Abstract
Antibody mimics have significant scientific and therapeutic utility for the disruption of protein–protein interactions inside cells; however, their delivery to the cell cytosol remains a major challenge. Here we show that protective antigen (PA), a component of anthrax toxin, efficiently transports commonly used antibody mimics to the cytosol of mammalian cells when conjugated to the N-terminal domain of LF (LFN). In contrast, a cell-penetrating peptide (CPP) was not able to deliver any of these antibody mimics into the cell cytosol. The refolding and binding of a transported tandem monobody to Bcr-Abl (its protein target) in chronic myeloid leukemia cells were confirmed by co-immunoprecipitation. We also observed inhibition of Bcr-Abl kinase activity and induction of apoptosis caused by the monobody. In a separate case, we show disruption of key interactions in the MAPK signaling pathway after PA-mediated delivery of an affibody binder that targets hRaf-1. We show for the first time that PA can deliver bioactive antibody mimics to disrupt intracellular protein–protein interactions. This technology adds a useful tool to expand the applications of these modern agents to the intracellular milieu.
Keywords: anthrax toxin, antibodies, intracellular delivery, protein–protein interactions, sortases
Introduction
Antibodies have proved to be powerful tools in facilitating the elucidation of disease mechanisms and generating novel and effective therapeutics. However, the use of antibodies has been limited to outside the cell because of two major factors: antibodies containing disulfide bonds might be unstable in the reducing environment of cytosol, and antibodies are unable to cross the cell plasma membrane to reach the cytosol. There are numerous intracellular targets and protein–protein interactions with large flat contact areas; these are considered difficult to perturb by small molecules. We believe there would be great interest in the use antibody mimics to target the intracellular protein–protein interactions, provided that they can be transported into the cytosol by a straightforward delivery platform.
In recent years, certain robust, single-domain, cysteine-free scaffold proteins have emerged as antibody mimics. These include monobodies derived from the tenth type III domain of human fibronectin (10FN3), affibodies derived from the immunoglobulin binding protein A, DARPins based on ankyrin repeat modules, and the B1 domain of protein G (GB1).[1] These antibody mimics have been engineered to bind extracellular receptors such as EGFR, HER2, VEGFR and integrin, as well as various intracellular targets, including caspases, Raf, Erk, c-Jun N-terminal kinase (JNK), Abl-SH2, and c-Jun.[2] Advances in directed evolution and molecular display technologies, such as phage display, yeast display, and ribosome display, make it possible to routinely generate a wide variety of high-affinity binders for specific protein targets.[3]
Research effort is now focused on applying these antibody mimics inside the cell (“intrabodies”) to target cytosolic proteins.[4] To achieve this, strategies are needed to allow facile and reliable delivery of these bioactive antibody mimics into the cytosol of various cell types. Delivery methods based on lipid-derived compounds,[5] polymeric nanoparticles,[6] inorganic nanocarriers,[7] supercharged proteins,[8] and, most commonly, cell-penetrating peptides (CPPs) such as the transactivator of transcription (TAT) of HIV-1, oligoarginine, and penetratin peptide derived from the Drosophila Antennapedia,[9] have been developed to deliver proteins of interest to the cytosol of mammalian cells. In most of these cases, high concentrations of these agents are required to achieve even modest effects, often because of inefficient cargo escape from the endosome.
Nature has evolved a variety of mechanisms to transport proteins across membranes into the cytosol of mammalian cells.[10] One bacterial protein-transport nanomachine is protective antigen (PA; 83 kDa), a component of anthrax toxin. PA is a receptor-binding, pore-forming transporter that delivers the enzymatic moieties of the toxin from the external milieu to the cytosol of mammalian cells. PA binds to host-cell receptors and is cleaved by a furin-family protease to yield a 63 kDa species (PA63) (Figure 1 A; step 1)[11] that self-assembles to form ring-shaped heptamers[12] and octamers.[13] These oligomers then form complexes with the cargo proteins (Kd∼1 nm) and are endocytosed. (Figure 1 A; steps 2–4). In the endosome, acidification triggers conformational change of the PA63 oligomers to form a transmembrane pore that unfolds and translocates the bound cargo proteins to the cytosol (Figure 1 A; step 5).[14] PA63 oligomers recognize the N-terminal domain (LFN, ∼30 kDa) of the toxin enzyme lethal factor (LF, 90 kDa).[15] Studies have shown that cargo fused to the C terminus of LFN can be transported to the cytosol via PA; most effort has focused on the delivery of peptides for vaccine development,[16] enzymes such as β-lactamase,[17] and enzymatic domains from diphtheria toxin (DTA), Shiga toxin, Pseudomonas exotoxin A (PEIII), and RTX toxin (ACD).[18] More recently, the PA/LFN system was shown to deliver Legionella pneumophila flagellin into macrophages.[19] However, no study has investigated the ability of PA/LFN system to translocate antibody mimics for the perturbation of intracellular protein–protein interactions.
Figure 1.
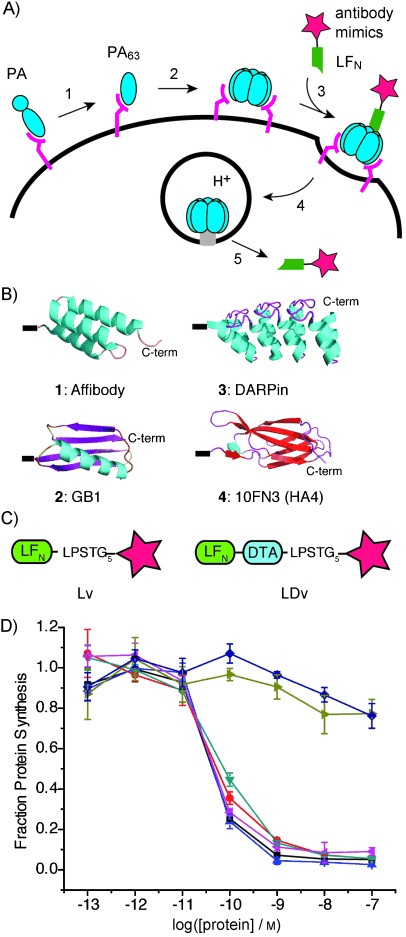
Delivery of antibody mimics into the cytosol by the LFN/PA system. A) Mechanism of entry of antibody mimic (star) into cells. B) Antibody mimics 1–4: affibody (PDB ID: 1Q2N), GB1 (1PGB), DARPin (adapted from 3ZU7), and HA4 (3K2M). C) Variants of antibody mimics (stars) attached to the C terminus of LFN (Lv) or LFN-DTA (LDv). D) Protein synthesis inhibition in CHO-K1 cells treated with LDv1–4 in the presence of 20 nm PA for 30 min. The cells were then washed three times with PBS and incubated with 1 μCi ml−1 3H-leucine for 1 h. Subsequently, the cells were washed three times with PBS and scintillation fluid was directly added to each well, and 3H incorporation into the cellular proteome was measured to determine the level of LDv inhibition of protein synthesis (n=3). Radioactive counts were normalized to those of cells treated with only PA (set to 1). •: LFN-DTA, •: LDv1, ▴: LDv2, ▾: LDv3, ◂: LDv4, ▸: DTA, ⧫: Lv1+DTA.
