Abstract
A matrix-assisted laser desorption/ionization time of flight/time of flight tandem mass spectrometer (MALDI TOF/TOF) has been used for high-speed precursor/fragment ion transition image acquisition. High throughput analysis is facilitated by a Nd:YLF solid state laser capable of pulse repetition rates up to 5 kHz, a high digitizer acquisition rate (up to 50 pixels/second), and continuous laser raster sampling. MS/MS experiments are enabled through the use of a precision timed ion selector, second source acceleration, and a dedicated collision cell. Continuous raster sampling is shown here to facilitate rapid MS/MS ion image acquisition from thin tissue sections for the drug rifampicin and of a common kidney lipid, SM4s(d18:1/24:1). The ability to confirm the structural identity of an analyte as part of the MS/MS imaging experiment is an essential part of the analysis. Additionally, the increase in sensitivity and specificity afforded by an MS/MS approach is highly advantageous, especially when interrogating complex chemical environments such as those in biological tissues. Herein, we report continuous laser raster sampling TOF/TOF imaging methodologies which demonstrate 8-14 fold increases in throughput compared to existing MS/MS instrumentation, an important advantage when imaging large areas on tissues.
INTRODUCTION
Imaging mass spectrometry (IMS) has emerged as a powerful analytical tool enabling direct chemical interrogation and is ideally suited to the in situ measurement of the complex array of molecular species present in cellular tissue.1 Specifically, the rapid development of matrix-assisted laser desorption/ionization (MALDI) IMS technology in recent years has focused on advanced instrumentation, sample preparation, and data acquisition methods to improve the throughput, specificity, and sensitivity of these types of measurements.1-9
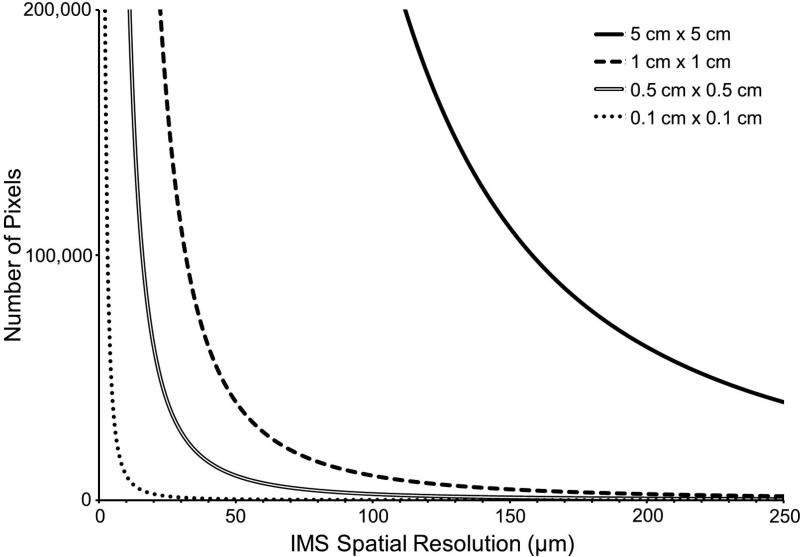
The importance of these types of improvements is especially evident in experiments involving biological tissue specimens. In MALDI tissue imaging experiments, a thinly sectioned tissue specimen is first mounted onto a target and then coated with a MALDI matrix in a manner which preserves the spatial integrity of the analytes of interest. A raster of the tissue surface is performed to generate a mass spectrum at each x, y coordinate (i.e., an individual pixel in ‘microprobe’ imaging mode).10 Ion intensity maps are then constructed as a function of x, y position for any single ion of interest. The total number of pixels in an image is thus a function of both the raster step size (i.e., the step resolution of the image) and the imaged area. As this is a squared relationship, the number of pixels required to sample a large tissue area11-14 and/or a tissue area at high spatial resolution3, 15-18 can be quite large (Figure 1). For example, a 1 cm2 tissue section imaged at 100 μm spatial resolution requires a 10,000 pixel image, whereas the same area imaged at 10 μm spatial resolution results in a 1,000,000 pixel image. Typical spectral acquisition rates (~2,000 to ~7,000 pixels/hour) can then require many hours or days to analyze these types of samples.
Figure 1.
The relationship between the number of pixels required to image a specified area is inversely related to the square of the spatial resolution defined across that area.
With the recent advances in laser technology, the analysis time is typically no longer limited by the laser repetition rate,17, 19-20 but rather by the instrument configuration, acquisition mode, and type of mass analyzer. Traditional IMS experiments are performed by moving the sample stage in discrete steps under a stationary laser. At each step, the stage is stopped to fire the laser a set number of times within the defined pixel area before moving to the next raster step. A second acquisition mode, termed continuous laser raster sampling, has also been described wherein the laser is fired continuously as the sample stage is moved at a constant velocity across an area of interest.9, 21-25 In this mode of acquisition, the lateral spatial resolution is then defined by:
| (1) |
where frep is the laser repetition rate, vstage is the sample stage velocity, and H.A. (hardware average) is the number of laser shots averaged for a single pixel.21 Continuous raster sampling has been demonstrated to give 5 to 10 fold improvements in throughput and is very well suited to imaging platforms for which the laser acquisition mode is a potential time-limiting segment of the experiment (e.g., time-of-flight [TOF] mass spectrometers).
In addition to throughput, specificity is extremely important when analyzing biological tissue specimens. Especially for small metabolites, there exist many isomeric, isobaric, and nearly-isobaric species that contribute to spectral complexity and cloud interpretation. In an imaging experiment, it is especially important to consider these potentially interfering species, as plotting the spatial distribution of a single peak may not represent only the spatial distribution of a single ion; rather, such an image may represent the confluence of several ions which have overlapping m/z values, giving a distorted picture of molecular distribution. In instances where a potential lack of specificity is due to nearly-isobaric species (i.e., ions which are extremely close, but still different, in m/z), the use of a high mass resolving power instrument platform (i.e., an orbitrap or Fourier transform ion cyclotron resonance [FTICR] mass spectrometer) may provide enhanced molecular specificity.26-27 In instances where isomeric and truly isobaric compounds are a concern, ion mobility (IM)12, 28 and tandem mass spectrometry (MS/MS or MSn)12, 24, 29-30 approaches have been successfully employed. In addition to enhancing specificity, these gas phase approaches can also improve sensitivity by eliminating chemical noise due to biological contaminants and/or the MALDI matrix. The ability of tandem imaging mass spectrometry approaches to also simultaneously identify the chemical structure of an analyte is particularly attractive. As such, imaging MS/MS methods have been successfully employed on several trap-based instrument platforms, including quadrupole-TOF (Q-TOF),12 linear ion trap (LIT), and LIT-orbitrap24, 29-30 mass spectrometers. However, trap-based MS/MS methods require lengthier experimental times, decreasing throughput. So-called beam-type MS/MS methods (i.e., where the ion beam is transmitted through a collision cell without trapping) are inherently faster (e.g., triple quadrupole [QQQ]23, 31 as well as in source decay [ISD] TOF32 and TOF/TOF33 imaging setups), but have seen far less use in imaging experiments.
