
Extended Figure 9.
a, Quantitative ChIP analysis of GR and PPARα occupancy at Kit and Pparα loci in mouse BFU-E cells following the indicated treatments. Units are arbitrary; signals using rabbit IgG are represented by gray dot lines across the plots (* p < 0.05; ** p < 0.01; Student t test. Error bars represent mean ± S.D. from three independent experiments.). b, Co-immunoprecipitation measuring interaction between GR and PPARα in mouse BFU-E cells isolated from E14.5 fetal livers in wild-type or PPARα−/− mice. BFU-E cells were untreated, or treated with DEX with or without GW7647 with or without GW6471. Whole cell lysates were incubated with anti-GR antibody and immunoprecipitates probed with specific antibodies as indicated. c, Colony forming assays to determine BFU-E colony numbers from 100 mouse BFU-E cells cultured with the indicated treatments. (* p < 0.05; Student t test. Error bars represent mean ± S.D. from three independent experiments.)
Extended Figure 10. Model of synergism between PPARα and GR to promote BFU-E self-renewal.
BFU-E cells normally undergo limited self-renewal to generate CFU-Es, which can differentiate into erythroblasts. GR is sequestered in the cytoplasm without GCs such as DEX. Upon GC treatment, liganded GR will be translocated into nucleus and bind to chromatin to regulate gene transcription important for BFU-E self-renewal. PPARα is often recruited to chromatin sites that are in close proximity to GR by GC treatment alone without any function on BFU-E self-renewal. Upon GCs and PPARα agonist co-treatment, activated PPARα interacts with GR to modulate GR transcriptional activity. This leads to enhanced BFU-E self-renewal, and over time generates more CFU-Es and erythroblasts.
Supplementary Material
Acknowledgments
We thank the Whitehead Institute Flow Cytometry Facility, Genome Technology Core and Bioinformatics & Research Computing (BARC) Facility, as well as MIT Koch Institute Flow Cytometry Core. We appreciate the help of Dr. Vijay Sankaran (Boston Children’s Hospital) for hemoglobin HPLC and Dr. Johan Flygare (Lund Universitet) for the plasmid encoding the RPS19 shRNA. We are very grateful to animal technicians Ferenc Reinhardt and Tony E. Chavarria for their assistance. This study was supported by grants to H.F.L. (DARPA #HR0011-14-2-0005; DOD/U.S. Army Medical Research and Materiel Command #W81WH-12-1-0449, NIH/NHLBI 2 P01 HL032262-25; as well as research support from the Diamond-Blackfan Anemia Foundation and Diamond Blackfan Anemia Canada. L.L.P. was supported by NIH grant DK100692. X.G. was supported by a postdoctoral fellowship from the Leukemia and Lymphoma Society.
Footnotes
Author Contributions
H.Y.L., X.G., L.L.P. and H.F.L. designed the experiments. H.Y.L., X.G. and R.R.E. performed the experiments. M.I.B. and H.L. conducted bioinformatic analyses of ChIP-Seq and RNA-Seq. H.Y.L., X.G. and H.F.L. wrote the manuscript with input from M.I.B.. All authors discussed the results and commented on the manuscript.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
RNA-Seq and ChIP-Seq data have been deposited in the Gene Expression Omnibus under accession numbers GSE63836 and GSE63837, respectively.
The authors declare no competing financial interests.
