Abstract
Corpus luteum (CL) regression is required during the estrous cycle. During CL regression, luteal cells stop producing progesterone and are degraded by apoptosis. However, the detailed mechanism of CL regression in cattle has not been fully elucidated. The aim of this study was to evaluate autophagy, lysosome activity, and apoptosis during CL regression in cattle. The expression of autophagy-related genes (LC3α, LC3β, Atg3, and Atg7) and the protein LC3-II was significantly higher in the late CL than in the mid CL. In addition, autophagy activity was significantly increased in the late CL. Moreover, gene expression of the autophagy inhibitor mammalian target of rapamycin (mTOR) was significantly lower in the late CL than in the mid CL. Lysosome activation and expression of cathepsin-related genes (CTSB, CTSD, and CTSZ) showed significant increases in the late CL and were associated with an increase in cathepsin B protein. In addition, mRNA expression and activity of caspase 3 (CASP3), an apoptotic enzyme, were significantly higher in the late CL than in the mid CL. These results suggest simultaneous upregulation of autophagy-related factors, lysosomal enzymes and apoptotic mediators, which are involved in regression of the bovine CL.
Keywords: Apoptosis, Autophagy, Cathepsin, Corpus luteum regression, LC3, Lysosome
The corpus luteum (CL), a biological clock of the estrous cycle and pregnancy, is a temporary endocrine structure responsible for the production of high levels of progesterone [1]. If pregnancy does not occur, the CL regresses until the next estrous cycle [2]. During regression, the CL first undergoes functional changes, i.e., the inhibition of progesterone production [3], followed by structural changes to decrease its size and weight and finally transformation into a scar tissue known as the corpus albicans [4]. However, the mechanism by which luteal regression is initiated and controlled is still not fully understood.
Recent studies have suggested that luteal cell death in humans and primates is induced by autophagy [5, 6]. Autophagy is a basic cellular mechanism associated with the degradation of unnecessary cytoplasmic organelles by isolating targeted components to form an autophagosome [7], which fuses with lysosomes to degrade the engulfed materials [8]. Autophagy is regulated by autophagy-related genes (Atg) and their associated proteins [9]. During the process of autophagy, mammalian target of rapamycin (mTOR) inhibits initiation of the autophagy pathway and autophagosome formation [10].
In ewes, luteal cells in the regressing CL are correlated with characteristics of lysosome activation such as the increase in lysosomal content and release of lysosomal enzymes into the cytosol [11]. Lysosomes are membrane-bound organelles involved in endocytosis, phagocytosis and autophagy [12]. In addition, lysosomes play an important role in the execution of apoptosis via lysosomal proteases, especially cysteine cathepsins [13]. Among the cysteine cathepsins, cathepsins B, L and D are the most abundant and can be used as apoptosis markers [13]. Cathepsin B is involved in apoptosis via activation of the pro-apoptotic protein BID, a member of the Bcl-2 family [14], which stimulates the mitochondrial apoptotic pathway [15]. Cathepsin B is found in various organs, including the liver, ovaries, and cumulus-oocyte complexes [16]. Cathepsin D, which is mainly involved in the proteolysis of endocytosed or autophagocytosed proteins at low pH, is also associated with the induction and promotion of apoptosis [17].
Apoptosis is strongly implicated in structural CL regression, as evidenced by DNA fragmentation and the expression of apoptosis-related genes [4, 18]. The pivotal apoptotic mediator caspase 3 (CASP3) has been detected in the human CL [19], and it is highly involved in luteal regression in cows [20] and other species [21].
Although the role of apoptosis in CL degradation has been extensively investigated, the status of autophagy during regression of the bovine CL has not been fully elucidated. The aim of this study was to evaluate autophagy and lysosome activity with respect to regression of the bovine CL.
Materials and Methods
Collection of CL tissues
Bovine ovaries were collected from a local abattoir and transported to the laboratory. Stages of the estrous cycle were estimated based on the morphological features of the ovaries [22]. In brief, the mid and late CL stages were detected according to the internal and external morphology and the presence of a large follicle. The mid CL (1.6–2.0 cm in diameter) is a fully formed CL with visible vasculature in the periphery and internal parts; the apex is initially red or brown, and the remainder of the gland is yellow or orange. The late CL is smaller (< 1.0 cm in diameter) and is characterized by a large follicle (> 1.0 cm in diameter); all the tissue is yellow or orange [23, 24].
Measurement of tissue progesterone
Progesterone (P4) was measured in luteal tissue homogenates as previously described [25] with some modifications. In brief, CL tissue samples were weighed and homogenized with a BioMasher I (Nippi, Tokyo, Japan) to prepare tissue homogenates. The homogenates were ultracentrifuged (12,000 rpm), and the supernatant was collected and adjusted to the required dilution ratio (100 µl/100 mg tissue homogenate) with phosphate-buffered saline (PBS). A 90-µl portion of each supernatant was stored at –20 C until analysis. P4 levels were measured by enzyme immunoassay as described previously [26]. The standard curve was constructed in the range of 0.391–100 ng/ml, and the effective dose for 50% inhibition (ED50) was 4.5 ng/ml.
