Summary
Platelets are generated from nucleated precursors referred to as megakaryocytes. The formation of platelets is one of the most elegant and unique developmental processes in eukaryotes. Because they enter the circulation without nuclei, platelets are often considered simple, non-complex cells that have limited functions beyond halting blood flow. However, emerging evidence over the past decade demonstrates that platelets are more sophisticated than previously considered. Platelets carry a rich repertoire of messenger RNAs (mRNAs), microRNAs (miRNAs), and proteins that contribute to primary (adhesion, aggregation, secretion) and alternative (immune regulation, RNA transfer, translation) functions. It is also becoming increasing clear that the “genetic code” of platelets changes with race, genetic disorders, or disease. Changes in the “genetic code” can occur at multiple points including megakaryocyte development, platelet formation, or in circulating platelets. This review focuses on regulation of the “genetic code” in megakaryocytes and platelets and its potential contribution to health and disease.
Keywords: megakaryocytes, platelets, genes, human
Introduction
Despite lacking nuclei, there is growing evidence that platelets have complex molecular signatures that influence their function in health and disease. This includes a rich portfolio of mRNAs and miRNAs. We are just beginning to understand the importance of RNAs in platelets, and how the megakaryocyte contributes to mRNA and miRNA expression patterns in circulating platelets. This review focuses on regulation of the genetic code in megakaryocytes and platelets where the genetic code is defined as: transfer of genetic information in the form of mRNAs and miRNAs from megakaryocytes to platelets.
Megakaryopoiesis and Thrombopoiesis: Key Determinants of the Genetic Code
Megakaryocytes descend from pluripotent stem cells and undergo multiple DNA replications without cell divisions by a unique process referred to as endomitosis [1–2]. The process of megakaryocyte maturation and differentiation is called megakaryopoiesis [3–4]. Committed megakaryocytes eventually produce hundreds if not thousands of circulating platelets. Thrombopoiesis is the term ascribed to the generation of platelets (i.e., thrombocytes) [4–5]. Although the exact duration of the combined processes in vivo are not known, megakaryopoiesis and thrombopoiesis occurs over a two-week period in ex vivo human culture systems[6]. Although shorter in duration, the process of megakaryopoiesis and thrombopoiesis in murine fetal-liver cell-derived megakaryocytes is similar to humans [7].
Megakaryopoiesis and thrombopoiesis are controlled by multiple cytokines, although thrombopoietin (TPO) is the key regulator of megakaryocyte maturation and proliferation[8]. Different matrices and exposure to flow also regulate proplatelet formation [9–13]. Genetic studies indicate that transcriptional responses driven by GATA-1 and Nuclear Factor-Erythroid 2 (NF-E2) control megakaryocyte development and differentiation [1, 14–16]. Transcription of mRNA and subsequent translation of the message into protein is essential for producing requisite cytoskeletal, granule, intracellular signaling, adhesion, and other proteins that are packaged into platelets[1]. These proteins guide and control the primary functions of platelets, which involve thrombus formation and cessation of blood flow.
A primary thrust of investigations focused on megakaryopoiesis and thrombopoiesis has been to identify culture mediums, cytokine cocktails, and extracellular matrices that yield high numbers of platelets with morphologic characteristics and functions similar to platelets that are freshly-isolated from the bloodstream. Results from these studies indicate that slight alterations in the in vitro culture conditions and/or constituents can significantly influence the generation and phenotype of newly-formed platelets[17–23]. This strongly suggests that changes in the bone marrow milieu can alter transcriptional, translational, and post-translational processes throughout megakaryocyte development, differentiation, and proplatelet formation. It also infers that megakaryocytes transfer unique genetic codes to platelets that are fluid, especially in disease situations.
The Genetic Code of Megakaryocytes
As described above, megakaryopoiesis and thrombopoiesis are complex processes where gene expression is under strict regulatory control[1]. As a result, there is ample opportunity for the genetic code to change as progenitor cells differentiate into mature megakaryocytes that produce platelets. Unfortunately, the exact genetic code of megakaryocytes is not known because megakaryocytes constitute less than 1% of all marrow cells[1]. Thus, it is extremely difficult to isolate sufficient numbers of purified bone marrow megakaryocytes, especially from human subjects. To overcome these challenges, investigators utilize immortalized or primary progenitor cell culture systems to study proplatelet-producing megakaryocytes. These culture systems are designed to reflect the in vivo situation as much as possible, but results need to be viewed with caution since in vitro megakaryocyte cultures do not completely replicate the bone marrow milieu.
