Abstract
Histone H2B monoubiquitination (H2Bub1) plays an important role in developmental regulation in various vertebrate species. However, the role of H2Bub1 in mammalian preimplantation development remains unclear. In the present study, we examined the role of H2Bub1 in the regulation of mouse preimplantation development. Based on immunocytochemical analysis using an anti-H2Bub1 antibody, no H2Bub1 signal was detected in the metaphase chromosomes of unfertilized oocytes or the pronuclei of early 1-cell stage embryos, but a weak signal was observed in late 1-cell stage embryos. The signal increased after cleavage into the 2-cell stage, and thereafter a strong signal was observed until the blastocyst stage. To assess the significance of H2Bub1 in the regulation of preimplantation development, RNF20 (an H2B-specific ubiquitin E3 ligase) was knocked down using small interfering RNA (siRNAs). In embryos treated with siRNA, the levels of Rnf20 mRNA and H2Bub1 decreased at the 4-cell and morula stages. Although these embryos developed normally until the morula stage, only one-third developed into the blastocyst stage. These results suggested that H2Bub1 is involved in the regulation of preimplantation development.
Keywords: H2B monoubiquitination, Mouse embryo, Preimplantation development
Monoubiquitination of histone H2B at the 120th lysine residue (H2Bub1) in mammals alters the chromatin structure and plays a role in developmental regulation in various multicellular organisms. H2Bub1 is catalyzed by Rad6p and Bre1p, a ubiquitin-conjugating E2 enzyme and ubiquitin E3 ligase, respectively, in yeast [1,2,3,4,5]. Rad6p is involved in the ubiquitination of various proteins [6,7,8,9]. The substrate specificity of Rad6p towards histone H2B is determined by Bre1p, which recruits Rad6p to the target chromatin [6, 10,11,12]. In Drosophila, H2B is monoubiquitinated by dBre1, the ortholog of yeast Bre1p. The decrease in H2Bub1 levels caused by a mutation in dBre1 results in the typical phenotype of Notch signaling disruption [13, 14]. In addition, an increase in H2Bub1 levels caused by a mutation in the Drosophila H2B ubiquitin protease scrawny increased the expression of Notch target genes, resulting in premature differentiation and a reduction in the number of stem cells in the germ line, follicles and intestine [13, 15]. In Arabidopsis, a deficiency in H2Bub1 caused by a mutation in Hub1 and Hub2 (plant orthologs of yeast Bre1) decreased seed dormancy and plant size and caused precocious flowering [13, 16, 17]. The increase in H2Bub1 levels caused by a mutation in ubiquitin-specific protease 26 also resulted in the early-flowering phenotype and smaller leaves [18]. RNF20 and RNF40 are human orthologs of yeast Bre1. Reduction of H2Bub1 using small interfering RNA (siRNAs) targeting these two genes resulted in decreased expression of HOX genes [19]. On the other hand, increase of H2Bub1 by overexpression of RNF20 caused abnormally stimulated expression of HOX genes [19]. Knockdown of Rnf20 led to insufficient upregulation of differentiation-related genes during mouse ES cell differentiation, resulting in deficient neural differentiation [20]. Numerous reports have shown that H2Bub1, which is mediated by Bre1p and its orthologs, plays pivotal roles in developmental regulation. However, to the best of our knowledge, little is known about the role of H2Bub1 in the regulation of preimplantation development.
Preimplantation development is driven by dynamically altered gene expression. In mice, transcription from the zygotic genome is initiated at the mid-1-cell stage after fertilization [21]. Although the transcriptional activity is very low during this stage, it increases gradually until the early 2-cell stage, after which a burst of transcription occurs at the mid to late 2-cell stage [21, 22]. Subsequently, active transcription occurs constantly until the blastocyst stage. The gene expression profile is also significantly altered as preimplantation development progresses [23,24,25]. Since H2Bub1 plays an important role in the regulation of active transcription in various types of cells and tissues [11, 20, 26,27,28], we hypothesized that H2Bub1 is involved in the regulation of preimplantation development through the control of gene expression.
In this report, we investigated the roles and dynamics of H2Bub1 in the regulatory mechanisms of preimplantation development. Our results showed that H2Bub1 was absent in unfertilized oocytes and early 1-cell stage embryos but present at low levels in late 1-cell stage embryos. It was detected at high levels during subsequent preimplantation development. Knockdown of Rnf20 using siRNAs decreased the rate of development to the blastocyst stage. In addition, the number of cells in embryos that could develop to the blastocyst stage decreased significantly. These results suggested that H2Bub1 is involved in the regulatory mechanisms of preimplantation development.
Materials and Methods
Collection and culture of oocytes and embryos
To collect unfertilized oocytes, cumulus-oocyte complexes (COCs) were obtained from 3-week-old BDF1 (B6D2F1) mice that had been injected with equine chorionic gonadotropin (ASKA Pharmaceutical, Tokyo) followed 48 h later by 5 IU human chorionic gonadotropin (ASKA Pharmaceutical). The COCs were transferred into human tubal fluid (HTF) medium [29] supplemented with 10 mg/ml BSA. Hyaluronidase was added to HTF medium at a final concentration of 300 μg/ml (Sigma-Aldrich, St. Louis, MO, USA) to remove cumulus cells. After being washed, the denuded oocytes were used for reverse transcription polymerase chain reaction (RT-PCR) or immunocytochemistry.