RNA isolation and quantitative real-time reverse transcription polymerase chain reaction (RT-PCR)
Total RNA from each sample was isolated and treated with DNase using a NucleoSpin RNA II Kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s instructions. The extracted RNA was then immediately used for RT-PCR or stored at –80 C until analysis. After standardizing the RNA quantity using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA), cDNA was synthesized with ReverTra Ace qPCR RT Master Mix (Toyobo, Osaka, Japan). Conventional PCR was performed using GoTaq Hot Start Green Master Mix (Promega, Madison, WI, USA). To assess gene expression, quantitative (q) PCR was used. Primers specific for autophagy-related genes (LC3α, LC3β, ATG3, and ATG7), cathepsin B (CTSB) and cathepsin D (CTSD) were designed and commercially synthesized (Eurofins Genomics, Tokyo, Japan). Primers for the genes cathepsins Z (CTSZ) [16], caspase 3 (CASP3) [27], mTOR [28], steroidogenic cytochrome P450, family 11, subfamily A, polypeptide 1 (CYP11A1), and 3-beta-hydroxysteroid dehydrogenase (HSD3β) [29] were synthesized based on the reported sequences. All primer sequences are presented in Table 1. The reactions were carried out in 96-well PCR plates, in a total volume of 10 µl containing 1 µl of 10 pmol/µl of each primer, 5 µl of Thunderbird SYBR qPCR Mix (Toyobo), and 3 µl of cDNA. After centrifugation, the plates were placed into a Roche LightCycler 480 instrument II (Roche, Basel, Switzerland) and subjected to the following cycling conditions: a denaturation step at 95 C for 30 sec, an amplification step of 50 cycles at 95 C for 10 sec, 57 C for 15 sec and 72 C for 30 sec, a melting-curve step at a gradient of 55–95 C with an increment of 2.2 C/sec and continuous fluorescence acquisition, and a cooling step at 4 C. The expression levels of the target genes were determined relative to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Table 1. List of oligonucleotide primers used for RT-PCR.
| Target gene | Primer sequence | GenBank accession number |
| CYP11A1 | 5’-AGGCAGAGGGAGACATAAGCA-3’ | (Atli et al. 2012) |
| 5’-GTGTCTTGGCAGGAATCAGGT-3’ | NM_176644.2 | |
| HSD3β | 5’-CCAAGCAGAAAACCAACGAC-3’ | (Atli et al. 2012) |
| 5’-ATGTCCACGTTCCCATCATT-3’ | NM_174343.3 | |
| LC3α | 5’-TGTCAACATGAGCGAGTTGGT-3’ | NM_001046175.1 |
| 5’-AGGAAGCCATCCTCGTCCTT-3’ | ||
| LC3β | 5’- CGAGAGCAGCATCCTACCAA -3’ | NM_001001169.1 |
| 5’- TGAGCTGTAAGCGCCTTCTT -3’ | ||
| ATG3 | 5’-AAGGGAAAGGCACTGGAAGT-3’ | NM_001075364.1 |
| 5’-GTGATCTCCAGCTGCCACAA-3’ | ||
| ATG7 | 5’- ATTGCTGCATCAAGAGACCCA -3’ | NM_001083795.2 |
| 5’- CCTTCTGGCGATTATGGTCA -3’ | ||
| CTSB | 5’- CACTTGGAAGGCTGGACACA-3’ | NM_174031.2 |
| 5’- GCATCGAAGCTTTCAGGCAG-3’ | ||
| CTSD | 5’- CCCGTGGAACACCTGATCGCCAA-3’ | NM_001166521.1 |
| 5’- CCCGATGCCGATCTCCCCGTA -3’ | ||
| CTSZ | 5’- GGGAGAAGATGATGGCAGAAAT -3’ | (Bettegowda et al. 2008) |
| 5’- TCTTTTCGGTTGCCATTATGC -3’ | NM_001077835.1 | |
| CASP3 | 5’-AGCCATGGTGAAGAAGGAATCA-3’ | (Ushizawa et al. 2006) |
| 5’-GGTACTTTGAGTTTCGCCAGGA-3’ | AY575000 | |
| mTOR | 5’-ATGCTGTCCCTGGTCCTTATG-3’ | (Nan et al. 2014) |
| 5’-GGGTCAGAGAGTGGCCTTCAA-3’ | XM_001788228.1 | |
| GAPDH | 5’-ACAGTCAAGGCAGAGAACGG-3’ | NM_001034034.2 |
| 5’-CCACATACTCAGCACCAGCA-3’ |
Immunodetection of LC3-II
The CL samples (100 mg each) were homogenized with a BioMasher (Nippi) and ultracentrifuged according to the manufacturer’s instruction. Then, 10 µl of homogenate was lysed as previously described [30] in buffer containing 90 µl of 2% sodium dodecyl sulfate (SDS; Wako Pure Chemical Industries, Osaka, Japan), 5% 2-mercaptoethanol (Sigma-Aldrich, St. Louis, MO, USA), 25% glycerol (Kanto Chemical, Tokyo, Japan), 0.5 M Tris-HCl (pH 6.8), and 0.5% bromophenol blue, and heated at 95 C for 5 min. Ten microliters of the sample was separated by SDS-polyacrylamide gel electrophoresis in a 15% polyacrylamide gel using an electrophoresis unit (Bio-Rad, Hercules, CA, USA). The separated proteins were transferred onto polyvinylidene fluoride membranes using an iBlot Gel Transfer System (Invitrogen, Carlsbad, CA, USA). The membrane was incubated with blocking solution containing 5% bovine serum albumin (BSA) (Sigma-Aldrich) in Tris-buffered saline containing 0.1% Tween 20 (TBS-T) for 1 h at room temperature and washed three times in TBS-T. The membranes were then incubated overnight at 4 C with an anti-LC3 rabbit polyclonal antibody (PM036Y, Medical & Biological Laboratories, Tokyo, Japan) and anti-GAPDH mouse monoclonal antibody (NB300-221SS; Novus Biologicals LLC, Littleton, CO, USA), both diluted 1:2000 with immunoreaction enhancer (Can Get Signal, Toyobo). After washing three times with TBS-T, the membranes were incubated with horseradish peroxidase (HRP)-labeled donkey anti-rabbit immunoglobulin G (IgG) (NA934VS; GE Healthcare Life Sciences, Little Chalfont, UK) diluted 1:25,000 in TBS-T to detect LC3. To detect GAPDH, HRP-labeled goat anti-mouse IgM (H+L) (bs-0368G; Bioss USA Antibodies, Woburn, MA, USA) was used (1:4000 dilution in TBS-T). After washing the membrane with TBS-T, chemifluorescence detection was performed with Immobilon Western Chemiluminescent HRP Substrate (P36599A; Merck Millipore, Billerica, MA, USA) and analyzed using a ChemiDoc System (Bio-Rad).
