INTRODUCTION
R2 elements exclusively insert into 28S rRNA genes (Figure 1). As a result of this specificity, R2 is one of the more tractable mobile elements to study and, thus, is now among the best understood elements both in terms of its mechanism and its population dynamics. The R2 element was first identified in the rDNA loci of Drosophila melanogaster in the early 1980's (1,2), when little was known of the structure or abundance of mobile elements in eukaryotes. In fact, the exclusive residence of the element at a specific site in the 28S gene initially suggested that it might be an intron. However, the findings that only a fraction of the genes contained the insertion, that 28S genes containing the insertion did not appear to be transcribed, and that many of the insertions had a sizeable deletion at the 5’ end all argued against its role as an intron. Insertions were soon identified at the same position of the 28S rRNA gene in many other species of insects (3,4,5). The complete sequence of the insertions in both D. melanogaster and Bombyx mori revealed a large open reading frame (ORF) encoding a reverse transcriptase that had greatest sequence similarity to that of non-LTR retrotransposons (6,7). R2 differed from most non-LTR retrotransposons, however, in that it only contained a single ORF. Furthermore, rather than an encoded apurinic endonuclease (APE) located amino-terminal to the reverse transcriptase (8), R2 encoded carboxyl terminal to the reverse transcriptase an endonuclease with an active site more similar to that of certain restriction enzymes (9).
Figure 1.
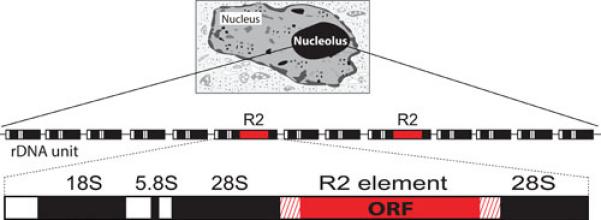
R2 elements insert within the 28S rRNA genes. The nucleolus, the site of rRNA transcription and processing, is organized around the hundreds of tandem units (rDNA units) that comprise the rDNA locus. Each rDNA unit is composed of a single transcription unit containing the 18S, 5.8S, and 28S genes (black boxes) and external and internal transcribed spacers (white boxes). The transcription units are separated by intergenic spacers (thin lines). A subset of the 28S genes in many animals contain R2 insertions near the middle of the gene (red box). R2 elements encode a single open reading frame (ORF).
The search for R2 in additional species was simple because the 28S gene sequences to either side of the R2 insertion site have undergone almost no substitutions in the entire evolution of eukaryotes. Thus it was straightforward to determine whether a species contained R2 insertions by direct cloning of 28S genes, PCR amplification of the insertion region, or computer searches of whole genome shotgun sequences. Such analyses have revealed R2 elements in most lineages of insects and arthropods (10,11) and in many other taxa of animals including nematodes, tunicates, and birds (12,13,14; unpublished data, DE Stage); however, there have been no reports of R2 elements in plants, fungi, or protozoans. The presence of R2 elements within a group can be spotty, for example only 4 out of 7 fish species examined have R2. Thus the apparent absence of R2 from some animal taxa may simply reflect the small numbers of species whose genomes have been tested. The large number of mammalian species examined without detecting R2 insertions does suggest with some confidence, however, that R2 is not present in this group.
The 3’ junctions of the 28S gene with the R2 insertions in all but two species are identical suggesting that the R2 endonuclease is highly specific and that it has rarely changed the specificity of the initial DNA cleavage since its origin. The two exceptions are the R2 elements of hydra, named R8, which insert into a specific sequence of the 18S rRNA gene (13), and the R2 elements of rotifer, named R9, which insert into a different site in the 28S rRNA gene (15). The ORF of all R2 elements is also very similar in coding capacity; the only significant difference is the number of zinc-finger motifs associated with DNA binding at the amino-terminal end of the protein (11,13,16). As described by Fujiwara in this volume (17), many other lineages of non-LTR retrotransposons have evolved sequence specificity for the rRNA genes or for other repeated sequences in the genomes of eukaryotes (18-22). Some of these site-specific elements are like R2 and encode a carboxyl-terminal restriction-like endonuclease, while others contain an amino-terminal APE domain. Among the latter, R1 elements insert in the 28S rRNA gene 74 bp downstream of the R2 insertion site. R1 elements were first identified along with the R2 elements of D. melanogaster (1,2) and subsequently in most lineages of arthropods (10). The turnover and evolution of R1 elements in the rDNA loci of Drosophila species is similar in most respects to that of the R2 elements (23-25).
Reconstructing the evolutionary history of R2 elements based on the sequence of their ORF, first in the genus Drosophila (26,27), then in all of arthropods (28), and finally in all animals (12,13,14), has suggested the R2 elements have evolved entirely by vertical descent. The absence of horizontal jumps between species has enabled the divergence of R2 elements to be used as a molecular clock to time the age of its various lineages as well as a guide to estimate the time of divergence of other retrotransposons (29). Unfortunately, because the R2 protein sequence eventually reaches maximal divergence, this dating can only be done with confidence for a time frame of less then 200 - 300 million years. Remarkably, multiple lineages of R2 have propagated in some animal lineages for the entire 200-300 million year time estimate (10,12,30,31). Why some animals are able to maintain multiple R2 lineages and other animals only one R2 lineage is unknown.
The long history and wide distribution of R2 is remarkable for a mobile element. Its success has been interpreted by some to indicate that R2 provides a useful function to the host. One possibility is that the R2 endonuclease initiates the recombinations that give rise to the concerted evolution of the locus (32). However, in instances where R2 elements are known to be active, the DNA cleavages generated by the endonuclease appear to lead to large deletions of the rDNA locus which are detrimental to the host (33). A second premise is that the inactivation of rDNA units by insertions and the subsequent reduction in 28S rRNA synthesis could influence the rate of development (34). However, the original findings leading to this conclusion have been challenged (35). The many species containing either multiple lineages of R2 or multiple classes of non-LTR retrotransposons inserted into the rDNA locus (10,12,28,31) are more consistent with the propagation of selfish genetic elements than a means used by animals to regulate gene expression. Finally, the frequently suggested argument that mobile elements provide useful genetic diversity seems unlikely for R2; comparisons of species with or without R2 elements reveal no sequence differences in 28S rRNA genes near the insertion site and no detectable difference in the mechanism of rRNA regulation. Thus R2 remains one of the best examples of an element that endures because it simply has the ability to make copies of itself (i.e. a selfish genetic element). R2 has likely been present since the origin of the metazoan radiation, est. 500-800 million years ago, making it one of the oldest known mobile genetic elements. Perhaps most remarkable is that throughout this long period R2 has undergone essentially no changes in its insertion site or in its mechanism of insertion. Clearly, R2 has found a niche in which it can hold its own in the genomic battle between element and host.
MECHANISM OF R2 INTEGRATION
Studies of the R2 integration mechanism have been conducted with the ORF of the R2 element from Bombyx mori expressed in and purified from E. coli (36,37). The purified protein (120 kilodaltons) was found to have all the RNA and DNA binding properties as well as enzymatic activities needed to complete a retrotransposition reaction. The critical first step in this reaction is the ability of the protein to nick one strand of the DNA target site and use the 3’ end of the DNA exposed by this cleavage to prime reverse transcription of the R2 RNA. The integration reaction was termed target DNA-primed reverse transcription, or TPRT (37), to distinguish it from the retrotransposition mechanism that had been previously discovered for retroviruses and LTR retrotransposons (reviewed in 38,39). The TPRT mechanism can explain three unusual properties revealed by the initial sequencing of non-LTR retrotransposons. First, the absence of the integrase domain usually seen in mobile elements can be explained because the new copies of the element are synthesized onto the target site rather than inserted into the target site. Second, the absence of the integrase also explains why many non-LTR retrotransposons generate variable in length or no target site duplications. Third, the uniform presence of a complete 3’ end but frequent presence of truncations at the 5’ end of non-LTR retrotransposons can be explained by the reverse transcriptase falling off before reaching the 5’ end of the RNA or reaching the end of a degraded RNA. Characterizations of the initial steps in the R2 TPRT reaction were previously summarized in Mobile DNA II (40). Here, experiments conducted since that publication are the primary focus.
Properties of the R2 reverse transcriptase
R2 elements from all lineages of animals encode a protein with a domain structure similar to that shown in Figure 2. The central domain of the R2 protein corresponds to the reverse transcriptase (RT). R2 RT has a number of properties that differentiate it from the RTs encoded by LTR retrotransposons and retroviruses. Clearly the most distinctive property is the ability of R2 RT to use the 3’ end of DNA to prime reverse transcription (41,42). This reaction is most efficient at the 28S gene insertion site with RNA containing the 3’ UTR of the R2 element as template. In the absence of these components and at a lower efficiency, R2 RT can prime the reverse transcription of any RNA using the 3’ end of any other RNA or single-stranded DNA as the primer. This priming also occurs in the absence of complementarity between the template and primer (43)
Figure 2.
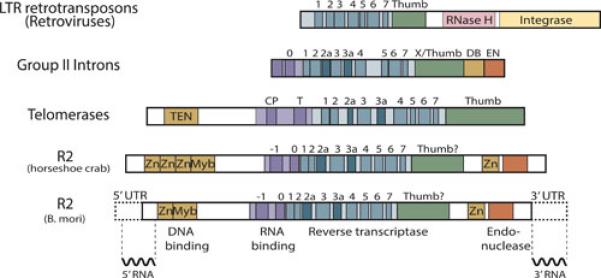
Domain structure of the R2 protein and its similarity to other elements. At the bottom is the R2 element from B. mori with the 5’ and 3’ untranslated regions indicated by dotted lines. The central region of the encoded protein contains the reverse transcriptase domain. The various conserved motifs within the fingers and palm regions (motifs 1 −7) and the predicted thumb are indicated. An RNA binding domain is immediately N-terminal to the reverse transcriptase and conserved motifs within this domain are labeled 0 and −1. The N-terminal region of the protein contains zinc finger (Zn) and c-myb (Myb) motifs, while the C-terminal region encodes a putative zinc-binding domain and the R2 endonuclease. Shown below the R2 diagram are the 5’ and 3’ regions of the R2 RNA that are bound by the R2 protein during a retrotransposition reaction (see Figure 4). The major difference among R2 elements from different species is the presence of one, two, or three zinc finger domains at the N-terminal end. The R2 element from horseshoe crab is an example of the latter. Comparison of the R2 protein with the pol gene of LTR retrotransposons (and retroviruses) reveals little in common except for 7 out of the 9 motifs in the reverse transcriptase domain. Most LTR retrotransposon pol genes also encode an RNase H and integrase not found in R2. The R2 protein has greater similarity to the proteins encoded by group II introns and telomerases. These three groups share all nine motifs of the reverse transcriptase. In the case of telomerase, these motifs are frequently termed 1, 2, 3, A, IFD, B, C, D, and E (from left to right) (69). Group II introns, telomerases, and R2 also share an RNA binding domain upstream of the reverse transcriptase (purple segment). Group II introns and R2 both encode an endonuclease domain at the 3’ end, while R2 and some telomerases have DNA binding domains (TEN) at the N-terminal end.