Herein, we describe a high speed tandem TOF imaging platform. Utilizing continuous raster sampling for the first time on a TOF/TOF platform, a 100 μm lipid ion image of a transverse mouse brain section is acquired in just over 20 minutes and allows identification of the lipid as part of the imaging experiment [lipid identified as SM4s(d18:1/24:1)]. A demonstration of the enhanced specificity of MS/MS imaging is shown for the detection of the drug rifampicin (RIF) from a rat kidney dosed in vitro. A demonstration of the enhanced sensitivity of MS/MS imaging is also shown for the detection of RIF from a rabbit liver dosed in vivo.
EXPERIMENTAL
Sample Preparation
Hand-spotted leucine enkephalin (YGGFL) was prepared via the dried-droplet method.34 Briefly, a ~5 μM analyte solution was mixed with an equal volume of 5 mg/mL α-cyano-4-hydroxycinnamic acid (CHCA) matrix solution (50/50/0.1 acetonitrile/water/trifluoroacetic acid), of which ~1 μL was then hand-spotted onto a stainless steel MALDI target.
10 μm thick transverse mouse brain sections were cryosectioned and then thaw-mounted onto conductive indium-tin oxide (ITO) slides. Application of a 1,5-diaminonapthalene (DAN) matrix layer was performed using a custom-built sublimation apparatus (110°C, 8 minutes, 50 mtorr).35-36 Following analysis, the MALDI matrix was removed using an ethanol wash and then stained using hematoxylin and eosin (H&E).
A rat kidney (637 mg) was dosed in vitro by immersion with agitation in a solution of rifampicin (200 μM) for a total of 100 hours. The kidney was then flash-frozen and stored at −80°C until analysis. 12 μm thick coronal sections were cryosectioned and then thaw-mounted onto a gold-coated stainless steel target. Matrix (20 mg/mL 2,4,6-trihydroxyacetophenone [THAP] in 50/50 ethanol/water) was applied via a hand-spraying protocol using a thin-layer chromatography (TLC) sprayer.37 A serial tissue section was stained using H&E.
For in vivo experiments, a New Zealand white rabbit infected with Mycobacterium tuberculosis was orally administered a cocktail of pharmaceuticals (rifampicin/isoniazid/pyrazinamide/moxifloxacin at 30/50/125/25 mg/kg) once daily for one week with the approval of the institutional animal care and use committee (IACUC) of the NIH/NIAID and was sacrificed 65 minutes after the final dose. The control tissue in this experiment was dosed with 50 mg/kg isoniazid only. The tissue was irradiated with UV light for disinfection, flash-frozen, and stored at −80°C until analysis. 12 μm sections were cryosectioned and thaw-mounted onto a gold-coated stainless steel target. Matrix (20 mg/mL 2,4,6- THAP in 50/50 ethanol/water) was manually applied using a TLC reagent sprayer. A serial section was also obtained for H&E staining.
YGGFL, angiotensin II, [glu1]-fibrinopeptide B, trifluoroacetic acid, CHCA, and DAN were all purchased from Sigma Aldrich (St. Louis, MO). C24:1-OH sulfatide lipid standard was purchased from Avanti Polar Lipids (Alabaster, AL). THAP was purchased from Fluka (Sigma Aldrich, St. Louis, MO). Acetonitrile and ethanol were purchased from Fisher Scientific (Pittsburg, PA). Mouse brain and rat kidney were purchased from Pel-Freez Biologicals (Rogers, AR).
Imaging Mass Spectrometry
Peptide standard analysis as well as lipid and RIF imaging experiments were performed on a MALDI TOF/TOF mass spectrometer designed and built by Marvin Vestal and his team at SimulTOF Systems (Sudbury, MA).38-40 Briefly, this instrument is a dual polarity TOF/TOF system operated at 8 kV in reflectron MS mode and operated at 4 kV in MS/MS mode. This system is equipped with a 349 nm, diode-pumped, frequency-tripled Nd:YLF laser (Spectra-Physics, Santa Clara, CA) capable of laser repetition rates up to 5 kHz. The laser beam is oriented at a 90° angle with respect to the target surface and the laser energy is controlled by adjusting the current applied to the diode. All laser energies reported here (μJ/pulse) are measured prior to attenuation (the attenuation is kept constant). In MS/MS mode, a precision timed ion selector (TIS) located at the velocity focal distance is used to isolate a precursor ion of interest. Following TIS isolation, the ions enter a collision cell and are then reaccelerated to 2 kV in the second source region. Fragmentation can be achieved using either laser induced dissociation (LID) (i.e., post-source decay [PSD] using no collision gas) or high energy collision induced dissociation (CID) (i.e., using collision gas). In MS mode, the TIS, collision cell, and second source acceleration regions are not active. A specified number of spectra are ‘hardware averaged’ on the acquisition card prior to writing the data to the hard disk. Instrument operation and data acquisition are controlled using the SimulTOF Controller and data analysis and ion image visualization were performed using the SimulTOF Viewer (SimulTOF Systems, Sudbury, MA). External calibration was performed using either a mixture of standard peptides (YGGFL, angiotensin II, and [glu1]-fibrinopeptide B for positive mode calibration) or matrix clusters (DAN and THAP for negative mode calibration).