Here we used transpeptidase sortase (SrtA)[20] to conjugate several commonly used antibody mimics to the C terminus of LFN and found that PA can mediate their transport into the cytosol of several different cell lines. We confirmed the refolding and binding of a tandem monobody to its protein target Bcr-Abl inside cells by co-immunoprecipitation. We observed inhibition of Abl kinase activity and subsequent cell death caused by the PA-delivered monobody. We show that the PA system can deliver an affibody that binds hRaf-1 to disrupt the MAPK signaling pathway.
Results and Discussion
Our antibody mimics consisted of scaffolds widely used to generate highly specific and potent binders: affibody, protein GB1, DARPin, and monobody (Figure 1 B). These scaffolds are disulfide-free, thus avoiding possible interference with passage through the PA translocase and potential stability problems in the reducing environment of the cytosol. Our chemoenzymatic bioconjugation route is based on SrtA*, an evolved SrtA, and is shown in Figure S1 in the Supporting Information.[21] SrtA* catalyzes the formation of covalent conjugates (designated Lv, Figure 1 C) between LFN containing the C-terminal LPXTG recognition motif and antibody mimics containing N-terminal oligoglycine.
We also prepared a series of conjugates (designated LDv, Figure 1 C) between LFN-DTA and each antibody mimic, in order to measure PA-mediated translocation into the cytosol. In anthrax toxin translocation studies, the A chain of diphtheria toxin (DTA), which catalyzes the ADP-ribosylation of EF-2 and inhibits protein synthesis, has been frequently used as a straightforward measure—the “gold standard” assay of PA-mediated translocation into the cytosol. Therefore, LFN-DTA variants (LDvs) allowed us to compare our findings with previous reports that used the same assay.[18a, c, 22] For each antibody mimic, after confirming translocation of the LDv, we also carried out studies with Lvs that lack the toxic DTA protein, thus avoiding interference with further characterization of antibody mimic function and delivery into the cytosol.
Each purified LDv (LDv1–4, Figure S2) was added to CHO-K1 cells in the presence of 20 nm PA. After 30 min, the cells were washed and incubated with medium supplemented with 3H-leucine. The efficiency of antibody mimic translocation was measured by the incorporation of 3H-Leu in the cellular proteome, as the level of protein synthesis inhibition is determined by the amount of LDv-containing variant in the cytosol. Despite their structural differences, all four variants (LDv1–4) translocated efficiently into the cytosol at levels comparable to that of the positive control, LFN-DTA (Figure 1 D). To confirm that the full-length protein was required for translocation to the cytosol, we performed control experiments by treating cells with DTA plus PA, or DTA together with LFN-affibody (Lv1) plus PA; we observed no protein synthesis inhibition in either case (Figure 1 D).
To investigate whether LDvs translocate to the cytosol by the same mechanism as for LFN-DTA, we carried out a series of control experiments with LDv1 containing affibody as the cargo (Figure 2 A). We investigated the role of LFN in LDv1 during the translocation process by using excess LFN (1 μm) to outcompete LDv1 in binding to PA. We found that 1 μm LFN significantly abolished translocation of LDv1, thus confirming the importance of LFN-mediated binding to PA (step 3, Figure 1 A). We then studied the role of endocytosis in LDv1 translocation by incubating cells with PA and LFN-DTA or LDv1 at 4 °C (instead of 37 °C) to arrest endocytosis. The translocation of both LFN-DTA and LDv1 was abolished under these conditions, thus indicating that endocytosis is a necessary step in the translocation process (step 4, Figure 1 A). To test the role of endosome acidification and active translocation (step 5, Figure 1 A), we performed two additional control experiments. First, we treated cells with PA and LFN-DTA or LDv1 in the presence of 200 nm bafilomycin A1, a specific inhibitor of vacuolar H+-ATPase, in order to block endosome acidification. We found that inhibition of endosome acidification abolished translocation of both LFN-DTA and LDv1, thus confirming the importance of the pH gradient for translocation. An additional control experiment was performed with a PA mutant (PA[F427H]) that binds and delivers the cargo to endosomes but arrests translocation through PA pore.[23] We found that PA[F427H] completely arrested translocation of LDv1, thus indicating that functional PA was required for LDv1 to reach the cytosol. These controls indicate that the translocation of LDv1 follows the same mechanism as that for LFN-DTA.
Figure 2.
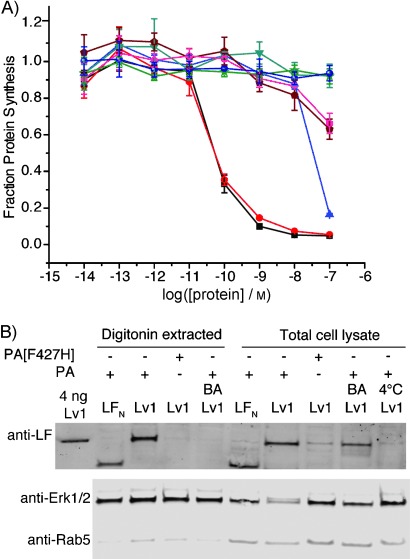
Control experiments validating the translocation mechanism of LDv1 and Lv1. A) Protein synthesis inhibition in CHO-K1 cells treated with variants in the presence of 20 nm PA for 30 min. BA=addition of 200 nm bafilomycin A1; 4 °C=incubation at 4 °C (instead of 37 °C); PA[F427H]=mutant PA instead of PA. ▪: LFN-DTA, •: LDv1, ▴: LDv1+LFN, •: LFN-DTA 4 °C, •: LDv1 4 °C, ★: LFN-DTA+BA, •: Lv1+BA, ▾: LDv1+PA[F427H]. B) CHO-K1 cells were treated with 250 nm Lv1 in the presence of 40 nm PA or PA[F427H] for 12 h (conditions/modifications as above). Total lysate and digitonin-extracted cytosolic proteins were prepared separately (∼300 000 cells per lane).
After confirming the key steps responsible for the translocation of antibody mimics, we investigated the delivery of Lvs into the cytosol by digitonin extraction and western blotting. Digitonin is a weak, nonionic detergent, and at low concentrations selectively permeabilizes the plasma membrane, thereby releasing cytosolic components from cells while the nuclear envelope and other major membrane organelles remain intact.[24] CHO-K1 cells were incubated with Lv1 and PA overnight (to allow multiple rounds of receptor-mediated endocytosis and translocation), washed, and trypsin digested to remove cell surface receptors and bound protein. In order to obtain the cytosolic fraction, cells were incubated with a buffer containing 50 μg mL−1 digitonin and 250 mm sucrose. Immunoblot analysis with antibodies against Rab5 (an early endosome marker) and Erk1/2 (a cytosolic marker) was carried out to confirm that only the cytosolic fraction was present. Minor amounts of Rab5 were detected in the digitonin-extracted cytosolic fractions, thus indicating little contamination from early endosomes (Figure 2 B). We observed a significant amount of Lv1 in the cytosolic fraction, similar to that in total cell lysate obtained by lysis buffer containing 1 % NP-40.