Studies in cultured systems do indicate that megakaryocytes transcribe thousands of mRNAs and express a diverse and rich repertoire of miRNAs, which regulate platelet biogenesis (reviewed by Edelstein and colleagues[24–25]). RNA expression studies in cultured megakaryocytes have several features including: proteins known to be present in platelets are generally detected at the mRNA level in megakaryocytes[26]; differentiated megakaryocytes have remarkably different mRNA expression patterns than non-megakaryocytic cells[27]; RNA expression patterns are fluid throughout megakaryocyte development and platelet biogenesis[28–29], and; by inference, RNAs present in megakaryocytes are thought to be transferred to circulating platelets[30].
mRNA and miRNA expression profiling in cultured megakaryocytes has primarily been used to identify key genes that regulate megakaryopoiesis and thrombopoiesis. Numerous studies have shown that miRNA regulation of mRNA stability and translation is critical for megakaryopoiesis and megakaryocyte differentiation [24–25]. However, very few studies in megakaryocytes have attempted to dissect how mRNAs and miRNAs are shuttled into platelets. Cecchetti et al.[26] has shown that proplatelet producing megakaryocytes differentially sort mRNAs for matrix metalloproteinases (MMPs) and tissue inhibitors of matrix metalloproteinases (TIMPs) into platelets. The mechanisms by which MMP and TIMP mRNAs are differentially transferred to platelets, however, remain unknown. There is also a paucity of studies examining whether changes in the megakaryocyte milieu alters the types and amounts of RNAs that are transferred to platelets.
In summary, the genetic code of megakaryocytes is incompletely understood and just recently being appreciated. However, there is strong circumstantial evidence that megakaryocytes transfer a rich and diverse repertoire of mRNAs and miRNAs to circulating platelets and that this transferable genetic code changes in disease (see below).
Megakaryocyte Investment of the Genetic Code into Platelets
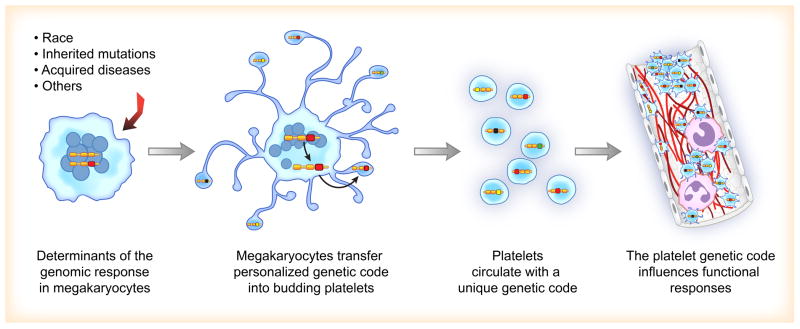
It is becoming increasing more clear that variations in race, genetic mutations, and acute disease affect the types of transcripts transferred from megakaryocytes to platelets. Changes in the megakaryocyte genetic code are thought to be reflected in the platelet transcriptome (Figure 1). In regards to race, platelet miRNA levels, which are capable of destabilizing mRNA, have recently been shown to inversely correlate with the expression of target transcripts in black subjects [31–32]. In the Platelet RNA and eXpression-1 (PRAX1) study, Edelstein and coworkers[31] found that a number of platelet mRNAs and miRNAs were differentially expressed in black versus white subjects. One of these mRNAs encoded for phosphatidylcholine transfer protein (PCTP, a regulator of lipids). Compared to white subjects, black subjects had significantly higher expression of PCTP mRNA and protein, which correlated with increased PAR4-mediated platelet aggregation. These investigators also identified that platelets isolated from black subjects had a network of differentially expressed miRNA-mRNA pairs, compared to white subjects. Prediction analyses and subsequent validation identified one of these miRNAs as miR-376c. miR-376c targets the PCTP gene, was inversely correlated with PTCP mRNA and protein levels, and was inversely correlated with PAR4 reactivity in humans. Further work identified the locus mapping to chromosome 14q32.2, which contained a large cluster of miRNA genes (commonly called the DLK1-DIO3 genomic region), the expression of which was significantly associated with race (black versus white race, P= 1.09 × 10−5), even after controlling for potential covariates. Follow-up studies by this same group of investigators also identified that platelets from blacks had 3 common single nucleotide polymorphisms in the PAR4 gene that were correlated with PAR4-induced platelet aggregation[32]. Together these data highlight that the genetic code of platelets – and presumably megakaryocytes, although this remains to be demonstrated – differs between black and white subjects and influences key platelet functional responses such as aggregation.
Figure 1. Megakaryocyte Investment of the Genetic Code into Platelets.
Variations in race, genetic mutations, or acute disease are associated with changes in the genetic code (i.e., mRNA and miRNA expression patterns) in megakaryocytes. Changes in the megakaryocyte genetic code are then transferred into developing platelets that are released into the circulation. Alterations in the platelet transcriptome and corresponding proteins influence functional responses in both health and disease.
Inherited gene mutations in the megakaryocyte also contribute to an altered genetic code in released platelets. In this regard, mutations in just one gene may have profound effects on other mRNAs (and their protein products and associated functions) in megakaryocytes and platelets. One example in humans is gray platelet syndrome (GPS), for which the causative mutation was recently identified[33]. GPS, an autosomal recessive inherited platelet disorder, is characterized by thrombocytopenia and large platelets that lack α granules. Kahr et al.[33] used next-generation genome-wide RNA sequencing (RNA-seq) of platelets to identify that mutations in the NBEAL2 are responsible for GPS. These RNA-seq studies also revealed that several transcripts (>100) were differentially expressed in platelets from GPS patients compared to platelets from unaffected individuals. The platelet transcriptome is also altered in patients with germline mutations in ETV6 (ets variant 6) that have thrombocytopenia and leukemia predisposition [34]. Together, these studies in humans suggest that targeted deletion of genes in mice is likely to have marked effects on the genetic code of megakaryocytes and platelets and their functional responses. They also raise the possibility that mouse models may be used to produce genetically-modified platelets through engineering of the genetic code.