This instrument is designed to use continuous laser raster sampling. Ion images were acquired at 100 μm spatial resolution in typewriter mode (i.e., acquisition in one lateral direction only) in order to eliminate differences in ion signal intensity dependent on the direction of sample stage motion.21 The vertical spatial resolution is determined by the motor step size between continuously rastered rows and the horizontal spatial resolution is determined via Equation 1. Laser repetition rate, stage speed, and number of hardware averages were optimized for the fastest acquisition rate while not exceeding a spectral acquisition speed of 50 pixels/second (to maintain a high digitizer efficiency and minimize data loss), not exceeding a laser shot overlap of 50 laser shots per unit area (to ensure that severe oversampling does not diminish sensitivity21), and not exceeding the maximum current output of the laser (at faster laser repetition rates, a higher diode current is required to achieve the same pulse energy compared to slower laser repetition rates).
Kidney imaging experiments were also performed on a Bruker Autoflex TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) and a linear ion trap mass spectrometer (LTQ XL, Thermo Scientific, San Jose, CA) equipped with a MALDI source. The Autoflex TOF/TOF provides a TOF/TOF platform for comparison which does not utilize continuous raster sampling. For the Autoflex TOF/TOF experiments, MS/MS images were acquired in LIFT “fragment only” MS/MS mode (using no collision gas). The precursor ion for RIF (m/z 821.4) was isolated using an isolation window of ±8 Da. A 150% detector boost, 60% laser power boost, and 1% (of parent ion mass) precursor ion selector (PCIS) window were used. 50 laser shots per pixel were averaged using no precursor filters (to ensure only a single ion was imaged using only 50 laser shots at each pixel). Data analysis and ion image visualization were performed using FlexImaging. For the ion trap experiments, the precursor ion for RIF (m/z 821.4) was isolated using an isolation width of 2 Da and fragmented using a normalized collision energy of 45%. Three microscans of five laser shots each were analyzed for each pixel. Data analysis and ion image visualization were performed using ImageQuest (Thermo Scientific, San Jose, CA).
Lipid ion identification was performed by searching fragment ion spectra against the LIPID MAPS (Lipidomics Gateway, www.lipidmaps.org) and METLIN Metabolite (http://metlin.scripps.edu) databases. Chemical structures and cleavage sites were confirmed using ChemBioDrawUltra 14.0 (CambridgeSoft Corporation, PerkinElmer Inc., Waltham, MA).
RESULTS
MS/MS Performance
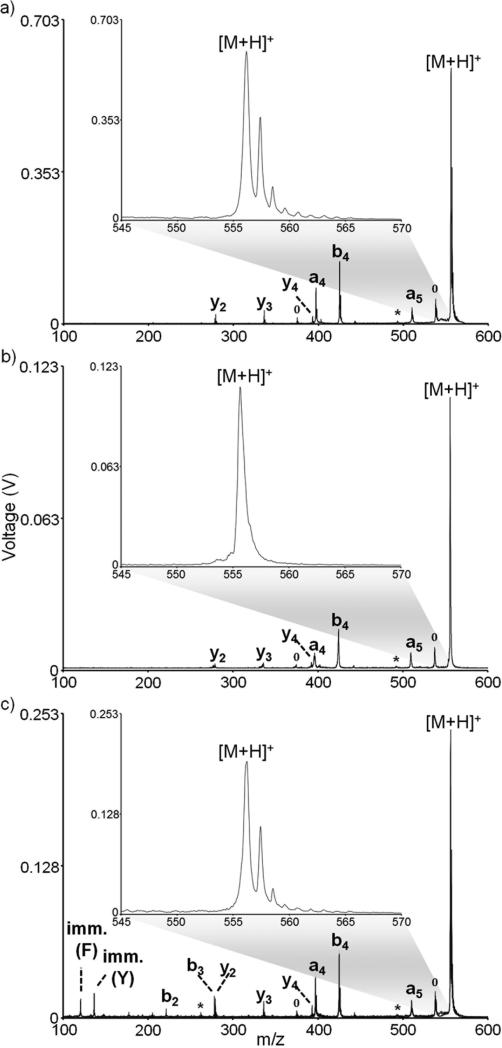
Important to any MS/MS experiment is the selectivity with which a precursor ion is chosen for fragmentation. In a TOF/TOF configuration such as the one described here, MS/MS selectivity is afforded by the precursor ion selection and the second source reacceleration. The resolution of the precursor ion selectivity can be enhanced by either narrowing the TIS precursor ion isolation window or by shortening the length of the second source reacceleration pulse, though manipulation of the isolation window is the more common approach. For example, by narrowing the width of the TIS gate from 40 ns to 15 ns in the positive ion mode MS/MS analysis of protonated YGGFL, the precursor ion window is narrowed from several (~5) Daltons to a single Dalton, allowing for the isolation of the primary 12C isotope of the [M+H]+ ion (Figure 2a inset and 2b inset). Although the isolation resolution is improved by narrowing the TIS window, the overall sensitivity is diminished by over a factor of 5. Varying isolation resolutions can be employed, but for isolation windows less than roughly 40 ns, ion signal intensity obtained from tissue is diminished. This is likely due to the velocity distribution of the ion packet, the fall time of the TIS power supply, and any fringe field effects from the TIS gate. Additionally, at the higher laser fluences typically required for efficient MS/MS performance (especially on biological tissue sections), precursor mass resolution is diminished, especially at higher mass. Thus, for the imaging experiments described herein, the TIS isolation window was maintained at ~40 ns.
Figure 2.
MS/MS analysis of [YGGFL+H]+ using (a) PSD and a 40 ns TIS window, (b) PSD and a 15 ns TIS window, and (c) high energy CID and a 40 ns TIS window. All spectra represent an average of 1500 laser shots. Degree symbols (°) denote water losses and asterisks (*) denote ammonia losses.
Fragmentation can be achieved using either PSD (no collision gas) or high energy CID (using collision gas). In PSD, residual energy from the MALDI process results in fragmentation after the ions have exited the source region of the instrument.41-45 In a TOF/TOF configuration, the observed fragments have dissociated after the initial source region but prior to the second source region.46-47 As fragmentation occurs in a field free region of the instrument, fragments from PSD have the same velocities (but different kinetic energies) as the intact parent ion, meaning they will arrive at the TIS gate at the same time and will be isolated as a common velocity family. The second source reacceleration then allows for separation of parent and fragment ions in the second TOF region (this separation of metastable ions has also been historically achieved in TOF configurations using a reflectron41-45). PSD spectra are typically equivalent to more conventional low energy collisional activation processes.46-47 For example, an MS/MS spectrum of YGGFL utilizing PSD for fragmentation produces many low-energy sequence ions from peptide backbone amide bond cleavages (Figure 2a and 2b). The formation of the a-, b-, and y-type ions are all consistent with relatively low energy processes, as is the b4/a4 ion ratio (i.e., a smaller b4/a4 ion ratio is typically consistent with a lower energy fragmentation process48-52). Conversely, the introduction of a collision gas induces high energy (keV) ion/neutral collisions in the collision cell, resulting in the generation of ions from high energy fragmentation channels, such as the tryptophan and phenylalanine immonium ions (Figure 2c).46, 53 The abundances of the low energy fragmentation channels (e.g., the a4 and b4 ions) are relatively unchanged with collision gas, indicating that high energy CID of the parent contributes relatively little to these fragmentation pathways. While high energy CID and consecutive fragmentation processes have proven useful in some analyses (e.g., using peptide side chain fragmentation to differentiate between isobaric leucine and isoleucine amino acid residues),46, 53 PSD fragmentation is sufficient for the analytes studied herein.