Control experiments were performed to validate translocation of Lv1 into the cytosol. No Lv1 was observed when the cells were incubated at 4 °C (endocytosis arrested), even in the total cell lysate. The absence of detectable Lv1 at 4 °C also validated that the wash and trypsin digestion protocol was sufficient to eliminate potential contamination from surface-bound protein. When the cells were treated with bafilomycin A1, a significant band for Lv1 was observed in the total cell lysate, whereas this band was not detectable in the cytosolic fraction, thus indicating that Lv1 was endocytosed but trapped in the non-acidified endosome. In addition, the absence of Lv1 in the cytosolic fraction for cells treated with bafilomycin A1 served as further evidence that our digitonin extraction protocol was sufficient to separate the cytosolic proteins from the rest of the cell lysate, including endosomes. Finally, when the cells were treated with Lv1 and PA[F427H] (instead of Lv1 and PA), some Lv1 was observed in the total cell lysate but none in the cytosolic fraction (Figure 2 B). Under these conditions, endocytosed Lv1 is trapped in the endosome because of the non-functional pore (mutant PA), and subsequently sorted to the lysosome for degradation, thus leaving only a small amount of Lv1 in the total cell lysate and none in the cytosolic fraction. These western blot results further corroborated the protein synthesis inhibition results with LFN-DTA (vide supra, Figure 1 D) and confirmed the translocation mechanism of the antibody mimic cargo. PA-mediated translocation of the other analogues (Lv2–4) was also studied by digitonin extraction and western blotting (Figure S3); bands at different molecular weights by anti-LF antibody confirmed the presence of these antibody mimic conjugates in the cytosol.
To compare the efficiency of the PA/LFN system with the CPP system, we first used a protein synthesis inhibition assay. We prepared DTA with a TAT peptide covalently attached at the N terminus. This construct showed at least 1000 times lower efficiency in protein synthesis inhibition than LFN-DTA and PA (Figure 3 A); this is consistent with a previous report[25] and indicates that the PA/LFN system is more efficient in delivering DTA to the cytosol. Next, we compared the efficiencies of the PA/LFN and CPP systems in delivering antibody mimics. We conjugated oligoglycine-containing antibody mimics 1–4 to a TAT peptide containing an HA tag and the LPSTGG motif to generate TAT-HA-antibody mimic constructs (Figure S4).[26] Translocation was studied by digitonin extraction and western blot analysis with anti-HA antibody. We treated cells with 2.5 μm TAT-HA-antibody mimic constructs (tenfold higher than for Lvs) for 4 h in serum-free medium and observed minor amounts of material in total-cell lysate (Figure S5 A). Then, when using the trypsin digestion and digitonin extraction protocol as described above, we detected no TAT-HA-antibody mimic protein by anti-HA immunoblotting with the cytosolic fractions, whereas a significant amount of Lv1 with HA tag (Lv1-HA) was detected for cells treated with 250 nm Lv1-HA and 40 nm PA for 4 h (Figure 3 B). To test if a different incubation time would improve translocation by TAT peptide, we treated cells with 2.5 μm TAT-HA-antibody mimics for 16 h; again, no material was evident in the total cell lysate (Figure S5 B). These results demonstrate the higher efficiency of the PA/LFN system in delivering antibody mimics over the TAT peptide method. Additionally, the amount of Lv1-HA detected by anti-HA immunoblotting was similar to that of Lv1 detected by anti-LF. The presence of a C-terminal HA-tag further validated the presence of full-length Lv1-HA in the cytosol.
Figure 3.
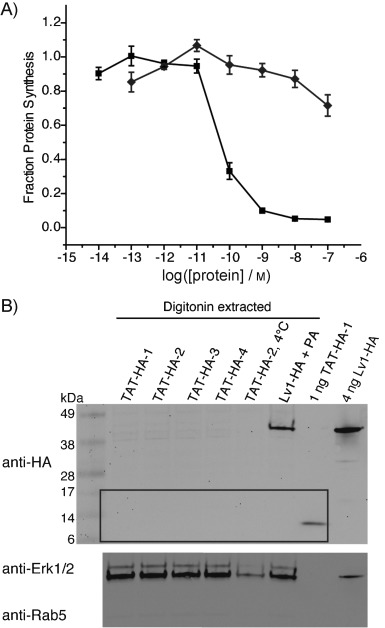
TAT peptide-mediated translocation of DTA and antibody mimics. A) Protein synthesis inhibition of CHO-K1 cells treated with varying concentrations of TAT-DTA (⧫) for 30 min, in comparison to LFN-DTA plus 20 nm PA (▪); assay conditions as in Figure 1. B) Western blot of cytosolic fractions extracted by digitonin from CHO-K1 treated with 2.5 μm TAT-HA-1–4 (see Figure 1 B) for 4 h (4 °C=incubation at 4 °C instead of 37 °C), in comparison to treatment with 250 nm Lv1-HA and 40 nm PA for 16 h. The cytosolic fraction was extracted using digitonin (∼280 000 cells per lane). The corresponding immunoblots of total lysates from cells treated with TAT-HA-1–4 for 4 h and 16 h are shown in Figure S5.
Our next question was whether the delivered antibody mimics can refold and bind to their targets inside the cytosol. Based on the protein synthesis inhibition assay, we confirmed that DTA could correctly refold in the cytosolic environment after translocation. For the antibody mimics, we chose to study the tandem 10FN3 monobody (HA4-7c12), which binds with nanomolar affinity to the Src homology 2 (SH2) domain of the oncoprotein Bcr-Abl.[2f] First, we sortagged this tandem monobody to LFN-DTA (LDv5) and used the protein synthesis inhibition assay to study the variant's translocation efficiency in chronic myeloid leukemia (CML) K562 cells. The assay showed that PA translocated LDv5 as efficiently as the control (LFN-DTA, Figure S6). After confirming that the PA system efficiently translocated the tandem monobody HA4-7c12, we sortagged it to LFN (Lv5, Figure 4 A), which lacks the cytotoxic DTA protein, and tested whether Lv5 could bind to Bcr-Abl in K562 cells after translocation to the cytosol. To ensure that the binding affinity of the monobody was not affected by the presence of LFN, which (apart from initiating translocation) provides an important epitope for detection in the cytosol by western blotting, we first measured the binding affinity of Lv5 for the Abl SH2 domain by SPR. We obtained a Kd value of 12 nm, similar to that for the monobody alone (6 nm, Figure S7). Next, we investigated whether the delivered Lv5 could refold and bind to Abl kinase inside the cell. K562 cells were treated with Lv5 and PA, and then subjected to immunoprecipitation with an anti-Abl antibody linked to agarose beads. Proteins eluted from the beads were subjected to immunoblot analysis with an anti-LF antibody. We observed a pull-down band corresponding to Lv5 when cells were treated with Lv5 and PA (Figure 4 B), thus confirming that at least some of the monobody properly folded after reaching the cytosol and was bound to intracellular Abl kinase. In control experiments, PA[F427H] was used instead of PA, in order to arrest translocation from the endosome to cytosol. No band was observed, thus confirming that cytosolic access of Lv5 was critical for the binding. Finally, the cells were treated with the binding mutant Lv5mut (HA4: Y87A; 7c12: Y62E/F87K) instead of Lv5. Similar amounts of Lv5 and Lv5mut were delivered to the cell; the pull-down band was absent for Lv5mut (Figure 4 B), thus indicating that the binding interaction only occurred when the LFN variant contained the functional binder. These results demonstrate that PA-delivered tandem monobody can refold and bind to its target inside the cell.
Figure 4.
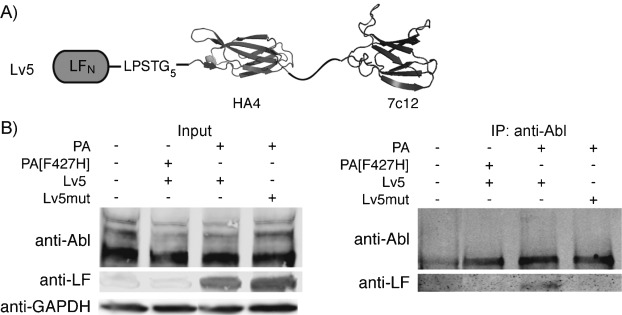
Delivery of Lv5 and binding of Lv5 to Abl kinase in K562 cells. A) Construct of Lv5 containing HA4 (PDB ID: 3K2M), GS-rich linker, and 7c12 (PDB ID: 3T04). B) Left: western blot of total cell lysate from K562 cells treated for 24 h with 50 nm Lv5 or Lv5mut in the presence of 20 nm PA or PA[F427H]. Right: the lysate was then subjected to co-immunoprecipitation (co-IP) with anti-Abl agarose beads. Cells treated with Lv5mut/PA or Lv5/PA[F427H] served as negative controls for co-IP.