For in vitro fertilization, the COCs were transferred into HTF medium and inseminated with spermatozoa that had been collected from the cauda epididymis of mature male ICR mice (SLC, Shizuoka, Japan) and incubated in HTF medium supplemented with 1% BSA at 38 C for 2 h. Six hours after insemination, the fertilized oocytes were washed and then cultured in potassium simplex optimized medium (KSOM [30]) containing 3 mg/ml BSA.
All of the procedures using animals were reviewed and approved by the University of Tokyo Institutional Animal Care and Use Committee and were performed in accordance with the Guiding Principles for the Care and Use of Laboratory Animals.
Immunocytochemistry
Oocytes and embryos were fixed with 3.7% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 15 min at room temperature. After being washed with PBS containing 1% BSA (1% BSA/PBS), the cells were permeabilized with 0.2% Triton X-100 in PBS for 30 min and then washed with PBS containing 0.05% Tween 20. The cells were incubated with 4 N HCl containing 0.1% Triton X-100 for 10 min at room temperature and then transferred into 0.1 M Tris-HCl (pH 8.5) containing 0.02% Triton X-100. After a 30-min incubation, the cells were washed in 1% BSA/PBS and then incubated with an antibody against H2Bub1 (Medimabs, Mont-Royal, QC, Canada; #MM-0029-p; 1:200 dilution with 1% BSA/PBS) overnight at 4 C. After washing with 1% BSA/PBS, cells were incubated with a fluorescent-labeled secondary antibody, namely, Alexa Fluor 488 conjugated anti-mouse IgG (Life Technologies, Carlsbad, CA, USA, #A11001; 1:100 dilution). Cells were mounted on a glass slide in Vectashield antibleaching solution (Vector Laboratories, Burlingame, CA, USA) containing 3 μg/ml 4’,6-diamino-2-phenylindole (DAPI; Dojindo, Kumamoto). The fluorescence signals were detected using an LSM 5 Exciter confocal microscope system (Carl Zeiss, Oberkochen, Germany).
Semi-quantitative RT-PCR
Total RNA was extracted from 12 oocytes and embryos using ISOGEN (Nippon Gene, Tokyo, Japan) and subjected to reverse transcription using an Oligo dT primer and PrimeScript RT-PCR kit according to the manufacturer’s instructions (Takara, Ohtsu, Japan). As an external control, 50 pg rabbit α-globin mRNA purchased from Invitrogen was added prior to total RNA isolation. The obtained cDNA was used for PCR with Ex Taq DNA polymerase (Takara). The primers used for PCR were as follows: Rnf20, 5′-tgcagatgacctcaaagcac-3′ (forward) and 5′-ttcatcacacttgggcacat-3′ (reverse), and rabbit α-globin, 5′-gtgggacaggagcttgaaat-3′ (forward) and 5′-gcagccacggtggcgagtat-3′ (reverse). PCR was performed by denaturation at 95 C for 2 min followed by 37 (Rnf20) or 24 cycles (rabbit α-globin) of 95 C for 20 sec (Rnf20) or 30 sec (rabbit α-globin) (denaturation), 60 C for 20 sec (annealing) and 72 C for 30 sec (elongation). To confirm knockdown of Rnf20, the primer pair 5’-gagtttgagcagacccttgc-3’ (forward) and 5’-gcagacgtgtcttgttcagg-3’ (reverse) or 5’- gcatcacaccatgtctcagg-3’ (forward) and 5’-cacccgctctaggacttcag-3’ (reverse) was used. These primers were designed to amplify the region near the target sites of siRNAs against Rnf20. The PCR conditions for these primer pairs were as follows: 95 C for 2 min, followed by 32 cycles of 95 C for 30 sec (denaturation), 63 C for 30 sec (annealing) and 72 C for 30 sec (elongation). To analyze the effects of Rnf20 knockdown on the expression of genes encoding histones, RNA was extracted from 25 embryos at the morula stage. The isolated RNA was treated with DNase I, after which the cDNA was reverse transcribed using random six-mer primers. After reverse transcription, cDNA was subjected to PCR using Ex Taq. The primers for PCR were as follows: hist1h2ab, 5’-ctaaggccaagacccgctc-3’ (forward) and 5’-tcgccagattacttccccttg-3’ (reverse); h2afy, 5’- gctagcgaagaagcgaggat-3’ (forward) and 5’-cccctttcttgcctcctgtc-3’ (reverse); hist1h2bb, 5’-tcgtgaacgacatcttcgag-3’ (forward) and 5’-ccctacgagctcacttggag-3’ (reverse); hist1h2bg, 5’- gcttgtttctaccatgcccg-3’ (forward) and 5’- atggtcgagcgcttgttgta-3’ (reverse); and hist2h2bb, 5’-atcacttcccgggagatcca-3’ (forward), 5’-agccttttgggtaaagccga-3’ (reverse). PCR was performed by denaturation at 95 C for 2 min followed by 32 (h2afy), 35 (hist1h2bb, hist1h2bg and hist2h2bb) or 38 cycles (hist1h2ab) of 95 C for 30 sec (denaturation), 60 C (hist1h2ab, hist1h2bb, hist1h2bg and hist2h2bb) or 63 C (h2afy) for 30 sec (annealing) and 72 C for 30 sec (elongation). PCR products were separated on 2% agarose gels and stained with ethidium bromide. The gel image was obtained using a DT-20MP UV illuminator (ATTO, Tokyo).