Drug Imaging
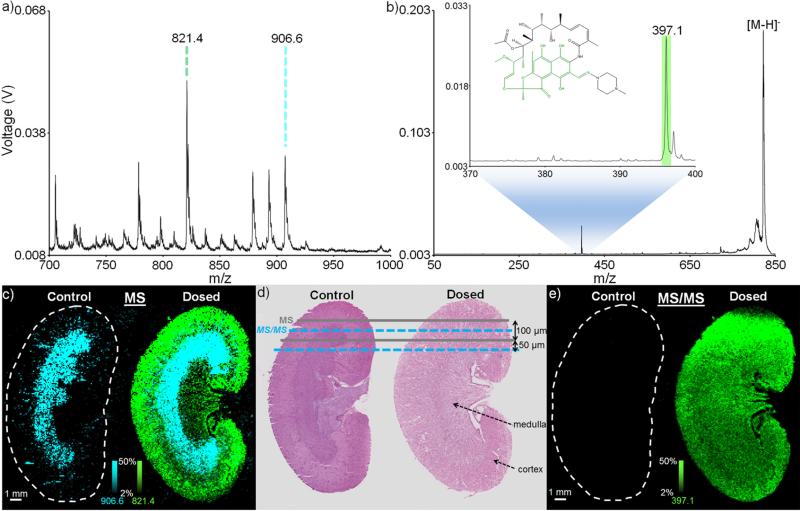
Especially for low molecular weight drugs and metabolites, there exist many isomeric, isobaric, and nearly-isobaric species that contribute to spectral complexity. This can be particularly important when examining, for example, the biodistribution of a drug.27 On instrument platforms where high resolution MS analysis is not an option, MS/MS is often used to maintain a high level of molecular specificity and ensure accuracy. In instances where large tissue areas are being examined, the high throughput capabilities of a MALDI TOF/TOF platform equipped with continuous laser raster sampling are attractive. This is exemplified here by imaging in negative ion mode the distribution of RIF, an antibiotic frequently used in the treatment of tuberculosis, in a rat kidney dosed in vitro (Figure 3). In MS mode, the singly deprotonated RIF ion, [M-H]− (m/z 821.4), is observed to localize primarily in the cortex of only the dosed kidney (Figure 3c). Conversely, a common kidney lipid, SM4s(d18:1/h24:0),54 is localized to the medulla region in both the control (non-dosed) and dosed kidneys (the identity of this lipid was later confirmed using MS/MS analysis, Supplemental Figure 1). Using continuous laser raster sampling, this 100 μm MS image consisted of 48,921 pixels and was acquired in 90 minutes. As the laser beam diameter is ~50 μm, a second image can then be acquired, with the continuously rastered rows interleaved within the first image (Figure 3d). Selecting the [M-H]− RIF ion to perform a 100 μm MS/MS image allows for observation of the distribution of the primary RIF fragment (m/z 821.4 → m/z 397.1), providing molecular confirmation of the drug distribution (Figure 3e). This MS/MS image contained 46,925 pixels and was acquired in 91 minutes. For both the MS and MS/MS images shown here, the stage velocity was 2 mm/s, the laser frequency was 1000 Hz, and each pixel represents 50 laser shot hardware averages. Laser pulse energies of 36.0 μJ and 63.7 μJ were used for the MS and MS/MS images, respectively. The latter pulse energy is near the maximum pulse energy of the laser at a 1000 Hz laser frequency, preventing the use of faster laser repetition rates (which would require the use of diode currents higher than those available on the current system to maintain the same pulse energy). In this configuration, though the digitizer is acquiring pixels (i.e., summed spectra) at a rate of 20 Hz, the overall effective acquisition rate is slightly lower. At the end of each line scan, the system is briefly paused to flush the acquired spectra from the acquisition card to the hard disk (this is done to minimize data loss). This brief flush period, combined with the delay time required to return the stage to the beginning of the next continuous raster line (since these images are required in typewriter mode), results in an effective pixel acquisition rate of ~9 Hz (46,925 pixels/91 minutes). Nevertheless, this represents a ~10 fold improvement in throughput over an analogous MS/MS image acquired on a TOF/TOF instrument with no continuous raster scanning (46,226 pixels/861 minutes, or an overall effective pixel acquisition speed of ~0.9 Hz, data not shown) and an ~8 fold improvement in throughput over an analogous MS/MS image acquired on an ion trap system (48,807 pixels/752 minutes, or an overall effective pixel acquisition speed of ~1 Hz, data not shown). On occasions such as this where an image requires a large number of pixels to effectively sample an area, the use of continuous raster sampling, coupled with TOF/TOF analysis, provides an efficient and specific means by which to perform analysis.
Figure 3.
IMS of kidney dosed in vitro: Average spectra for (a) MS analysis and (b) MS/MS analysis of the [M-H]− of rifampicin from the dosed kidney (each spectrum is an average of a 100 pixel region of interest). The inset of (b) shows a zoom-in of the primary rifampicin fragment (see RIF structure inset). (c) A 48,921 pixel MS mode image (normalized to total ion current [TIC]) acquired in 90 minutes shows rifampicin to localize only to the dosed kidney (green), while a common sulfatide, SM4s(d18:1/h24:0), localizes to the medulla region of both the control and dosed kidney sections (blue). (d) Hematoxylin and eosin (H&E) stains of serial kidney sections are used to indicate the manner by which the interleaved MS and MS/MS scans are performed. (e) A 46,925 pixel MS/MS mode image acquired in 91 minutes showing the primary fragment of rifampicin (821.4 → 397.1) confirms the drug localization. All ion images are plotted using a ±0.4 m/z tolerance. Due to the difference in experimental modes (MS and MS/MS), ion (and image) intensities are not comparable between a (c) and b (e).