Immunohistochemistry of cathepsin B
Small pieces of CL tissue were collected, embedded in Optimal Cutting Temperature compound (Sakura Finetek, Tokyo, Japan), and stored at –80 C until use. Cross sections were prepared using a Leica CM3050 S Research Cryostat (Leica Biosystems Nussloch, Nussloch, Germany) and fixed in 4% (w/v) paraformaldehyde (pH 7.4). Then, the slides were washed in PBS and permeabilized by incubation with 0.25% (v/v) Triton-X100 in PBS (PBS-T) for 10 min. After washing with PBS-T, the samples were blocked with 1% (w/v) BSA in PBS-T for 30 min and incubated for 1 h at room temperature with the primary mouse monoclonal antibody to cathepsin B (ab58802; Abcam, Cambridge, UK) diluted 1:200 in 1% BSA-PBS-T. Slides were then washed three times in PBS-T for 5 min and incubated for 30 min at room temperature with the fluorescein-conjugated secondary antibody ( CAPPELTM Research Products, Durham, NC, USA ) diluted 1:300 in 1% BSA-PBS-T:goat anti-mouse IgM (µ chain). The slides were washed in PBS-T three times for 5 min, covered with 10 µl of mounting solution (Vectashield with DAPI, Vector Laboratories, Burlingame, CA, USA), and examined under a BZ-9000 Biorevo fluorescence microscope (Keyence, Osaka, Japan) using a 550-nm excitation filter.
Detection of autophagy activity
Autophagy activity was detected as described previously [31] with some modifications. Cyto-ID Green Autophagy Detection Reagent (Enzo Life Sciences, Farmingdale, NY, USA), a novel amphiphilic autophagosome tracer dye that measures the autophagic vacuoles and monitors autophagic flux in live cells, co-localizes with LC3 and has negligible nonspecific staining of lysosomes [32], was used according to the manufacturer’s protocol. In brief, small pieces (less than 2 × 2 × 2 mm) of CL tissue samples were incubated in 1× Assay Buffer (Enzo) at 37 C for 30 min in the presence of 2 μl/ml reaction mix. After rinsing in PBS, the stained samples were mounted onto a glass slide and observed immediately under a BZ-9000 Biorevo fluorescence microscope (Keyence) using a 550-nm excitation filter. Images were captured, and fluorescence intensity was analyzed using the ImageJ Software (National Institutes of Health, Bethesda, MD, USA). For the image analysis, the tissues were selected by setting the threshold above the background level as constant for all analyzed images, and the average fluorescence intensity was calculated to evaluate the autophagy activity.
Detection of cathepsin B and lysosomal activities
Cathepsin B activity was detected by using a Magic Red Cathepsin B Assay Kit (MR-RR) 2 (Immunochemistry Technologies, LLC, Minneapolis, MN, USA) according to the manufacturer’s protocol with a slight modification. In brief, small pieces (less than 2 × 2 × 2 mm) of each sample were incubated in 300 µl of serum-free Dulbecco’s modified Eagle medium (DMEM) containing 1 µl reaction mix in a humidified atmosphere of 5% CO2 at 38.5 C for 30 min. Active lysosomes were detected by incubating the CL tissues with 1 µM of LysoTracker Red (L-7528, Molecular Probes, Eugene, OR, USA) in DMEM supplemented with 10% fetal calf serum in a humidified atmosphere of 5% CO2 at 38.5 C for 30 min. After rinsing in PBS, the stained samples were mounted onto a glass slide and observed immediately under a fluorescence microscope using a 590-nm excitation filter. Images were captured and analyzed as described above by measuring the fluorescent intensity of cathepsin B (active form) and the average size of lysosomes.
Detection of caspase 3 activity in CL tissues
To evaluate the apoptotic status, caspase 3 activity in the CL tissues was detected using caspase 3 Magic Red-(DEVD)2 substrate reagent (Immunochemistry Technologies, LLC) according to the manufacturer’s protocol. Small pieces (less than 2 × 2 × 2 mm) of CL tissue were incubated in 300 µl of serum-free DMEM containing 2 µl reaction mix in a humidified atmosphere of 5% CO2 at 38.5 C for 30 min. After rinsing in PBS, the stained samples were mounted onto a glass slide and observed under a fluorescence microscope using a 590-nm excitation filter. Images were captured and analyzed as described above.
Statistical analysis
Each experiment was performed using more than 3 different CL tissues for each analysis, and the data are expressed as means ± SEM. The statistical significance of differences between the tissues was analyzed with Student’s t-tests for independent samples using the SPSS version 16.0 software (SPSS, Chicago, IL, USA); P < 0.05 was considered statistically significant.