Acute and chronic diseases also alter the platelet genetic code. Seminal work by Healy and colleagues[35] identified significant quantitative differences in the platelet transcriptome between patients with acute ST-segment-elevation myocardial infarction (STEMI) and patients with stable coronary disease. Specifically, these investigators found that by profiling platelet mRNA, they could identify that the expression of CD69 (involved in platelet aggregation) and myeloid-related protein-8 (MRP-8/14, involved in calcium signaling, arachidonic acid metabolism, and cytoskeletal reorganization) was significantly increased in isolated platelets from STEMI patients. These findings suggested that the platelet transcriptome is dynamic and predicts functional alterations of clinical significance.
Another human disease state where the platelet transcriptome appears to be altered is in patients with systemic lupus erythematosis (SLE). SLE is an autoimmune disease known for injurious inflammatory responses in both solid organs and the circulatory system. Much of this inflammation is thought to be driven by an ongoing type I interferon response. SLE is also associated with an increased risk of cardiovascular disease and atherothrombosis. Recent work by Lood and colleagues[36] used real-time PCR to investigate whether platelets isolated from patients with SLE displayed a type I interferon (IFN) gene signature. These investigators found that platelets from SLE patients, compared to healthy controls, had highly increased expression of many type I IFN-regulated genes. Among the genes validated in more detail, the mRNA expression of CD69 in platelets from SLE patients was >6-fold higher relative to healthy controls. Expression of CD69 protein (and other type I IFN proteins) was also significantly increased in SLC patients compared to controls. Platelets from SLE patients also exhibited heightened platelet reactivity, as measured by binding of annexin V and the formation of platelet-monocyte aggregates, compared to platelets from control subjects [37–39]. In the immortalized megakaryocyte cell line MEG-01s, stimulation with IFNα led to significant increases in the expression of CD69 and other IFN-regulated proteins. Taken together, the authors concluded that platelets from SLE patients have a type I IFN transcriptional signature with increased levels of corresponding proteins, including CD69[36]. These findings also support the hypothesis that in SLE patients, circulating IFNα production by plasmacytoid dendritic cells may alter the genetic code of megakaryocytes within the bone marrow niche, leading to the production of platelets enriched for IFN-regulated proteins.
Body mass index and circulating levels of C-reactive protein (CRP) and interleukin-6 are also associated with changes in platelet gene expression [40–41], and there are many other published lines of evidence consistent with the supposition that human diseases are associated with changes in the genetic code of platelets[30, 42–45]. While an in-depth discussion of these established and emerging findings are beyond the scope of the current brief review, the reader is encouraged to keep an eye on this rapidly evolving area of investigation. It is also important to note that the aforementioned studies infer that differential transfer of mRNA occurs during proplatelet formation. While this may be true, there is evidence that tumor cells transfer RNA into platelets [46]. The study by Nilsson and colleagues [46] suggests that RNA profiling in blood platelets may serve as a diagnostic platform for cancer, but it is unknown if this type of RNA transfer has functional consequences in platelets.
Regulation of the Genetic Code in Circulating Platelets
When mRNAs reach platelets their structural features mimic their expression in megakaryocytes. In other words, they are of predicted length (i.e., not degraded) and each mRNA is capped and polyadenylated on the 5′- and 3′-untranslated region, respectively [47–50]. Platelet miRNAs also resemble miRNAs found in megakaryocytes and nucleated cells [51–52]. Surprisingly, platelets have functional machinery that allows them to splice pre-mRNAs into mature mRNAs and process pre-miRNAs into miRNAs [51, 53–54]. These unexpected splicing/processing functions provide anucleate platelets with alternative paths of gene regulation that can affect their genetic code.
RNAs are presumably fairly stable in platelets, but in vivo studies of RNA stability have not been performed. In situ hybridization studies from freshly-isolated human platelets suggest that all subpopulations of platelets express mRNA [53]. However, these studies are limited to select candidates and their qualitative nature does not distinguish whether mRNA expression levels vary among platelet subpopulations (i.e., older versus younger platelets). These caveats need to be considered as one evaluates the genetic code of platelets.
Studies over the last 50 years have demonstrated that platelets are capable of translating a subset mRNAs into proteins and this list continues to grow. Translation of platelet mRNAs is reviewed in detail elsewhere [50, 55]. Recent studies not included in these reviews, however, demonstrate that dengue infected platelets synthesize interleukin-1β that is packaged into MPs and induces endothelial permeability [56]. Corduan and colleagues[57] also recently demonstrated rapid dissociation of Ago2 protein complexes from serpine1 mRNA, which codes for plasminogen activator inhibitor-1 (PAI-1) that is known to be translated by activated platelets [58–59]. This disassembly process is another exquisite example of platelets possessing unexpected, complex gene expression pathways.