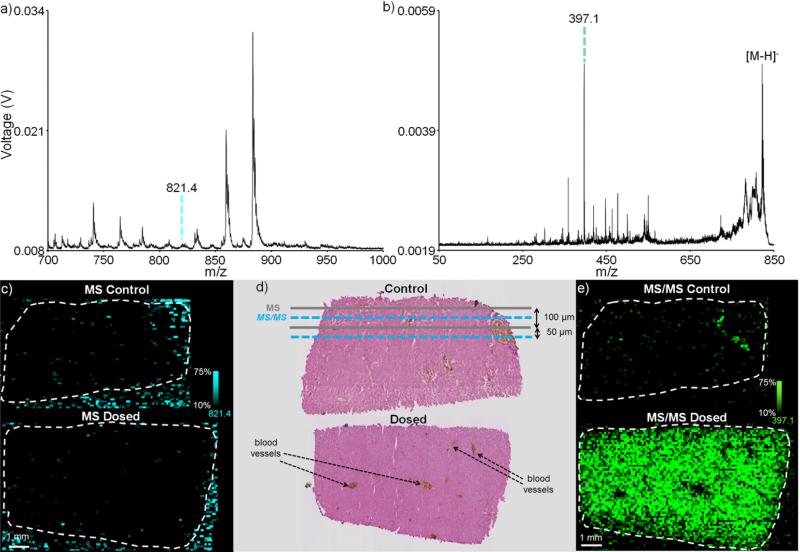
In addition to enhancing specificity and providing molecular confirmation, MS/MS imaging can also improve sensitivity by eliminating chemical noise due to biological contaminants and/or the MALDI matrix. This is shown here by imaging in negative ion mode the distribution of RIF in a rabbit liver dosed in vivo (Figure 4). In MS mode, the singly deprotonated RIF ion, [M-H]− (m/z 821.4), is not observed (Figure 4a). An ion image plotted showing the distribution of m/z 821.4 ± 0.4 Da shows no rifampicin localization as the drug signal is overwhelmed by the chemical background of the tissue (Figure 4c). Using continuous laser raster sampling, this 100 μm MS image consisted of 14,387 pixels and was acquired in 35 minutes. As with the kidney imaging experiment, a second ion image can be acquired by interleaving the acquisition rows within the first image (Figure 4d). Selecting the [M-H]− RIF ion to perform a 100 μm MS/MS image allows for observation of the distribution of the primary RIF fragment (m/z 821.4 → m/z 397.1). RIF is observed to localize to the hepatocytes in the liver, but not in the blood vessels (Figure 4e). This is consistent with a steady-state dosing regimen where RIF would no longer be present in the blood, but would have been taken up by the hepatocytes. The improvement in signal-to-noise afforded by the MS/MS mode image (13,822 pixels acquired in 49 minutes) is due to the elimination of chemical noise and now provides an image of drug distribution. For both the MS and MS/MS images shown here, the stage velocity was 1 mm/s, the laser frequency was 1000 Hz, and each pixel represents 100 laser shot hardware averages. Laser pulse energies of 36.0 μJ and 63.7 μJ were used for the MS and MS/MS images, respectively.
Figure 4.
IMS of liver dosed in vivo: Average spectra for (a) MS analysis and (b) MS/MS analysis of the [M-H]− of rifampicin from the dosed liver (each spectrum is an average of a 100 pixel region of interest). (c) A 14,387 pixel MS mode image acquired in 35 minutes shows no rifampicin localization as the drug signal is overwhelmed by the chemical background (blue). (d) Hematoxylin and eosin (H&E) stains of serial liver sections are used to indicate the manner by which the interleaved MS and MS/MS scans are performed. (e) The improvement in signal-to- noise afforded by the MS/MS mode image (13,822 pixels acquired in 49 minutes) allows for the detection of the primary fragment of rifampicin (821.4 → 397.1) and provides an image of drug distribution. All ion images are plotted using a ±0.4 m/z tolerance. Due to the difference in experimental modes (MS and MS/MS), ion (and image) intensities are not comparable between a (c) and b (e).
Lipid Imaging
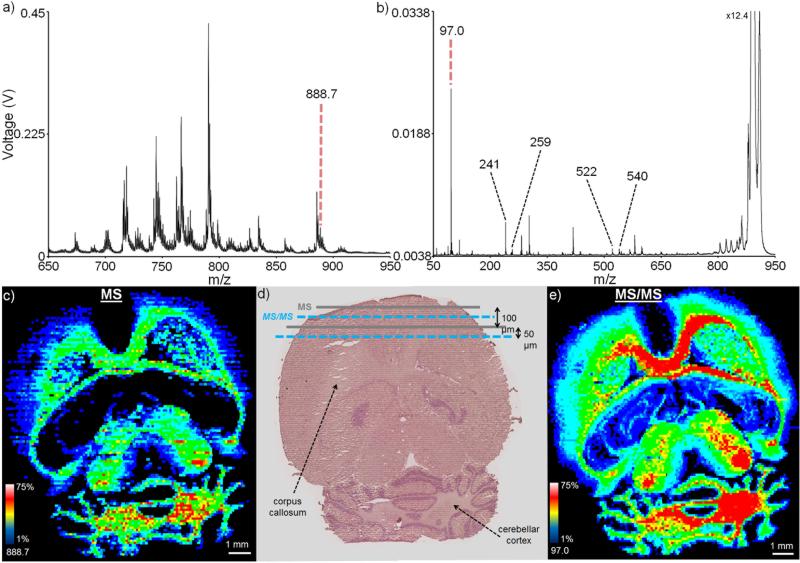
In addition to using MS/MS to improve the molecular specificity of an imaging experiment, the ability to use a MS/MS fragmentation pattern to simultaneously image and identify an ion of interest can be useful. This is exemplified here by imaging the distribution of a sulfatide in a transverse mouse brain section in negative ion mode (Figure 5). In MS mode, the singly deprotonated lipid ion, [M-H]− (m/z 888.7), is observed to localize to regions of the corpus callosum and cerebellar cortex (Figure 5c). As with the drug imaging experiment, a second image can be acquired by interleaving the laser rows between those of the first image (Figure 5d). Selecting the precursor ion at m/z 888.7 for MS/MS imaging provides a series of lipid fragment ions, allowing for identification of the lipid as SM4s(d18:1/24:1) (Figure 5b).55-56 For example, the fragment ion at m/z 259 corresponds to galactose 3-sulfate, with fragment ion at m/z 241 representing a dehydration product of the galactose-sulfate fragment (see Supplementary Figure 2 for MS/MS of a SM4s(d18:1/24:1) lipid standard). Cleavage of the NH-CO bond is also observed, resulting in a direct loss of the fatty acyl group and giving a m/z 540, b1 fragment ion (the dehydrated version of this ion at m/z 522 is also observed). An ion image representing the bisulfate fragment ion (m/z 888.7 → m/z 97.0) gives a more specific and sensitive image than the MS image, showing a similar spatial distribution to the m/z 888.7 parent ion (Figure 5e). Other fragment ions are also present in the MS/MS spectrum and are likely derived from other isobaric or nearly-isobaric species (see Supplementary Figure 3 for an example of how spatial localization can be used to exclude these interfering fragment ions). This MS/MS image contained 16,904 pixels and was acquired in 22 minutes (4 mm/s stage speed, 4000 Hz laser, 22.5 μJ pulse energy, 100 hardware averages per pixel, 40 Hz digitizer pixel acquisition rate), resulting in an overall effective pixel acquisition rate of ~13 Hz. This MS/MS image represents the maximum laser current (and thus pulse energy) available at this laser repetition frequency and nears the maximum digitizer acquisition rate at high efficiency, resulting ~14 and ~12 fold improvements in throughput compared to the MS/MS acquisition speeds mentioned earlier for the (discrete raster) TOF/TOF and ion trap systems, respectively.