A second interesting property of R2-RT is its higher processivity than that of most RTs. Processivity refers to the product length that can be catalyzed by the enzyme before it dissociates from the template. In single cycle reactions on RNA and DNA templates, R2 RT synthesizes cDNA that is 2 to 5 times the length of that synthesized by retroviral RTs (44,45). This difference in processivity is likely a result of the different demands placed on these enzymes. DNA synthesis by retroviral or LTR RTs occurs within virus-like particles within the cytoplasm. In these particles, the RT is able to undergo rounds of dissociation from and reassociation to the template before giving rise to full-length products (38,39). In contrast, DNA synthesis by R2 RT occurs in the nucleus at the DNA target site. If R2 RT dissociates from the RNA template then reassociation could be difficult, and the result is a truncated (dead) copy. Consistent with this model, R2 RT has been shown to initiate only poorly at the 3’ end of long DNA primers annealed to RNA templates (43). Thus there appears to have been selective pressure on R2 and likely other non-LTR retrotransposons to evolve RTs with high processivity.
Another unusual ability of R2 RT is that it can jump from the 5’ end of one RNA template to the 3’ end of another RNA template. These “end-to-end template jumps” do not require sequence identity between the templates (43,46). Instead, R2 RT adds up to five non-templated nucleotides to the cDNA when it reaches the end of a template. Microhomologies between these overhanging nucleotides and sequences near the 3’ end of the acceptor template enable the polymerase to jump between templates (46). R2 end-to-end template jumps are similar to the template jumps observed for viral RNA directed RNA polymerases (47) as well as for the Mauriceville retroplasmid and group II intron RTs (48,49) but differ from the template switching reaction associated with retroviral DNA synthesis, which does require sequence identity between the donor and acceptor RNA templates (50).
The R2 protein has no identifiable RNase H domain (9,51) and no RNase H activity has been detected in vitro (37,45) suggesting the template for second strand DNA synthesis in a retrotransposition reaction is an RNA:DNA duplex. Consistent with this model R2 RT has the ability to displace an annealed RNA or DNA strand as it uses an RNA or DNA strand as template (43,45). Remarkably, the processivity of R2 RT is not reduced by the presence of an annealed strand. Retroviral RTs, on the other hand, do possess RNase H activity and show only limited ability to displace RNA annealed to DNA (52,53). Because most non-LTR retrotransposons do not have an RNase H domain, strand displacement may be a common property of non-LTR RTs. However, the acquisition of an RNase H domain by some elements (51) and the finding that group II intron RTs have a strong strand displacement activity but depend on the host RNase H for mobility (54) leave the question open as to the extent non-LTR retrotransposons may rely on host RNase H activity.
Recently, it was shown that R2 RT has a level of nucleotide misincorporation (mutation rate) similar to that of HIV-1 RT (55,56). Like HIV-1 the low fidelity of R2 RT is a result of its ability to extend a mismatch if the wrong nucleotide is incorporated into the product. For HIV-1 the high error rate has been suggested to enable the virus to escape the immune system of the host (57). In the case of R2 RT, the high error rate could be a consequence of the unusual flexibility needed at the active site to enable priming in the absence of sequence complementarity. However, because R2 retrotransposition is infrequent relative to the many germ line replications that occur each generation, the long-term nucleotide substitution rate for R2 is not significantly above that associated with typical genes (55).
DNA and RNA binding domains of the R2 protein
The C-terminal end of the R2 protein contains the endonuclease domain (Figure 2). A similar domain appears to be present in all non-LTR retrotransposons that do not encode an APE-like endonuclease at the N-terminal end of their protein (29). The active site of the R2 endonuclease was found to be similar to that of type IIs restriction enzymes (9). The catalytic and DNA binding domains of type IIs restriction enzymes are separate, thus, these enzymes bind the DNA a short distance from the cleavage site. The separation of cleavage from binding is also suggested for R2 as protein foot-print analyses indicted that most DNA contacts by the R2 protein are located upstream and downstream of the insertion site (58, 59). The C-terminal end of the R2 protein also encodes a potential zinc-binding domain that could play a role in DNA binding (59,60). Whether this motif is involved in nucleic acid binding or simply involved in protein folding remains unresolved as mutations in this motif of R2 gave rise to an unstable protein, precluding further analysis (unpublished data, SM Christensen). A potential zinc-binding domain downstream of the RT domain is found in many non-LTR retrotransposons. In vivo integration assays have revealed that mutations in this putative zinc-binding domain eliminate retrotransposition activity in L1 elements of mammals (61).
At the N-terminal end of the R2 protein (Figure 2) are classic C2-H2 zinc-finger and Myb-like nucleic acid binding motifs (9,62). DNA-binding and DNase footprint analyses of wild type and mutant polypeptides spanning the 140 amino acid N-terminal end revealed that the Zn-finger motif binds the DNA target from 1 to 3 bases upstream of the cleavage site while the Myb-motif binds DNA sequences from 10 to 15 base pair downstream of the insertion site (62). Because the complete R2 protein also protects a region of DNA from 10-40 bp upstream of the cleavage site, the C-terminal domain of the protein was postulated to be responsible for this upstream binding (62). Recently, however, the N-terminal domain of the R2 protein from the horseshoe crab was shown to bind this upstream region (16). The horseshoe crab R2 protein differs from the B. mori R2 in having three zinc-finger domains instead of one (figure 2). It is possible that both the N-terminal and C-terminal domains of all R2 proteins contribute to protein binding 10-40 bp upstream of the insertion site, but the isolated N-terminal domain of the B. mori protein binds too weakly to be detected in vitro. Analysis of the DNA-binding motifs that are N-terminal to the RT domain in other non-LTR retrotransposons show considerable flexibility in their binding to the target site (63).
An RNA binding domain of the R2 protein was recently identified immediately N-terminal to the RT domain (64). This domain comprises two conserved sequence motifs that have been termed 0 and −1 because they are encoded before motifs 1 through 7 of the RT domain (Figure 2). Mutations within either motif affect all properties of the R2 protein that require RNA binding. These include the ability of the protein to conduct the TPRT reaction, the ability of the protein to bind RNA in gel mobility shift assays, and the ability of the protein to cleave the second strand of the target site. Sequence similarity to the 0 motif can be found in all lineages of non-LTR retrotransposons (29,65,66), suggesting a similar RNA binding domain. As diagramed in Figure 2, an RNA binding domain of telomerase and of the group-II introns is also located upstream of the RT domain (67-69). Indeed, sequence similarity exists between the 0 motifs of R2 and group-II introns (11,70,71). The similar location of these RNA binding domains provides support to the model, originally based on the sequences of the reverse transcriptase domain, for a close evolutionary relationship among these three groups of genetic elements (70,72,73). This common origin and structural similarity suggests that future studies of non-LTR retrotransposition mechanisms should use telomerase and group II introns as guides.
Nature of the RNA template: the R2 ribozyme
An understanding of the R2 integration reaction requires an understanding of the RNA template that is used for reverse transcription. The exact 3’ end of the RNA did not appear critical to the TPRT mechanism, as in vitro experiments showed that similar TPRT integrated products were formed whether the RNA templates ended at the precise 3’ end of the R2 element or contained downstream 28S sequences (41,42). On the other hand, the exact 5’ end of the RNA did appear important, as in vivo integration results (74,75) indicated the products differed depending on whether 28S sequences were present or absent at this end of the transcript.
An analysis of Drosophila simulans stocks that supported frequent R2 retrotransposition events (described in greater detail below) revealed R2 transcripts of the approximate length of a full-length R2 element (76). Several lines of evidence suggested these transcripts were derived by co-transcription with the 28S rRNA gene (Figure 3A). First, previous studies could not identify a promoter at the 5’ end of full-length D. melanogaster elements using transient transcription assays in tissue culture cells (77). Second, the level of full-length R2 transcripts usually correlated with the level of transcription of extensively 5’ truncated elements (78). Third, in stocks with significant levels of R2 transcripts low levels of 28S/R2 co-transcripts could also be observed (76). Detailed RT-PCR analysis of these stocks revealed that the co-transcripts were derived from inserted rDNA units containing R2 5’ junctions with small deletions. Thus not only was processing of the R2 transcript from a 28S co-transcript suggested, but processing at the 5’ end was dependent upon the sequence at the 28S/R2 5’ junction (79).
Figure 3.
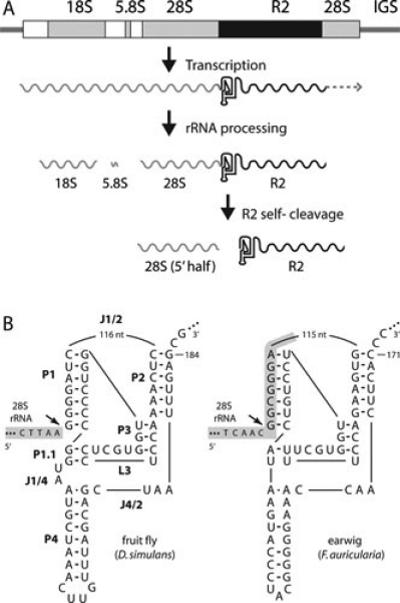
The R2 ribozyme. (A) An rDNA transcription unit is diagramed with 18S, 5.8S, and 28.S genes (gray boxes), transcribed spacers (white boxes), and R2 insertion (black box). All three rRNAs are normally processed from the single primary transcript. When a unit contains an R2 insertion, a self-cleaving ribozyme encoded at the 5’ end of the element releases the 5’ end of the R2 transcript from the upstream 28S rRNA sequence. It is not known if transcription ends at the 3’ end of the R2 element, or if this end is processed from downstream 28S gene sequences. (B) On the left is the D. simulans R2 ribozyme folded in a structure similar to that of the hepatitis delta virus (HDV) ribozyme (80,81). The various components of the ribozyme are labeled as in the HDV ribozyme: P, base-paired region; L, loop; J, nucleotides joining paired regions. 28S gene sequences are shaded with gray. On the right is the R2 ribozyme from Forficula auricularia (earwig). Self-cleavage (arrow) occurs at the precise junction of the R2 element with the 28S gene in the case of the D. simulans element and upstream of the junction in the 28S gene sequences in the case of the R2 element from F. auricularia.