Our next goal was to investigate the possibility of using the delivered tandem monobody to perturb protein function and related signaling pathways in cancer cells. Overexpression of HA4-7c12 in K562 cells has been reported to strongly inhibit kinase activity and induce apoptosis by disrupting a critical intramolecular SH2-kinase domain–domain interaction.[2f] Despite the high affinity and specificity of the monobody in targeting the allosteric module in Bcr-Abl and its utility in fighting therapy resistance of Bcr-Abl mutants, Hantschel et al. determined that intracellular delivery was the biggest hurdle to achieving its practical application.[2f, 27] In order to test if PA-mediated delivery could overcome this, we used PA to translocate Lv5 into K562 cells. Based on the linear relationship between the amount of protein loaded and the signal intensity of each band detected by anti-LF antibody (Figure S8), we estimated a total of 1 ng Lv5 delivered into ∼100 000 cells (Figure S9); this is ∼10 fg (110 000 Lv5 molecules) in each cell (cytosolic concentration ∼80 nm). Although this concentration is above the Kd for the Abl SH2 domain, we did not expect strong inhibition of Bcr-Abl kinase, because of the high concentration of Bcr-Abl inside K562 cells. As detected by western blotting, monobody binding resulted in a modest reduction in activation loop (Tyr412) phosphorylation of Bcr-Abl (Figure S9). We then investigated the effect of this activity inhibition on inducing apoptosis of K562 cells. By TUNEL staining (detects DNA fragmentation by labeling the termini), we observed apoptosis after K562 cells were treated with Lv5 and PA (Figure 5 A). This was not observed when PA[F427H] or Lv5mut was used. The proportion of apoptotic cells after delivery of Lv5 was 20 % of that caused by the small molecule imatinib (1 μm, Figure S10), which inhibits kinase activity by binding close to the ATP binding site of Bcr-Abl. The different inhibition mechanism and the much higher concentration of cytosolic imatinib might explain the difference in the proportion of apoptotic cells as compared to the Lv5 and PA treatment. In addition, the extent of apoptosis by Lv5 was also lower than that induced by overexpression of HA4-7c12.[2f] The high concentration of monobody achieved by overexpression and selection for only transfection-positive cells met the challenge of requiring high local concentration to interfere with the intramolecular interactions between the SH2 domain and kinase domain in Bcr-Abl. The modest inhibition of kinase activity and induction of apoptosis by PA-mediated delivery of Lv5 serves as a proof-of-concept that intracellularly delivered monobodies can perturb the activity of an oncoprotein.
Figure 5.
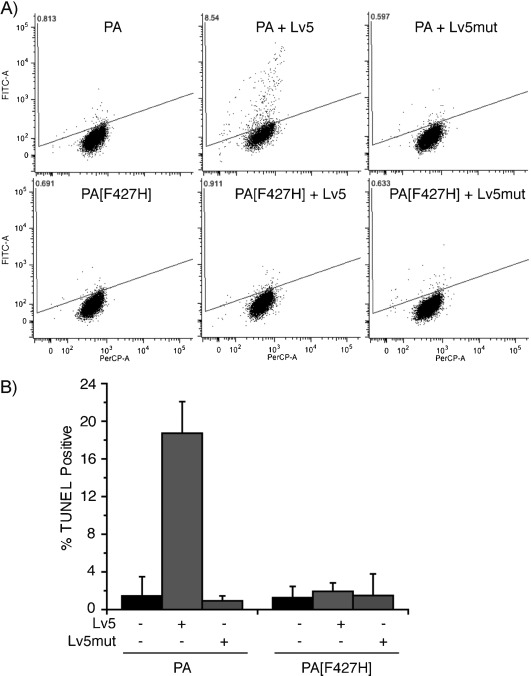
Monitoring of apoptosis of K562 cells treated with Lv5 and PA by the TUNEL assay. A) K562 cells treated with the indicated analogues (500 nm Lv5 or Lv5mut in the presence of 80 nm PA or PA[F427H]) for three days and analyzed for apoptosis; 1 μm imatinib served as a positive control. Representative dot plots from flow cytometry show terminal deoxynucleotidyl transferase (TdT) catalyzed BrdUTP incorporation into the DNA strand breaks of apoptotic cells; this is detected by Alexa Fluor 488-labeled anti-BrdU antibody (FITC). PerCP indicates the DNA fraction. B) Quantification of TUNEL-positive cells: intensities normalized to K562 cells treated with imatinib (100 %) and non-treated cells (0 %). The dot-plots of imatinib-treated and untreated cells are shown in Figure S10. Data are averages of three independent experiments.
We also investigated PA-mediated delivery of a binder based on affibody (ABRaf), which was evolved to bind to human Raf-1 (hRaf-1, Kd=100 nm),[2c] a protein kinase of central importance in the MAPK/ERK signaling pathway. Although ABRaf has been shown to inhibit the Ras/Raf interaction in vitro, it has not been tested for inhibition of MAPK signaling pathway in cells. We transfected human embryonic kidney 293T (HEK293T) cells with ABRaf, and observed an average of 37 % reduction in phosphorylation levels of MAPK (pErk1/2, downstream of Ras/Raf pathway) upon epidermal growth factor (EGF) activation Figure 6 A), thus validating the cellular function of this binder in blocking the MAPK signaling pathway. We then conjugated ABRaf to LFN (Lv6), and found that PA delivered ∼240 nm Lv6 (∼5 fg or 79 000 molecules per cell) in HEK293T cells (Figure 6 B). We measured the phosphorylation level of MAPK (pErk1/2) upon EGF activation in cells treated with Lv6 and PA or PA[F427H]. Cells treated with Lv6 and PA showed ∼25 % reduction of pErk1/2 relative to EGF-activated untreated cells (P<0.01, Figure 6 C). PA[F427H] (negative control) showed a similar level of pErk1/2 as for EGF-activated untreated cells (P=0.27, Figure 6 C). This experiment is another example of using PA to translocate functional binders to disrupt protein–protein interactions and the associated signaling pathway in cells.
Figure 6.
