Abstract
Purpose
Recruitment of histone deacetylases (HDAC) is a mechanism of transcriptional repression implicated in the differentiation block in acute myeloid leukemia (AML). We hypothesized that the HDAC inhibitor romidepsin could cause transcriptional derepression, up-regulation of specific target genes in AML, and differentiation of the leukemic clone. The primary objectives of the study were to evaluate the safety and efficacy of romidepsin in advanced AML.
Experimental Design
Twenty patients were stratified into cohort A or B based on the absence or presence of chromosomal abnormalities known to recruit HDACs, including those involving core binding factor (CBF). Romidepsin was administered i.v. at 13 mg/m2/d on days 1,8, and 15 of a 28-day cycle. Pharmacodynamic endpoints were evaluated at serial time points.
Results
Common adverse effects noted were grade 1 to 2 nausea, anorexia, and fatigue. No objective evidence of antileukemic activity was seen in cohort A. In cohort B, although there were no clinical responses by standard criteria, antileukemic activity was observed in 5 of 7 patients. Two patients had clearance of bone marrow blasts and 3 patients had a >50% decrease in bone marrow blasts. Furthermore, in cohort B, at 24 h, there was a significant increase in MDR1 (P = 0.005), p15 (P = 0.01), and p14 (P < 0.0001) expression. In cohort A, although there was a trend toward up-regulation of MDR1, p15, and p14 expression, these changes were not statistically significant.
Conclusion
Romidepsin has differential antileukemic and molecular activity in CBF AML. Development of this agent in CBF AML should focus on combinations that target related mechanisms of gene silencing such as DNA methylation.
Recruitment of histone deacetylases (HDAC) is an important epigenetic mechanism of transcriptional dysregulation and gene silencing in acute myeloid leukemia (AML). Acetylation of the lysine tails of histones, a process mediated by histone acetyl transferases, results in loosening of DNA-histone interactions and facilitates binding of specific transcriptional coregulators, resulting in transcriptionally active chromatin. HDACs cleave acetyl groups from histones and block this DNA conformational change, resulting in transcriptional repression of specific genes, including those important for cellular differentiation, cell cycle regulation, and apoptosis (1).
AML is a biologically heterogeneous disease; both epigenetic and genetic alterations have been implicated in disease pathogenesis. Approximately 30% to 40% of AML is associated with recurring chromosomal translocations that result in the generation of chimeric fusion genes. We and others have shown that histone acetyl transferases such as CBP and p300 (2–4) are disrupted in chromosomal translocations in leukemia. In addition, transcriptional repression by several chimeric fusion proteins in AML occurs via recruitment of HDACs either directly or in cooperation with corepressors (5–8). These findings provide strong evidence for transcriptional dysregulation involving the histone acetylation/deace-tylation pathway in leukemogenesis. For example, the fusion proteins AML1-ETO and core binding factor β (CBFβ)-MYH11 resulting from the t(8;21)(q22;q22) and (inv 16)(pl3;q22) or t(16;16)(pl3;q22), respectively, have been shown to act as transcriptional repressors. Either directly or through its hetero-dimeric partner CBFβ, AML1 is the target of all three chromosomal rearrangements and acts as a transcriptional activator, whereas the fusion proteins repress transcription of AML1 target genes such as pl4ARF, MDR1, interleukin-3, and CSF1-R (5, 9, 10).
Unlike gene deletions that are irreversible, transcriptional repression by epigenetic mechanisms can be reversed for therapeutic benefit by chromatin remodeling agents such as HDAC inhibitors (HDACI). Inhibitors of HDACs are now in clinical trials and have been shown to exert antitumor effects in vitro and in vivo. Romidepsin (depsipeptide) is a potent HDACI currently in development that inhibits class I HDACs. In addition, the agent has been shown to induce G1 and G2-M cell cycle arrest, differentiation, and morphologic reversion of transformed cells in parallel with accumulation of highly acetylated histones within cells (11–15). Significant antileuke-mia effects, including cellular differentiation coupled with reexpression of AML1 target genes such as interleukin-3, have been noted with HDACIs including romidepsin in cell lines and primary leukemia samples harboring the t(8;21) (7, 16–19). This raises the possibility that this cytogenetic subset of AML could be particularly amenable to therapy with HDACIs. The finding that myeloid leukemia cell lines that lack this specific chromosomal translocation are also susceptible in vitro to treatment with HDACIs, however, suggests that treatment with HDACIs could be more widely applicable in AML (17, 20).
We report here our results from a phase II trial of romidepsin in AML, including a cohort of patients with CBF AML. The primary objectives of the trial were to determine the efficacy and safety of romidepsin in AML. Secondary objectives included an evaluation of the effects of romidepsin in vivo on various biologic markers including global and promoter-specific histone acetylation and expression of specific target genes.
Patients, Materials and Methods
Patients and eligibility criteria
Patients were eligible for this study if they had relapsed or refractory AML. Patients with relapsed AML who were ages <60 years had to be in second relapse or greater, whereas patients who were ≥60 years could be in first relapse. Patients with previously untreated AML, ages ≥60 years and who were not candidates for or who refused conventional chemotherapy, were also eligible for this trial. Patients enrolled on this trial could not be candidates for allogeneic stem cell transplantation at the time of enrollment. Patients had to be aged at least 18 years, have a CALGB performance status of ≥2, and give informed consent. There was no limitation to the number or types of prior therapies received, including autologous or allogeneic stem cell transplantation. No cytotoxic therapy or radiation therapy was permitted within 2 weeks (6 weeks for mitomycin C and nitrosoureas and 4 weeks if prior therapy was with an investigational agent). The exception was hydroxyurea, which was permitted during the first cycle of therapy to control hyperleukocytosis (defined as WBC count over 20,000/μL). Patients could not have received prior therapy with romidepsin and could not be receiving concurrent therapy with drugs known to have HDACI activity such as sodium valproate (21).