Results
Evaluation of CL regression status
To evaluate the stage of CL regression, the CL tissue samples were analyzed for P4 levels, expression of the genes involved in progesterone synthesis and the activity of caspase-3, an apoptotic indicator. P4 production was significantly lower in the late CL than in the mid CL (P < 0.05; Supplementary Fig. 1A: online only), and this decrease was correlated with a decrease in the expression of the steroidogenic CYP11A1 and HSD3β genes (P < 0.05; Supplementary Fig. 1B and C). To further confirm the CL regression status, the expression and enzymatic activity of the apoptotic mediator caspase 3 was analyzed in the mid and late CL. The results revealed that caspase 3 activity was significantly higher in the late CL stage than in the mid CL stage (Supplementary Fig. 2A and B: online only); strong signals of caspase 3 activity were observed in late CL sample (Supplementary Fig. 2B). Quantitative assessment showed that the fluorescent signal indicative of the cleaved caspase 3 substrate was significantly stronger in the late CL stage (P < 0.05; Supplementary Fig. 2C). These results were substantiated by markedly higher CASP3 expression in the late CL stage compared with the mid CL stage (P < 0.01; Supplementary Fig. 2D).
Assessment of expression and activity of autophagy-related genes and protein
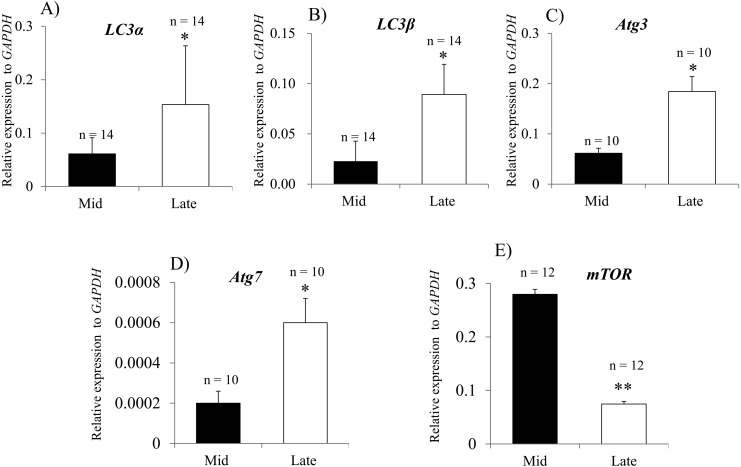
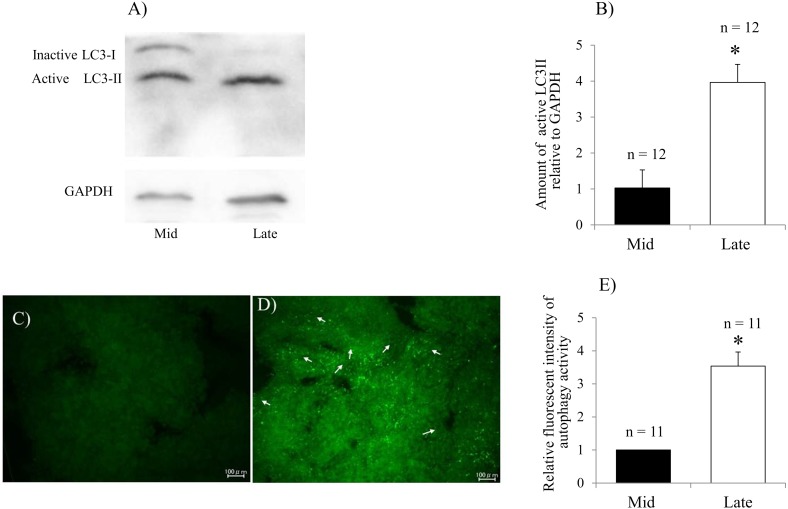
To evaluate autophagy status during regression of the bovine CL, the expression levels of LC3α, LC3β, Atg3, Atg7 and mTOR were analyzed by qPCR. The expression levels of all genes were significantly higher in the late CL stage than in the mid CL stage (Fig. 1 A–D). Expression of mTOR, an autophagy inhibitory factor, was significantly decreased in late CL stage (Fig. 1E). LC3 has inactive LC3-I and active LC3-II forms. LC3-II localizes inside and outside the mature autophagosome and can therefore be considered an autophagy marker [33]. To assess the active membrane-bound LC3 form, LC3-II protein was detected by western blot analysis. Significantly higher LC3-II expression was observed in the late CL than the mid CL (Fig 2A and B). For further confirmation, autophagy activity was evaluated in mid and late CLs by detecting the autophagosomes with Cyto-ID dye. Clear and strong spots indicating autophagosomes were observed in the late CL (Fig. 2C and D). The fluorescence intensity of active autophagosomes was significantly higher in the late CL than in the mid CL (Fig. 2E).
Fig. 1.
Expression of autophagy-related genes during bovine CL regression. Relative mRNA levels of LC3α (A), LC3β (B), Atg3 (C), Atg7 (D) and mTOR (E). All data are normalized to GAPDH expression and shown as means ± SEM. *P < 0.05. **P < 0.01. n, total number of analyzed tissues.
Fig. 2.
Evaluation of LC3 proteins and autophagy activity during bovine CL regression. Expression of the active membrane-bound LC3-II protein and inactive LC3-I protein detected by western blotting analysis (A) and the relative level of active LC3-II protein to GAPDH in the mid and late CL (B). Fluorescence microscopy of the autophagic vacuole accumulation in mid (C) and late (D) CL; Strong and diffuse green signals were observed in the late CL (white arrows). Quantification of autophagy activity was performed by calculating the fluorescence intensity (E). The data are expressed as means ± SEM. *P < 0.05. n, total number of analyzed tissues.