To examine processing, RNA templates comprising different lengths of the consensus D. simulans 28S/R2 5’ junction were generated in vitro with T7 RNA polymerase (79). In addition to the expected RNA product, these simple RNA synthesis reactions also produced RNA fragments resulting from cleavage at the exact 5’ junction of the R2 element with the upstream 28S rRNA sequences. The autocatalysis was efficient with up to 98% of the RNA cleaved. RNA self-cleavage required the first 184 nucleotides of the R2 element but was not dependent upon the upstream 28S sequences. As shown in Figure 3B, the first 184 nucleotides of the R2 RNA could be folded into a secondary structure containing a double pseudoknot and five base paired regions that was similar to the self-cleaving ribozyme previously characterized from the hepatitis delta virus (HDV) (80,81). Remarkably, 21 of the 27 nucleotide positions of the HDV ribozyme that had been shown to generate the active site of the HDV ribozyme were identical to the R2 sequence (80-82). Using an entirely structure-based bioinformatics approach, the R2 ribozyme was independently identified by Ruminski and co-workers (83).
An analysis of R2 from many other animals spanning its entire host range revealed that the 5’ end of all R2 elements could be folded into an HDV-like ribozyme structure (84). Comparison of these different R2 ribozymes revealed considerable flexibility in some aspects of the structure and in the sequence. Surprisingly, however, several distantly related R2 lineages had sequences comprising the active site that were highly similar to those observed for the D. simulans R2 and HDV ribozymes. Presumably the limited parameter space afforded by using only four nucleotides to make the ribozyme has resulted in the R2 ribozyme converging upon the same sequences on multiple occasions. HDV-like ribozymes have also been found in other non-LTR retrotransposon lineages including several that are not site-specific (83-86).
The vast majority of the 28S/R2 5’ junction RNAs that were directly tested for activity in vitro showed detectable levels of self-cleavage. The major difference in the activity of the R2 ribozyme from diverse animals to that observed in Drosophila was the location of RNA self-cleavage. Unlike the D. simulans R2 ribozyme, which cleaves at the precise 28S/R2 5’junction, many R2 ribozymes cleaved in GC-rich regions of the 28S rRNA either 13 or 28 nucleotides upstream of the R2 insertion site (Figure 3B). As described below in the discussion of the R2 integration mechanism, self-cleavage by the ribozyme in the upstream 28S sequences in some species gives rise to insertions with uniform 5’ junctions while self-cleavage at the 5’ end of R2 in other species gives rise to variable junctions (11,84,87).
The discovery of the R2 ribozyme also helped to explain the presence of several non-autonomous parasites of R2 in the rDNA locus of some Drosophila species (88). For example, a short (530 bp) element propagates at the precise R2 insertion site of the 28S rRNA genes in D. willistoni. Divergent but clearly derived from the typical R2 elements found in this species, this short element had lost the entire ORF but had retained the 5’ ribozyme to enable processing from the 28S co-transcript and the structure of the 3’ UTR to enable binding by the R2 integration machinery. The elements were termed SIDEs for Short Internally Deleted Elements. Surprisingly, a more often encountered type of SIDE contained an R2 ribozyme at the 5’ end and the 3’ UTR from an R1 element (88). These R2/R1 hybrid SIDEs were located in the typical R1 insertion site suggesting that while their RNA was processed by the R2 ribozyme it was the R1 integration machinery generating the new insertions. In many respects these SIDEs are similar to the non-autonomous SINE elements that parasitize other non-LTR retrotransposons (LINEs) (89,90). The presence of R2s, R1s, and their SIDEs suggest that in some species there can be intense competition between selfish elements for the limited number of 28S rRNA insertion sites.
Current model of R2 integration
The initiation of the R2 integration reaction, i.e. using the initial nick at the target site to prime reverse transcription, was deduced soon after the R2 protein of B. mori was purified (37,41,42). The key to understanding the second half of the integration reaction was revealed over 10 years later when it was discovered that in addition to binding RNA from the 3’ end of the R2 transcript, R2 protein could also bind a segment of RNA from near the 5’ end of the R2 element (60). This 300 nt segment of RNA starts within the 5’ UTR and ends just before the sequences encoding the N-terminal zinc-finger (Figure 2). A distinctive structure of this RNA is a pseudoknot that is conserved across silk moths (91,92). The segment of R2 RNA that is bound by the protein determines its function in the integration reaction. In the presence of the 3’ RNA, the R2 protein binds the 28S gene upstream of the insertion site. In the presence of the 5’ RNA the R2 protein binds the 28S gene downstream of the insertion site.
In the complete model for R2 retrotransposition, these two protein/RNA complexes are proposed to perform symmetric reactions as diagramed in Figure 4. The subunit bound upstream initiates the retrotransposition reaction by both cleaving the bottom (first) strand of the DNA target and polymerizing the first DNA strand onto the released 3’ OH. The R2 subunit bound downstream of the insertion site appears to remain inactive until after the bound 5’ RNA is removed (60). Presumably this RNA is ‘pulled’ from the subunit as the RNA is used for first strand DNA synthesis. The downstream subunit then initiates the second half of the reaction by cleaving the top (second) DNA strand. The protein again utilizes the released 3’ end of the DNA as a primer and polymerizes the second DNA strand. Second strand synthesis involves displacement of the annealed RNA strand by the R2 protein (43,45). Gel shift experiments suggest single R2 protein subunits separately bind the 3’ and 5’ ends of the RNA (59,93). However the stoichiometry of a complete integration reaction is not known.
Figure 4.
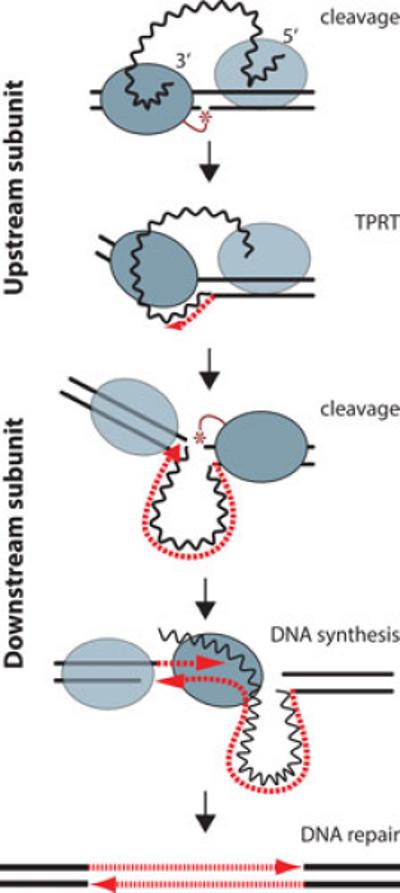
The R2 retrotransposition model. An R2 integration reaction is proposed to involve symmetric cleavage/DNA synthesis steps by R2 proteins bound upstream and downstream of the insertion site. From top to bottom, protein bound upstream of the insertion site is associated with the 3’ end of the R2 transcript. This protein both cleaves the bottom stand of DNA and catalyzes the reverse transcription of the R2 RNA using the cleaved DNA target as primer, target primed reverse transcription (TPRT). R2 protein bound downstream of the insertion site is associated with the 5’ end of the R2 transcript. When the reverse transcription reaction catalyzed by the upstream protein dislodges the 5’ RNA, the downstream protein cleaves the top DNA strand and again uses the cleaved DNA to prime second strand DNA synthesis. Second strand synthesis requires the polymerase to displace the R2 RNA. Because in the absence of bound RNA the downstream protein does not bind tightly to the DNA target, it is shown dissociated from the target site during polymerization. The integration reaction is completed by the host repair machinery which fills in the single stranded gaps at the target site. Blue oval, protein subunit (dark, active; light, inactive); wavy black line, R2 RNA; dashed red lines, synthesized DNA; solid black lines; target DNA.
The initiation of second strand DNA synthesis has not yet been documented in the in vitro retrotransposition reaction (37,45,46,59). Because this step is so inefficient in vitro, we can not exclude the possibility that second strand synthesis is accomplished in vivo by a host polymerase as appears to be the case for group II intron retrohoming (54). Based on the variation observed at the 5’ junction of endogenous R2 insertions in different species, it also appears to be a step that has evolved. In B. mori and many other animals, the R2 5' junctions within a species show little variation except for the occasional direct duplication of 28S gene sequences of a characteristic length for each species (11,84). However, in species of Drosophila and other animals, most 5’ junctions contain variable deletions of the 28S target site and variable additions of non-templated nucleotides (87,94). As described above in the discussion of the R2 ribozyme, this difference is correlated with the location of the R2 self-cleavage.
A model to explain the difference in the uniformity of the 5’ junction in different animals is diagramed in Figure 5. If the RNA template contains upstream 28S sequences, then the cDNA that is generated by reverse transcription can anneal to the upper strand of the DNA target. This annealing allows efficient and precise priming of second-strand synthesis and uniform 5’ junctions. However, if the 5’ end of the transcript contains only R2 sequences, then the cDNA generated by reverse transcription has no sequence identity with the upper strand of the DNA target. Second strand DNA synthesis is then postulated to initiate at a region of microhomology between the DNA strand upstream of the insertion site and the extra nucleotides added to the cDNA strand as the reverse transcriptase runs off the RNA template (46). Because these extra nucleotides are random, the microhomology used in each integration event can vary giving rise to the different length deletions of 28S sequences and/or non-templated nucleotide additions observed in some species.
Figure 5.
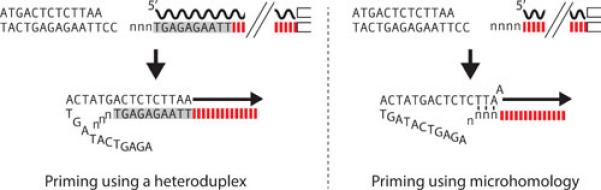
Variation in the priming of second-strand DNA synthesis. R2 elements differ in whether the 5’ end of the RNA template used in the integration reaction ends at the boundary between R2 and the 28S gene or extends a short distance upstream in the 28S rRNA sequence. This difference is dependent upon the location of the self-cleavage site by the R2 ribozyme (see text). Left panel. If self-cleavage by the R2 ribozyme is upstream in the 28S gene sequences, the resulting cDNA strand can form a heteroduplex with the upstream target DNA. This heteroduplex can stabilize the integration intermediate resulting in precise initiation of second strand synthesis (arrow) and uniform 5’ ends for different R2 copies. Right panel. If self-cleavage is at the 28S/R2 junction, there are no 28S sequences on the DNA strand (cDNA) generated by reverse transcription. As a consequence, the R2 protein must use regions of microhomology to initiate second strand synthesis (arrow). Priming frequently involves the 3-5 non-templated nucleotides added to the cDNA strand as the enzyme ran off the RNA template (lower case n's). This use of chance microhomologies to prime second strand DNA synthesis gives rise to sequence variation at the 5’ junctions of different integrated copies of R2. Wavy black line, RNA with 5’ end denoted; red dashed line, first strand DNA composed of R2 sequences; gray box, first strand DNA sequences complementary to upstream DNA target sequences.