Because of the potential for QTc prolongation, a class effect of HDACIs, patients had to have a left ventricular ejection fraction of ≥40% and QTc interval of <500 ms on electrocardiogram to be eligible. In addition, patients could not have had a myocardial infarct within the preceding 3 months or have symptomatic heart disease or left ventricular hypertrophy on electrocardiogram.
Patients were ineligible if the serum creatinine or serum bilirubin were greater than twice the upper limit of normal or the serum transaminases were greater than three times the upper limit of normal. Patients were also excluded if they had an uncontrolled intercurrent illness such as an active infection, symptomatic heart failure, unstable angina, cardiac arrhythmia, pregnancy, HIV infection, or any other medical condition that would make the administration of romidepsin hazardous. The protocol was reviewed and approved at each institution's institutional review board, and all subjects enrolled gave written informed consent.
Treatment and response evaluation
This was an open-label, multicenter phase II study conducted through the University of Chicago Phase II Consortium. Patients were stratified into two cohorts based on the absence (cohort A) or presence (cohort B) of chromosomal aberrations such as the t(8;21), inv(16), and t(15;17) that encode proteins known to recruit HDACs. Treatment was identical for both cohorts. Romidepsin was administered i.v. over 4 h through a central or peripheral venous catheter at a dose of 13 mg/m2/d on days 1, 8, and 15 with cycles repeated every 28 days. Prophylactic antiemetics (ondansetron and prochlorperazine or the equivalent) were administered before and 24 to 72 h after therapy. During the course of the study, the protocol was amended to require that potassium and magnesium levels be maintained at a minimum level of 4 mmol/L and 2 mg/dL, respectively, to minimize the likelihood of QTc prolongation and/or cardiac arrhythmias arising from romidepsin therapy. Cardiac monitoring was employed with serial electrocardiograms and multigated acquisition scans before and after therapy. Treatment could continue indefinitely in the absence of disease progression or unacceptable adverse events. Toxicity was graded using National Cancer Institute Common Toxicity Criteria version 3.0. Patients experiencing a drug-related grade 3 or 4 nonhematologic toxicity were eligible for retreat-ment at a reduced dose of 10 mg/m2/d provided the toxicity had resolved to a grade ≤2. Response was evaluated at day 21 of each cycle of therapy using previously described criteria for AML (22).
Pharmacodynamic studies
Peripheral blood mononuclear cells were collected immediately before therapy (hour 0) and at 4 and 24 h following therapy with romidepsin on days 1 and 8 of the initial cycle of treatment for the pharmacodynamic assays described below.
Gene expression analysis—pl4ARF, MDR1, and pl5INK4B
Total RNA was extracted from the purified cells using RNA STAT-60 (Tel-Test “B”). Total RNA (1-5 μg) was converted into cDNA according to the Transcriptor Reverse Transcriptase (Roche Diagnostics) manufacturer's instructions.
Real-time reverse transcription-PCR was done with individual primer and probe sets designed for detection of pl4ARF, MDR1, and pl5INK4B (pl5) transcripts. Patient specimens, serial 5-fold dilutions of cell line cDNA, and controls lacking template were amplified within a LightCycler instrument (Roche Diagnostics). For each assay, a standard curve was generated using cDNA derived from the appropriate cell line. Each reaction was done in a 20 μL reaction mix including cDNA, Amplitaq Gold DNA polymerase (PE Biosvstems), primers, and TaqMan probe. Similarly, the ABL transcript was amplified to quantitate the target gene (pl4ARF, MDR1, and pl5) transcript copy number relative to an endogenous control. The MDR1, pl5, and ABL transcripts were amplified using previously published primer and probe sequences (23–25). The pl4ARF transcript was detected using the following primer and probe sequences: [p14ARF 5¶ TCGTGCTGATGC-TACTG, [p14ARF 3¶ CAGCGTGTCCAGGAAG, and [p14ARF probe) FAM-TTCCTAGAAGACCAGGTCATGATGATGG-TAMRA.
All patient and negative control samples were evaluated in triplicate, and absolute transcript quantities were obtained by comparing CT (threshold cycle) values to respective standard curves following PCR amplification of pl4ARF, MDR1, and pl5 transcripts. To adjust for differences in RNA integrity and cDNA synthesis efficiency, the absolute pl4ARF, MDR1, and pl5 transcript copy number was normalized to the absolute transcript copy number of ABL. The normalized transcript copy number was expressed as the ratio of the absolute pl4ARF, MDR1, and p15 transcript copy number over the ABL transcript copy number.
Chromatin immunopwdpitation/promoter-speciflc histone acetylation
Chromatin immunoprecipitation analysis was done using the Chromatin Immunoprecipitation Assay Kit (Upstate Biotechnology). Formaldehyde was added to cells to crosslink associated proteins to DNA and after sonicating extracted chromatin to an average length of 500 bp, protein-DNA complexes were immunoprecipitated with commercial (Upstate Biotechnology) antibodies to acetylated histone H3 and H4. After immunoprecipitation, DNA was extracted, purified, and analyzed by quantitative PCR using specific primer pairs for regions p15, MDR1, and ABL (control) promoters.