Expression of cathepsin genes and cathepsin B protein
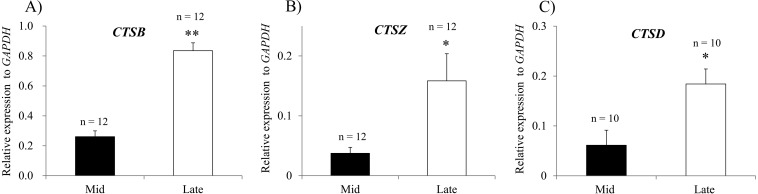
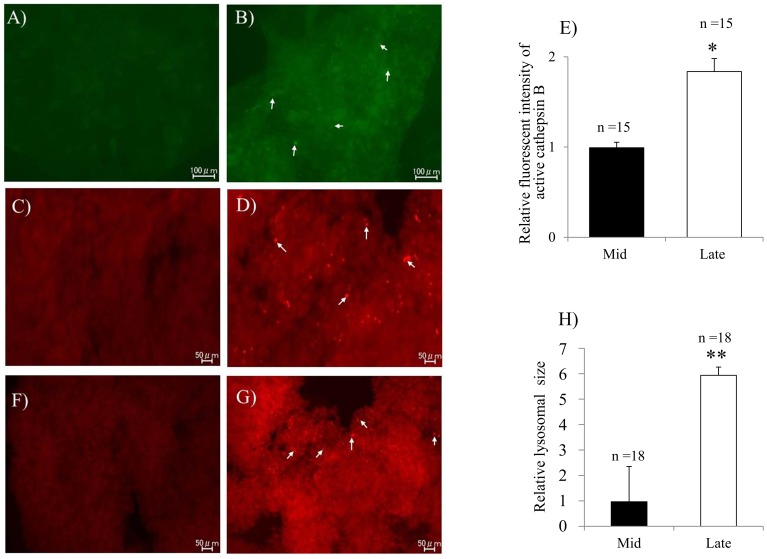
To evaluate the expression of the CTSB, CTSD, and CTSZ genes during CL regression, qPCR analysis was performed. Significantly higher mRNA levels of CTSB, CTSD and CTSZ were detected in the late CL than in the mid CL (Fig 3A–C). The increase in CTSB expression was further confirmed by evaluating the cathepsin B protein using immunohistochemistry (Fig. 4A and B) and cathepsin B activity (Fig. 4C and D) in the late CL. The relative florescent intensity of cathepsin B was significantly higher in the late CL tissues than that in the mid CL tissues (Fig. 4E)
Fig. 3.
Expression of cathepsin-encoding genes during bovine CL regression. Relative mRNA levels of CTSB (A), CTSZ (B) and CTSD (C). The data are normalized to GAPDH expression and are shown as means ± SEM. *P < 0.05. **P < 0.01. n, total number of analyzed tissues.
Fig. 4.
Detection of lysosomal cathepsin B protein, activity and active lysosomes in the regressing bovine CL. Immunohistochemical localization of cathepsin B protein in the mid (A) and late (B) CL. Green fluorescent dots of cathepsin B in the cytoplasm of late CL (white arrows) cells. Cathepsin B activity in the mid (C) and late (D) CL. The activity was indicated by diffuse red fluorescence in the cytoplasm of late luteal cells (white arrows). Relative fluorescence intensity of cathepsin B (E). Intracellular localization of lysosomes in the mid (F) and late (G) CL; Active lysosomes appeared as diffuse red fluorescence spots in the cytoplasm of luteal cells especially in the late CL (white arrows). Lysosomal size was performed (H). The data are expressed as means ± SEM. *P < 0.05. **P < 0.01. n, total number of analyzed tissues.
Evaluation of active lysosomes
To evaluate the relationship between lysosomal activity and CL regression, detection of active lysosomes was performed. Red fluorescent signals indicative of active lysosomes were clearly observed in the late CL tissue (Fig. 4F and G). The average lysosomal size was significantly larger in the late CL tissues than that in the mid CL tissues (Fig. 4H).
Discussion
In the present study, we performed a) a molecular evaluation of autophagy, b) investigated the association between the expression of autophagy markers and apoptotic mediators, and c) investigated the possible role of lysosomal activity and proteases expressions during CL regression. The overall results strongly suggest that the expression of autophagy-specific genes and lysosomal activity associated with the lysosome cathepsin expression pattern are highly related to apoptosis in the process of CL regression.
In the molecular evaluation of autophagy during CL regression, higher expression of Atg3 and Atg7 was observed in the late CL, possibly owing to their role in the formation of an LC3-phosphatidylethanolamine complex, which plays an essential role in membrane dynamics during autophagy [34, 35]. In addition, we detected increases in expression of LC3α and LC3β, which encode two LC3 forms, and in the expression of the activated membrane-bound derivative LC3-II (an autophagy marker [33]) in the regressing CL tissues. These findings are in agreement with a previous study in rats that showed upregulation of LC3 and the membrane-bound LC3-II in the late-luteal CL stage [36]. Upregulation of autophagy-related genes and proteins in the late CL is supported by the significant decrease in mTOR expression, which has an inverse relationship with autophagy [10]. The inhibitory effect of mTOR on autophagy depends on prevention of the induction stage [37, 38]. In addition, autophagy activity was significantly increased in the late CL after autophagosomes were detected. The increases in both activity and mRNA expressions of autophagy and autophagy-related factors were accompanied by a decrease in expression of mTOR in the late CL, suggesting that autophagy participates in the process of bovine luteolysis.
We also investigated the association between the expression of autophagy markers and apoptotic mediators. Upregulation of autophagy-related factors was associated with higher expression and activity of the apoptotic initiator caspase 3. The correlation between autophagy and apoptosis has been reported previously [39]. It has been demonstrated in a rat model in which an increase in apoptotic death in granulosa cells paralleled the accumulation of autophagosomes in luteal cells during CL regression [36]. In bovine CL regression, the extrinsic apoptosis pathway has been shown to be particularly important for the onset of regression [40], at which point caspase 3 activation and transduction of cell death signals occur via the tumor necrosis factor alpha (TNFα) pathway [41, 42]. TNF-α was identified as the predominant immune regulatory factor for macroautophagy in the skeletal muscle [43]. In addition, TNF-α treatment has been shown to induce macroautophagy in Ewing sarcoma cells [44]. Taken together, it can be concluded that TNF-α can promote autophagy and may act as a link between extrinsic apoptosis and autophagy during CL regression, thus explaining the synchronized elevation of LC3 expression and caspase 3 activity observed in our study.