Consistent with this model, in all animals the junctions of 5’ truncated R2 insertions, which are generated when the reverse transcriptase doesn't reach the 5’ end of the RNA template, are found to involve microhomologies and non-templated additions (84,87,94). Direct support for this model has also been provided by in vivo R2 injection experiments in Drosophila, which yielded integrations with precise 5’ junctions when the injected R2 RNA contained upstream 28S sequences and highly variable junctions when the injected R2 RNA did not contain upstream 28S sequences (74). Similarly a tissue culture R2 integration system based on a baculovirus vector required the R2 transcript to contain upstream 28S sequence to give rise to precise R2 integrations into the 28S rRNA gene (75). Models for the formation of a heteroduplex between the cDNA strand and the DNA template have also been hypothesized during initiation of second strand synthesis for other non-LTR retrotransposons (87,95).
EXPRESSION OF R2 ELEMENTS
R2 copies are rapidly gained and lost from rDNA loci
Theoretical studies and computer simulations have suggested that repeated crossovers over time are responsible for the nearly identical sequence of the rRNA genes within a species, but allow these rRNA sequences to evolve in unison over time (concerted evolution) (reviewed in 96). These crossovers result in a wide range in the number of rDNA units per loci for the individuals in a population. This concerted evolution of the rDNA locus would also affect the sequence and number of R2 elements residing within the rDNA units. Thus, while copies of mobile elements that insert throughout a genome can remain for long periods at locations where they do no harm, all R2 elements would be predicted to be like the rDNA units themselves and undergo turnover. Evidence for this turnover was found in the low levels of nucleotide sequence divergence (range 0.0 - 0.6 %) between R2 copies within a species (26,27,87).
It would appear technically difficult to monitor the turnover of R2 elements because all elements insert into the same site within the 28S genes and all R2 copies and 28S rRNA genes are nearly identical in sequence (87,97). Fortunately in species of Drosophila, individual R2 insertions can be monitored because the RNA transcript does not contain upstream 28S sequences (79), and thus as diagramed in Figure 5, have highly variable 5’ junctions. These variants include small deletions of the 28S gene and/or additional non-templated sequences as well as R2 5’ truncations ranging from less than 100 bp to more than 3 kb.
A PCR based assay was developed to generate profiles of the distinctive “lengths” associated with the 5’ end of the R2 elements in individual flies (24). The assay utilizes a forward primer that anneals to the 28S gene about 75 bp upstream of the R2 insertion site as well as a series of reverse primers that anneal to R2 sequences spaced at 200 to 400 bp intervals throughout the R2 element. The PCR amplification products are separated on 5% acrylamide gels to monitor the R2 5’ truncations or on high resolution sequencing gels to obtain the single base pair resolution needed to differentiate between the full-length R2 inserts. Such assays revealed most stocks of D. melanogaster and D. simulans contained 15 to 25 different 5’ truncated R2 copies and 20 to 30 full-length R2 copies of which 10-15 copies contained junctions that differed in length (23,24,98). The “5’ profiles” for flies from different populations or even from individuals within the same population could completely differ suggesting old copies were lost and new copies gained; that is, the R2 elements in these species were turning over (24,98). A similar analysis of 5’ truncated elements in the tadpole shrimp likewise revealed R2 copies were being gained and lost (99). PCR assays were also conducted to monitor the R1 non-LTR retrotransposons that are also present in the rDNA of the two Drosophila species. While most full-length R1 copies are of identical length, and thus cannot be monitored, the profiles of 5’ truncated copies of R1 from individual flies suggested a level of turnover similar to that of R2 (24).
To directly monitor the rate of turnover, the R2 and R1 elements were examined in the Harwich mutation accumulation lines of D. melanogaster generated by Mackay and co-workers (100). Those flies had first undergone 45 generations of inbreeding to remove any variation before multiple sublines were started (generation 0) and maintained by mass mating a small number of flies at each generation. At generation 350, the R2 and R1 5’ profiles were determined in 19 sublines (25,101). Over this ~17 year period, each subline was found to have gained 0 to 2 new R2 5’ variants (mean 0.8 insertions/line) and to have lost 0 to 8 of the 34 ancestral R2 variants (mean 2.9 deletions/line) indicating the R2 elements were slowly being lost from these lines. In contrast, a mean of 9.5 new insertions and 2.1 deletions of ancestral R1 elements were detected in the sublines indicating that R1 elements were increasing in number. Equally important, the sizes of the rDNA loci in the sublines after 350 generations varied from 140 to 310 units, indicating that many crossover events must have occurred. However, almost all of the R2 and R1 length variants, whether new or ancestral, were present at one copy per locus indicating that these crossovers had seldom duplicated individual insertions (102). The variation in rDNA number among sublines was almost exclusively associated with the number of uninserted rDNA units. Thus copies of R1 and R2 were lost from the rDNA locus, presumably by recombination, but copies of these insertions were seldom duplicated by recombination.
Active R2 retrotransposition and its developmental timing
The question thus became: are new R2 insertions generated at a low rate in possibly all individuals, or can R2 elements undergo higher rates of retrotransposition in individuals of a specific genetic composition or under certain physiological conditions? The search for flies with R2 activity was easier to conduct in D. simulans than in D. melanogaster. Unlike D. melanogaster where rDNA loci are on both the X and Y chromosomes, D. simulans encodes an rDNA locus only on the X chromosome. Therefore, D. simulans males will have a single rDNA locus that can be scanned for new insertions. A survey of laboratory stocks originally derived from a single population in California revealed that the males in most of the D. simulans stocks showed only minimal differences in their R2 5’ profiles, consistent with little or no retrotransposition and slow rates of deletion (98). However, the males in a few stocks showed extensive variation among their R2 5’ profiles. Indeed, in two stocks virtually every male had a 5’ profile that differed from every other tested male suggesting very high rates of R2 turnover.
To directly monitor the rates of new insertions in these lines, the R2 5’ profiles were determined for the male progeny of single pair crosses (33). New R2 insertions, typically only 1 or 2 per locus, were observed in about 10% of the sons from each cross. Two lines examined in detail showed R2 insertion rates of 0.12 and 0.15 insertions per locus per generation. Surprisingly, large numbers of parental R2 copies were also being deleted in these crosses with an average loss of over four R2 copies (range 1 to 15 copies) per deletion event. Thus the rates of R2 deletions in these R2 active lines based on the single generation experiments (0.22 and 0.44 deletions/locus/generation) were actually higher than the rates of new insertions. High R2 deletion rates were never seen in lines in which new insertions were not detected suggesting that R2 activity itself was causing the deletions. Presumably, the presence of multiple R2 endonucleases attempting to initiate integration in multiple rDNA units results in the deletion of large segments of the rDNA locus.
To determine if the high rates of R2 activity seen in these stocks could be maintained over multiple generations, the progeny of individual pairs were monitored after 30 generations (33). The rates of new insertions per generation in these 30-generation experiments were similar to those found after the one-generation experiment, however, the rates of R2 deletions determined from these long-term experiments was significantly less. In fact, the ~ 0.10 deletions per locus per generation found in these long term experiments was now similar to the rate of new insertions. Thus many of the large deletions of the rDNA locus that were seen in the one generation experiment presumably gave rise to less viable flies which were then lost from the small populations being maintained.
The ability to detect new R2 insertions in a single generation made it possible to ask when and where R2 retrotranspositions occurred. The above studies detected retrotranspositions in the female germ line since sons were compared to their mother. To look at the timing of the retrotransposition events in the development of the germ cells over 200 male progeny from a single female were screened for new R2 insertions (33). Thirty-one of the 32 new R2 insertions that were scored were only found in one or two males indicating that most retrotransposition events occurred late in the development of the egg, i.e. during oogenesis rather than during germ cell propagation. To look for activity in the male germ line, males from each of the R2 active stocks were crossed to attached-X (XXY) females. The only surviving male progeny from this cross inherit their Y chromosome from their mother and their X chromosome from their father. New R2 insertions were also detected through the male germ line but at 3-4 fold lower levels than observed through the female germ line.
Finally, R2 insertions were also assayed in somatic tissues (103). Somatic insertions were defined as an R2 variant not inherited from either the father or mother and present in only a subset of the tissues tested from an animal. The tissues examined were individual imaginal discs dissected from third instar larvae as well as various adult tissues (e.g. leg, wing, antenna, proboscis). Somatic R2 insertions were detected in about one quarter of the animals tested from the D. simulans stocks that contained highly active R2 elements. Remarkably in one third of these somatic events, the same new insertion was detected in more than one body segment. Because determination of body segments occurs at the blastoderm stage in 2-3 hour embryos, this implied that the R2 retrotransposition events were occurring in the first two hours of embryo development. During this time, embryonic nuclei are entering the pole plasma to become the germ line. Therefore, an R2 insertion event that occurred at this time could give rise to germ line mosaics, which at the next generation would be scored as a germ line event. Thus the low number of R2 retrotransposition events that were originally scored in the male germ line as well as a fraction of the retrotranspositions scored in the female germ line (33) may actually represent somatic events rather than true germ line events.
Control over R2 activity is at the level of transcription
Since the first discovery of R2 insertions in D. melanogaster a question frequently asked has been when are they transcribed (78,104,105). Even though they reside in a significant fraction of the genes in arguably the most actively transcribed loci, the levels of R2 transcripts were found to be extremely low to nonexistent. Nuclear run-on experiments (106) as well as the direct microscopic observation of transcribing rDNA loci (107) indicated that the low level of detectable R2 transcripts was due to transcription repression and not the rapid degradation of RNA transcripts. The chromatin associated with R2 insertions contained the typical epigenetic marks linked to heterochromatin (106). These studies revealed that most of the uninserted rDNA units also appeared to be packaged into heterochromatin. These findings are consistent with numerous studies conducted in many eukaryotic taxa that found only a fraction of the total number of rDNA units are actively transcribed (108-110). The fraction of transcriptionally active rDNA units in D. melanogaster has been estimated at only 30 – 40 units of the several hundred units typically present in the rDNA loci (32,106,111). Thus in most flies the rDNA units containing an R2 insertion are not transcribed.
The transcriptional state of the R2-inserted units could be changed, however, by manipulating the composition of the rDNA locus (78). R2 protein/RNA complexes of B. mori (similar to those used in the in vitro studies of the TPRT reaction) were injected into D. melanogaster embryos to give rise to flies with B. mori R2 insertions. Several of these flies also contained large deletions of the rDNA loci which was presumably the result of the injected R2 endonuclease cleaving multiple rDNA units within a locus and the subsequent loss of large sections. In these stocks, the level of transcript from the B. mori R2 insertion was inversely correlated with the total number of uninserted units in the locus. For example, a stock that contained about 20 uninserted units on the X chromosome rDNA locus had a level of B. mori R2 transcription in females that was about 100-fold higher than stocks that had greater than 80 uninserted units (78). Interestingly, transcription of specific endogenous D. melanogaster R2 insertions was also inversely correlated with the number of uninserted units.