Figure 5.
Mouse brain imaging: Average spectra for (a) MS analysis and (b) MS/MS analysis of the m/z 888.7 (each spectrum is an average of a 100 pixel region of interest). (c) A 17,346 pixel MS mode image acquired in 43 minutes shows the m/z 888.7 lipid to localize to specific regions of the brain, including the corpus callosum and cerebellar cortex. (d) H&E stain of a serial brain section following IMS shows the interleaved MS and MS/MS analyses. (e) A 16,904 pixel MS/MS mode image acquired in 22 minutes showing bisulfate fragment ion (888.7 → 97.0) confirms the parent m/z 888.7 lipid localization. Using other fragment ions, the lipid of interest can be identified as SM4s(d18:1/24:1). All ion images are plotted using a ±0.4 m/z tolerance. Due to the difference in experimental modes (MS and MS/MS), ion (and image) intensities are not comparable between a (c) and b (e).
CONCLUSIONS
The use of continuous raster sampling on a MALDI TOF/TOF system has been demonstrated here to improve the throughput ~8-14 fold compared to conventional TOF/TOF and ion trap systems, resulting in overall effective pixel acquisition rates of up to 13 Hz. This has been shown to facilitate drug imaging where other isobaric species may complicate spectral and image interpretation, improving the molecular specificity and sensitivity of the resulting MS/MS ion image and dramatically reducing the analysis time. This method of MS/MS image acquisition has also been shown to facilitate lipid identification while decreasing the time required for image acquisition. As IMS analyses are extended to smaller spatial resolutions and larger tissue areas, throughput becomes an important analytical figure of merit. Coupled with the benefits of MS/MS methodology (i.e., improved sensitivity and chemical specificity), continuous laser raster sampling offers one method by which to maintain high throughput for large imaging datasets.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Marvin Vestal, Kevin Hayden, and George Mills at SimulTOF Systems for their instrument support and Clifton E. Barry III and Laura E. Via for providing the rabbit livers. This work was sponsored by the National Institutes of Health/National Institute of General Medical Sciences under Award 5P41 GM103391-03 and, in part, by the Bill and Melinda Gates Foundation Accelerator grant N01 HD23342. B.M.P. was supported by the National Institutes of Health under Award T32 ES007028. C.W.C. acknowledges receipt of a fellowship from Aegis Sciences Corporation.
REFERENCES
- 1.Norris JL, Caprioli RM. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem. Rev. 2013;113(4):2309–2342. doi: 10.1021/cr3004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaurand P, Norris JL, Cornett DS, Mobley JA, Caprioli RM. New developments in profiling and imaging of proteins from tissue sections by MALDI mass spectrometry. J. Proteome Res. 2006;5(11):2889–2900. doi: 10.1021/pr060346u. [DOI] [PubMed] [Google Scholar]
- 3.Zavalin A, Todd EM, Rawhouser PD, Yang J, Norris JL, Caprioli RM. Direct imaging of single cells and tissue at sub-cellular spatial resolution using transmission geometry MALDI MS. J. Mass Spectrom. 2012;47(11):1473–1481. doi: 10.1002/jms.3108. [DOI] [PubMed] [Google Scholar]
- 4.Norris JL, Caprioli RM. Imaging Mass Spectrometry: A New Tool for Pathology in a Molecular Age. Proteomics. Clinical Applications. 2013 doi: 10.1002/prca.201300055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas A, Charbonneau J, Fournaise E, Chaurand P. Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: enhanced information in both positive and negative polarities after 1,5-diaminonapthalene deposition. Anal. Chem. 2012;84(4):2048–2054. doi: 10.1021/ac2033547. [DOI] [PubMed] [Google Scholar]
- 6.Harris GA, Nicklay JJ, Caprioli RM. Localized in situ hydrogel-mediated protein digestion and extraction technique for on-tissue analysis. Anal. Chem. 2013;85(5):2717–2723. doi: 10.1021/ac3031493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amstalden van Hove ER, Smith DF, Heeren RMA. A concise review of mass spectrometry imaging. Journal of Chromatography. A. 2010;1217(25):3946–3954. doi: 10.1016/j.chroma.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 8.Aleš S. Mass spectrometric imaging of small molecules. Trends Biotechnol. 2010;28 doi: 10.1016/j.tibtech.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 9.McDonnell LA, Heeren RMA. Imaging mass spectrometry. Mass Spectrom. Rev. 2007;26(4):606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 10.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 1997;69 doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 11.Stoeckli M, Staab D, Schweitzer A, Gardiner J, Seebach D. Imaging of a beta-peptide distribution in whole-body mice sections by MALDI mass spectrometry. J. Am. Soc. Mass Spectrom. 2007;18(11):1921–1924. doi: 10.1016/j.jasms.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Trim PJ, Henson CM, Avery JL, McEwen A, Snel MF, Claude E, Marshall PS, West A, Princivalle AP, Clench MR. Matrix-assisted laser desorption/ionization-ion mobility separation-mass spectrometry imaging of vinblastine in whole body tissue sections. Anal. Chem. 2008;80(22):8628–8634. doi: 10.1021/ac8015467. [DOI] [PubMed] [Google Scholar]
- 13.Solon EG, Schweitzer A, Stoeckli M, Prideaux B. Autoradiography, MALDI-MS, and SIMS-MS imaging in pharmaceutical discovery and development. The AAPS journal. 2010;12(1):11–26. doi: 10.1208/s12248-009-9158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reyzer ML, Chaurand P, Angel PM, Caprioli RM. Direct molecular analysis of whole-body animal tissue sections by MALDI imaging mass spectrometry. Methods in Molecular Biology. 2010;656:285–301. doi: 10.1007/978-1-60761-746-4_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaurand P, Schriver KE, Caprioli RM. Instrument design and characterization for high resolution MALDI-MS imaging of tissue sections. J. Mass Spectrom. 2007;42(4):476–489. doi: 10.1002/jms.1180. [DOI] [PubMed] [Google Scholar]