Transfer of the Platelet Genetic Code to Target Cells
Although RNAs are typically considered intrinsic regulators of cellular function, emerging evidence suggests that mRNAs and miRNAs can be transferred from one cell to another. In this regard, several studies have demonstrated that platelets are capable of transferring RNAs to recipient cells (reviewed by Clancy and Freedman[60]). Risitano and colleagues [61] found that labeled RNA from platelet-like particles is transferred to leukocytes and endothelial cells, and this transfer alters gene expression patterns in the target cells. Using a green fluorescent-based system, they also showed that RNA from the platelet-like particles is translated by the recipient cells. This same group also demonstrated that platelet RNA transfer to leukocytes occurs in vivo.
In addition to mRNA transfer, platelets transfer miRNAs to endothelial cells. Laffont and coworkers[62] found that platelets transfer miR-223 to human umbilical vein endothelial cells (HUVECs). miR-233 was transferred in platelet microparticles (MPs), which contained a functional Argonaute-2 (Ago2)-miR233 complex that regulated the expression of a reporter construct in recipient HUVEC’s. Similarly, Gidlöf et al.[63] found that platelets isolated from patients with myocardial infarction release specific miRNAs that can regulate endothelial cell gene expression. Gidlöf and colleagues also found that activated platelets can transfer fluorescently-labeled miRNA and endogenously released miR-39 to endothelial cells.
An important consideration is that intercellular transfer of platelet RNAs and proteins to recipient cells can occur via MPs. Given that the vast majority of circulating MPs are derived from platelets, and the numbers of MPs increases significantly in disease situations due to their increased uptake and/or decreased consumption, it is plausible that platelet MPs transfer significant amounts of genetic information to recipient cells [64–66]. This transfer has widespread implications for several fields, especially if the genetic code bestowed into MPs changes in disease situations.
Conclusion
There is no doubt that the basic functions of platelets are similar between individuals. However, it is becoming increasingly clear that platelets from different individuals and races have distinct genetic codes that personalize and predict how they react and respond. Furthermore, variations in the platelet genetic code become more pronounced in acute and chronic disease situations. Identifying what these differences are, how they occur, and the means by which the platelet genetic code affects and/or causes human disease will be an exciting element of future studies. In addition, disease-specific interrogation of the megakaryocyte and platelet genetic code will undoubtedly reveal surprises that broaden our understanding of how circulating platelets perform in sickness and in health.
Acknowledgments
We thank Ms. Diana Lim for her creative input and outstanding preparation of the figure. Portions of the work reviewed in this report were funded by NHLBI and NIA to MTR and ASW (grants HL112311, HL126547, and AG048022).
Footnotes
Authors state no conflicts of interests.
References
- 1.Italiano J, Jr, Hartwig JH. Megakaryocyte Development and Platelet Formation. In: Michelson AD, editor. Platelets. 3. San Diego: Elsevier; 2013. pp. 27–49. [Google Scholar]
- 2.Ebbe S. Biology of megakaryocytes. Prog Hemost Thromb. 1976;3:211–29. [PubMed] [Google Scholar]
- 3.Deutsch VR, Tomer A. Advances in megakaryocytopoiesis and thrombopoiesis: from bench to bedside. Br J Haematol. 2013;161:778–93. doi: 10.1111/bjh.12328. [DOI] [PubMed] [Google Scholar]
- 4.Smith BW, Murphy GJ. Stem cells, megakaryocytes, and platelets. Curr Opin Hematol. 2014;21:430–7. doi: 10.1097/MOH.0000000000000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaushansky K. Thrombopoiesis. Semin Hematol. 2015;52:4–11. doi: 10.1053/j.seminhematol.2014.10.003. S0037-1963(14)00077-8 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Foulks JM, Marathe GK, Michetti N, Stafforini DM, Zimmerman GA, McIntyre TM, Weyrich AS. PAF-acetylhydrolase expressed during megakaryocyte differentiation inactivates PAF-like lipids. Blood. 2009;113:6699–706. doi: 10.1182/blood-2008-11-186312. blood-2008-11-186312 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thon JN, Montalvo A, Patel-Hett S, Devine MT, Richardson JL, Ehrlicher A, Larson MK, Hoffmeister K, Hartwig JH, Italiano JE., Jr Cytoskeletal mechanics of proplatelet maturation and platelet release. J Cell Biol. 2010;191:861–74. doi: 10.1083/jcb.201006102. jcb.201006102 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaushansky K. Thrombopoietin: the primary regulator of platelet production. Blood. 1995;86:419–31. [PubMed] [Google Scholar]