The recovery of naturally occurring stable stocks of D. simulans that exhibited frequent R2 retrotranspositions (98) provided a means to directly study control over R2 activity. Northern blots of RNA from stocks with frequent R2 retrotranspositions (R2 active) and stocks with infrequent/no retrotranspositions (R2 inactive) revealed a clear correlation between the level of full-length R2 transcripts and the level of R2 retrotransposition events (76). Consistent with the studies of when and where R2 retrotranspositions were occurring (33,103) R2 transcripts were detected in both males and females and in both germ line and somatic tissues of larvae and adults. Nuclear run-on experiments again suggested that control was at the level of transcription rather than at a post-transcription step (76).
To assess the genetic control over R2 transcription, crosses were conducted between the R2 active and R2 inactive stocks (76). In both the F1 and F2 progeny of these crosses, the pattern of high versus low R2 transcript level suggested that control over R2 transcription mapped to the single rDNA locus on the X chromosome and was otherwise little influenced by the genetic background of the fly. For example, monitoring individual male progeny from crosses between animals with R2 active loci (XR2-A) and animals with R2 inactive loci (XR2-I) revealed that even after many generations of random mating all males with the XR2-A locus had a high R2 transcript level, and all males with the XR2-I locus had low levels of R2 transcript. Surprisingly, females that contained both an XR2-I and an XR2-A locus showed nucleolar dominance (112). These heterozygous females supported only low R2 transcript levels suggesting the XR2-I locus showed dominance over the XR2-A locus. Microscopic analysis of the secondary constrictions in these heterozygous females revealed a transcriptionally active rDNA locus on only one X chromosome, directly demonstrating nucleolar dominance. The dominance of an XR2-I locus over an XR2-A locus only occurred when the latter had very high levels of R2 transcription. Females with two rDNA loci that each supported more intermediate levels of R2 transcription in males typically had a transcriptionally active rDNA locus on both X chromosomes and R2 transcript levels near the average of the two loci (76).
While the extremely high levels of R2 transcription seen in some D. simulans stocks have to date not been observed in D. melanogaster, crosses between D. melanogaster stocks with moderate and extremely low levels of R2 transcription again revealed intermediate levels of transcription in the female progeny (113). However, in D. melanogaster an rDNA locus is also present on the Y chromosome. Surprisingly, the level of R2 transcripts detected in the male progeny was found to be dependent upon the level of R2 transcription in the father and independent of the level of R2 transcription in the mother. This suggested that the Y chromosome rDNA locus showed nucleolar dominance over the X chromosome rDNA locus (113,114). Microscopic analysis confirmed that only the rDNA locus on the Y chromosome was transcriptionally active. While the key to R2 activity, it is interesting to note that control over rDNA locus expression also appears to have an influence on the regulation of genes throughout the genome. The size of the rDNA locus and thus the number of genes that must be turned-off has been shown to affect the level of expression of many genes, in particular those that are influenced by position-effect variegation (113,115,116).
A model for the regulation of R2 activity
Little is known about the factors affecting the long-term stability of a retrotransposable element. Do the defensive mechanisms used by the host to control the element occasionally breakdown, thus, the element experiences windows of opportunity to replenish its number? Or, are the host's defensive mechanisms not completely effective, thus, there are continuous but low levels of element activity? In the case of R2, the former appears more likely as most laboratory stocks exhibited no R2 activity, while a few stocks demonstrated high levels of R2 activity over many generations. Because higher numbers of R2 elements were found associated with the rDNA loci of these active stocks and because host control over R2 activity mapped to the rDNA locus itself, one plausible model was that the many R2 elements were simply overwhelming the cellular defensive machinery (76,98). However the alternative model in which the activation of R2 elements gave rise to the higher numbers of copies was also a viable possibility. To directly address this issue, it was necessary to obtain a snapshot of the dynamic nature of the rDNA locus in natural populations.
To this end, ~100 D. simulans lines were generated that contained individual rDNA loci (iso-rDNA lines) from each of two populations (Atlanta, San Diego) (117). For each population, about one half of the iso-rDNA lines were found to have no detectable level of R2 transcription. The remaining lines had levels of R2 transcription that varied over a factor of 100. Indeed, each population gave rise to one or two lines with levels of R2 transcripts that equaled or even exceeded that seen in our most active laboratory stocks. A subset of the lines encompassing the full range of R2 transcript levels were tested for their ability to generate new R2 insertions. As found in the laboratory stocks, the R2 transcript level correlated with the frequency of new retrotranspositions events, again suggesting that regulation was at the level of transcription.
To determine the differences among these lines that could be responsible for R2 activity, 100 lines were selected for a more detailed analysis of the physical properties of their rDNA loci (118). The mean number of R2 elements in the lines was 50 (range 25 to 80), and the mean locus size was 250 units (range 150 to 400). A small but significant trend towards higher R2 transcription was associated with higher numbers of R2 copies and with a smaller rDNA locus size (i.e. fewer rDNA units), however, there was a wide range of R2 transcript levels associated with all values of locus size and R2 copy number. Thus, a simple model in which R2 number or the fraction of inserted rDNA units in the locus would determine whether a stock contained active R2 elements did not appear valid.
The property of the rDNA loci that could be directly linked to R2 activity was revealed by genomic blots which monitored the distribution of R2 elements in the rDNA locus. A NotI restriction enzyme site is located in the R2 element but not in either the rDNA unit or its spacer region. Therefore, a pulsed-field gel of NotI-digested genomic DNA probed with 18S sequences reveals the spacing between R2-inserted rDNA units in the locus, with the largest NotI fragment representing the most extensive region of the rDNA locus that is free of R2 insertions. Those lines without detectable R2 transcription, both iso-rDNA lines from natural populations as well as laboratory stocks, had a region greater than 40-units (>400 kb) of the their rDNA loci free of R2 insertions. For those stocks with measurable levels of R2 transcription, the largest R2-free region was less (frequently much less) than 40 units in size (76,117). Thus the critical property predicting R2 activity was whether the R2-inserted rDNA units were distributed uniformly throughout the rDNA locus or were clustered, the latter producing at least one large region free of R2 insertions.
These findings gave rise to the “transcription domain” model as shown in Figure 6A. Fundamental to this model were the previous findings that only 30 to 40 rDNA units within the rDNA loci of Drosophila are transcribed (106,111) and the electron microscopic observations that actively transcribed rDNA units are contiguous (107,111). The transcription domain model suggests that the host activates for transcription a block of ~40 rDNA units that contains the fewest R2 insertions. The remaining rDNA units are packaged into transcriptionally inactive heterochromatin. If the area activated for transcription is entirely free of R2 insertions, then R2 transcripts are not produced. If a large region without R2 insertions is unavailable in the locus, the cell is required to include R2-inserted units in the transcription domain. Loci with transcribed R2-inserted units give rise to the generation of new R2 copies by retrotransposition. How the host is able to differentiate inserted and uninserted units is not known. However, small RNA silencing pathways (119,120) which induce heterochromatin formation of the R2-inserted units, potentially spreading into flanking units, would seem a likely mechanism. The finding that an XR2-I locus can be dominant over an XR2-A locus suggests that in extreme instances heterochromatin formation can spread through the whole ribosomal array.
Figure 6.
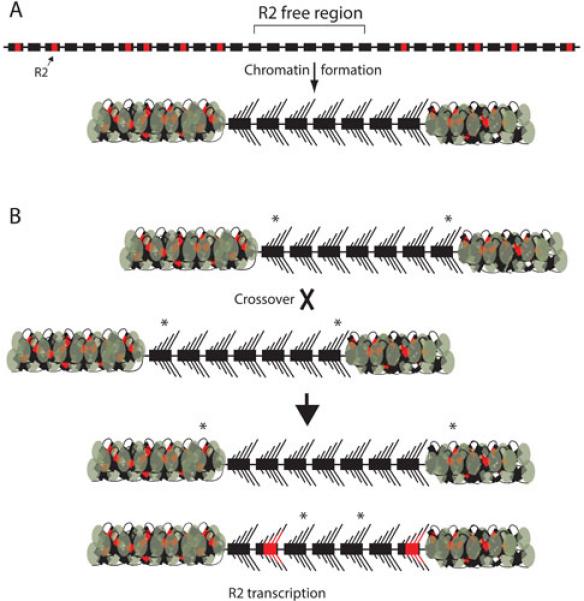
The transcription domain model of the rDNA locus and the long-term stability of R2 elements. (A) Uninserted (black boxes) and R2-inserted (black boxes with red insert) rDNA units are interspersed throughout the tandem array of rRNA genes. In Drosophila, a contiguous region of the rDNA locus with the lowest level of R2 insertions is selected for transcription. For simplicity this region is drawn as only seven units, but in D. simulans it is believed to be about 40 units. The remainder of the locus is packaged into heterochromatin (the compacted DNA plus protein flanking the active region). If the region selected as the transcription domain is free of R2-inserted units, then there is no R2 transcription and no R2 retrotransposition. (B) The driving force in the concerted evolution of the rDNA locus is crossovers between chromosomes. Most of these crossovers occur within the transcription domain (see text). The diagramed crossover produces one chromosome with an expanded R2 free region. Because the same number of rDNA units is still activated for transcription, some of the units that were transcribed before the crossover are packaged into heterochromatin after the crossover. Asterisks marking the original boundary of the transcription domain show this shift. The other chromosome product of the recombination contains an rDNA locus with a smaller R2-free region. In this case, rDNA units originally flanking the transcription domain are now activated for transcription. These flanking units contain R2 inserted units and thus copies of the R2 element are transcribed and retrotranspositions result.
Interestingly, variation in the ability of an organism to transcribe a region of the rDNA locus free of R2 insertions can explain why species have very different percentages of their rDNA units inserted with R2 (10,27,31,121,122). If an organism is less capable at differentiating between inserted and uninserted rDNA units, than all R2 inserts are potentially harmful and will be selected against. This selection against all copies means that the level of R2 elements in the locus will remain low. However, if the organism is adept at differentiating between inserted and uninserted units, than larger numbers of R2 elements can accumulate in the locus with minimal effects on the host.
Simulating rDNA loci and a stable population of R2 elements
Many studies have suggested that the driving force in the concerted evolution of the rRNA genes are random crossovers between rDNA loci (123-125). The presence of R2 elements, each inactivating a rDNA unit, were incorporated into computer simulations (126). By varying the crossover rate, the rate of R2 retrotransposition, and the number of uninserted units needed for full viability of the host, it was possible to simulate stable populations that contained from low to high mean levels of R2 elements. In all simulations, with or without the R2 element, the mean number of functional rDNA units per locus for the population far exceeded the minimum number required for full viability. The presence of R2-inserted units simply added to the total number of rDNA units present in each locus. The large excess in the number of uninserted rDNA units observed in organisms (106,108-110) was explained by the wide range in rDNA locus size that is generated by the crossovers. In any population, these crossovers generate a small number of individuals (the extreme low end of the range) that have insufficient numbers of rDNA units for peak fitness and are therefore lost from the population. The loss of those individuals with the smallest loci results in a continual increase in the mean number of rDNA units per locus over generations. This gradual increase in mean locus size was previously noted by Lyckegaard and Clark (125). To avoid this increase, all simulations of the rDNA locus assume either selection against very large locus size or a low rate of chromosomal loop-deletion (i.e. recombination between rDNA units on the same chromosome).