Global histone acetylation
Global histone H3 acetylation was analyzed by flow cytometry for detection of acetylated histone H3 as described previously (26). Briefly, f 0.5 × 106 cells were processed with permeabilization reagents according to the manufacturer's instructions (Caltag). The cells were then incubated with anti-acetylated H3 antibodies (Upstate Biotechnology), washed, and subsequently incubated with FITC-conjugated secondary antibody (Biosource International). Samples were analyzed (30,000 events) by flow cytometry (Coulter system II software). The assay was validated using Kasumi-1 cells, an AMLl-ETO-expressing cell line.
Statistical methods
For patients in cohort A, patients without t(8:21), inv(16) or other chromosomal abnormality involving AML1, a Simon's optimal two-stage design was employed. Enrollment to this cohort would be stopped for lack of efficacy if no responses were observed in the first 12 evaluable patients. Otherwise, another 25 evaluable patients would be enrolled for a total of 37 patients, and if ≥4 responses were observed, the treatment would be considered sufficiently active to warrant further study. The sample size was calculated to test a null hypothesis of 5% response rate against an alternative hypothesis of 20% response rate, with 90% power at one-sided significance level of 0.10.
A second, smaller cohort of patients (cohort B, maximum number = 10) with t(8;21), invl6, or t(15;17) or other translocations encoding proteins known to recruit HDACs would also be recruited and their responses were analyzed separately.
A random-effects linear model was used to examine whether there was a significant change in gene expression over the time points analyzed within the first 24 h of treatment (hour effect: hour 0 versus hours 4 and 24) and whether there was a significant change in the kinetics of gene expression between days and 1 and 8 of therapy (day effect: day 8 versus day 1). Logarithms of gene expression values were analyzed, because these made the values more normally distributed.
Results
Patient characteristics
Twenty patients were enrolled between June 2002 and October 2006, including 13 patients on cohort A and 7 patients on cohort B. Pretreatment patient characteristics for the study as a whole are summarized in Table 1. Baseline characteristics for patients enrolled on cohort B are summarized in Table 2. Despite the fact that these cytogenetic subgroups of AML have been shown traditionally to have a good prognosis, all patients enrolled in cohort B had multiply relapsed and/or refractory disease. In addition, almost all patients within this cohort had additional chromosomal abnormalities or other features predictive of poor risk disease, in addition to translocations involving AML1 or CBFb. One patient (patient 7) with a novel t(4;21)(q31.3;q22) was stratified into cohort B, because interphase cytogenetics (fluorescence in situ hybridization analysis) with a TEL/AML1 probe showed involvement of AML1 on chromosome band 21q22.
Table 1.
Baseline patient characteristics
| Characteristics | No. patients (%) |
|---|---|
| Age (y) | |
| < 60 | 9 (45) |
| ≥60 | 11 (55) |
| Sex | |
| Male | 8 (40) |
| Female | 12 (60) |
| Performance status | |
| 0 | 7 (35) |
| 1 | 11 (55) |
| 2 | 2 (10) |
| Diagnosis | |
| AML | |
| De novo | 14 (70) |
| Therapy related/antecedent hematologic disorder | 6 (30) |
| Bone marrow cytogenetics | |
| t(8;21)/inv(16) or t(4;21) | 7 (35) |
| Normal karyotype | 3 (15) |
| 11 q23, −5, −7, complex | 6 (30) |
| Other | 4 (20) |
| Stage of disease | |
| Primary refractory | 6 (30) |
| First relapse | 2 (10) |
| Beyond first relapse | 10 (50) |
| Previously untreated (and age > 60 y) | 2 (10) |
| Prior therapy | |
| Cytarabine | 18 (90) |
| High-dose cytarabine | 16 (80) |
| Autologous stem cell transplant | 7 (35) |
| Duration of first complete remission | |
| ≥12 mo | 2 (10) |
Table 2.
Cohort B: baseline characteristics and response to therapy
| Patient no. | Age | Stage of disease | Duration of first complete remission (mo) | Prior high-dose cytarabine | Prior autologous hematopoietic stem cell transplant | Bone marrow cytogenetics | No. cycles | Response |
|---|---|---|---|---|---|---|---|---|
| 2 | 60 | Relapse 3 | 2 | Yes | Yes | 46, XY, t(8;21) (q22;q22)/ t(1;5)(q32;q13) |
1 | Progressive disease |
| 5 | 39 | Relapse 5, refractory |
16 | Yes | Yes | 46,XX,t(8;21) (q22;q22),del(9) (q13q22) |
2 | Bone marrow blast clearance (from 68% to < 5%) after 1 cycle |
| 7 | 75 | Relapse 3 | 8 | No | No | 46,XX,t(4;21)(q31.3; q22)/46,XX |
3 | Bone marrow blast clearance (from 70% to < 5%) after 2 cycles |
| 17 | 25 | Relapse 1, refractory |
5 | Yes | Yes | 45,X,-X,t(8;21)(q22; q22)/46,XX |
1 | Progressive disease |
| 19 | 60 | Relapse 2 | 12 | Yes | No | 45X,-Yadd(1)(p36.3), t(8;21)(q22; q22) /46XY |
2 | > 50% decrease in marrow blasts (from 63% to 10%) after 1 cycle |
| 20 | 35 | Relapse 2 | 7 | Yes | Yes | 47,XY,+8,inv(16) (p13.1q22)/46XY |
2 | > 50% decrease (from 74% to 31%) in marrow blasts after 1 cycle |
| 21 | 51 | Relapse 1, refractory twice |
10 | Yes | No | 46,XX,add(7)(p22), t(8;21)(q22;q22); 47,idem,+der(21) t(8;21)(q22; q22)/46XX |
1 | > 50% decrease (from 76% to 27% blasts) in marrow blasts |
NOTE: HiDAC: high dose cytarabine. Auto HCT: autologous hematopoietic stem cell transplant. CR: complete remission.