- 9.Nakagawa Y, Nakamura S, Nakajima M, Endo H, Dohda T, Takayama N, Nakauchi H, Arai F, Fukuda T, Eto K. Two differential flows in a bioreactor promoted platelet generation from human pluripotent stem cell-derived megakaryocytes. Exp Hematol. 2013;41:742–8. doi: 10.1016/j.exphem.2013.04.007. S0301-472X(13)00165-3 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Thon JN, Mazutis L, Wu S, Sylman JL, Ehrlicher A, Machlus KR, Feng Q, Lu S, Lanza R, Neeves KB, Weitz DA, Italiano JE., Jr Platelet bioreactor-on-a-chip. Blood. 2014;124:1857–67. doi: 10.1182/blood-2014-05-574913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Buduo CA, Wray LS, Tozzi L, Malara A, Chen Y, Ghezzi CE, Smoot D, Sfara C, Antonelli A, Spedden E, Bruni G, Staii C, De Marco L, Magnani M, Kaplan DL, Balduini A. Programmable 3D silk bone marrow niche for platelet generation ex vivo and modeling of megakaryopoiesis pathologies. Blood. 2015 doi: 10.1182/blood-2014-08-595561. blood-2014-08-595561 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawaguchi T, Hatano R, Yamaguchi K, Nawa K, Hashimoto R, Yokota H. Fibronectin promotes proplatelet formation in the human megakaryocytic cell line UT-7/TPO. Cell Biol Int. 2012;36:39–45. doi: 10.1042/CBI20110383. CBI20110383 [pii] [DOI] [PubMed] [Google Scholar]
- 13.Leven RM, Tablin F. Extracellular matrix stimulation of guinea pig megakaryocyte proplatelet formation in vitro is mediated through the vitronectin receptor. Exp Hematol. 1992;20:1316–22. [PubMed] [Google Scholar]
- 14.Vyas P, Ault K, Jackson CW, Orkin SH, Shivdasani RA. Consequences of GATA-1 deficiency in megakaryocytes and platelets. Blood. 1999;93:2867–75. [PubMed] [Google Scholar]
- 15.Visvader JE, Elefanty AG, Strasser A, Adams JM. GATA-1 but not SCL induces megakaryocytic differentiation in an early myeloid line. EMBO J. 1992;11:4557–64. doi: 10.1002/j.1460-2075.1992.tb05557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shivdasani RA, Rosenblatt MF, Zucker-Franklin D, Jackson CW, Hunt P, Saris CJ, Orkin SH. Transcription factor NF-E2 is required for platelet formation independent of the actions of thrombopoietin/MGDF in megakaryocyte development. Cell. 1995;81:695–704. doi: 10.1016/0092-8674(95)90531-6. [DOI] [PubMed] [Google Scholar]
- 17.Hatami J, Andrade PZ, Alves de Matos AP, Djokovic D, Lilaia C, Ferreira FC, Cabral JM, da Silva CL. Developing a co-culture system for effective megakaryo/thrombopoiesis from umbilical cord blood hematopoietic stem/progenitor cells. Cytotherapy. 2015 doi: 10.1016/j.jcyt.2014.12.010. S1465–3249(15)00006-7 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Mills JA, Paluru P, Weiss MJ, Gadue P, French DL. Hematopoietic differentiation of pluripotent stem cells in culture. Methods Mol Biol. 2014;1185:181–94. doi: 10.1007/978-1-4939-1133-2_12. [DOI] [PubMed] [Google Scholar]
- 19.Pineault N, Boucher JF, Cayer MP, Palmqvist L, Boyer L, Lemieux R, Proulx C. Characterization of the effects and potential mechanisms leading to increased megakaryocytic differentiation under mild hyperthermia. Stem Cells Dev. 2008;17:483–93. doi: 10.1089/scd.2007.0149. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi K, Monzen S, Yoshino H, Abe Y, Eguchi-Kasai K, Kashiwakura I. Effects of a 2-step culture with cytokine combinations on megakaryocytopoiesis and thrombopoiesis from carbon-ion beam-irradiated human hematopoietic stem/progenitor cells. J Radiat Res. 2008;49:417–24. doi: 10.1269/jrr.07132. JST.JSTAGE/jrr/07132 [pii] [DOI] [PubMed] [Google Scholar]
- 21.Vanikar AV, Mishra VV, Firoz A, Shah VR, Dave SD, Patel RD, Kanodia KV, Patel JV, Patel CN, Trivedi HL. Successful generation of donor specific hematopoietic stem cell lines from co-cultured bone marrow with human embryonic stem cell line: a new methodology. Transplant Proc. 2007;39:658–61. doi: 10.1016/j.transproceed.2007.01.048. S0041-1345(07)00097-8 [pii] [DOI] [PubMed] [Google Scholar]