More sophisticated simulations were next attempted that could reproduce the structure of the rDNA loci and the dynamics of the R2 elements observed for the natural populations of D. simulans (118). The characterized properties of the rDNA loci that had to be reproduced included: a) the frequency range of R2 elements in the loci (118), b) the frequency range of all rDNA units (118), c) the infrequent duplication of individual copies of R2 by crossover events (102,117), d) the more rapid change in the number of uninserted units in a locus than the number of inserted units (102), e) the finding that about 45% of the individuals in a population have some level of R2 transcription (117), f) the linkage of R2 activity to the distribution of the R2 inserted units in the rDNA locus (i.e. the transcription domain model) (76,117), and g) the size of the transcription domain at about 40 units (76,111,117). Simulated populations that duplicated the natural populations could be generated and most importantly required R2 retrotransposition rates only slightly higher than that which had been estimated from direct observations (33,117).
The simulations suggested two other properties of the system. First, the only means found to minimize the number of duplicated R2 copies in the simulations was to localize the crossover events to the transcription domain. Clustering crossovers within the transcription domain is consistent with studies that suggested transcription increased crossover rates while heterochromatin typically inhibited recombination (127,128) and with predictions that regions free of insertions would allow multiple contiguous rDNA units to align and thus more likely to be involved in chromosome pairing. Second, in order to match the natural populations the simulations suggested that selection against individuals with active R2 elements had to be quite low (118). Even a 1% reduction in fitness for individuals that were transcribing a single R2-inserted unit was sufficient to rapidly drive R2 elements out of the simulated populations. Such a low affect on fitness would suggest that those individuals in natural populations forced to transcribe R2-inserted units could readjust the size of the transcription domain and compensate for any potential imbalance in 28S, 18S and 5.8S rRNA levels.
As shown in Figure 6B, the key to the long-term survival of R2 in the rDNA loci of many organisms is the ability of fully functional R2 copies to reside in rDNA units outside the transcription domain for many generations with essentially no effect on the host. Invariably, however, crossovers within the transcription domain increase and decrease the area free of R2 insertions and generate rDNA loci in which the organism is forced to place R2-inserted units within the transcription domain. The resultant production of R2 transcripts and subsequent R2 retrotransposition events can then repopulate the locus. Thus it is the stochastic nature of crossover within the rDNA locus that enables R2 elements to survive in many lineages.
Ultimately, it has been the exclusive niche of R2 elements for 28S rRNA genes that has made possible the wide range of studies described here. These long-term hitchhikers of eukaryotic genomes have provided valuable insights into the origin of non-LTR retrotransposons, their mechanism of integration, and their mechanism of expression. In studying R2 activity and control over that activity, R2 elements have also served as a useful tool to better understand both the evolution and transcriptional control of the rDNA locus itself.
ACKNOWLEDGEMENTS
The authors thank William Burke for his comments and assistance on this manuscript. We also want to thank Alan Lambowitz for his comments. We especially thank all the past and present members of the Eickbush lab whose invaluable contributions over the years have furthered our understanding of the R2 element. This work was made possible by the many years of support through National Institutes of Health Grant Number R01 GM42790.
REFERENCES
- 1.Dawid IB, Rebbert ML. Nucleotide sequence at the boundaries between gene and insertion regions in the rDNA of D. melanogaster. Nucleic Acids Res. 1981;9:5011–5020. doi: 10.1093/nar/9.19.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roiha H, Miller JR, Woods LC, Glover DM. Arrangements and rearrangements of sequence flanking the two types of rDNA insertion in D. melanogaster. Nature. 1981;290:749–753. doi: 10.1038/290749a0. [DOI] [PubMed] [Google Scholar]
- 3.Smith VL, Beckingham K. The intron boundaries and flanking rRNA coding sequences of Calliphora erythrocephala rDNA. Nucleic Acids Res. 1984;12:1707–1724. doi: 10.1093/nar/12.3.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujiwara H, Orgura T, Takada N, Miyajima N, Ishikawa H, Maekawa H. Introns and their flanking sequences of B. mori rDNA. Nucleic Acids Res. 1984;12:6861–6869. doi: 10.1093/nar/12.17.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eickbush TH, Robins B. B. mori 28S genes contain insertion elements similar to the type I and type II elements of D. melanogaster. EMBO J. 1985;4:2281–2285. doi: 10.1002/j.1460-2075.1985.tb03927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burke WD, Calalang CC, Eickbush TH. The site-specific ribosomal insertion element type II of Bombyx mori (R2Bm) contains the coding sequence for a reverse transcriptase-like enzyme. Mol Cell Biol. 1987;7:2221–2230. doi: 10.1128/mcb.7.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jakubczak JL, Xiong Y, Eickbush TH. Type I (R1) and Type II (R2) ribosomal DNA insertions of Drosophila melanogaster are retrotransposable elements closely related to those of Bombyx mori. J Mol Biol. 1990;212:37–52. doi: 10.1016/0022-2836(90)90303-4. [DOI] [PubMed] [Google Scholar]
- 8.Feng Q, Moran JV, Kazazian HH, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 9.Yang J, Malik HS, Eickbush TH. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc Natl Acad Sci USA. 1999;96:7847–7852. doi: 10.1073/pnas.96.14.7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakubczak JL, Burke WD, Eickbush TH. Retrotransposable elements R1 and R2 interrupt the rRNA genes of most insects. Proc Natl Acad Sci USA. 1991;88:3295–3299. doi: 10.1073/pnas.88.8.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burke WD, Malik HS, Jones JP, Eickbush TH. The domain structure and retrotransposition mechanism of R2 elements are conserved throughout arthropods. Mol Biol Evol. 1999;16:502–511. doi: 10.1093/oxfordjournals.molbev.a026132. [DOI] [PubMed] [Google Scholar]
- 12.Kojima KK, Fujiwara H. Cross-genome screening of novel sequence-specific non-LTR retrotransposons: various multicopy RNA genes and microsatellites are selected as targets. Mol Biol Evol. 2004;21:207–217. doi: 10.1093/molbev/msg235. [DOI] [PubMed] [Google Scholar]
- 13.Kojima KK, Kuma K, Toh H, Fujiwara H. Identification of rDNA-specific non-LTR retrotransposons in Cnidaria. Mol Biol Evol. 2006;23:1984–1993. doi: 10.1093/molbev/msl067. [DOI] [PubMed] [Google Scholar]
- 14.Luchetti A, Mantovani B. Non-LTR R2 element evolutionary patterns: phylogenetic incongruences, rapid radiation and the maintenance of multiple lineages. PLoS ONE. 2013;8:e57076. doi: 10.1371/journal.pone.0057076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gladyshev EA, Arkhipova IR. Rotifer rDNA-specific R9 retrotransposable elements generate an exceptionally long target site duplication upon insertion. Gene. 2009;448:145–150. doi: 10.1016/j.gene.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 16.Thompson BK, Christensen SM. Independently derived targeting of 28S rDNA by A- and D-clade R2 retrotransposons. Mobile Genetic Elements. 2011;1:29–37. doi: 10.4161/mge.1.1.16485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiwara H. Mobile DNA III. 2014 [to be completed] [Google Scholar]
- 18.Burke WD, Müller F, Eickbush TH. R4, a non-LTR retrotransposon specific to the large subunit rRNA gene of nematodes. Nucleic Acids Res. 1995;23:4628–4634. doi: 10.1093/nar/23.22.4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malik HS, Eickbush TH. NeSL-1, an ancient lineage of site-specific non-LTR retrotransposons from Caenorhabditis elegans. Genetics. 2000;154:193–203. doi: 10.1093/genetics/154.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burke WD, Singh D, Eickbush TH. R5 retrotransposons insert into a family of infrequently transcribed 28S rRNA genes of Planaria. Mol Biol Evol. 2003;20:1260–1270. doi: 10.1093/molbev/msg141. [DOI] [PubMed] [Google Scholar]
- 21.Burke WD, Malik HS, Rich SM, Eickbush TH. Ancient lineages of non-LTR retrotransposons in the primitive eukaryote, Giardia lamblia. Mol Biol Evol. 2002;19:619–630. doi: 10.1093/oxfordjournals.molbev.a004121. [DOI] [PubMed] [Google Scholar]
- 22.Kojima KK, Fujiwara H. Evolution of target specificity in R1 clade non-LTR retrotransposons. Mol Biol Evol. 2003;20:351–361. doi: 10.1093/molbev/msg031. [DOI] [PubMed] [Google Scholar]
- 23.Jakubczak JL, Zenni MK, Woodruff RC, Eickbush TH. Turnover of R1 (Type I) and R2 (Type II) retrotransposable elements in the ribosomal DNA of Drosophila melanogaster. Genetics. 1992;131:129–142. doi: 10.1093/genetics/131.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pérez-González CE, Eickbush TH. Dynamics of R1 and R2 Elements in the rDNA locus of Drosophila simulans. Genetics. 2001;158:1557–1567. doi: 10.1093/genetics/158.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pérez-González CE, Eickbush TH. Rates of R1 and R2 retrotransposition and elimination from the rDNA locus of Drosophila melanogaster. Genetics. 2002;162:799–811. doi: 10.1093/genetics/162.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eickbush DG, Eickbush TH. Vertical transmission of the retrotransposable elements R1 and R2 during the evolution of the Drosophila melanogaster species subgroup. Genetics. 1995;139:671–684. doi: 10.1093/genetics/139.2.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lathe WC, III, Eickbush TH. A single lineage of R2 retrotransposable elements is an active, evolutionarily stable component of the Drosophila rDNA locus. Mol Biol Evol. 1997;14:1232–1241. doi: 10.1093/oxfordjournals.molbev.a025732. [DOI] [PubMed] [Google Scholar]
- 28.Burke WD, Malik HS, Lathe WC, Eickbush TH. Are retrotransposons long term hitchhikers? Nature. 1998;239:141–142. doi: 10.1038/32330. [DOI] [PubMed] [Google Scholar]
- 29.Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 30.Burke WD, Eickbush DG, Xiong Y, Jakubczak JL, Eickbush TH. Sequence relationship of retrotransposable elements R1 and R2 within and between divergent insect species. Mol Biol Evol. 1993;10:163–185. doi: 10.1093/oxfordjournals.molbev.a039990. [DOI] [PubMed] [Google Scholar]