Microinjection of siRNAs
Stealth RNAiTM small interfering RNAs (siRNAs) against Rnf20 (siRnf20 #1: 5’-cagucacaguucucuguccuguaua, and siRnf20 #2: 5’-caggaguucuguaaguugcagggua-3’) or GFP (siGFP: 5’-ccacuaccugagcacccaguccgcc-3’) were purchased from Life Technologies. Microinjection was performed on an inverted microscope (Eclipse TE300; Nikon, Tokyo, Japan) using a micromanipulator (Narishige) and microinjector (IM300; Narishige). To remove surrounding cumulus cells at 2 h post insemination, the zygotes were treated with hyaluronidase in HTF for 5 min, followed by repeated pipetting using narrow glass capillaries. Approximately 5 pl siRNA (10 μM) was microinjected into the cytoplasm of zygotes in Hepes-buffered KSOM using narrow glass capillaries (Harvard Apparatus; GC100 TF-10).
Immunosurgery of blastocysts
Embryos were collected at the blastocyst stage 96 h after insemination and treated with anti-mouse red blood cells (Rockland, Boyertown, PA, USA, #110-4139; 1:100) in α-MEM (Life Technologies) for 30 min at 37 C. The cells were washed and incubated in α-MEM for 5 min. To stain DNA in the trophectoderm, the embryos were incubated in α-MEM containing guinea pig complement sera (Sigma-Aldrich, Cat# S-1639; diluted 1:100) and propidium iodide (PI; Sigma-Aldrich, P4864; 1:100) for 10 min, after which the embryos were washed in α-MEM. Embryos were treated with hypotonic solution (0.9% sodium citrate containing 0.3% polyvinylpyrrolidone) and then fixed in a mixture of methanol, acetic acid, and sterilized water (5:1:4). The fixed embryos were adhered to the surface of the glass slide by dripping a solution of methanol and acetic acid (3:1) on them. The glass slides were dried and consecutively soaked in mixtures of methanol and acetic acid (3:1) and methanol, acetic acid and sterilized water (3:3:1) for 30 min and 1 min, respectively. The embryos were enclosed in Vectashield containing 3 µg/ml DAPI with cover slips. Images of the embryos were obtained using an LSM 5 Exciter confocal microscope system, after which the numbers of nuclei stained purple (trophectoderm) and blue (ICM) were counted using an LSM Image Browser (Carl Zeiss).
Results
Alteration of H2Bub1 during preimplantation development
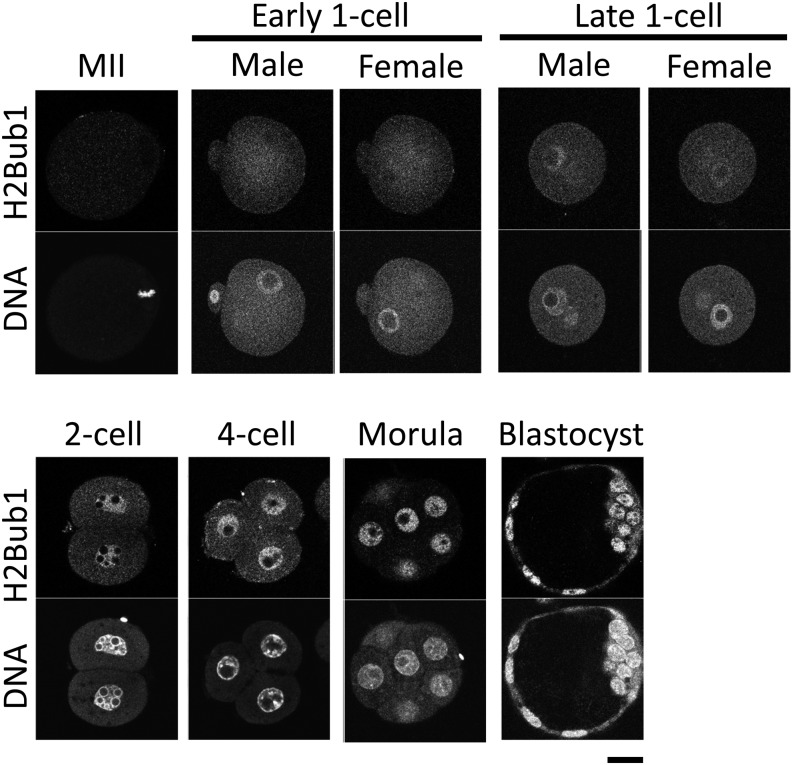
To examine the dynamics of H2Bub1 during preimplantation development, we performed immunocytochemistry using an anti-H2Bub1 antibody. H2Bub1 was not detected in the metaphase chromosomes of MII oocytes or the pronuclei of early 1-cell stage embryos (Fig. 1). However, a weak fluorescent signal of H2Bub1 was observed at the late 1-cell stage, and the signal intensity increased after cleavage into the 2-cell stage. The intense H2Bub1 signal was maintained during subsequent preimplantation development until the blastocyst stage (Fig. 1).
Fig. 1.
H2Bub1 in mouse preimplantation embryos. The MII stage oocytes (MII) and preimplantation embryos were immunostained with anti-ubiquitinated H2B (H2Bub1) antibody. Embryos at the early 1-cell, late 1-, 2- and 4-cell, morula and blastocyst stages were collected 6, 12, 28, 45, 70 and 96 h after insemination, respectively. Images of two confocal planes are shown for 1-cell stage embryos to clearly reveal the male and female pronuclei. More than three independent experiments (in which more than 20 oocytes/embryos were observed) were performed for each stage of oocyte and preimplantation embryo. DNA was stained with 4’,6-diamino-2-phenylindole (DAPI). Similar results were obtained in each experiment, and representative images are shown. Bar = 20 μm.