We also evaluated lysosomal activity and protease expression levels. Our findings demonstrate an increased number of active lysosomes in the late CL. Lysosomes contain enzymes that break down unused intracellular materials, such as acid hydrolases synthesized in the endoplasmic reticulum and packaged in the Golgi complex into so-called primary lysosomes. These lysosomes fuse with endosomes to form endolysosomes or secondary lysosomes; hydrogen ions then create an acidic environment and activate the enzymes. Thus, a mature lysosome characterized by a highly acidic pH and activated hydrolytic enzymes is formed [45]. In our experiments, a higher intensity of the acidic probe in the late CL tissues indicates an increased number and size of mature lysosomes with low pH and activated hydrolytic enzymes.
Lysosomal number and enzymatic activity are strongly related to CL regression; lysosomes appear to be more numerous in small luteal cells than in large luteal cells before the initiation of luteolysis in sheep [46]. Moreover, the activity of lysosomal enzymes increases at the beginning of the progesterone decline and regression of the CL in ewes [47]. During structural luteolysis, luteal cells undergo apoptosis [4, 18], and lysosomes participate in the execution of apoptosis via lysosomal enzymes, including cysteine proteases and the aspartic protease cathepsin D [13]. Lysosomes play an important role in macroautophagy via formation of autophagolysosomes [8], which are increased in response to PGF2α in sheep luteal cells [48]. These findings may explain the lysosome accumulation and upregulation of the associated protease cathepsin B in bovine luteal cells during CL regression.
Higher expression of the CTSB gene may be related to the induction of apoptosis via the activation of a pro-apoptotic protein, BID. Truncated BID participates in the mitochondrial apoptotic pathway involved in the activation of procaspase 9 [49]. The simultaneous increase in cathepsin B and LC3 has been observed in a previous study in which excessive activation of autophagy led to the elevation of cathepsin B production, which could stimulate intracellular inflammasomes, activate NOD-like receptors and subsequently activate proinflammatory caspases [50]. These results suggest that cathepsin B activity during mitochondrial apoptosis may link autophagy with a proinflammatory response [51]. In our study, the observed high expression and activity of cathepsin B in the late CL stage may be related to its role in apoptosis and autophagy.
Similarly, higher expression of the CTSD gene in the late CL stage may be attributed to its role in the regulation of both intrinsic and extrinsic apoptotic pathways. It was suggested that the increase in CTSD in the regressing CL is related to the redox events that lead to cell death and suggests a caspase-independent apoptotic pathway [52]. A synchronous increase in CTSD expression and autophagy activity may be attributed to the role of cathepsin D in the proteolysis of endocytosed or autophagocytosed proteins at low pH [17].
The high level of CTSZ in the late CL might be due to the presence of macrophages in the regressing CL observed in many species [53]. Macrophages play a role in luteolysis via phagocytosis of decaying luteal cells or secretion of cytokines and reactive oxygen species [54]. Given that cathepsin Z is expressed predominantly by monocytes and macrophages, the accumulation of these cells in late CL tissues may explain elevated CTSZ expression.
In conclusion, our study suggests that autophagy and apoptosis are involved in the regression of the bovine CL, as evidenced by a simultaneous increase in the expression of apoptotic caspase 3 and ubiquitin-like LC3. Additional evidence includes the upregulation of lysosomal activity, synthesis of lysosomal cathepsins, and formation of autophagolysosomes. Therefore, this study suggests cross talk between lysosomal function, autophagy, and apoptosis during regression of the CL in bovine.
Supplementary
Acknowledgments
This study was supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (KAKENHI, 24580439 and 24780265).
References
- 1.Fields MJ, Fields PA. Morphological characteristics of the bovine corpus luteum during the estrous cycle and pregnancy. Theriogenology 1996; 45: 1295–1325. [DOI] [PubMed] [Google Scholar]
- 2.Stocco C, Telleria C, Gibori G. The molecular control of corpus luteum formation, function, and regression. Endocr Rev 2007; 28: 117–149. [DOI] [PubMed] [Google Scholar]
- 3.Ireland JJ, Mihm M, Austin E, Diskin MG, Roche JF. Historical perspective of turnover of dominant follicles during the bovine estrous cycle: key concepts, studies, advancements, and terms. J Dairy Sci 2000; 83: 1648–1658. [DOI] [PubMed] [Google Scholar]
- 4.Juengel JL, Garverick HA, Johnson AL, Youngquist RS, Smith MF. Apoptosis during luteal regression in cattle. Endocrinology 1993; 132: 249–254. [DOI] [PubMed] [Google Scholar]
- 5.Morales C, García-Pardo L, Reymundo C, Bellido C, Sánchez-Criado JE, Gaytán F. Different patterns of structural luteolysis in the human corpus luteum of menstruation. Hum Reprod 2000; 15: 2119–2128. [DOI] [PubMed] [Google Scholar]