Treatment outcome
Antileukemic activity
Of the 13 patients enrolled in cohort A 6 received 1 cycle of therapy, 1 patient received 1.66 cycles (two of the three planned doses during cycle 2), and 2 patients received 2 cycles. Four patients received <1 cycle of therapy (two of the three planned doses of therapy). All 13 patients were evaluable for toxicity, and 12 patients were evaluable for response. One patient with therapy-related leukemia and a background history of breast cancer failed to complete 1 cycle of therapy due to recurrence of a malignant pleural effusion from her breast cancer and was considered inevaluable for response. Treatment was discontinued in three other patients before completion of cycle 1 because of disease progression. There was no objective evidence of antileukemic activity in cohort A.
Of the 7 patients enrolled in group B, 1 patient received 3 cycles of therapy, 3 patients received 2 cycles of therapy, and 4 patients received 1 cycle of therapy. There were no objective responses by classic response criteria in part due to the failure of normal hematopoiesis. Antileukemic activity was observed with disappearance of bone marrow blasts after 1 and 2 cycles of therapy, respectively, in 2 patients, in the setting of a relatively cellular marrow (marrow cellularity of ≥20%). An additional 3 patients had a >50% decrease in bone marrow blasts (Table 2). None of these 5 patients had received hydroxyurea while being treated on this study. In addition, none of these 5 patients had any evidence of an intervening period of marrow aplasia, and in 3 of these patients, an increasing percentage of promyelocytes, myelocytes, and monocytes was observed in the marrow aspirates concurrent with the reduction in blast count. These findings suggest that the antileukemic effects of romidepsin in these patients were mediated through a differentiating, rather than a nonspecific cytotoxic, effect. The antileukemic response was short-lived, however, with all 5 patients developing disease progression within 30 days of achieving their best response to treatment with romidepsin.
Toxicity
The treatment was well tolerated. The most common treatment-related toxicities were mild (grade 1-2) nausea, anorexia, and fatigue occurring in 35%, 25%, and 20% of patients, respectively. Twenty-five percent of patients had grade 1 cardiac changes consisting mainly of asymptomatic nonspecific ST and T wave abnormalities (Table 3). Grade 1 and 2 liver function abnormalities occurred in 10% of patients.
Table 3.
Incidence of commonly encountered toxicities (%)
| Toxicity | Grade 1 | Grade 2 | Grade 3 | Grade 4 |
|---|---|---|---|---|
| Vomiting | 25 | 10 | 5 | 0 |
| Anorexia | 20 | 5 | 0 | 0 |
| Cardiac | 25 | 0 | 0 | 0 |
| Fatigue | 10 | 10 | 0 | 0 |
| LFT | 5 | 5 | 5 | 0 |
Pharmacodynamic studies
Target gene expression
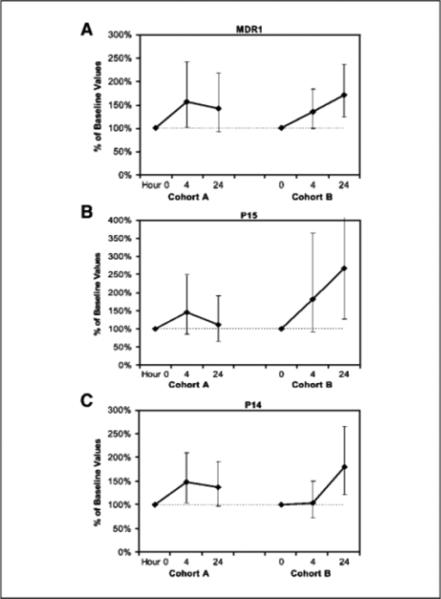
Kasumi-1 cells (an AML1-ETO-expressing cell line) were used for assay validation. This cell line was treated with romidepsin for 4 h at concentrations ranging from 1 to 100 nmol/L. A dose-dependent up-regulation in the expression of the MDR1 gene, a known target of AML1 -ETO mediated transcriptional repression, was observed. Similarly, up-regulation of pl4ARF, which is also a target of AMLl-ETO-mediated repression, was shown. To determine whether the use of romidepsin in AML in vivo results in up-regulation of specific target genes at the doses evaluated in this study, quantitative real-time reverse transcription-PCR analysis for MDR1, pl4ARF, and pl5 in patient-derived samples was done in 11 patients in cohort A and 6 patients in cohort B. These revealed a trend toward up-regulation of MDR1, pl5, and p14ARF expression by hour 24 in cohort A (42%, 12%, and 36% mean increase in expression, respectively); however, these changes were not statistically significant (Fig. 1A-C). In contrast, in cohort B, at 24 h, there was a 75% mean increase in MDR1 expression (P = 0.005), a 162% mean increase in pl5 (P = 0.01), and a 106% mean increase in pl4ARF (P < 0.0001; Fig. 1A-C). The kinetics of the changes in gene expression were similar on both days 1 and 8 of therapy in both cohorts of patients.