References
- 1.Kaushansky KLAM, Beutler E, Kipps JT, Seligsohn U, Prchal TJ. Title of book: Williams Hematology. 8. 2010. [Google Scholar]
- 2.Livingston DH, et al. Bone marrow failure following severe injury in humans. Annals of surgery. 2003;238:748–753. doi: 10.1097/01.sla.0000094441.38807.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones KB, Anderson DW, Longmore GD. Effects of recombinant hematopoietins on blood-loss anemia in mice. The Iowa orthopaedic journal. 2005;25:129–134. [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson Y, et al. Impaired erythropoiesis after haemorrhagic shock in mice is associated with erythroid progenitor apoptosis in vivo. Acta anaesthesiologica Scandinavica. 2008;52:605–613. doi: 10.1111/j.1399-6576.2008.01656.x. [DOI] [PubMed] [Google Scholar]
- 5.Zimmerman JL. Use of blood products in sepsis: an evidence-based review. Critical care medicine. 2004;32:S542–547. doi: 10.1097/01.ccm.0000145906.63859.1a. [DOI] [PubMed] [Google Scholar]
- 6.Fiorillo A, et al. Unresponsiveness to erythropoietin therapy in a case of Blackfan Diamond anemia. American journal of hematology. 1991;37:65. doi: 10.1002/ajh.2830370121. [DOI] [PubMed] [Google Scholar]
- 7.Nathan DG, Clarke BJ, Hillman DG, Alter BP, Housman DE. Erythroid precursors in congenital hypoplastic (Diamond-Blackfan) anemia. The Journal of clinical investigation. 1978;61:489–498. doi: 10.1172/JCI108960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vlachos A, Muir E. How I treat Diamond-Blackfan anemia. Blood. 2010;116:3715–3723. doi: 10.1182/blood-2010-02-251090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niemeyer CM, et al. Treatment trial with recombinant human erythropoietin in children with congenital hypoplastic anemia. Contributions to nephrology. 1991;88:276–280. doi: 10.1159/000419537. discussion 281. [DOI] [PubMed] [Google Scholar]
- 10.Flygare J, Rayon Estrada V, Shin C, Gupta S, Lodish HF. HIF1alpha synergizes with glucocorticoids to promote BFU-E progenitor self-renewal. Blood. 2011;117:3435–3444. doi: 10.1182/blood-2010-07-295550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, et al. ZFP36L2 is required for self-renewal of early burst-forming unit erythroid progenitors. Nature. 2013;499:92–96. doi: 10.1038/nature12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willig TN, et al. Identification of new prognosis factors from the clinical and epidemiologic analysis of a registry of 229 Diamond-Blackfan anemia patients. DBA group of Societe d’Hematologie et d’Immunologie Pediatrique (SHIP), Gesellshaft fur Padiatrische Onkologie und Hamatologie (GPOH), and the European Society for Pediatric Hematology and Immunology (ESPHI) Pediatric research. 1999;46:553–561. doi: 10.1203/00006450-199911000-00011. [DOI] [PubMed] [Google Scholar]
- 13.Lipton JM, Atsidaftos E, Zyskind I, Vlachos A. Improving clinical care and elucidating the pathophysiology of Diamond Blackfan anemia: an update from the Diamond Blackfan Anemia Registry. Pediatric blood & cancer. 2006;46:558–564. doi: 10.1002/pbc.20642. [DOI] [PubMed] [Google Scholar]
- 14.Bauer A, et al. The glucocorticoid receptor is required for stress erythropoiesis. Genes & development. 1999;13:2996–3002. doi: 10.1101/gad.13.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Lindern M, et al. The glucocorticoid receptor cooperates with the erythropoietin receptor and c-Kit to enhance and sustain proliferation of erythroid progenitors in vitro. Blood. 1999;94:550–559. [PubMed] [Google Scholar]
- 16.O’Malley B. The steroid receptor superfamily: more excitement predicted for the future. Molecular endocrinology. 1990;4:363–369. doi: 10.1210/mend-4-3-363. [DOI] [PubMed] [Google Scholar]
- 17.Moore JT, Collins JL, Pearce KH. The nuclear receptor superfamily and drug discovery. ChemMedChem. 2006;1:504–523. doi: 10.1002/cmdc.200600006. [DOI] [PubMed] [Google Scholar]
- 18.Yang LP, Keating GM. Fenofibric acid: in combination therapy in the treatment of mixed dyslipidemia. American journal of cardiovascular drugs : drugs, devices, and other interventions. 2009;9:401–409. doi: 10.2165/11203920-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 19.Muoio DM, et al. Peroxisome proliferator-activated receptor-alpha regulates fatty acid utilization in primary human skeletal muscle cells. Diabetes. 2002;51:901–909. doi: 10.2337/diabetes.51.4.901. [DOI] [PubMed] [Google Scholar]
- 20.Lee SS, et al. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Molecular and cellular biology. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flygare J, et al. Deficiency of ribosomal protein S19 in CD34+ cells generated by siRNA blocks erythroid development and mimics defects seen in Diamond-Blackfan anemia. Blood. 2005;105:4627–4634. doi: 10.1182/blood-2004-08-3115. [DOI] [PubMed] [Google Scholar]
- 22.Ebert BL, et al. An RNA interference model of RPS19 deficiency in Diamond-Blackfan anemia recapitulates defective hematopoiesis and rescue by dexamethasone: identification of dexamethasone-responsive genes by microarray. Blood. 2005;105:4620–4626. doi: 10.1182/blood-2004-08-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong P, et al. Gene induction and repression during terminal erythropoiesis are mediated by distinct epigenetic changes. Blood. 2011;118:e128–138. doi: 10.1182/blood-2011-03-341404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.