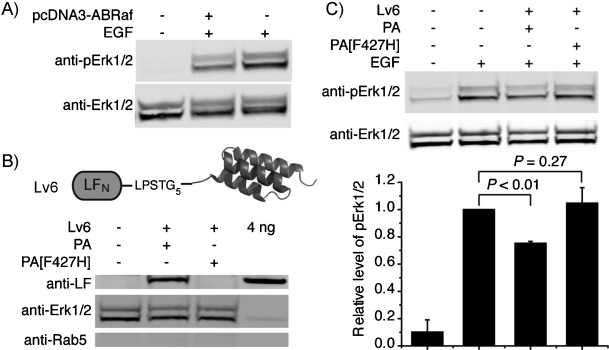
Perturbation of the MAPK signaling pathway by PA-mediated delivery of an affibody (Lv6) that targets Raf. A) HEK293T cells were transfected with pcDNA3-ABRaf for 24 h, starved overnight, treated with 5 ng mL−1 EGF for 7 min, lysed with buffer containing 1 % NP-40, and subjected to anti-pErk1/2 immunoblotting. The membrane was stripped and re-blotted with anti-Erk1/2 antibody to serve as loading control. B) Cytosolic fractions extracted by digitonin from HEK293T cells treated with 500 nm Lv6 and 80 nm PA or PA[F427H] in serum-free medium for 12 h (∼400 000 cells per lane). C) HEK293T cells treated with 500 nm Lv6 and 80 nm PA or PA[F427H] in serum-free medium for 12 h, then with 5 ng mL−1 EGF for 7 min, lysed, and analyzed as in (A). Bar graph: quantification of phosphorylation of Erk1/2 (pErk1/2; n=3); bars correspond to the lanes above. pErk1/2 bands normalized to that of Erk1/2, and compared to cells treated with EGF (set to 1). Data are averages of three experiments. P values: Student's t-Test.
Conclusion
We have demonstrated the ability of PA/LFN to deliver antibody mimics to the cytosol of cells and also the possibility of using the delivered binders to perturb critical protein–protein interactions in cancer cells. Prior efforts with this system have mainly focused on the delivery of enzymes that can exert a strong biological effect at very low concentrations, as has been demonstrated here by the LFN-DTA activity assays. However, for an antibody mimic binder to exert inhibitory effects on protein–protein interactions inside cells, the concentration of the antibody mimic binder needs to reach a level above Kd and close to the concentration of competing endogenous binding proteins. We aimed to answer the questions as to whether PA could both efficiently deliver the antibody mimics and transport sufficient amounts of cargo to perturb protein–protein interactions.
We first systematically investigated translocation mediated by PA of four different antibody mimics: all α-helical (affibody, DARPin), all β-sheet (monobody), and α-helical and β-sheet proteins (GB1). We did not assume these scaffolds would translocate efficiently, because prior investigations have indicated that certain C-terminal modifications of LFN or LFN-DTA can abrogate translocation through PA. In particular, imaging studies indicated that fluorescent proteins fused to the C terminus of LF significantly attenuate translocation.[28] Other studies have shown that DTA with an artificial disulfide or dihydrofolate reductase (DHFR) complexed with methotrexate can arrest translocation when attached to the C terminus of LFN.[29] Using the DTA-based protein synthesis inhibition assay, we found that all four antibody mimics translocated as efficiently as the positive control LFN-DTA. We are unaware of any system that permits the facile delivery of antibody mimics ranging in structural diversity as shown here.
Our translocation studies began with LDvs and the protein synthesis inhibition assay, because this is commonly used to probe anthrax toxin entry into the cytosol and thus allowed us to compare our findings to other reports. However, in order to use antibody mimics to perturb protein–protein interactions, we needed to eliminate the interference of DTA. Therefore, we also investigated the translocation of LFN-antibody mimics (Lvs) in the presence of PA. To facilitate the analysis of Lv delivery, we used a reliable western blot approach to assess full-length cargo in the cytosolic fraction (obtained by using digitonin to permeabilize the plasma membrane). For all experiments, we confirmed successful extraction of cytosolic proteins from the rest of the cellular components by staining to check for the presence of Erk1/2 and absence of Rab5. The successful extraction of cytosolic proteins was further validated by the absence of Lv1 in the digitonin-extracted fractions from cells treated with bafilomycin A1 or PA[F427H]. Additionally, we observed no Lv1 in the total cell lysate under 4 °C treatment conditions, in order to validate the effectiveness of trypsin digestion in removing surface-bound Lv (leaving only intracellular proteins for detection). We detected the conjugates of antibody mimics to LFN by the anti-LF antibody, and confirmed the presence of the antibody mimics attached to LFN by their different molecular weights. Further evidence that full-length material translocated into the cytosol was obtained by detection of Lv1-HA with the C-terminal HA-tag, which gave a similar amount of material detected as that by the anti-LF antibody.
We also used digitonin extraction and western blot to gauge the amount of material delivered into the cell cytosol. Based on the linear relationship between the amount of loaded protein and the signal intensity on the anti-LF immunoblot, we were able to estimate the amount of material delivered, by comparing the immunoblot signal to that of a known amount of protein loaded on the same blot. As the antibody mimics do not interfere with translocation through the PA pore, the amount delivered to the cytosol is dependent on the number of anthrax receptors on cells; these are present on most human cells (2000– 50 000 per cell, higher in certain cancer cells).[30] Theoretically, one round of translocation for cells harboring 50 000 receptors would give ∼20 000 molecules in one cell, if seven receptors deliver three copies of LFN-cargo variant. Thus we estimated, by western blot quantification, that after multiple rounds of translocation we achieved mid-nanomolar concentrations of cargo in the cytosol.
We next studied whether a functional antibody mimic binder can properly refold after translocation, as translocation through the PA pore requires protein unfolding. Based on co-immunoprecipitation of the monobody with the anti-Abl antibody, we confirmed that the monobody correctly refolds in the cytosolic environment after translocation. The efficiency of refolding and the portion of functional protein is presumed to depend on the cytosolic stability and degradation rates of the variants. For the binders based on monobody and affibody, we observed inhibition of Bcr-Abl kinase and disruption of the MAPK signaling pathway, respectively, thus providing evidence that the antibody mimics properly fold and function after translocation into the cytosol. As the amount of tandem monobody delivered was not significantly higher than the Kd for the Bcr-Abl target, the extent of apoptosis caused by Lv5 was not as great as that achieved by overexpression of the tandem monobody (where its cytosolic concentration reaches a much higher level). The lower biological effect of Lv5 was probably due to the presence of high concentration of endogeneous Abl kinase, as well as the difficulty in interfering with the intramolecular domain–domain interaction of Abl kinase. In contrast, even though the delivered amount of affibody binder for hRaf-1 was not much higher than the Kd for its target, the inhibition of the MAPK signaling pathway by Lv6 was close to that achieved by overexpression of the affibody binder. This could be attributable to the lower target concentration and the higher efficiency in disrupting Ras/Raf intermolecular interactions, compared to the Bcr-Abl target. The delivery of more material and/or increased potency of the cargo for its target are additional challenges in biomolecular delivery.
The high adaptability and promiscuity of the PA transporter enabled delivery without the need for protein engineering or screening, unlike delivery methods such as CPPs where a number of sequences or linkers need to be screened for efficient delivery of each cargo. In the case of CPPs, the stability and identity of the peptide transduction sequence can lead to significant differences in delivery efficiency, and strong adherence to the cell surface and endosomal membranes often blocks cargo escape into the cytosol. For example, it was reported that CPP fusions to DTA were not able to achieve delivery into the cell,[25] even at concentrations a thousand times higher than that required for PA-mediated translocation; we confirmed this result. We also investigated the efficiency of the TAT peptide in the delivery of the antibody mimics and found that, even after treatment of the same cell lines with tenfold more material, the TAT peptide was not able to deliver any of the four antibody mimics. These results demonstrate the significantly better delivery efficiency by PA over the TAT peptide. Additionally, because of highly inefficient endosomal escape for delivery into the cytosol,[31] most delivery methods use high concentrations of components, and this can give rise to cellular toxicity. We studied whether the delivery components PA and LFN (by themselves or in combination) are toxic to cells. Under the conditions tested, we did not see any toxicity, based on a TUNEL apoptosis assay (Figure 5) or an MTS assay (Figure S11).