Fig. 1.
Romidepsin induced an up-regulation of MDR1, p15, and p14ARF genes. Quantitative PCR analysis of (A) MDR1, (B) p15, and (C) p14ARF at baseline, 4 h, and 24 h following romidepsin treatment in patients in cohort A revealed a trend toward increased gene expression, but these changes were not statistically significant. In cohort B, at 24 h, there was a 75% mean increase in (A) MDR1 expression (P = 0.005), a 162% mean increase in (B) p15 (P = 0.01), and a (C) 106% mean increase in p14 (P < 0.0001). Statistical significance was determined using a random-effects linear model. Geometric means and corresponding 95% confidence intervals are presented.
Global histone protein acetylation
This assay was optimized using Kasumi-1 cells treated with romidepsin at a dose of 10 nmol/L for 48 h. An increase in histone H3 acetylation was observed following romidepsin treatment (Fig. 2A). Similarly, an increase in histone H3 acetylation was seen in 6 of 7 (86%) patients in cohort B, including all 5 patients with evidence of antileukemic activity (Fig. 2B and C). Four of 13 (31%) patients in cohort A also showed an increase in H3 acetylation. These results confirm that romidepsin induces genome-wide histone acetylation in AML, as reported previously, and suggest that this effect may be more pronounced in patients with translocations involving AML1.
Fig. 2.
Romidepsin induced an increase in global histone (H3) acetylation. A, an increase in H3 acetylation is shown after incubation of Kasumi-1 cells with 10 nmol/L romidepsin for 48 h (red curve) compared with untreated sample (blue curve). B, representative M3 acetylation histogram on a patient (patient 17) in cohort B. Blue curve, hour 0 (pretreatment) sample. Red curve, 24 h post-therapy sample and demonstrates an increase in H3 acetylation. C, an increase in global histone H3 acetylation after therapy is shown in 6 of 7 (86%) patients in cohort B.
Promoter-specific histone acetylation
In 4 patients with sufficient material preserved, chromatin immunoprecipitation analysis was carried out for promoter-specific changes in histone H3 and H4 acetylation. An induction in H3 acetylation was observed at both pl5 and MDR1 promoters (Fig. 3A and B), in keeping with the gene expression data that showed an up-regulation of pl5 and MDR1 expression in the same 4 patients.
Fig. 3.
Romidepsin-induced histone acetylation H3 acetylation at the (A) p15 promoter and (B) MDR1 promoter in 4 patients analyzed at 4 h after therapy compared with pre-therapy samples.
Discussion
This phase II study of romidepsin in relapsed and refractory AML shows that this HDACI has a differential antileukemic activity in patients with CBF AML [predominantly t(8;21)] and that this was associated with an up-regulation of AML1 target genes. These findings are in keeping with observations in vitro that have established that t(8;21) AML is sensitive to therapy with various HDACIs including romidepsin (7, 16–19, 27). Despite the in vitro evidence suggesting an inherent sensitivity of this cytogenetic subset of AML to this class of agents, this is the first study to actually show antileukemic activity in vivo in a cohort of patients with t(8;21) AML and other translocations involving the AML1 gene.
Romidepsin has been investigated in a phase I trial (28) conducted at this dose and schedule in two groups of patients with chronic lymphocytic leukemia and AML, respectively. In that trial that involved 10 AML patients, there were no patients with CBF AML. There were no objective responses reported, nor was there evidence of significant antileukemic activity (such as clearance of bone marrow blasts). The phase I experience mirrors our experience in the 13 patients enrolled in cohort A of this study. The authors of the phase I trial also observed an increase in global H3 and H4 acetylation as well as acetylation at the p21 promoter in the patients studied. Another recendy published pilot trial of romidepsin in myeloid neoplasms included 9 patients with AML and 3 with MDS, none of whom had CBF AML. Of these, 1 patient with previously untreated AML and a del (7q) achieved a complete remission after one infusion of romidepsin administered at a higher dose (18 mg/m2) than used in our study. The response duration was brief, however, with recurrent cytopenias being documented 29 days after the achievement of CR (29). Other clinical trials investigating HDACIs in AML have recruited very few patients with CBF AML and have seldom enrolled patients with t(8;21) (30–36).
In our own trial, an antileukemic effect was seen in the majority of CBF AML patients studied, and global histone H3 acetylation was observed following romidepsin therapy in several patients, particularly those with CBF AML. In addition, up-regulation of MDRl and pl4ARF (both targets of AML1-ETO mediated transcriptional repression; refs. 9, 10) were observed, and these effects were most pronounced in the CBF AML subgroup as we had originally hypothesized. Interestingly, although p15, a well-described target of epigenetic silencing in AML, was also up-regulated in several patients in both subgroups, the increase in expression after romidepsin treatment was most striking, as a whole, in patients with CBF AML.