- 22.Pastos KM, Slayton WB, Rimsza LM, Young L, Sola-Visner MC. Differential effects of recombinant thrombopoietin and bone marrow stromal-conditioned media on neonatal versus adult megakaryocytes. Blood. 2006;108:3360–2. doi: 10.1182/blood-2006-04-018036. blood-2006-04-018036 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cortin V, Garnier A, Pineault N, Lemieux R, Boyer L, Proulx C. Efficient in vitro megakaryocyte maturation using cytokine cocktails optimized by statistical experimental design. Exp Hematol. 2005;33:1182–91. doi: 10.1016/j.exphem.2005.06.020. S0301-472X(05)00303-6 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Edelstein LC, Bray PF. MicroRNAs in platelet production and activation. Blood. 2011;117:5289–96. doi: 10.1182/blood-2011-01-292011. blood-2011-01-292011 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edelstein LC, McKenzie SE, Shaw C, Holinstat MA, Kunapuli SP, Bray PF. MicroRNAs in platelet production and activation. J Thromb Haemost. 2013;11 (Suppl 1):340–50. doi: 10.1111/jth.12214. [DOI] [PubMed] [Google Scholar]
- 26.Cecchetti L, Tolley ND, Michetti N, Bury L, Weyrich AS, Gresele P. Megakaryocytes differentially sort mRNAs for matrix metalloproteinases and their inhibitors into platelets: a mechanism for regulating synthetic events. Blood. 2011;118:1903–11. doi: 10.1182/blood-2010-12-324517. blood-2010-12-324517 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F, He J, Zhu FM, Liu JH, Qin F, Chen S, Xu G, Lu XJ, Yan LX. Analysis of mRNA expression profiles of megakaryocytes from human cord blood CD34+ cells ex vivo expanded using Solexa sequencing. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2011;33:529–32. [PubMed] [Google Scholar]
- 28.Opalinska JB, Bersenev A, Zhang Z, Schmaier AA, Choi J, Yao Y, D’Souza J, Tong W, Weiss MJ. MicroRNA expression in maturing murine megakaryocytes. Blood. 2010;116:e128–38. doi: 10.1182/blood-2010-06-292920. blood-2010-06-292920 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bluteau O, Langlois T, Rivera-Munoz P, Favale F, Rameau P, Meurice G, Dessen P, Solary E, Raslova H, Mercher T, Debili N, Vainchenker W. Developmental changes in human megakaryopoiesis. J Thromb Haemost. 2013;11:1730–41. doi: 10.1111/jth.12326. [DOI] [PubMed] [Google Scholar]
- 30.Massimi I, Guerriero R, Lotti LV, Lulli V, Borgognone A, Romani F, Barilla F, Gaudio C, Gabbianelli M, Frati L, Pulcinelli FM. Aspirin influences megakaryocytic gene expression leading to up-regulation of multidrug resistance protein-4 in human platelets. Br J Clin Pharmacol. 2014;78:1343–53. doi: 10.1111/bcp.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edelstein LC, Simon LM, Montoya RT, Holinstat M, Chen ES, Bergeron A, Kong X, Nagalla S, Mohandas N, Cohen DE, Dong JF, Shaw C, Bray PF. Racial differences in human platelet PAR4 reactivity reflect expression of PCTP and miR-376c. Nat Med. 2013;19:1609–16. doi: 10.1038/nm.3385. nm.3385 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edelstein LC, Simon LM, Lindsay CR, Kong X, Teruel-Montoya R, Tourdot BE, Chen ES, Ma L, Coughlin S, Nieman M, Holinstat M, Shaw CA, Bray PF. Common variants in the human platelet PAR4 thrombin receptor alter platelet function and differ by race. Blood. 2014;124:3450–8. doi: 10.1182/blood-2014-04-572479. blood-2014-04-572479 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kahr WH, Hinckley J, Li L, Schwertz H, Christensen H, Rowley JW, Pluthero FG, Urban D, Fabbro S, Nixon B, Gadzinski R, Storck M, Wang K, Ryu GY, Jobe SM, Schutte BC, Moseley J, Loughran NB, Parkinson J, Weyrich AS, Di Paola J. Mutations in NBEAL2, encoding a BEACH protein, cause gray platelet syndrome. Nat Genet. 2011;43:738–40. doi: 10.1038/ng.884. ng.884 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noetzli L, Lo RW, Lee-Sherick AB, Callaghan M, Noris P, Savoia A, Rajpurkar M, Jones K, Gowan K, Balduini CL, Pecci A, Gnan C, De Rocco D, Doubek M, Li L, Lu L, Leung R, Landolt-Marticorena C, Hunger S, Heller P, Gutierrez-Hartmann A, Xiayuan L, Pluthero FG, Rowley JW, Weyrich AS, Kahr WH, Porter CC, Di Paola J. Germline mutations in ETV6 are associated with thrombocytopenia, red cell macrocytosis and predisposition to lymphoblastic leukemia. Nat Genet. 2015 doi: 10.1038/ng.3253. ng.3253 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Healy AM, Pickard MD, Pradhan AD, Wang Y, Chen Z, Croce K, Sakuma M, Shi C, Zago AC, Garasic J, Damokosh AI, Dowie TL, Poisson L, Lillie J, Libby P, Ridker PM, Simon DI. Platelet expression profiling and clinical validation of myeloid-related protein-14 as a novel determinant of cardiovascular events. Circulation. 2006;113:2278–84. doi: 10.1161/CIRCULATIONAHA.105.607333. [DOI] [PubMed] [Google Scholar]