H2Bub1 plays a role in preimplantation development
As described above, H2Bub1 appeared after fertilization and was maintained at a high level until the blastocyst stage. Based on this result, we assessed the significance of H2Bub1 for the progression of preimplantation development. To accomplish this, we knocked down Rnf20, a histone H2B-specific E3 ubiquitin ligase, and examined whether the decrease in H2Bub1 caused by Rnf20 knockdown affected preimplantation development. In previous studies, knockdown of Rnf20 significantly decreased H2Bub1 levels in several different types of cells [20, 31,32,33].
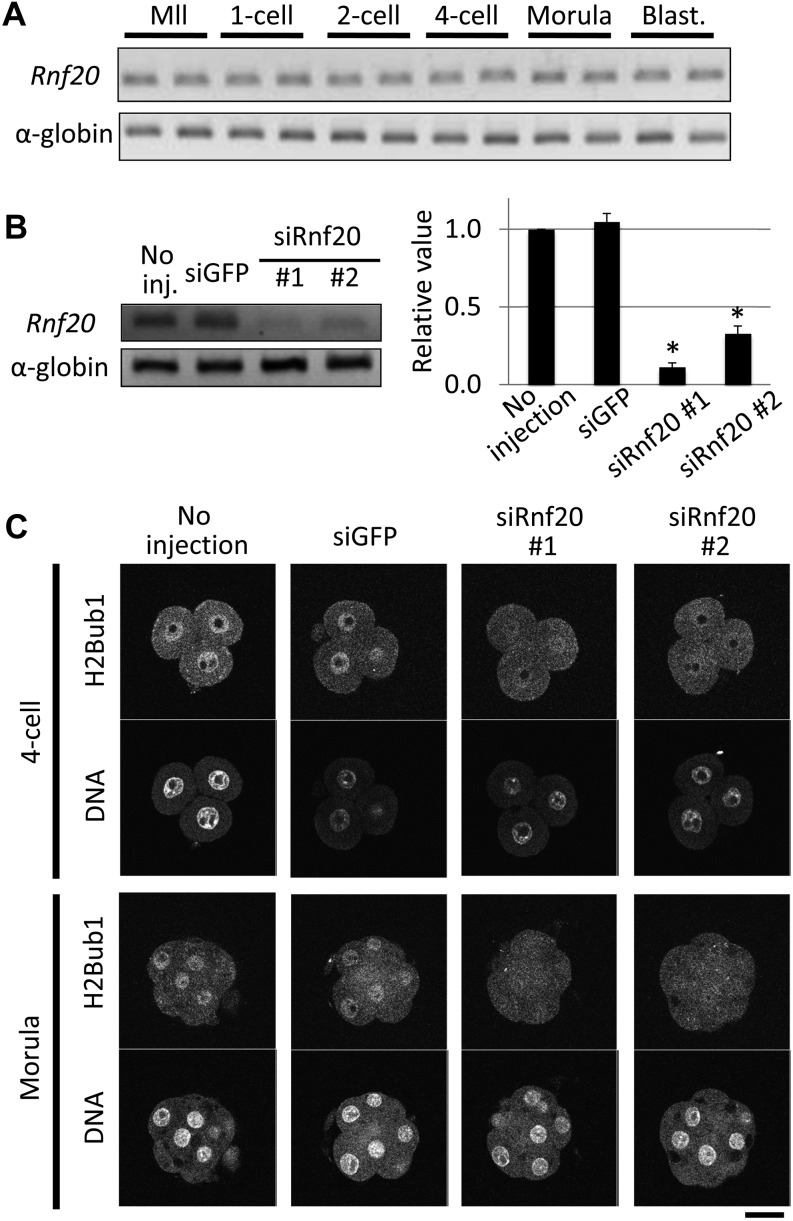
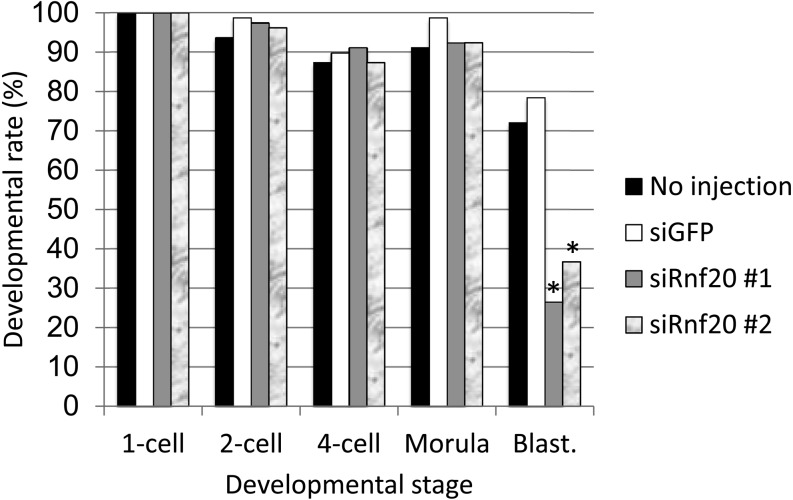
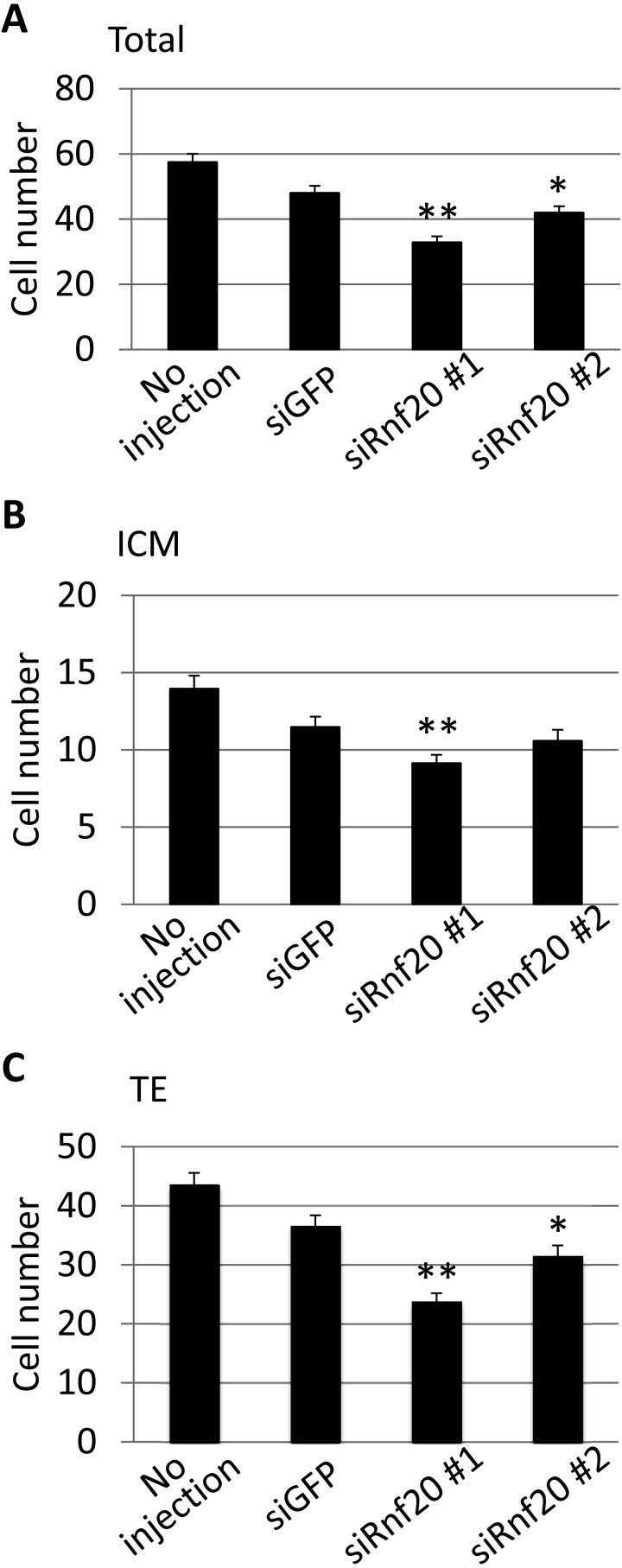
First, we examined the expression of Rnf20 in preimplantation embryos. RT-PCR analysis revealed that Rnf20 mRNA was expressed at a constant level throughout preimplantation development (Fig. 2A). To suppress Rnf20 expression, siRNAs against Rnf20 were injected into 1-cell stage embryos. The level of Rnf20 mRNA was reduced efficiently by the two siRNAs (siRnf20 #1 and #2) at the 4-cell stage, and the level of H2Bub1 was decreased in 4-cell and morula stage embryos (Fig. 2B and C), although the siRNAs could not reduce the H2Bub1 level at the 1- and 2-cell stages (data not shown). These results indicate that Rnf20 is an essential factor regulating H2Bub1 in preimplantation embryos, as observed in several types of cells. The Rnf20-knocked down embryos developed normally until the morula stage. However, only one-third developed to the blastocyst stage, whereas more than 70% of control embryos injected with GFP siRNA developed to the blastocyst stage (Fig. 3). Among the embryos that had developed to the blastocyst stage, the number of cells was significantly smaller in the embryos injected with Rnf20 siRNAs than in the control embryos (P < 0.05 and < 0.01 for siRnf20 #1 and siRnf20 #2, respectively, based on the Student’s t-test) (Fig. 4A). The decrease in cell number was observed both in the inner cell mass and trophectoderm, which were discriminated based on differential staining of nuclei after immunosurgery (Fig. 4B and C). Taken together, these results suggested that H2Bub1 is involved in preimplantation development.
Fig. 2.
Knockdown of Rnf20 by siRNAs decreased H2Bub1 levels in preimplantation embryos. (A) Expression of ring finger protein 20 (Rnf20) was examined using RT-PCR in MII stage oocytes (MII) and 1-, 2- and 4-cell, morula and blastocyst (Blast.) stage embryos, which were collected 0, 12, 28, 45, 70 and 96 h after insemination, respectively. Three independent experiments with duplicate samples were performed, and a representative image is shown. Rabbit α-globin was used as an external control. (B) Knockdown of Rnf20 using siRNAs. Small interfering RNAs against Rnf20 (siRnf20 #1 and #2) or green fluorescent protein (siGFP) were injected into the cytoplasm of the embryos 2 h after insemination. The Rnf20 transcript was examined by RT-PCR at the 4-cell stage 45 h after insemination. The intensities of the bands were quantified and shown in the bar graph. The value of no injection was set as 1, and the values of other groups were expressed relative to this value. Asterisks indicate statistically significant difference compared with control siGFP (P < 0.05; Student’s t-test). Four independent experiments were performed. Error bars represent the SEM. (C) Decreases in H2Bub1 in the Rnf20 knock-down embryos. Embryos injected with siRNAs against Rnf20 or GFP were collected 45 and 70 h after insemination (4-cell and morula stages, respectively) and immunostained with anti-H2Bub1 antibody. DNA was stained with DAPI. Three independent experiments were conducted, and representative images are shown. As a control, embryos without injection and those injected with siGFP were examined.