Although we observed an antileukemic effect of romidepsin in several patients with CBF AML, including a patient with a novel t(4;21) shown by fluorescence in situ hybridization analysis to involve AML1, these effects were short in duration. The reason for this finding remains to be determined. One likely explanation is that the patients enrolled on this study had advanced disease and the majority had evidence of clonal evolution with additional chromosomal aberrations present besides those involving AML1 or CBFb, implicating other pathways. Because romidepsin up-regulates MDR1 in vitro and in vivo (37, 38), a finding that has been described with several other HDACIs and observed in our study in both subgroups of patients, and romidepsin is a known substrate for MDR1, (39) another possibility is that the drug may mediate its own resistance in vivo. In vitro, acquired resistance to romidepsin has been shown from reversible MDR1 induction caused by histone hyperacetylation at the MDR1 promoter (37), and MDR1-mediated drug efflux has also been shown in cell lines following romidepsin exposure (40). Although we were able to show for the first time that romidepsin induced histone acetylation at the MDR1 promoter in vivo, insufficient material was available to assess the level and function of MDR1 and other MDR-related proteins. Future clinical trials investigating this agent should, therefore, evaluate the possibility of up-regulation of MDR1 and MDRl-mediated drug efflux as a potential mechanism of chemoresistance to romidepsin. If true, this may provide a potential rationale for therapeutic combinations with MDR inhibitors.
In summary, our experience in this trial provides in vivo evidence to support a proof of principle for further clinical investigation of HDACIs in CBF AML and particularly t(8;21) AML. Although CBF AML has a better prognosis than other subtypes of AML, f 50% of patients relapse, with the prognosis of patients with the t(8;21) apparently worse than those with the inv(16) (41). Recently, the AML1-ETO fusion protein has been shown to associate with DNA methyltransferase 1 (27) in addition to recruiting HDACs. Therefore, future studies in this area should consider combining HDACIs with agents such as DNA methyltransferase inhibitors that have been shown to be synergistic with HDACIs in vitro in a variety of malignancies including t(8;21) AML (17, 27, 42, 43). Our pharmacodynamic studies raise the possibility of MDR1 up-regulation as a potential mechanism of chemoresistance to romidepsin in AML and underscore the need for additional investigation of this issue in preclinical studies and clinical trials investigating romidepsin and similar agents for the treatment of AML.
Translational Relevance.
This phase II multicenter study was based on the hypothesis that the HDACI romidepsin could cause transcriptional derepression, up-regulation of specific target genes in AML, and differentiation of the leukemic clone. The study was designed to include a cohort of patients with CBF AML, a subgroup that we hypothesized may be particularly amenable to therapy with HDACI based on the presence of chromosome abnormalities such as t(8;21), known to recruit HDACs. The article provides the first in vivo evidence of clinical and molecular effects of a HDACI in a cohort with CBF AML. These results serve as a proof of principle for further clinical investigation of HDACIs in CBF AML, particularly t(8;21) AML.
Acknowledgments
Grant support: National Cancer Institute/Cancer Therapy Evaluation Program contract N01-CM-62201 in collaboration with Gloucester Pharmaceuticals, American Society of Oncology Career Development Award (O.M. Odenike), American Cancer Society Institutional Grant (O. M. Odenike), and National Cancer InstituteTranslational Research funds (O.M. Odenike and S. Alkan).
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with18 U.S.C. Section 1734 solely to indicate this fact.
Disclosure of Potential Conflicts of Interest
O. Odenike has received research support from MGI, Curagen, and TopoTarget, is on the advisory board for MGI, and has an ownership interest in Gloucester Pharmaceuticals. O. Odenike is an inventor on a patent that relates to the use of HDACIs for treating AML. The patent is owned by the University of Chicago.
References
- 1.Redner RL. Chromatin remodeling and leukemia: new therapeutic paradigms. Blood. 1998;94:417–28. [PubMed] [Google Scholar]
- 2.Sobulo OM, Borrow J, Tomek R, et al. MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t(11;16)(q23;p13.3). Proc Natl Acad Sci U S A. 1997;94:8732–7. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borrow J, Stanton VP, Jr., Andresen JM, et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 4.Taki T, Sako M, Tsuchida M, Hayashi Y. The t(11;16)(q23;p13) translocation in myelodysplastic syndrome fuses the MLL gene to the CBP gene. Blood. 1997;89:3945–50. [PubMed] [Google Scholar]