In summary, we report for the first time PA-mediated delivery of antibody mimics into the cytosol and successful disruption of critical protein–protein interactions inside cells. As an example, we found that an SH2-binding tandem monobody can be delivered and functioned as an inhibitor of the oncoprotein Bcr-Abl inside cancer cells. We also demonstrated inhibition of the Ras/Raf interaction by an affibody thereby blocking the MAPK signaling pathway, which plays a central role in the control of cell proliferation, survival, and growth. The delivery of antibody mimics to the cytosol of cells as indicated by DTA activity and western blotting, together with the observation of functional tandem monobody binder to the oncoprotein Bcr-Abl and affibody binder to hRaf-1, supports our belief that the PA-mediated protein delivery system (designed by nature) will significantly expand the biomolecular delivery toolbox. Our future efforts will focus on increasing the amount of material delivered and the delivery of more potent antibody mimics. This platform provides new possibilities to apply modern intrabody technology to disrupt processes inside cells.
Experimental Section
Materials: All reagents were purchased from Sigma–Aldrich and Life Technologies except where otherwise indicated. The following primary and secondary antibodies were used: goat anti-LF (bD-17; Santa Cruz Biotechnology, Dallas, TX), rabbit anti-HA (Sigma–Aldrich), rabbit anti-Abl (agarose conjugate, K-12; Santa Cruz Biotechnology), rabbit anti-pAbl (pTyr412; Cell Signaling Technology, Danvers, MA), rabbit anti-Erk1/2 and rabbit anti-phospho-Erk1/2 (Thr202/Tyr204, Cell Signaling), rabbit anti-Rab5 (Cell Signaling), goat anti-rabbit (IRdye 800CW; LI-COR Biosciences, Lincoln, NE), donkey anti-goat (IRdye 680LT; LI-COR Biosciences). Additional methods including expression and purification of the proteins used in our studies are described in detail in the Supporting Information.
One-pot sortagging reaction using Staphylococcus aureus SrtA*: SrtA*-mediated ligation was used to ligate proteins to His6-SUMO-LFN-DTA-LPSTGG-His5 or His6-SUMO-LFN-LPSTGG-His6 as previously described (Figure S1).[21b] LFN-DTA-LPSTGG-His5 or LFN-LPSTGG-His6 (50 μm), SrtA* (5 μm), and G5-protein (100–500 μm) were incubated with Ni-NTA beads in sortase buffer (Tris⋅HCl (50 mm, pH 7.5), CaCl2 (10 mm), NaCl (150 mm)) for 30 min at RT rocking. Ni-NTA beads that have bound unreacted starting material, SrtA*-His6, His6-SUMO, and GG-His6 were centrifuged (16 000 g, 4 °C) to minimize the formation of hydrolyzed side product. The supernatant (containing sortagged product) was collected. The beads were washed three times. The supernatant and the three washes were subjected to gel filtration to separate excess oligoglycine protein reactants. SDS-PAGE and LCMS were used to assess the purity of the products (Figure S2, Table S2, and LCMS Appendix; isolated yields in Table S3; protein variants are listed in Table S5).
Protein synthesis inhibition assay: CHO-K1 cells were maintained in F-12K medium supplemented with FBS (10 %, v/v) at 37 °C with 5 % CO2. The cells were plated in a 96-well plate (∼30 000 per well) 16 h prior to the assay. LDvs were prepared in tenfold serial dilutions followed by the addition of protective antigen (PA83; 20 nm). The samples were added to CHO-K1 cells for 30 min at 37 °C and 5 % CO2. The cells were washed three times with PBS, then incubated with leucine-free F-12K medium (100 μL) supplemented with 3H-leucine (1 μCi mL−1, PerkinElmer) for 1 h at 37 °C with 5 % CO2. The cells were washed three times with PBS and suspended in scintillation fluid (150 μL). 3H-Leu incorporation into cellular proteins was measured to determine the inhibition of protein synthesis by LFN-DTA. The scintillation counts from cells treated with PA only (control) were used for normalization. Each experiment was done in triplicate. The data were fitted by using OriginLab software (Northhamptown, MA) with a sigmoidal Boltzmann fit using Equation (1):
| (1) |
where x0 is the log EC50 value (Table S4).
Uptake of Lv and TAT-HA-1–4 in CHO-K1 cells: CHO-K1 cells were plated in 12-well plate 16 h prior to treatment. For Lvs, cells were treated with Lv (250 nm) in the presence of PA (40 nm) in F-12K with FBS overnight at 37 °C with 5 % CO2. For TAT-HA-antibody mimic constructs, cells were treated with TAT-HA-1–4 (2.5 μm) in F-12K without FBS for 4 h at 37 °C and 5 % CO2. Translocation controls included PA[F427H] (instead of PA), addition of bafilomycin A1 (200 nm), and incubation at 4 °C instead of 37 °C.
Cytosolic protein extraction and whole-cell lysate preparation: After uptake of the antibody mimics, cells were washed with PBS, detached and digested with Trypsin-EDTA (0.25 %) for 5 min at 37 °C to remove surface-bound protein, then washed twice with PBS. The cell pellets were then subjected to cytosolic extraction or whole-cell lysate preparation. For cytosolic protein extraction, cells (∼106) were suspended in buffer (100 μL: digitonin (50 μg mL−1) in NaCl (75 mm), NaH2PO4 (1 mm), Na2HPO4 (8 mm), sucrose (250 mm)) supplemented with Protease Inhibitor Cocktail (Roche) for 10 min on ice, then centrifuged (16 000 g, 5 min). For whole-cell lysate, cells were lysed in IP lysis buffer (Tris⋅HCl (25 mm, pH 7.5), NaCl (150 mm), NP-40 (1 %, v/v)) supplemented with Protease Inhibitor Cocktail on ice for 30 min, then centrifuged (16 000 g, 10 min). Supernatants were collected for blotting and analysis.
Western blot: Transfer was done with a TE 70 Semi-Dry Transfer Unit (GE Healthcare) and nitrocellulose membranes (Whatman) in Tris⋅HCl (48 mm) containing glycine (39 mm), SDS (0.0375 %, v/v), and methanol (20 %, v/v). The membrane was blocked (RT, 2 h) with blocking buffer (LI-COR) and then incubated with goat anti-LF or rabbit anti-HA antibody in LI-COR blocking buffer overnight at 4 °C. The membranes were washed with TBST (Tris⋅HCl (50 mm), NaCl (150 mm), Tween 20 (0.1 %, v/v)) then blotted with secondary antibody conjugated to IRdye (LI-COR) and imaged with an Odyssey Infrared Imaging System (LI-COR). The western blot images were analyzed and quantified with the Image Studio Lite program (LI-COR).
Co-immunoprecipitation of Lv5 with Abl kinase: K562 cells were treated with PA or PA[F427H] (40 nm) and LFN-antibody mimic (50 nm) for 24 h. Cells (∼11×106) were trypsinized, washed with PBS, frozen at −80 °C, and then lysed in IP lysis buffer (500 μL) supplemented with Protease Inhibitor Cocktail (Roche) on ice for 30 min. After centrifugation (16 000 g, 15 min), the lysate (450 μL) was incubated with 12.5 μg anti-Abl agarose beads for 4 h. The resulting immune complexes were washed three times with lysis buffer and once with lysis buffer without NP-40. The bound proteins were eluted with SDS (0.2 %) and Tween 20 (0.1 %) and subjected to SDS-PAGE separation. Immunoblotting was performed with anti-Abl or anti-LF antibody.