- 36.Lood C, Amisten S, Gullstrand B, Jonsen A, Allhorn M, Truedsson L, Sturfelt G, Erlinge D, Bengtsson AA. Platelet transcriptional profile and protein expression in patients with systemic lupus erythematosus: up-regulation of the type I interferon system is strongly associated with vascular disease. Blood. 2010;116:1951–7. doi: 10.1182/blood-2010-03-274605. blood-2010-03-274605 [pii] [DOI] [PubMed] [Google Scholar]
- 37.Michelson AD, Barnard MR, Krueger LA, Valeri CR, Furman MI. Circulating monocyte-platelet aggregates are a more sensitive marker of in vivo platelet activation than platelet surface P-selectin: studies in baboons, human coronary intervention, and human acute myocardial infarction. Circulation. 2001;104:1533–7. doi: 10.1161/hc3801.095588. [DOI] [PubMed] [Google Scholar]
- 38.Rondina MT, Brewster B, Grissom CK, Zimmerman GA, Kastendieck DH, Harris ES, Weyrich AS. In vivo platelet activation in critically ill patients with primary 2009 influenza A(H1N1) Chest. 2012;141:1490–5. doi: 10.1378/chest.11-2860. chest.11-2860 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rondina MT, Weyrich AS, Zimmerman GA. Platelets as cellular effectors of inflammation in vascular diseases. Circ Res. 2013;112:1506–19. doi: 10.1161/CIRCRESAHA.113.300512. 112/11/1506 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freedman JE, Larson MG, Tanriverdi K, O’Donnell CJ, Morin K, Hakanson AS, Vasan RS, Johnson AD, Iafrati MD, Benjamin EJ. Relation of platelet and leukocyte inflammatory transcripts to body mass index in the Framingham heart study. Circulation. 2010;122:119–29. doi: 10.1161/CIRCULATIONAHA.109.928192. CIRCULATIONAHA.109.928192 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McManus DD, Beaulieu LM, Mick E, Tanriverdi K, Larson MG, Keaney JF, Jr, Benjamin EJ, Freedman JE. Relationship among circulating inflammatory proteins, platelet gene expression, and cardiovascular risk. Arterioscler Thromb Vasc Biol. 2013;33:2666–73. doi: 10.1161/ATVBAHA.112.301112. ATVBAHA.112.301112 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raghavachari N, Xu X, Harris A, Villagra J, Logun C, Barb J, Solomon MA, Suffredini AF, Danner RL, Kato G, Munson PJ, Morris SM, Jr, Gladwin MT. Amplified expression profiling of platelet transcriptome reveals changes in arginine metabolic pathways in patients with sickle cell disease. Circulation. 2007;115:1551–62. doi: 10.1161/CIRCULATIONAHA.106.658641. CIRCULATIONAHA.106.658641 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reilly SJ, Li N, Liska J, Ekstrom M, Tornvall P. Coronary artery bypass graft surgery up-regulates genes involved in platelet aggregation. J Thromb Haemost. 2012;10:557–63. doi: 10.1111/j.1538-7836.2012.04660.x. [DOI] [PubMed] [Google Scholar]
- 44.Calverley DC, Phang TL, Choudhury QG, Gao B, Oton AB, Weyant MJ, Geraci MW. Significant downregulation of platelet gene expression in metastatic lung cancer. Clin Transl Sci. 2010;3:227–32. doi: 10.1111/j.1752-8062.2010.00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colombo G, Gertow K, Marenzi G, Brambilla M, De Metrio M, Tremoli E, Camera M. Gene expression profiling reveals multiple differences in platelets from patients with stable angina or non-ST elevation acute coronary syndrome. Thromb Res. 2011;128:161–8. doi: 10.1016/j.thromres.2011.02.012. S0049-3848(11)00093-4 [pii] [DOI] [PubMed] [Google Scholar]
- 46.Nilsson RJ, Balaj L, Hulleman E, van Rijn S, Pegtel DM, Walraven M, Widmark A, Gerritsen WR, Verheul HM, Vandertop WP, Noske DP, Skog J, Wurdinger T. Blood platelets contain tumor-derived RNA biomarkers. Blood. 2011;118:3680–3. doi: 10.1182/blood-2011-03-344408. blood-2011-03-344408 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schubert S, Weyrich AS, Rowley JW. A tour through the transcriptional landscape of platelets. Blood. 2014;124:493–502. doi: 10.1182/blood-2014-04-512756. blood-2014-04-512756 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rowley JW, Oler AJ, Tolley ND, Hunter BN, Low EN, Nix DA, Yost CC, Zimmerman GA, Weyrich AS. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood. 2011;118:e101–11. doi: 10.1182/blood-2011-03-339705. blood-2011-03-339705 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rowley JW, Schwertz H, Weyrich AS. Platelet mRNA: the meaning behind the message. Curr Opin Hematol. 2012;19:385–91. doi: 10.1097/MOH.0b013e328357010e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weyrich AS, Lindemann S, Tolley ND, Kraiss LW, Dixon DA, Mahoney TM, Prescott SM, McIntyre TM, Zimmerman GA. Change in protein phenotype without a nucleus: translational control in platelets. Seminars in Thrombosis and Hemostasis. 2004;30:493–500. doi: 10.1055/s-2004-833484. [DOI] [PubMed] [Google Scholar]