- 16.Jun JH, Song Z, Liu Z, Nikolau BJ, Yeung ES, Lee YJ. High-spatial and high-mass resolution imaging of surface metabolites of Arabidopsis thaliana by laser desorption-ionization mass spectrometry using colloidal silver. Anal. Chem. 2010;82(8):3255–3265. doi: 10.1021/ac902990p. [DOI] [PubMed] [Google Scholar]
- 17.Sabine G, Martin K, Oliver S, Bernhard S. Laser spot size and laser power dependence of ion formation in high resolution MALDI imaging. Int. J. Mass Spectrom. 2010;294 [Google Scholar]
- 18.Thiery-Lavenant G, Zavalin AI, Caprioli RM. Targeted multiplex imaging mass spectrometry in transmission geometry for subcellular spatial resolution. J. Am. Soc. Mass Spectrom. 2013;24(4):609–614. doi: 10.1007/s13361-012-0563-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holle A, Haase A, Kayser M, Höhndorf J. Optimizing UV laser focus profiles for improved MALDI performance. J. Mass Spectrom. 2006;41(6):705–716. doi: 10.1002/jms.1041. [DOI] [PubMed] [Google Scholar]
- 20.Trim PJ, Djidja M-C, Atkinson SJ, Oakes K, Cole LM, Anderson DMG, Hart PJ, Francese S, Clench MR. Introduction of a 20 kHz Nd:YVO4 laser into a hybrid quadrupole time-of-flight mass spectrometer for MALDI-MS imaging. Anal. Bioanal. Chem. 2010;397(8):3409–3419. doi: 10.1007/s00216-010-3874-6. [DOI] [PubMed] [Google Scholar]
- 21.Spraggins JM, Caprioli RM. High-speed MALDI-TOF imaging mass spectrometry: rapid ion image acquisition and considerations for next generation instrumentation. J. Am. Soc. Mass Spectrom. 2011;22(6):1022–1031. doi: 10.1007/s13361-011-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simmons DA. Improved MALDI-MS imaging performance using continuous laser rastering. Applied Biosystems Technical Note. 2008:1–5. [Google Scholar]
- 23.Hopfgartner G, Varesio E, Stoeckli M. Matrix-assisted laser desorption/ionization mass spectrometric imaging of complete rat sections using a triple quadrupole linear ion trap. Rapid communications in mass spectrometry : RCM. 2009;23(6):733–736. doi: 10.1002/rcm.3934. [DOI] [PubMed] [Google Scholar]
- 24.Lanekoff I, Burnum-Johnson K, Thomas M, Short J, Carson JP, Cha J, Dey SK, Yang P, Prieto Conaway MC, Laskin J. High-speed tandem mass spectrometric in situ imaging by nanospray desorption electrospray ionization mass spectrometry. Anal. Chem. 2013;85(20):9596–9603. doi: 10.1021/ac401760s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffiths R, Sarsby J, Guggenheim E, Race A, Steven R, Fear J, Lalor P, Bunch J. Formal lithium fixation improves direct analysis of lipids in tissue by mass spectrometry. Anal. Chem. 2013;85(15):7146–7153. doi: 10.1021/ac400737z. [DOI] [PubMed] [Google Scholar]
- 26.Cornett DS, Frappier SL, Caprioli RM. MALDI-FTICR imaging mass spectrometry of drugs and metabolites in tissue. Anal. Chem. 2008;80(14):5648–5653. doi: 10.1021/ac800617s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castellino S, Groseclose MR, Wagner D. MALDI imaging mass spectrometry: bridging biology and chemistry in drug development. Bioanalysis. 2011;3(21):2427–2441. doi: 10.4155/bio.11.232. [DOI] [PubMed] [Google Scholar]
- 28.McLean JA, Ridenour WB, Caprioli RM. Profiling and imaging of tissues by imaging ion mobility-mass spectrometry. J. Mass Spectrom. 2007;42(8):1099–1105. doi: 10.1002/jms.1254. [DOI] [PubMed] [Google Scholar]
- 29.Landgraf RR, Prieto Conaway MC, Garrett TJ, Stacpoole PW, Yost RA. Imaging of lipids in spinal cord using intermediate pressure matrix-assisted laser desorption-linear ion trap/Orbitrap MS. Anal. Chem. 2009;81(20):8488–8495. doi: 10.1021/ac901387u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perdian DC, Lee YJ. Imaging MS methodology for more chemical information in less data acquisition time utilizing a hybrid linear ion trap-orbitrap mass spectrometer. Anal. Chem. 2010;82(22):9393–9400. doi: 10.1021/ac102017q. [DOI] [PubMed] [Google Scholar]
- 31.Prideaux B, Dartois V, Staab D, Weiner D, Goh A, Via L, Barry C, Stoeckli M. High-sensitivity MALDI-MRM-MS imaging of moxifloxacin distribution in tuberculosis-infected rabbit lungs and granulomatous lesions. Anal. Chem. 2011;83(6):2112–2118. doi: 10.1021/ac1029049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Debois D, Bertrand V, Quinton L, De Pauw-Gillet M-C, De Pauw E. MALDI-in source decay applied to mass spectrometry imaging: a new tool for protein identification. Anal. Chem. 2010;82(10):4036–4045. doi: 10.1021/ac902875q. [DOI] [PubMed] [Google Scholar]
- 33.Nilsson A, Fehniger TE, Gustavsson L, Andersson M, Kenne K, Marko-Varga G, Andre PE. Fine mapping the spatial distribution and concentration of unlabeled drugs within tissue micro-compartments using imaging mass spectrometry. PLoS One. 2010;5(7):e11411. doi: 10.1371/journal.pone.0011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 1988;60(20):2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 35.Hankin JA, Barkley RM, Murphy RC. Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 2007;18(9):1646–1652. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J, Caprioli RM. Matrix sublimation/recrystallization for imaging proteins by mass spectrometry at high spatial resolution. Anal. Chem. 2011;83(14):5728–5734. doi: 10.1021/ac200998a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwartz SA, Reyzer ML, Caprioli RM. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J. Mass Spectrom. 2003;38(7):699–708. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- 38.Vestal M, Hayden K. High performance MALDI-TOF mass spectrometry for proteomics. Int. J. Mass Spectrom. 2007;268 [Google Scholar]
- 39.Vestal M. Modern MALDI time-of-flight mass spectrometry. J. Mass Spectrom. 2009;44(3):303–317. doi: 10.1002/jms.1537. [DOI] [PubMed] [Google Scholar]
- 40.Vestal ML. TOF-TOF with high resolution precursor selection and multiplexed MS-MS. 2010 US 7,838,824 B2.