- 31.Stage DE, Eickbush TE. Maintenance of multiple lineages of R1 and R2 retrotransposable elements in the ribosomal RNA gene loci of Nasonia. Insect Mol Biol. 2010;19(suppl.1):37–48. doi: 10.1111/j.1365-2583.2009.00949.x. [DOI] [PubMed] [Google Scholar]
- 32.Hawley RS, Marcus CH. Recombinational controls of rDNA redundancy in Drosophila. Annu Rev Genet. 1989;23:87–120. doi: 10.1146/annurev.ge.23.120189.000511. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Zhou J, Eickbush TH. Rapid R2 retrotransposition leads to the loss of previously inserted copies via large deletions of the rDNA locus. Mol Biol Evol. 2008;25:229–237. doi: 10.1093/molbev/msm250. [DOI] [PubMed] [Google Scholar]
- 34.Hollocher H, Templeton AR. The molecular through ecological genetics of abnormal abdomen in Drosophila mercatorum. VI. The non-neutrality of the Y chromosome rDNA polymorphism. Genetics. 1994;136:1373–1384. doi: 10.1093/genetics/136.4.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malik HS, Eickbush TH. Retrotransposable elements R1 and R2 in the rDNA units of Drosophila mercatorum: abnormal abdomen revisited. Genetics. 1999;151:653–665. doi: 10.1093/genetics/151.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong Y, Eickbush TH. Functional expression of a sequence-specific endonuclease encoded by the retrotransposon R2Bm. Cell. 1988;55:235–246. doi: 10.1016/0092-8674(88)90046-3. [DOI] [PubMed] [Google Scholar]
- 37.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 38.Craigie R. Retroviral DNA integration. In: Craig NL, Craige R, Gellert M, Lambowitz AM, editors. Mobile DNA 11. ASM Press; Washington, DC: 2002. pp. 613–630. [Google Scholar]
- 39.Voytas DF, Boeke JD. Ty1 and Ty5 of Saccharomyces cerevisiae. In: Craig NL, Craige R, Gellert M, Lambowitz AM, editors. Mobile DNA 11. ASM Press; Washington, DC: 2002. pp. 631–662. [Google Scholar]
- 40.Eickbush TH. R2 and related site-specific non-long terminal repeat retrotransposons. In: Craig NL, Craige R, Gellert M, Lambowitz AM, editors. Mobile DNA 11. ASM Press; Washington, DC: 2002. pp. 813–835. [Google Scholar]
- 41.Luan DD, Eickbush TH. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol Cell Biol. 1995;15:3882–3891. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luan DD, Eickbush TH. Downstream 28S gene sequences on the RNA template affect the choice of primer and the accuracy of initiation by the R2 reverse transcriptase. Mol Cell Biol. 1996;16:4726–4734. doi: 10.1128/mcb.16.9.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bibillo A, Eickbush TH. The reverse transcriptase of the R2 non-LTR retrotransposon: continuous synthesis of cDNA on non-continuous RNA templates. J Mol Biol. 2002;316:459–473. doi: 10.1006/jmbi.2001.5369. [DOI] [PubMed] [Google Scholar]
- 44.Bibillo A, Eickbush TH. High processivity of the reverse transcriptase from a non-long terminal repeat retrotransposon. J Biol Chem. 2002;277:34836–34845. doi: 10.1074/jbc.M204345200. [DOI] [PubMed] [Google Scholar]
- 45.Kurzynska-Kokorniak A, Jamburuthugoda VK, Bibillo A, Eickbush TH. DNA-directed DNA polymerase and strand displacement activity of the reverse transcriptase encoded by the R2 retrotransposon. J Mol Biol. 2007;374:322–333. doi: 10.1016/j.jmb.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bibillo A, Eickbush TH. End-to-end template jumping by the reverse transcriptase encoded by the R2 retrotransposon. J Biol Chem. 2004;279:14945–14953. doi: 10.1074/jbc.M310450200. [DOI] [PubMed] [Google Scholar]
- 47.Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol) is sufficient for template switching in vitro. J Biol Chem. 1999;274:2706–2716. doi: 10.1074/jbc.274.5.2706. [DOI] [PubMed] [Google Scholar]
- 48.Chen B, Lambowitz AM. De novo and DNA primer-mediated initiation of cDNA synthesis by the Mauriceville retroplasmid reverse transcriptase involve recognition of a 3’ CCA sequence. J Mol Biol. 1997;271:311–332. doi: 10.1006/jmbi.1997.1185. [DOI] [PubMed] [Google Scholar]
- 49.Mohr S, Ghanem E, Smith W, Sheeter D, Qin Y, King O, Polioudakis D, Iyer VR, Hunicke-Smith S, Swamy S, Kuersten S, Lambowitz AM. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA. 2013;19:958–970. doi: 10.1261/rna.039743.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peliska JA, Benkovic SJ. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992;258:1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- 51.Malik HS, Eickbush TH. Phylogenetic analysis of Ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 2001;11:1187–1197. doi: 10.1101/gr.185101. [DOI] [PubMed] [Google Scholar]
- 52.Kelleher CD, Champoux JJ. Characterization of RNA strand displacement synthesis by Moloney Murine Leukemia virus reverse transcriptase. J Biol Chem. 1998;273:9976–9986. doi: 10.1074/jbc.273.16.9976. [DOI] [PubMed] [Google Scholar]
- 53.Lanciault C, Champoux JJ. Single unpaired nucleotides facilitate HIV-1 reverse transcriptase displacement synthesis through duplex RNA. J Biol Chem. 2004;279:32252–32261. doi: 10.1074/jbc.M404117200. [DOI] [PubMed] [Google Scholar]
- 54.Yao J, Truong DM, Lambowitz AM. Genetic and biochemical assays reveal a key role for replication restart proteins in group II intron retrohoming. PLOS Genetics. 2013;9:e1003469. doi: 10.1371/journal.pgen.1003469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jamburuthugoda VK, Eickbush TH. The reverse transcriptase encoded by the non-LTR retrotransposon R2 is as error-prone as that encoded by HIV-1. J Mol Biol. 2011;407:661–672. doi: 10.1016/j.jmb.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim B, Ayran JC, Sagar SG, Adman ET, Fuller SM, Tran NH, Horrigan J. New human immunodeficiency virus, type 1 reverse transcriptase (HIV-1 RT) mutants with increased fidelity of DNA synthesis. Accuracy, template binding, and processivity. J Biol Chem. 1999;274:27666–27673. doi: 10.1074/jbc.274.39.27666. [DOI] [PubMed] [Google Scholar]
- 57.Preston BD, Poiesz BJ, Loeb LA. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 58.Christensen S, Eickbush TH. Footprint of the R2Bm protein on its target site before and after cleavage in the presence and absence of RNA. J Mol Biol. 2004;336:1035–1045. doi: 10.1016/j.jmb.2003.12.077. [DOI] [PubMed] [Google Scholar]
- 59.Christensen SM, Eickbush TH. R2 target primed reverse transcription: ordered cleavage and polymerization steps by protein subunits asymmetrically bound to the target DNA. Mol Cell Biol. 2005;25:6617–6628. doi: 10.1128/MCB.25.15.6617-6628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Christensen SM, Ye J, Eickbush TH. RNA from the 5’ end of the R2 retrotransposon controls R2 protein binding to and cleavage of its DNA target site. Proc Natl Acad Sci USA. 2006;104:17602–17607. doi: 10.1073/pnas.0605476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 62.Christensen SM, Bibillo A, Eickbush TH. Role of the R2 element amino-terminal domain in the target-primed reverse transcription reaction. Nucleic Acids Res. 2005;33:6461–6468. doi: 10.1093/nar/gki957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shivram H, Cawley D, Christensen SM. Targeting novel sites: the N-terminal DNA binding domain of non-LTR retrotransposons is an adaptable module that is implicated in changing site specificities. Mob Genet Elements. 2011;1:169–178. doi: 10.4161/mge.1.3.18453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jamburuthugoda VK, Eickbush TH. Identification of RNA binding motifs in the R2 retrotransposon-encoded reverse transcriptase. Nuc Acids Res. 2014 doi: 10.1093/nar/gku514. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clements AP, Singer MF. The human LINE-1 reverse transcriptase: effects of deletions outside the common reverse transcriptase domain. Nucleic Acids Res. 1998;26:3528–3535. doi: 10.1093/nar/26.15.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moran JV, Gilbert N. Mammalian LINE-1 retrotransposons and related elements. In: Craig NL, Craige R, Gellert M, Lambowitz AM, editors. Mobile DNA 11. ASM Press; Washington, DC: 2002. pp. 836–869. [Google Scholar]
- 67.Gu SQ, Cui X, Mou S, Mohr S, Yao J, Lambowitz AM. Genetic identification of potential RNA-binding regions in a group II intron-encoded reverse transcriptase. RNA. 2010;16:732–747. doi: 10.1261/rna.2007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rouda S, Skordalakes E. Structure of the RNA binding domain of telomerase: implications for RNA recognition and binding. Structure. 2007;15:1403–1412. doi: 10.1016/j.str.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 69.Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17:513–518. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- 70.Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blocker FJ, Mohr G, Conlan LH, Qi L, Belfort M, Lambowitz AM. Domain structure and three-dimensional model of a group II intron-encoded reverse transcriptase. RNA. 2005;11:14–28. doi: 10.1261/rna.7181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eickbush TH. Telomerase and retrotransposons: which came first? Science. 1997;277:911–912. doi: 10.1126/science.277.5328.911. [DOI] [PubMed] [Google Scholar]
- 73.Arkhipova IA, Pyatkov KI, Meselson M, Evgenev MB. Retroelements containing introns in diverse invertebrate taxa. Nature Genetics. 2003;33:123–124. doi: 10.1038/ng1074. [DOI] [PubMed] [Google Scholar]
- 74.Eickbush DG, Luan DD, Eickbush TH. Integration of Bombyx mori R2 sequences into the 28S ribosomal DNA loci of D. melanogaster. Mol Cell Biol. 2000;20:213–223. doi: 10.1128/mcb.20.1.213-223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fujimoto H, Hirukawa Y, Tani H, Matsuura Y, Hashido K, Tsuchida K, Takada N, Kobayashi M, Maekawa H. Integration of the 5’ end of the retrotransposon, R2Bm, can be complemented by homologous recombination. Nucleic Acids Res. 2004;32:1555–1565. doi: 10.1093/nar/gkh304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eickbush DG, Ye J, Zhang X, Burke WD, Eickbush TH. Epigenetic regulation of retrotransposons within the nucleolus of Drosophila. Mol Cell Biol. 2008;28:6452–6461. doi: 10.1128/MCB.01015-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.George JA, Eickbush TH. Conserved features at the 5' end of Drosophila R2 retrotransposable elements: implications for transcription and translation. Insect Mol Biol. 1999;8:3–10. doi: 10.1046/j.1365-2583.1999.810003.x. [DOI] [PubMed] [Google Scholar]