- 5.Hiebert SW, Lutterbach B, Amann J. Role of compressors in transcriptional repression mediated by thet(8;21), t(16;21), t(12;21), and inv(16) fusion proteins. Curr Opin Hematol. 2001;8:197–200. doi: 10.1097/00062752-200107000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci U S A. 1998;95:10860–5. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Saunthararajah Y, Redner RL, Liu JM. Inhibitors of histone deacetylase relieve ETO-mediated repression and induce differentiation of AML1-ETO leukemia cells. Cancer Res. 1999;59:2766–9. [PubMed] [Google Scholar]
- 8.Lutterbach B, Hou Y, Durst KL, Hiebert SW. The inv(16) encodes an acute myeloid leukemia 1 transcriptional corepressor. Proc Natl Acad Sci U S A. 1999;96:12822–7. doi: 10.1073/pnas.96.22.12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linggi B, Muller-Tidow C, van de Locht L, et al. The t(8;21) fusion protein, AML1 ETO, specifically represses the transcription of the p14(ARF) tumor suppressor in acute myeloid leukemia. Nat Med. 2002;8:743–50. doi: 10.1038/nm726. [DOI] [PubMed] [Google Scholar]
- 10.Lutterbach B, Sun D, Schuetz J, Hiebert SW. The MYND motif is required for repression of basal transcription from the multidrug resistance 1 promoter by the t(8;21) fusion protein. Mol Cell Biol. 1998;18:3604–11. doi: 10.1128/mcb.18.6.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fecteau KA, Mei J, Wang HC. Differential modulation of signaling pathways and apoptosis of ras-transformed 10TI/2 cells by the depsipeptide FR901228. J Pharmacol Exp Ther. 2002;300:890–9. doi: 10.1124/jpet.300.3.890. [DOI] [PubMed] [Google Scholar]
- 12.Nakajima H, Kim YB, Terano H, Yoshida M, Horinouchi S. FR901228, a potent antitumor antibiotic, is a novel histone deacetylase inhibitor. Exp Cell Res. 1998;241:126–33. doi: 10.1006/excr.1998.4027. [DOI] [PubMed] [Google Scholar]
- 13.Sandor V, Bakke S, Robey RW, et al. Phase I trial of the histone deacetylase inhibitor, depsipeptide (FR901228, NSC 630176), in patients with refractory neoplasms. Clin Cancer Res. 2002;8:718–28. [PubMed] [Google Scholar]
- 14.Sandor V, Senderowicz A, Mertins S, et al. P21-dependent g (1) arrest with downregulation of cyclin D1 and upregulation of cyclin E by the histone deacetylase inhibitor FR901228. Br J Cancer. 2000;83:817–25. doi: 10.1054/bjoc.2000.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueda H, Nakajima H, Hori Y, Goto T, Okuhara M. Action of FR901228, a novel antitumor bicyclic depsipeptide produced by Chromobacterium violaceum no. 968, on Ha-ras transformed NIH3T3 cells. Biosci Biotechnol Biochem. 1994;58:1579–83. doi: 10.1271/bbb.58.1579. [DOI] [PubMed] [Google Scholar]
- 16.Gozzini A, Rovida E, Dello Sbarba P, Galimberti S, Santini V. Butyrates, as a single drug, induce histone acetylation and granulocytic maturation: possible selectivity on core binding factor-acute myeloid leukemia blasts. Cancer Res. 2003;63:8955–61. [PubMed] [Google Scholar]
- 17.Klisovic MI, Maghraby EA, Parthun MR, et al. Depsipeptide (FR 901228) promotes histone acetylation, gene transcription, apoptosis and its activity is enhanced by DNA methyltransferase inhibitors in AML1/ETO-positive leukemic cells. Leukemia. 2003;17:350–8. doi: 10.1038/sj.leu.2402776. [DOI] [PubMed] [Google Scholar]
- 18.Liu S, Klisovic RB, Vukosavljevic T, et al. Targeting AML1/ETO-histone deacetylase repressor complex: a novel mechanism for valproic acid-mediated gene expression and cellular differentiation in AML1/ETO-positive acute myeloid leukemia cells. J Pharmacol ExpTher. 2007;321:953–60. doi: 10.1124/jpet.106.118406. [DOI] [PubMed] [Google Scholar]
- 19.Yang G, Thompson MA, Brandt SJ, Hiebert SW. Histone deacetylase inhibitors induce the degradation of the t(8;21) fusion oncoprotein. Oncogene. 2007;26:91–101. doi: 10.1038/sj.onc.1209760. [DOI] [PubMed] [Google Scholar]
- 20.Ferrara FF, Fazi F, Bianchini A, et al. Histone deacetylase-targeted treatment restores retinoic acid signaling and differentiation in acute myeloid leukemia. Cancer Res. 2001;61:2–7. [PubMed] [Google Scholar]
- 21.Gottlicher M, Minucci S, Zhu P, et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–78. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheson BD, Bennett JM, Kopecky KJ, et al. Revised recommendations of the international working group for diagnosis, standardization of response criteria, treatment outcomes, and reporting standards for therapeutic trials in acute myeloid leukemia. J Clin Oncol. 2003;21:4642–9. doi: 10.1200/JCO.2003.04.036. [DOI] [PubMed] [Google Scholar]
- 23.Olesen LH, Norgaard JM, Pallisgaard N, Bukh A, Hokland P. Validation and clinical implication of a quantitative real-time PCR determination of MDR1 gene expression: comparison with semi-quantitative PCR in 101 patients with acute myeloid leukemia. Eur J Haematol. 2003;70:296–303. doi: 10.1034/j.1600-0609.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 24.Tessema M, Langer F, Dingemann J, Ganser A, Kreipe H, Lehmann U. Aberrant methylation and impaired expression of the p15(INK4b) cell cycle regulatory gene in chronic myelomonocytic leukemia (CMML). Leukemia. 2003;17:910–8. doi: 10.1038/sj.leu.2402891. [DOI] [PubMed] [Google Scholar]