Fig. 3.
Effect of Rnf20 knockdown on the progression of preimplantation development. The small interfering RNAs against Rnf20 (siRnf20 #1 and #2) or green fluorescent protein (siGFP) were injected into the cytoplasm of the embryos 2 h after insemination. Development to the 1-, 2- and 4-cell, morula and blastocyst (Blast.) stages was observed at 12, 28, 45, 70 and 96 h after insemination, respectively. Four independent experiments were performed, and the data were accumulated. A total of 79 embryos were examined in each experimental group. Black, white, grey and marble bars indicate no injection, siGFP, siRnf20 #1 and siRnf20 #2, respectively. Values indicated by asterisks are significantly lower than the control values (siGFP) (P < 0.01; χ2-test).
Fig. 4.
Effect of Rnf20 knockdown on the cell numbers in embryos at the blastocyst stage. The embryos were injected with small interfering RNAs against Rnf20 (siRnf20 #1 and #2) or green fluorescent protein (siGFP) 2 h after insemination and collected at the blastocyst stage 96 h after insemination. The numbers of cells in the whole embryos (A), in the inner cell mass (ICM; B) and in the trophectoderm (TE; C) were determined. For the determination of cell numbers in the ICM and TE, the nuclei of the embryos were stained differentially after immunosurgery. Four independent experiments were performed, and the data were accumulated. In each experimental group, more than 44 embryos were analyzed in total. Error bars indicate standard errors (SE). Values indicated by asterisks are significantly lower than those of the control embryos (siGFP) (based on the Student’s t-test; *P < 0.05; **P < 0.01).
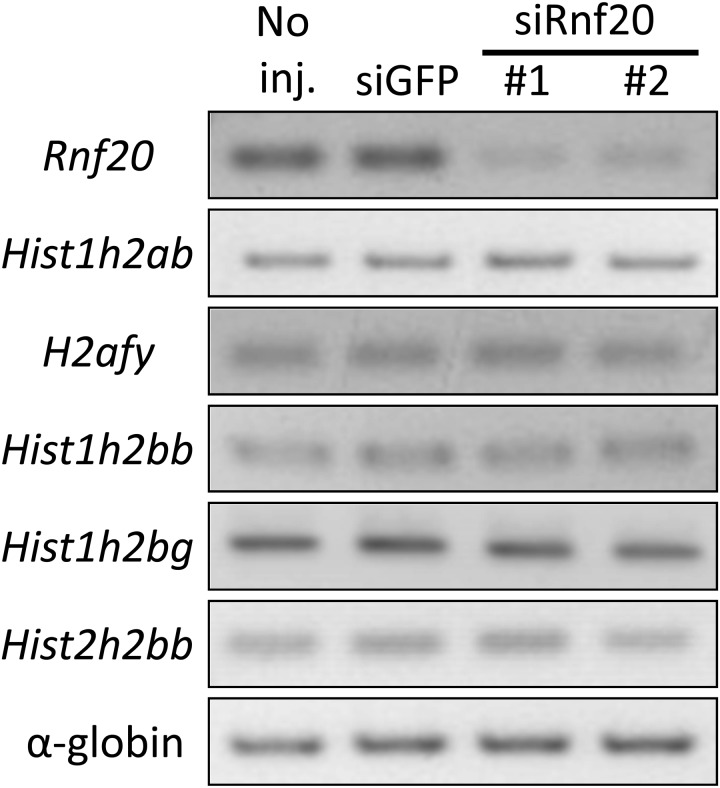
Effects of decreased H2Bub1 levels on gene expression in preimplantation embryos
It has been reported that H2Bub1 is involved in developmental regulation at various stages by regulating gene expression in several organisms. Based on the decrease in developmental rate caused by the knockdown of Rnf20, we investigated the effects of decreased H2Bub1 levels on gene expression in preimplantation embryos. Since it was reported that the expression of histone H2A and H2B genes was significantly decreased upon depletion of RNF20 in human somatic cells [33], we explored the expression of Hist1h2ab, H2afy, Hist1h2bb, Hist1h2bg and Hist2h2bb in Rnf20 knockdown embryos. However, no obvious change in the expression of these genes occurred after the H2Bub1 levels decreased in morula stage embryos (Fig. 5).
Fig. 5.
Effect of Rnf20 knockdown on the expression of histone H2A and H2B genes. The expression of histone H2A and H2B genes in the Rnf20 knockdown embryos was examined using RT-PCR. The embryos were injected with siRnf20 #1, siRnf20 #2 or siGFP 2 h after insemination and collected at the morula stage 70 h after insemination. Two independent experiments were performed, and similar results were obtained. A representative image is shown. Rabbit α-globin was used as an external control.