TUNEL assay with Lv5 in K562 cells: K562 cells were plated in a 24-well plate (150 000 cells per mL in each well) and treated with LFN-antibody mimic (500 nm) and PA or PA[F427H] (60 nm) in serum-free medium for one day. FBS (10 %) was added, and the cells were incubated for two days (positive control: imatinib (1 μm)). The cells were fixed with paraformaldehyde (3.2 %), then treated with methanol (90 %) on ice or stored at −20 °C, followed by TUNEL staining by using the Apo-BrdU TUNEL assay kit from Life Technologies.
Transfection of HEK293T with pcDNA3-ABRaf: HEK293T cells were plated in a 24-well plate overnight to reach ∼90 % confluency. The cells were then transfected with plasmid pcDNA3-ABRaf-GFP (see the Supporting Information for plasmid details) by using Lipofectamine 2000 (Life Technologies). The medium was changed after 5 h. The total transfection time was 24 h. For pErk1/2 analysis, the cells were starved for 12 h then treated with EGF (5 ng mL−1) for 7 min, washed with cold PBS, and lysed in-plate with IP lysis buffer containing NP-40 (1 %), Protease Inhibitor Cocktail (Roche), and phosphoSTOP (Roche). The lysate was subjected to SDS-PAGE separation, transferred to nitrocellulose membrane, and blocked with BSA (5 %), Na3VO4, and NaF. The immunoblotting with anti-pErk1/2 antibody (Cell Signaling) was performed overnight in BSA (3 %) with Na3VO4 and NaF. After stripping the membrane with Restore Plus Western Blot Stripping Buffer (Thermo Scientific), the membrane was blocked again and immunoblotted with anti-Erk1/2 antibody (Cell Signaling).
Delivery of Lv6 into HEK293T cells: HEK293T cells were plated in a 24-well plate and left overnight to reach ∼80 % confluency. The cells were then treated with PA (80 nm) and Lv6 (500 nm) in serum-free medium for 12 h. For detection of cytosolic Lv6, the cells were detached with trypsin, washed with PBS, and suspended in digitonin (100 μg mL−1) for 10 min (as above). For detection of pErk1/2, the cells were treated with EGF (5 ng mL−1) for 7 min, washed with cold PBS, lysed in-plate with IP lysis buffer containing NP-40 (1 %), Protease Inhibitor Cocktail (Roche), and phosphoSTOP (Roche). Immunoblotting with anti-pErk1/2 and anti-Erk1/2 was as described above.
Acknowledgments
This research was generously sponsored by MIT start-up funds, the MIT Reed Fund, NSF CAREER Award (CHE-1351807), and a Damon Runyon Cancer Research Foundation award for B.L.P, and a National Science Foundation Graduate Research Fellowship for A.E.R. We would like to thank Prof. R. John Collier (Harvard) for his encouragement, support, and some of the laboratory equipment. We are indebted to the NERCE facility (grant: U54 AI057159) for expressing the toxin proteins. Patent applications covering material reported in this manuscript were filed by MIT-TLO. We thank Jingjing Ling, Mike Lu, Dan Chinnapen, and Daphne van Scheppingen for their helpful discussions. We also thank Prof. Dane Wittrup for providing the fibronectin clone.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
miscellaneous_information
References
- 1a.Caravella J, Lugovskoy A. Curr. Opin. Chem. Biol. 2010;14:520–528. doi: 10.1016/j.cbpa.2010.06.175. [DOI] [PubMed] [Google Scholar]
- 1b.Gilbreth RN, Koide S. Curr. Opin. Struct. Biol. 2012;22 doi: 10.1016/j.sbi.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1c.Koide S, Koide A, Lipovšek D. Methods Enzymol. 2012;503 doi: 10.1016/B978-0-12-396962-0.00006-9. [DOI] [PubMed] [Google Scholar]
- 1d.Tamaskovic R, Simon M, Stefan N, Schwill M, Plückthun A. Methods Enzymol. 2012;503 doi: 10.1016/B978-0-12-396962-0.00005-7. [DOI] [PubMed] [Google Scholar]
- 1e.Löfblom J, Feldwisch J, Tolmachev V, Carlsson J, Ståhl S, Frejd FY. FEBS Lett. 2010;584 doi: 10.1016/j.febslet.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 1f.Mandal K, Uppalapati M, Ault-Riche D, Kenney J, Lowitz J, Sidhu SS, Kent SBH. Proc. Natl. Acad. Sci. USA. 2012;109 doi: 10.1073/pnas.1210483109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Schweizer A, Roschitzki-Voser H, Amstutz P, Briand C, Gulotti-Georgieva M, Prenosil E, Binz HK, Capitani G, Baici A, Plückthun A, Grütter MG. Structure. 2007;15:625–636. doi: 10.1016/j.str.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 2b.Schroeder T, Barandun J, Flutsch A, Briand C, Mittl PRE, Grütter MG. Structure. 2013;21 doi: 10.1016/j.str.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 2c.Grimm S, Salahshour S, Nygren P-Å. New Biotechnol. 2012;29 doi: 10.1016/j.nbt.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 2d.Parizek P, Kummer L, Rube P, Prinz A, Herberg FW, Plückthun A. ACS Chem. Biol. 2012;7 doi: 10.1021/cb3001167. [DOI] [PubMed] [Google Scholar]
- 2e.Kummer L, Parizek P, Rube P, Millgramm B, Prinz A, Mittl PRE, Kaufholz M, Zimmermann B, Herberg FW, Plückthun A. Proc. Natl. Acad. Sci. USA. 2012;109:E2248–E2257. doi: 10.1073/pnas.1205399109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2f.Grebien F, Hantschel O, Wojcik J, Kaupe I, Kovacic B, Wyrzucki AM, Gish GD, Cerny-Reiterer S, Koide A, Beug H, Pawson T, Valent P, Koide S, Superti-Furga G. Cell. 2011;147 doi: 10.1016/j.cell.2011.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2g.Wojcik J, Hantschel O, Grebien F, Kaupe I, Bennett KL, Barkinge J, Jones RB, Koide A, Superti-Furga G, Koide S. Nat. Struct. Mol. Biol. 2010;17 doi: 10.1038/nsmb.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2h.Lundberg E, Brismar H, Gräslund T. Biotechnol. Appl. Biochem. 2009;52 doi: 10.1042/BA20070178. [DOI] [PubMed] [Google Scholar]
- 3a.Methods Enzymol. 2012;502:1–344. [Google Scholar]
- 3b.Methods Enzymol. 2012;503 [Google Scholar]
- 3c.Moore SJ, Hayden Gephart MG, Bergen JM, Su YS, Rayburn H, Scott MP, Cochran JR. Proc. Natl. Acad. Sci. USA. 2013;110 doi: 10.1073/pnas.1311333110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marschall ALJ, Frenzel A, Schirrmann T, Schüngel M, Dubel S. mAbs. 2011;3:3–16. doi: 10.4161/mabs.3.1.14110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zelphati O, Wang Y, Kitada S, Reed JC, Felgner PL, Corbeil J. J. Biol. Chem. 2001;276:35103–35110. doi: 10.1074/jbc.M104920200. [DOI] [PubMed] [Google Scholar]