- 6.Dickson SE, Bicknell R, Fraser HM. Mid-luteal angiogenesis and function in the primate is dependent on vascular endothelial growth factor. J Endocrinol 2001; 168: 409–416. [DOI] [PubMed] [Google Scholar]
- 7.Berg TO, Fengsrud M, Strømhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem 1998; 273: 21883–21892. [DOI] [PubMed] [Google Scholar]
- 8.Bernard A, Klionsky DJ. Autophagosome formation: tracing the source. Dev Cell 2013; 25: 116–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010; 140: 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, Cecconi F. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 2013; 15: 406–416. [DOI] [PubMed] [Google Scholar]
- 11.Stacy BD, Gemmell RT, Thorburn GD. Morphology of the corpus luteum in the sheep during regression induced by prostaglandin F2ALPHA. Biol Reprod 1976; 14: 280–291. [DOI] [PubMed] [Google Scholar]
- 12.Eskelinen EL, Tanaka Y, Saftig P. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol 2003; 13: 137–145. [DOI] [PubMed] [Google Scholar]
- 13.Ivanova S, Repnik U, Bojic L, Petelin A, Turk V, Turk B. Lysosomes in apoptosis. Methods Enzymol 2008; 442: 183–199. [DOI] [PubMed] [Google Scholar]
- 14.Lamparska-Przybysz M, Gajkowska B, Motyl T. Cathepsins and BID are involved in the molecular switch between apoptosis and autophagy in breast cancer MCF-7 cells exposed to camptothecin. J Physiol Pharmacol 2005; 56(Suppl 3): 159–179. [PubMed] [Google Scholar]
- 15.Antonsson B. Bax and other pro-apoptotic Bcl-2 family “killer-proteins” and their victim the mitochondrion. Cell Tissue Res 2001; 306: 347–361. [DOI] [PubMed] [Google Scholar]
- 16.Bettegowda A, Patel OV, Lee KB, Park KE, Salem M, Yao J, Ireland JJ, Smith GW. Identification of novel bovine cumulus cell molecular markers predictive of oocyte competence: functional and diagnostic implications. Biol Reprod 2008; 79: 301–309. [DOI] [PubMed] [Google Scholar]
- 17.Kågedal K, Johansson U, Ollinger K. The lysosomal protease cathepsin D mediates apoptosis induced by oxidative stress. FASEB J 2001; 15: 1592–1594. [DOI] [PubMed] [Google Scholar]
- 18.Rueda BR, Tilly KI, Hansen TR, Hoyer PB, Tilly JL. Expression of superoxide dismutase, catalase and glutathione peroxidase in the bovine corpus luteum: evidence supporting a role for oxidative stress in luteolysis. Endocrine 1995; 3: 227–232. [DOI] [PubMed] [Google Scholar]
- 19.Khan SM, Dauffenbach LM, Yeh J. Mitochondria and caspases in induced apoptosis in human luteinized granulosa cells. Biochem Biophys Res Commun 2000; 269: 542–545. [DOI] [PubMed] [Google Scholar]
- 20.Rueda BR, Tilly KI, Botros IW, Jolly PD, Hansen TR, Hoyer PB, Tilly JL. Increased bax and interleukin-1beta-converting enzyme messenger ribonucleic acid levels coincide with apoptosis in the bovine corpus luteum during structural regression. Biol Reprod 1997; 56: 186–193. [DOI] [PubMed] [Google Scholar]
- 21.Rueda BR, Hendry IR, Tilly JL, Hamernik DL. Accumulation of caspase-3 messenger ribonucleic acid and induction of caspase activity in the ovine corpus luteum following prostaglandin F2alpha treatment in vivo. Biol Reprod 1999; 60: 1087–1092. [DOI] [PubMed] [Google Scholar]
- 22.Kawate N, Inaba T, Mori J. Gonadotropin-releasing hormone receptors in anterior pituitaries of cattle during the estrous cycle. Anim Reprod Sci 1991; 24: 185–191. [Google Scholar]
- 23.Ireland JJ, Murphee RL, Coulson PB. Accuracy of predicting stages of bovine estrous cycle by gross appearance of the corpus luteum. J Dairy Sci 1980; 63: 155–160. [DOI] [PubMed] [Google Scholar]
- 24.Kawate N, Akiyama M, Suga T, Inaba T, Tamada H, Sawada T, Mori J. Change in concentrations of luteinizing hormone subunit messenger ribonucleic acids in the estrous cycle of beef cattle. Anim Reprod Sci 2001; 68: 13–21. [DOI] [PubMed] [Google Scholar]
- 25.Wijayagunawardane MP, Miyamoto A, Cerbito WA, Acosta TJ, Takagi M, Sato K. Local distributions of oviductal estradiol, progesterone, prostaglandins, oxytocin and endothelin-1 in the cyclic cow. Theriogenology 1998; 49: 607–618. [DOI] [PubMed] [Google Scholar]
- 26.Okuda K, Uenoyama Y, Fujita Y, Iga K, Sakamoto K, Kimura T. Functional oxytocin receptors in bovine granulosa cells. Biol Reprod 1997; 56: 625–631. [DOI] [PubMed] [Google Scholar]
- 27.Ushizawa K, Takahashi T, Kaneyama K, Hosoe M, Hashizume K. Cloning of the bovine antiapoptotic regulator, BCL2-related protein A1, and its expression in trophoblastic binucleate cells of bovine placenta. Biol Reprod 2006; 74: 344–351. [DOI] [PubMed] [Google Scholar]
- 28.Nan X, Bu D, Li X, Wang J, Wei H, Hu H, Zhou L, Loor JJ. Ratio of lysine to methionine alters expression of genes involved in milk protein transcription and translation and mTOR phosphorylation in bovine mammary cells. Physiol Genomics 2014; 46: 268–275. [DOI] [PubMed] [Google Scholar]
- 29.Atli MO, Bender RW, Mehta V, Bastos MR, Luo W, Vezina CM, Wiltbank MC. Patterns of gene expression in the bovine corpus luteum following repeated intrauterine infusions of low doses of prostaglandin F2alpha. Biol Reprod 2012; 86: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aono A, Nagatomo H, Takuma T, Nonaka R, Ono Y, Wada Y, Abe Y, Takahashi M, Watanabe T, Kawahara M. Dynamics of intracellular phospholipid membrane organization during oocyte maturation and successful vitrification of immature oocytes retrieved by ovum pick-up in cattle. Theriogenology 2013; 79: 1146–1152.e1. [DOI] [PubMed] [Google Scholar]