- 25.Stock W, Yu D, Karrison T, et al. Quantitative real-time RT-PCR monitoring of BCR-ABL in chronic myelogenous leukemia shows lack of agreement in blood and bone marrow samples. Int J Oncol. 2006;28:1099–103. [PubMed] [Google Scholar]
- 26.Amin HM, Saeed S, Alkan S. Histone deacetylase inhibitors induce caspase-dependent apoptosis and downregulation of daxx in acute promyelocytic leukaemia with t(15;17). Br J Haematol. 2001;115:287–97. doi: 10.1046/j.1365-2141.2001.03123.x. [DOI] [PubMed] [Google Scholar]
- 27.Liu S, Shen T, Huynh L, et al. Interplay of RUNX1/MTG8 and DNA methyltransferase 1 in acute myeloid leukemia. Cancer Res. 2005;65:1277–84. doi: 10.1158/0008-5472.CAN-04-4532. [DOI] [PubMed] [Google Scholar]
- 28.Byrd JC, Marcucci G, Parthun MR, et al. A phase 1 and pharmacodynamic study of depsipeptide (FK228) in chronic lymphocytic leukemia and acute myeloid leukemia. Blood. 2005;105:959–67. doi: 10.1182/blood-2004-05-1693. [DOI] [PubMed] [Google Scholar]
- 29.Klimek VM, Fircanis S, Maslak P, et al. Tolerability, pharmacodynamics, and pharmacokinetics studies of depsipeptide (romidepsin) in patients with acute myelogenous leukemia or advanced myelodysplastic syndromes. Clin Cancer Res. 2008;14:826–32. doi: 10.1158/1078-0432.CCR-07-0318. [DOI] [PubMed] [Google Scholar]
- 30.Blum W, Klisovic RB, Hackanson B, et al. Phase I study of decitabine alone or in combination with valproic acid in acute myeloid leukemia. J Clin Oncol. 2007;25:3884–91. doi: 10.1200/JCO.2006.09.4169. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Manero G, Kantarjian HM, Sanchez-Gonzalez B, et al. Phase 1/2 study of the combination of 5-aza-¶deoxycytidine with valproic acid in patients with leukemia. Blood. 2006;108:3271–9. doi: 10.1182/blood-2006-03-009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Manero G, Yang H, Bueso-Ramos C, et al. Phase 1 study of the histone deacetylase inhibitor vorinostat (suberoylanilide hydroxamic acid [SAHA]) in patients with advanced leukemias and myelodysplastic syndromes. Blood. 2008;111:1060–6. doi: 10.1182/blood-2007-06-098061. [DOI] [PubMed] [Google Scholar]
- 33.Gojo I, Jiemjit A, Trepel JB, et al. Phase 1 and pharmacologic study of MS-275, a histone deacetylase inhibitor, in adults with refractory and relapsed acute leukemias. Blood. 2007;109:2781–90. doi: 10.1182/blood-2006-05-021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gore SD, Baylin S, Sugar E, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66:6361–9. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- 35.Kuendgen A, Schmid M, Schlenk R, et al. The histone deacetylase (HDAC) inhibitor valproic acid as monotherapy or in combination with all-trans retinoic acid in patients with acute myeloid leukemia. Cancer. 2006;106:112–9. doi: 10.1002/cncr.21552. [DOI] [PubMed] [Google Scholar]
- 36.Soriano AO, Yang H, Faderl S, et al. Safety and clinical activity of the combination of 5-azacytidine, valproic acid, and all-trans retinoic acid in acute myeloid leukemia and myelodysplastic syndrome. Blood. 2007;110:2302–8. doi: 10.1182/blood-2007-03-078576. [DOI] [PubMed] [Google Scholar]
- 37.Xiao JJ, Huang Y, Dai Z, et al. Chemoresistance to depsipeptide FK228 [(E)-(1S,4S,10S,21R)-7-[(Z)-ethylidene]-4,21-diisopropyl-2-oxa-12,13-dithia-5,8,20,23-tetraazabicyclo[8,7,6]-tricos-16-ene-3,6,9,22-pentanone] is mediated by reversible MDR1 induction in human cancer cell lines. J Pharmacol Exp Ther. 2005;314:467–75. doi: 10.1124/jpet.105.083956. [DOI] [PubMed] [Google Scholar]
- 38.Robey RW, Zhan Z, Piekarz RL, Kayastha GL, Fojo T, Bates SE. Increased MDR1 expression in normal and malignant peripheral blood mononuclear cells obtained from patients receiving depsipeptide (FR901228, FK228, NSC630176). Clin Cancer Res. 2006;12:1547–55. doi: 10.1158/1078-0432.CCR-05-1423. [DOI] [PubMed] [Google Scholar]
- 39.Lee JS, Paull K, Alvarez M, et al. Rhodamine efflux patterns predict P-glycoprotein substrates in the National Cancer Institute drug screen. Mol Pharmacol. 1994;46:627–38. [PubMed] [Google Scholar]
- 40.Xiao JJ, Foraker AB, Swaan PW, et al. Efflux of depsipeptide FK228 (FR901228, NSC-630176) is mediated by P-glycoprotein and multidrug resistance-associated protein 1. J Pharmacol Exp Ther. 2005;313:268–76. doi: 10.1124/jpet.104.072033. [DOI] [PubMed] [Google Scholar]
- 41.Marcucci G, Mrozek K, Ruppert AS, et al. Prognostic factors and outcome of core binding factor acute myeloid leukemia patients with t(8;21) differ from those of patients with inv(16): a Cancer and Leukemia Group B study. J Clin Oncol. 2005;23:5705–17. doi: 10.1200/JCO.2005.15.610. [DOI] [PubMed] [Google Scholar]
- 42.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–7. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 43.Gozzini A, Santini V. Butyrates and decitabine cooperate to induce histone acetylation and granulocytic maturation of t(8;21) acute myeloid leukemia blasts. Ann Hematol. 2005;84(Suppl 1):54–60. doi: 10.1007/s00277-005-0006-z. [DOI] [PubMed] [Google Scholar]