- 41.Xuejun T, Werner E, Kenneth GS, John BW. Daughter ion mass spectra from cationized molecules of small oligopeptides in a reflecting time-of-flight mass spectrometer. Anal. Chem. 1988;60 doi: 10.1021/ac00168a029. [DOI] [PubMed] [Google Scholar]
- 42.Tang X, Ens W, Mayer F, Standing KG, Westmore JB. Measurement of unimolecular decay in peptides of masses greater than 1200 units by a reflecting time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 1989;3(12):443–448. doi: 10.1002/rcm.1290031210. [DOI] [PubMed] [Google Scholar]
- 43.Boesl U, Weinkauf R, Schlag EW. Reflectron time-of-flight mass spectrometry and laser excitation for the analysis of neutrals, ionized molecules and secondary fragments. Int. J. Mass Spectrom. Ion Processes. 1992;112 [Google Scholar]
- 44.Kaufmann R, Kirsch D, Spengler B. Sequenching of peptides in a time-of-flight mass spectrometer: evaluation of postsource decay following matrix-assisted laser desorption ionisation (MALDI). Int. J. Mass Spectrom. Ion Processes. 1994;131 [Google Scholar]
- 45.Kaufmann R, Chaurand P, Kirsch D, Spengler B. Post-source decay and delayed extraction in matrix-assisted laser desorption/ionization-reflectron time-of-flight mass spectrometry. Are there trade-offs? Rapid Commun. Mass Spectrom. 1996;10(10):1199–1208. doi: 10.1002/(SICI)1097-0231(19960731)10:10<1199::AID-RCM643>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 46.Suckau D, Resemann A, Schuerenberg M, Hufnagel P, Franzen J, Holle A. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal. Bioanal. Chem. 2003;376(7):952–965. doi: 10.1007/s00216-003-2057-0. [DOI] [PubMed] [Google Scholar]
- 47.Bienvenut WV, Déon C, Pasquarello C, Campbell JM, Sanchez J-C, Vestal ML, Hochstrasser DF. Matrix-assisted laser desorption/ionization-tandem mass spectrometry with high resolution and sensitivity for identification and characterization of proteins. Proteomics. 2002;2(7):868–876. doi: 10.1002/1615-9861(200207)2:7<868::AID-PROT868>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 48.Goeringer DE, McLuckey SA. Relaxation of internally excited high-mass ions simulated under typical quadrupole ion trap storage conditions. Int. J. Mass Spectrom. 1998;177:163–174. [Google Scholar]
- 49.Asano KG, Goeringer DE, Butcher DJ, McLuckey SA. Bath gas temperature and the appearance of ion trap tandem mass spectra of high-mass ions. Int. J. Mass Spectrom. 1999;190-191:281–293. [Google Scholar]
- 50.Sztáray J, Memboeuf A, Drahos L, Vékey K. Leucine enkephalin-a mass spectrometry standard. Mass Spectrom. Rev. 2011;30(2):298–320. doi: 10.1002/mas.20279. [DOI] [PubMed] [Google Scholar]
- 51.Jue AL, Racine AH, Glish GL. The effect of ion trap temperature on the dissociation of peptide ions in a quadrupole ion trap. Int. J. Mass Spectrom. 2011;301:74–83. [Google Scholar]
- 52.Prentice BM, Santini RE, McLuckey SA. Adaptation of a 3-D quadrupole ion trap for dipolar DC collisional activation. J. Am. Soc. Mass Spectrom. 2011;22(9):1486–1492. doi: 10.1007/s13361-011-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ioannis AP. The interpretation of collision-induced dissociation tandem mass spectra of peptides. Mass Spectrom. Rev. 1995;14 [Google Scholar]
- 54.Grove KJ, Voziyan PA, Spraggins JM, Wang S, Paueksakon P, Harris RC, Hudson BG, Caprioli RM. Diabetic nephropathy induces alterations in the glomerular and tubule lipid profiles. J. Lipid Res. 2014;55(7):1375–1385. doi: 10.1194/jlr.M049189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu F, Bohrer A, Turk J. Electrospray ionization tandem mass spectrometric analysis of sulfatide. Determination of fragmentation patterns and characterization of molecular species expressed in brain and in pancreatic islets. Biochim. Biophys. Acta. 1998;1392(2-3):202–216. doi: 10.1016/s0005-2760(98)00034-4. [DOI] [PubMed] [Google Scholar]
- 56.Hsu F-F, Turk J. Studies on sulfatides by quadrupole ion-trap mass spectrometry with electrospray ionization: structural characterization and the fragmentation processes that include an unusual internal galactose residue loss and the classical charge-remote fragmentation. J. Am. Soc. Mass Spectrom. 2004;15(4):536–546. doi: 10.1016/j.jasms.2003.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.