- 31.Clarke AJ, Ellinghaus U, Cortini A, Stranks A, Simon AK, Botto M, Vyse TJ. Autophagy is activated in systemic lupus erythematosus and required for plasmablast development. Ann Rheum Dis 2014; 1: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JS, Lee GM. Monitoring of autophagy in Chinese hamster ovary cells using flow cytometry. Methods 2012; 56: 375–382. [DOI] [PubMed] [Google Scholar]
- 33.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 1999; 147: 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature 2000; 408: 488–492. [DOI] [PubMed] [Google Scholar]
- 35.Nath S, Dancourt J, Shteyn V, Puente G, Fong WM, Nag S, Bewersdorf J, Yamamoto A, Antonny B, Melia TJ. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat Cell Biol 2014; 16: 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi J, Jo M, Lee E, Choi D. The role of autophagy in corpus luteum regression in the rat. Biol Reprod 2011; 85: 465–472. [DOI] [PubMed] [Google Scholar]
- 37.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 2008; 181: 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 2009; 20: 1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.González-Polo RA, Boya P, Pauleau AL, Jalil A, Larochette N, Souquère S, Eskelinen EL, Pierron G, Saftig P, Kroemer G. The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J Cell Sci 2005; 118: 3091–3102. [DOI] [PubMed] [Google Scholar]
- 40.Kliem H, Berisha B, Meyer HH, Schams D. Regulatory changes of apoptotic factors in the bovine corpus luteum after induced luteolysis. Mol Reprod Dev 2009; 76: 220–230. [DOI] [PubMed] [Google Scholar]
- 41.Okuda K, Korzekwa A, Shibaya M, Murakami S, Nishimura R, Tsubouchi M, Woclawek-Potocka I, Skarzynski DJ. Progesterone is a suppressor of apoptosis in bovine luteal cells. Biol Reprod 2004; 71: 2065–2071. [DOI] [PubMed] [Google Scholar]
- 42.Korzekwa A, Murakami S, Wocławek-Potocka I, Bah MM, Okuda K, Skarzynski DJ. The influence of tumor necrosis factor alpha (TNF) on the secretory function of bovine corpus luteum: TNF and its receptors expression during the estrous cycle. Reprod Biol 2008; 8: 245–262. [DOI] [PubMed] [Google Scholar]
- 43.Keller CW, Fokken C, Turville SG, Lünemann A, Schmidt J, Münz C, Lünemann JD. TNF-alpha induces macroautophagy and regulates MHC class II expression in human skeletal muscle cells. J Biol Chem 2011; 286: 3970–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Djavaheri-Mergny M, Amelotti M, Mathieu J, Besançon F, Bauvy C, Souquère S, Pierron G, Codogno P. NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy. J Biol Chem 2006; 281: 30373–30382. [DOI] [PubMed] [Google Scholar]
- 45.Singh I. Textbook of human histology : (with colour atlas & practical guide). New Delhi; St. Louis: Jaypee Brothers Medical Publishers; 2011: 386 p. [Google Scholar]
- 46.Sawyer HR. Structural and functional properties of the corpus luteum of pregnancy. J Reprod Fertil Suppl 1995; 49: 97–110. [PubMed] [Google Scholar]
- 47.Niswender GD, Reimers TJ, Diekman MA, Nett TM. Blood flow: a mediator of ovarian function. Biol Reprod 1976; 14: 64–81. [DOI] [PubMed] [Google Scholar]
- 48.McClellan MC, Abel JH, Jr, Niswender GD. Function of lysosomes during luteal regression in normally cycling and PGF alpha-treated ewes. Biol Reprod 1977; 16: 499–512. [PubMed] [Google Scholar]
- 49.Sandes E, Lodillinsky C, Cwirenbaum R, Argüelles C, Casabé A, Eiján AM. Cathepsin B is involved in the apoptosis intrinsic pathway induced by Bacillus Calmette-Guérin in transitional cancer cell lines. Int J Mol Med 2007; 20: 823–828. [PubMed] [Google Scholar]
- 50.Niemi K, Teirilä L, Lappalainen J, Rajamäki K, Baumann MH, Öörni K, Wolff H, Kovanen PT, Matikainen S, Eklund KK. Serum amyloid A activates the NLRP3 inflammasome via P2X7 receptor and a cathepsin B-sensitive pathway. J Immunol 2011; 186: 6119–6128. [DOI] [PubMed] [Google Scholar]
- 51.Li S, Du L, Zhang L, Hu Y, Xia W, Wu J, Zhu J, Chen L, Zhu F, Li C, Yang S. Cathepsin B contributes to autophagy-related 7 (Atg7)-induced nod-like receptor 3 (NLRP3)-dependent proinflammatory response and aggravates lipotoxicity in rat insulinoma cell line. J Biol Chem 2013; 288: 30094–30104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.González-Fernández R, Martínez-Galisteo E, Gaytán F, Bárcena JA, Sánchez-Criado JE. Changes in the proteome of functional and regressing corpus luteum during pregnancy and lactation in the rat. Biol Reprod 2008; 79: 100–114. [DOI] [PubMed] [Google Scholar]
- 53.Kirsch TM, Friedman AC, Vogel RL, Flickinger GL. Macrophages in corpora lutea of mice: characterization and effects on steroid secretion. Biol Reprod 1981; 25: 629–638. [DOI] [PubMed] [Google Scholar]
- 54.Benyo DF, Pate JL. Tumor necrosis factor-alpha alters bovine luteal cell synthetic capacity and viability. Endocrinology 1992; 130: 854–860. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.