Discussion
In this study, we examined the role of H2Bub1 in the regulation of preimplantation development. Immunocytochemical analysis showed that H2Bub1 was absent from the pronuclei of early 1-cell stage embryos. However, a weak H2Bub1 signal was detected during the late 1-cell stage. The signal became stronger at the 2-cell stage and remained high until the blastocyst stage (Fig. 1). Knockdown of Rnf20 via siRNAs decreased H2Bub1 in the nuclei (Fig. 2), which caused a reduction in blastocyst stage development (Figs. 3 and 4). These results suggest that H2Bub1 mediated by RNF20 is involved in the regulation of preimplantation development.
H2Bub1 likely regulates preimplantation development through the regulation of gene expression, since H2Bub1 stimulates transcriptional elongation by RNA polymerase II (RNAPII) [26, 27]. The H2Bub1 levels changed in a manner to the transcriptional activity during preimplantation development. It was previously reported that transcriptional activity was not detected in the early 1-cell stage, appeared in the late stage but at a low level and then increased in the 2-cell stage [21]. The reduction of H2Bub1 by Rnf20 knockdown via siRNAs caused developmental arrest at the morula stage (Fig. 3), although it was reported that embryos arrested at the 2-cell stage when transcription was inhibited after fertilization [34]. Since the H2Bub1 level was not reduced by siRNAs before the 4-cell stage, the role of H2Bub1 in developmental regulation at the 1- and 2-cell stages remains unknown. It would be interesting to determine whether H2Bub1 is involved in zygotic gene activation during this period.
Previous reports suggested that H2Bub1 regulates histone H3K4 and H3K79 methylation, which are tightly associated with active transcription [3, 12, 35,36,37,38,39]. H2Bub1 may regulate changes in gene expression by altering methylation of these residues during preimplantation development. However, H3K79 dimethylation (H3K79me2) is maintained at low levels after fertilization and then increases at the blastocyst stage [40], although H2Bub1 was already detected in the late 1-cell stage and increased in the 2-cell stage in the present study (Fig. 1). The changes in H2Bub1 differed from those in H3K4 trimethylation (H3K4me3). H3K4me3 increased in the male pronuclei at the 1-cell stage. However, it decreased in the 2-cell stage, and the lower level of H3K4me3 was maintained until the blastocyst stage [41, 42]. Thus, H2Bub1 does not seem to act as an upstream mechanism for H3K79me2 and H3K4me3 during preimplantation development. Instead, other mechanisms may cause the dynamic changes in H3K79me2. Indeed, DOT1L (the sole H3K79 methyltransferase) was not localized in the nucleus at the 2-cell stage, and forced nuclear localization of DOT1L caused hypermethylation of H3K79me2 in 2-cell stage embryos [43], suggesting that H3K79me2 is regulated by the nuclear localization of DOT1L during this stage. On the other hand, H3K4me3 is catalyzed by several histone methyltransferases and demethylases [44, 45]. It remains unclear which of these enzymes is involved in the changes in H3K4me3 during preimplantation development.
We analyzed the expression of specific histone genes that are positively regulated by H2Bub1 in human somatic cells in Rnf20-deficient preimplantation embryos. No apparent effect on the expression of these genes was observed (Fig. 5). The results suggested that the roles of H2Bub1 in the regulation of gene expression are dependent on cell type. Rnf20-deficient ES cells did not show abnormal gene expression profiles, except when treated with retinoic acid to induce differentiation [20]. This may be important for understanding the role of H2Bub1 in preimplantation development and identifying genes that are misregulated in H2Bub1-deficient preimplantation embryos. A comprehensive analysis of the transcriptome via RNA sequencing may be used to identify these genes.
References
- 1.Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell 2003; 11: 261–266. [DOI] [PubMed] [Google Scholar]
- 2.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science 2000; 287: 501–504. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev 2011; 25: 1345–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev 2003; 17: 2733–2740. [DOI] [PubMed] [Google Scholar]
- 5.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 2006; 75: 243–269. [DOI] [PubMed] [Google Scholar]
- 6.Osley MA. Regulation of histone H2A and H2B ubiquitylation. Brief Funct Genomics Proteomics 2006; 5: 179–189. [DOI] [PubMed] [Google Scholar]
- 7.Watkins JF, Sung P, Prakash S, Prakash L. The extremely conserved amino terminus of RAD6 ubiquitin-conjugating enzyme is essential for amino-end rule-dependent protein degradation. Genes Dev 1993; 7: 250–261. [DOI] [PubMed] [Google Scholar]
- 8.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002; 419: 135–141. [DOI] [PubMed] [Google Scholar]
- 9.Sung P, Berleth E, Pickart C, Prakash S, Prakash L. Yeast RAD6 encoded ubiquitin conjugating enzyme mediates protein degradation dependent on the N-end-recognizing E3 enzyme. EMBO J 1991; 10: 2187–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, Johnston M, Shilatifard A. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell 2003; 11: 267–274. [DOI] [PubMed] [Google Scholar]
- 11.Shukla A, Chaurasia P, Bhaumik SR. Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability. Cell Mol Life Sci 2009; 66: 1419–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 2012; 81: 65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright DE, Wang C-Y, Kao C-F. Flickin’ the ubiquitin switch: the role of H2B ubiquitylation in development. Epigenetics 2011; 6: 1165–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bray S, Musisi H, Bienz M. Bre1 is required for Notch signaling and histone modification. Dev Cell 2005; 8: 279–286. [DOI] [PubMed] [Google Scholar]