- 51.Landry P, Plante I, Ouellet DL, Perron MP, Rousseau G, Provost P. Existence of a microRNA pathway in anucleate platelets. Nat Struct Mol Biol. 2009;16:961–6. doi: 10.1038/nsmb.1651. nsmb.1651 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ple H, Landry P, Benham A, Coarfa C, Gunaratne PH, Provost P. The repertoire and features of human platelet microRNAs. PLoS One. 2012;7:e50746. doi: 10.1371/journal.pone.0050746. PONE-D-12-24904 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Denis MM, Tolley ND, Bunting M, Schwertz H, Jiang H, Lindemann S, Yost CC, Rubner FJ, Albertine KH, Swoboda KJ, Fratto CM, Tolley E, Kraiss LW, McIntyre TM, Zimmerman GA, Weyrich AS. Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell. 2005;122:379–91. doi: 10.1016/j.cell.2005.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shashkin PN, Brown GT, Ghosh A, Marathe GK, McIntyre TM. Lipopolysaccharide is a direct agonist for platelet RNA splicing. J Immunol. 2008;181:3495–502. doi: 10.4049/jimmunol.181.5.3495. 181/5/3495 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zimmerman GA, Weyrich AS. Signal-dependent protein synthesis by activated platelets: new pathways to altered phenotype and function. Arterioscler Thromb Vasc Biol. 2008;28:s17–24. doi: 10.1161/ATVBAHA.107.160218. 28/3/s17 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hottz ED, Lopes JF, Freitas C, Valls-de-Souza R, Oliveira MF, Bozza MT, Da Poian AT, Weyrich AS, Zimmerman GA, Bozza FA, Bozza PT. Platelets mediate increased endothelium permeability in dengue through NLRP3-inflammasome activation. Blood. 2013;122:3405–14. doi: 10.1182/blood-2013-05-504449. blood-2013-05-504449 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corduan A, Ple H, Laffont B, Wallon T, Plante I, Landry P, Provost P. Dissociation of SERPINE1 mRNA from the translational repressor proteins Ago2 and TIA-1 upon platelet activation. Thromb Haemost. 2015;113:14–07-0622. doi: 10.1160/TH14-07-0622. [pii] [DOI] [PubMed] [Google Scholar]
- 58.Nylander M, Osman A, Ramstrom S, Aklint E, Larsson A, Lindahl TL. The role of thrombin receptors PAR1 and PAR4 for PAI-1 storage, synthesis and secretion by human platelets. Thromb Res. 2012;129:e51–8. doi: 10.1016/j.thromres.2011.12.021. S0049-3848(11)00686-4 [pii] [DOI] [PubMed] [Google Scholar]
- 59.Brogren H, Karlsson L, Andersson M, Wang L, Erlinge D, Jern S. Platelets synthesize large amounts of active plasminogen activator inhibitor 1. Blood. 2004;104:3943–8. doi: 10.1182/blood-2004-04-1439. [DOI] [PubMed] [Google Scholar]
- 60.Clancy L, Freedman JE. New paradigms in thrombosis: novel mediators and biomarkers platelet RNA transfer. J Thromb Thrombolysis. 2014;37:12–6. doi: 10.1007/s11239-013-1001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Risitano A, Beaulieu LM, Vitseva O, Freedman JE. Platelets and platelet-like particles mediate intercellular RNA transfer. Blood. 2012;119:6288–95. doi: 10.1182/blood-2011-12-396440. blood-2011-12-396440 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laffont B, Corduan A, Ple H, Duchez AC, Cloutier N, Boilard E, Provost P. Activated platelets can deliver mRNA regulatory Ago2*microRNA complexes to endothelial cells via microparticles. Blood. 2013;122:253–61. doi: 10.1182/blood-2013-03-492801. blood-2013-03-492801 [pii] [DOI] [PubMed] [Google Scholar]
- 63.Gidlof O, van der Brug M, Ohman J, Gilje P, Olde B, Wahlestedt C, Erlinge D. Platelets activated during myocardial infarction release functional miRNA, which can be taken up by endothelial cells and regulate ICAM1 expression. Blood. 2013;121:3908–17. S1–26. doi: 10.1182/blood-2012-10-461798. blood-2012-10-461798 [pii] [DOI] [PubMed] [Google Scholar]
- 64.Burnouf T, Goubran HA, Chou ML, Devos D, Radosevic M. Platelet microparticles: detection and assessment of their paradoxical functional roles in disease and regenerative medicine. Blood Rev. 2014;28:155–66. doi: 10.1016/j.blre.2014.04.002. S0268-960X(14)00033-2 [pii] [DOI] [PubMed] [Google Scholar]
- 65.Lynch SF, Ludlam CA. Plasma microparticles and vascular disorders. Br J Haematol. 2007;137:36–48. doi: 10.1111/j.1365-2141.2007.06514.x. BJH6514 [pii] [DOI] [PubMed] [Google Scholar]
- 66.Goubran HA, Burnouf T, Stakiw J, Seghatchian J. Platelet microparticle: A sensitive physiological “fine tuning” balancing factor in health and disease. Transfus Apher Sci. 2014 doi: 10.1016/j.transci.2014.12.015. S1473-0502(14)00253-5 [pii] [DOI] [PubMed] [Google Scholar]