- 6a.Hasadsri L, Kreuter J, Hattori H, Iwasaki T, George JM. J. Biol. Chem. 2009;284:6972–6981. doi: 10.1074/jbc.M805956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6b.Kamaly N, Xiao Z, Valencia PM, Radovic-Moreno AF, Farokhzad OC. Chem. Soc. Rev. 2012;41 doi: 10.1039/c2cs15344k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu Z, Biswas A, Zhao M, Tang Y. Chem. Soc. Rev. 2011;40:3638–3655. doi: 10.1039/c0cs00227e. [DOI] [PubMed] [Google Scholar]
- 8a.Cronican JJ, Thompson DB, Beier KT, McNaughton BR, Cepko CL, Liu DR. ACS Chem. Biol. 2010;5:747–752. doi: 10.1021/cb1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8b.Thompson DB, Cronican JJ, Liu DR. Methods Enzymol. 2012;203 doi: 10.1016/B978-0-12-396962-0.00012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Caron NJ, Torrente Y, Camirand G, Bujold M, Chapdelaine P, Leriche K, Bresolin N, Tremblay JP. Mol. Ther. 2001;3:310–318. doi: 10.1006/mthe.2001.0279. [DOI] [PubMed] [Google Scholar]
- 9b.Foerg C, Merkle HP. J. Pharm. Sci. 2008;97 doi: 10.1002/jps.21117. [DOI] [PubMed] [Google Scholar]
- 9c.Cai S-R, Xu G, Becker-Hapak M, Ma M, Dowdy SF, McLeod HL. Eur. J. Pharm. Sci. 2006;27 doi: 10.1016/j.ejps.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 9d.Heitz F, Morris MC, Divita G. Br. J. Pharmacol. 2009;157 doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alouf JE, Popoff MR, editors. The Comprehensive Sourcebook of Bacterial Protein Toxins. 3edrd ed. San Diego: Academic Press; 2006. [Google Scholar]
- 11.Klimpel KR, Molloy SS, Thomas G, Leppla SH. Proc. Natl. Acad. Sci. USA. 1992;89:10277–10281. doi: 10.1073/pnas.89.21.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milne JC, Furlong D, Hanna PC, Wall JS, Collier RJ. J. Biol. Chem. 1994;269:20607–20612. [PubMed] [Google Scholar]
- 13.Kintzer AF, Thoren KL, Sterling HJ, Dong KC, Feld GK, Tang II, Zhang TT, Williams ER, Berger JM, Krantz BA. J. Mol. Biol. 2009;392:614–629. doi: 10.1016/j.jmb.2009.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krantz BA, Trivedi AD, Cunningham K, Christensen KA, Collier RJ. J. Mol. Biol. 2004;344:739–756. doi: 10.1016/j.jmb.2004.09.067. [DOI] [PubMed] [Google Scholar]
- 15.Pannifer AD, Wong TY, Schwarzenbacher R, Renatus M, Petosa C, Bienkowska J, Lacy DB, Collier RJ, Park S, Leppla SH, Hanna P, Liddington RC. Nature. 2001;414:229–233. doi: 10.1038/n35101998. [DOI] [PubMed] [Google Scholar]
- 16.Ballard JD, Collier RJ, Starnbach MN. Proc. Natl. Acad. Sci. USA. 1996;93:12531–12534. doi: 10.1073/pnas.93.22.12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Hobson JP, Liu S, Rønø B, Leppla SH, Bugge TH. Nat. Methods. 2006;3:259–261. doi: 10.1038/nmeth862. [DOI] [PubMed] [Google Scholar]
- 17b.Bachran C, Morley T, Abdelazim S, Fattah RJ, Liu S, Leppla SH. mBio. 2013;4 doi: 10.1128/mBio.00201-13. e00201-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Arora N, Leppla SH. Infect. Immun. 1994;62:4955–4961. doi: 10.1128/iai.62.11.4955-4961.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18b.Arora N, Leppla SH. J. Biol. Chem. 1993;268 [PubMed] [Google Scholar]
- 18c.Milne JC, Blanket SR, Hanna PC, Collier RJ. Mol. Microbiol. 1995;15 doi: 10.1111/j.1365-2958.1995.tb02375.x. [DOI] [PubMed] [Google Scholar]
- 18d.Cordero CL, Kudryashov DS, Reisler E, Fullner Satchell KJ. J. Biol. Chem. 2006;281 doi: 10.1074/jbc.M605275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Moltke J, Trinidad NJ, Moayeri M, Kintzer AF, Wang SB, van Rooijen N, Brown CR, Krantz BA, Leppla SH, Gronert K, Vance RE. Nature. 2012;490:107–126. doi: 10.1038/nature11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Nat. Chem. Biol. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- 21a.Chen I, Dorr BM, Liu DR. Proc. Natl. Acad. Sci. USA. 2011;108:11399–11404. doi: 10.1073/pnas.1101046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21b.Ling JJ, Policarpo RL, Rabideau AE, Liao X, Pentelute BL. J. Am. Chem. Soc. 2012;134 doi: 10.1021/ja302354v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Collier RJ, Kandel J. J. Biol. Chem. 1971;246:1496–1503. [PubMed] [Google Scholar]
- 22b.Wilson BA, Collier RJ. Curr. Top. Microbiol. Immunol. 1992;175 doi: 10.1007/978-3-642-76966-5_2. [DOI] [PubMed] [Google Scholar]
- 23a.Sun J, Lang AE, Aktories K, Collier RJ. Proc. Natl. Acad. Sci. USA. 2008;105:4346–4351. doi: 10.1073/pnas.0800701105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23b.Krantz BA, Melnyk RA, Zhang S, Juris SJ, Lacy DB, Wu Z, Finkelstein A, Collier RJ. Science. 2005;309 doi: 10.1126/science.1113380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adam SA, Sterne Marr R, Gerace L. J. Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falnes PØ, Wesche J, Olsnes S. Biochemistry. 2001;40:4349–4358. doi: 10.1021/bi002443l. [DOI] [PubMed] [Google Scholar]
- 26.Nagahara H, Vocero-Akbani AM, Snyder EL, Ho A, Latham DG, Lissy NA, Becker-Hapak M, Ezhevsky SA, Dowdy SF. Nat. Med. 1998;4:1449–1452. doi: 10.1038/4042. [DOI] [PubMed] [Google Scholar]
- 27.Hantschel O, Grebien F, Superti-Furga G. Oncotarget. 2011;2:828–829. doi: 10.18632/oncotarget.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Zornetta I, Brandi L, Janowiak B, Dal Molin F, Tonello F, Collier RJ, Montecucco C. Cell. Microbiol. 2010;12:1435–1445. doi: 10.1111/j.1462-5822.2010.01480.x. [DOI] [PubMed] [Google Scholar]
- 28b.Zheng S, Zhang G, Li J, Chen PR. Angew. Chem. Int. Ed. 2014;53 doi: 10.1002/anie.201403945. [DOI] [PubMed] [Google Scholar]
- Angew. Chem. 2014;126 [Google Scholar]
- 29.Wesche J, Elliott JL, Falnes PØ, Olsnes S, Collier RJ. Biochemistry. 1998;37:15737–15746. doi: 10.1021/bi981436i. [DOI] [PubMed] [Google Scholar]
- 30.Abi-Habib RJ, Urieto JO, Liu S, Leppla SH, Duesbery NS, Frankel AE. Mol. Cancer Ther. 2005;4:1303–1310. doi: 10.1158/1535-7163.MCT-05-0145. [DOI] [PubMed] [Google Scholar]
- 31a.Wadia JS, Stan RV, Dowdy SF. Nat. Med. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 31b.Pirie CM, Hackel BJ, Rosenblum MG, Wittrup KD. J. Biol. Chem. 2011;286 doi: 10.1074/jbc.M110.186973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
miscellaneous_information