- 15.Buszczak M, Paterno S, Spradling AC. Drosophila stem cells share a common requirement for the histone H2B ubiquitin protease scrawny. Science 2009; 323: 248–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao Y, Dai Y, Cui S, Ma L. Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 2008; 20: 2586–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng J, Shen W-H. Dynamic regulation and function of histone monoubiquitination in plants. Front Plant Sci 2014; 5: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitz RJ, Tamada Y, Doyle MR, Zhang X, Amasino RM. Histone H2B deubiquitination is required for transcriptional activation of FLOWERING LOCUS C and for proper control of flowering in Arabidopsis. Plant Physiol 2009; 149: 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu B, Zheng Y, Pham A-D, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell 2005; 20: 601–611. [DOI] [PubMed] [Google Scholar]
- 20.Fuchs G, Shema E, Vesterman R, Kotler E, Wolchinsky Z, Wilder S, Golomb L, Pribluda A, Zhang F, Haj-Yahya M, Feldmesser E, Brik A, Yu X, Hanna J, Aberdam D, Domany E, Oren M. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol Cell 2012; 46: 662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 1997; 181: 296–307. [DOI] [PubMed] [Google Scholar]
- 22.Schultz RM. Regulation of zygotic gene activation in the mouse. BioEssays 1993; 15: 531–538. [DOI] [PubMed] [Google Scholar]
- 23.Hamatani T, Carter MG, Sharov AA, Ko MSH. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 2004; 6: 117–131. [DOI] [PubMed] [Google Scholar]
- 24.Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell 2004; 6: 133–144. [DOI] [PubMed] [Google Scholar]
- 25.Latham KE, Schultz RM. Embryonic genome activation. Front Biosci 2001; 6: d748–d759. [DOI] [PubMed] [Google Scholar]
- 26.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 2006; 125: 703–717. [DOI] [PubMed] [Google Scholar]
- 27.Fuchs G, Oren M. Writing and reading H2B monoubiquitylation. Biochim Biophys Acta 2014; 1839: 694–701. [DOI] [PubMed] [Google Scholar]
- 28.Chandrasekharan MB, Huang F, Sun Z-W. Histone H2B ubiquitination and beyond: Regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation. Epigenetics 2010; 5: 460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinn P, Begley AJ. Effect of human seminal plasma and mouse accessory gland extracts on mouse fertilization in vitro. Aust J Biol Sci 1984; 37: 147–152. [DOI] [PubMed] [Google Scholar]
- 30.Erbach GT, Lawitts JA, Papaioannou VE, Biggers JD. Differential growth of the mouse preimplantation embryo in chemically defined media. Biol Reprod 1994; 50: 1027–1033. [DOI] [PubMed] [Google Scholar]
- 31.Shema E, Kim J, Roeder RG, Oren M. RNF20 inhibits TFIIS-facilitated transcriptional elongation to suppress pro-oncogenic gene expression. Mol Cell 2011; 42: 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vethantham V, Yang Y, Bowman C, Asp P, Lee J-H, Skalnik DG, Dynlacht BD. Dynamic loss of H2B ubiquitylation without corresponding changes in H3K4 trimethylation during myogenic differentiation. Mol Cell Biol 2012; 32: 1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, Hublarova P, Moyal L, Gana-Weisz M, Shiloh Y, Yarden Y, Johnsen SA, Vojtesek B, Berger SL, Oren M. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev 2008; 22: 2664–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warner CM, Versteegh LR. In vivo and in vitro effect of alpha-amanitin on preimplantation mouse embryo RNA polymerase. Nature 1974; 248: 678–680. [DOI] [PubMed] [Google Scholar]
- 35.Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim J-E, Chen J, Lazar MA, Blobel GA, Vakoc CR. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol 2008; 28: 2825–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129: 823–837. [DOI] [PubMed] [Google Scholar]
- 37.Vermeulen M, Mulder KW, Denissov S, Pijnappel WWMP, van Schaik FMA, Varier RA, Baltissen MPA, Stunnenberg HG, Mann M, Timmers HTM. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 2007; 131: 58–69. [DOI] [PubMed] [Google Scholar]
- 38.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 2007; 25: 15–30. [DOI] [PubMed] [Google Scholar]
- 39.Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, Roeder RG. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 2013; 152: 1021–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ooga M, Inoue A, Kageyama S, Akiyama T, Nagata M, Aoki F. Changes in H3K79 methylation during preimplantation development in mice. Biol Reprod 2008; 78: 413–424. [DOI] [PubMed] [Google Scholar]
- 41.Lepikhov K, Walter J. Differential dynamics of histone H3 methylation at positions K4 and K9 in the mouse zygote. BMC Dev Biol 2004; 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bošković A, Bender A, Gall L, Ziegler-Birling C, Beaujean N, Torres-Padilla M-E. Analysis of active chromatin modifications in early mammalian embryos reveals uncoupling of H2A.Z acetylation and H3K36 trimethylation from embryonic genome activation. Epigenetics 2012; 7: 747–757. [DOI] [PubMed] [Google Scholar]
- 43.Ooga M, Suzuki MG, Aoki F. Involvement of DOT1L in the remodeling of heterochromatin configuration during early preimplantation development in mice. Biol Reprod 2013; 89: 145. [DOI] [PubMed] [Google Scholar]
- 44.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell 2007; 131: 633–636. [DOI] [PubMed] [Google Scholar]
- 45.Boland MJ, Nazor KL, Loring JF. Epigenetic regulation of pluripotency and differentiation. Circ Res 2014; 115: 311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]