- 78.Eickbush DG, Eickbush TH. Transcription of endogenous and exogenous R2 elements in the rDNA gene locus of Drosophila melanogaster. Mol Cell Biol. 2003;23:3825–3836. doi: 10.1128/MCB.23.11.3825-3836.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eickbush DG, Eickbush TH. R2 retrotransposons encode a self-cleaving ribozyme for processing from an rRNA co-transcript. Mol Cell Biol. 2010;30:3142–3150. doi: 10.1128/MCB.00300-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Been MD, Wickham GS. Self-cleaving ribozymes of hepatitis delta virus RNA. Eur J Biochem. 1997;247:741–753. doi: 10.1111/j.1432-1033.1997.00741.x. [DOI] [PubMed] [Google Scholar]
- 81.Ferré-D’Amaré AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 82.Nehdi A, Perreault J-P. Unbiased in vitro selection reveals the unique character of the self-cleaving antigenomic HDV RNA sequence. Nucleic Acids Res. 2006;34:584–592. doi: 10.1093/nar/gkj463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ruminski DJ, Webb C-HT, Riccitelli NJ, Lupták A. Processing and translation initiation of non-long terminal repeat retrotransposons by hepatitis delta virus (HDV)-like self-cleaving ribozymes. J Biol Chem. 2012;286:41286–41295. doi: 10.1074/jbc.M111.297283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eickbush DG, Burke WD, Eickbush TH. Evolution of the R2 retrotransposon ribozyme and its self-cleavage site. PLoS One. 2013;8:e66441. doi: 10.1371/journal.pone.0066441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Webb C-H, Riccitelli NJ, Ruminski DJ, Lupták A. Widespread occurrence of self-cleaving ribozymes. Science. 2009;326:953. doi: 10.1126/science.1178084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sánchez-Luque FJ, López MC, Macias F, Alonso C, Thomas MC. Identification of an hepatitis delta virus-like ribozyme at the mRNA 5’ end of the L1Tc retrotransposon from Trypanosoma cruzi. Nucleic Acids Res. 2011;39:8065–8077. doi: 10.1093/nar/gkr478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stage DE, Eickbush TH. Origin of nascent lineages and the mechanisms used to prime second-strand DNA synthesis in the R1 and R2 retrotransposons of Drosophila. Genome Biology. 2009;10:R49. doi: 10.1186/gb-2009-10-5-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eickbush DG, Eickbush TH. R2 and R1/R1 hybrid non-autonomous retrotransposons derived by internal deletions of full-length elements. Mobile DNA. 2012;3:10. doi: 10.1186/1759-8753-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ohshima K, Okada N. SINEs and LINEs: symbionts of eukaryotic genomes with a common tail. Cytogenet Genome Res. 2005;110:475–490. doi: 10.1159/000084981. [DOI] [PubMed] [Google Scholar]
- 90.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 91.Kierzek E, Kierzek R, Moss WN, Christensen SM, Eickbush TH, Turner DH. Isoenergetic penta- and hexanucleotide microarray probing and chemical mapping provide a secondary structure model for an RNA element orchestrating R2 retrotransposon protein function. Nucleic Acids Res. 2008;36:1770–1782. doi: 10.1093/nar/gkm1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kierzek E, Christensen SM, Eickbush TE, Kierzek R, Turner DH, Moss WN. Secondary structures for 5′ regions of R2 retrotransposon RNAs reveal a novel conserved pseudoknot and regions that evolve under different constraints. J Mol Biol. 2009;374:322–333. doi: 10.1016/j.jmb.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang J, Eickbush TH. RNA-induced changes in the activity of the endonuclease encoded by the R2 retrotransposable element. Mol Cell Biol. 1998;18:3455–3465. doi: 10.1128/mcb.18.6.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.George JA, Burke WD, Eickbush TH. Analysis of the 5' junctions of R2 insertions with the 28S gene: implications for non-LTR retrotransposition. Genetics. 1996;142:853–863. doi: 10.1093/genetics/142.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ostertag EM, Kazazian HH. Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 96.Eickbush TH, Eickbush DG. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics. 2007;175:477–485. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stage DE, Eickbush TH. Sequence variation within the rRNA gene loci of 12 Drosophila species. Genome Res. 2007;17:1888–1897. doi: 10.1101/gr.6376807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang X, Eickbush TH. Characterization of active R2 retrotransposition in the rDNA locus of Drosophila simulans. Genetics. 2005;170:195–205. doi: 10.1534/genetics.104.038703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mingazzini V, Luchetti A, Mantovani B. R2 dynamics in Triops cancriformis (Bosc, 1801) (Crustacea, Branchiopoda, Notostraca): turnover rate and 28S concerted evolution. Heredity. 2011;106:567–575. doi: 10.1038/hdy.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mackay TFC, Lyman RF, Jackson MS, Terzian C, Hill WG. Polygenic mutation in Drosophila melanogaster: estimates from divergence among inbred strains. Evolution. 1992;46:300–316. doi: 10.1111/j.1558-5646.1992.tb02039.x. [DOI] [PubMed] [Google Scholar]
- 101.Pérez-González CE, Burke WD, Eickbush TH. R1 and R2 retrotransposition and deletion in the rDNA loci on the X and Y chromosomes of Drosophila melanogaster. Genetics. 2003;165:675–685. doi: 10.1093/genetics/165.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Averbeck KT, Eickbush TH. Monitoring the mode and tempo of concerted evolution in the Drosophila melanogaster rDNA locus. Genetics. 2005;171:1837–1846. doi: 10.1534/genetics.105.047670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Eickbush MT, Eickbush TH. Retrotransposition of R2 elements in somatic nuclei during the early development of Drosophila. Mobile DNA. 2011;2:11. doi: 10.1186/1759-8753-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Long EO, Dawid IB. Expression of ribosomal DNA insertions in Drosophila melanogaster. Cell. 1979;18:1185–1196. doi: 10.1016/0092-8674(79)90231-9. [DOI] [PubMed] [Google Scholar]
- 105.Kidd SJ, Glover DM. D. melanogaster ribosomal DNA containing type II insertions is variably transcribed in different strains and tissues. J Mol Biol. 1981;151:645–662. doi: 10.1016/0022-2836(81)90428-9. [DOI] [PubMed] [Google Scholar]
- 106.Ye J, Eickbush TH. Chromatin structure and transcription of the R1- and R2-inserted rRNA genes of Drosophila melanogaster. Mol Cell Biol. 2006;23:8781–8790. doi: 10.1128/MCB.01409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jamrich M, Miller OL. The rare transcripts of interrupted rDNA genes in Drosophila melanogaster are processed or degraded during synthesis. EMBO J. 1984;3:1541–1545. doi: 10.1002/j.1460-2075.1984.tb02008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Conconi A, Widmer RM, Koller T, Sogo JM. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- 109.Conconi A, Sogo JM, Ryan CA. Ribosomal gene clusters are uniquely proportioned between open and closed chromatin structures in both tomato leaf cells and exponentially growing suspension cultures. Proc Natl Acad Sci USA. 1992;89:5256–5260. doi: 10.1073/pnas.89.12.5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dammann R, Lucchini R, Koller T, Sogo JM. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McKnight SL, Miller OL. Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell. 1976;8:305–319. doi: 10.1016/0092-8674(76)90014-3. [DOI] [PubMed] [Google Scholar]
- 112.Tucker S, Vitins A, Pikaard CS. Nucleolar dominance and ribosomal RNA gene silencing. Curr Opin Cell Biol. 2010;22:351–356. doi: 10.1016/j.ceb.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhou J, Sackton TB, Martinsen L, Lemos B, Eickbush TH, Hartl DL. Y chromosome mediates ribosomal DNA silencing and modulates the chromatin state in Drosophila. Proc Natl Acad Sci USA. 2012;109:9941–9946. doi: 10.1073/pnas.1207367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Greil F, Ahmad K. Nucleolar dominance of the Y chromosome in Drosophila melanogaster. Genetics. 2012;191:1119–1128. doi: 10.1534/genetics.112.141242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Paredes S, Branco AT, Hartl DL, Maggert KA, Lemos B. Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS Genetics. 2011;7:e1001376. doi: 10.1371/journal.pgen.1001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Paredes S, Maggert KA. Expression of I-CreI endonuclease generates deletions within the rDNA of Drosophila. Genetics. 2009;181:1661–1671. doi: 10.1534/genetics.108.099093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhou J, Eickbush TH. The pattern of R2 retrotransposon activity in natural populations of Drosophila simulans reflects the dynamic nature of the rDNA locus. PLoS Genetics. 2009;5:e1000386. doi: 10.1371/journal.pgen.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou J, Eickbush MT, Eickbush TH. A population genetic model for the maintenance of R2 retrotransposons in rRNA gene loci. PLoS Genetics. 2013;8:e1003179. doi: 10.1371/journal.pgen.1003179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Senti K-A, Brennecke J. The piRNA pathway: a fly's perspective on the guardian of the genome. Trends in Genetics. 2010;26:499–509. doi: 10.1016/j.tig.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ghesini S, Luchetti A, Marini M, Mantovani B. The non-LTR retrotransposon R2 in termites (Insecta, Isoptera): characterization and dynamics. J Mol Evol. 2011;72:296–305. doi: 10.1007/s00239-011-9430-y. [DOI] [PubMed] [Google Scholar]
- 122.Montiel EE, Cabrero J, Ruiz-Estévez M, Burke WD, Eickbush TH, Camacho JPM, López-León MD. Preferential occupancy of R2 retroelements on the B chromosome of the grasshopper Eyprepocnemis plorans. 2014 doi: 10.1371/journal.pone.0091820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ohta T. Evolution and variation of multigene families. Springer-Verlag; Berlin/Heidelberg, Germany/New York; 1980. [Google Scholar]
- 124.Ohta T, Dover GA. Population genetics of multigene families that are dispersed into two or more chromosomes. Proc Natl Acad Sci USA. 1983;80:4079–4083. doi: 10.1073/pnas.80.13.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lyckegaard EMS, Clark AG. Evolution of ribosomal RNA gene copy number on the sex chromosomes of Drosophila melanogaster. Mol Biol Evol. 1991;8:458–474. doi: 10.1093/oxfordjournals.molbev.a040664. [DOI] [PubMed] [Google Scholar]
- 126.Zhang X, Eickbush MT, Eickbush TH. Role of recombination in the long-term retention of transposable elements in rRNA gene loci. Genetics. 2008;180:1617–1626. doi: 10.1534/genetics.108.093716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Aguilera A, Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nature Rev Genetics. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 128.Voelket-Meiman K, Keil RL, Roeder GS. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987;48:1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]


