Abstract
Background
Smoking is a notorious risk factor for chronic mucus hypersecretion (CMH). CMH frequently occurs in Chronic Obstructive Pulmonary Disease (COPD). The question arises whether the same single nucleotide polymorphisms (SNPs) are related to CMH in smokers with and without COPD.
Methods
We performed two genome wide association studies on CMH under an additive genetic model in male heavy smokers (≥20 pack-years) with COPD (n=849, 39.9% CMH) and without COPD (n=1,348, 25.4% CMH), followed by replication and meta-analysis in comparable populations, and assessment of the functional relevance of significantly associated SNPs.
Results
GWA analysis on CMH in COPD and non-COPD yielded no genome wide significance after replication. In COPD, our top SNP (rs10461985, p=5.43×10−5) was located in the GDNF-antisense gene that is functionally associated with the GDNF gene. Expression of GDNF in bronchial biopsies of COPD patients was significantly associated with CMH (p=0.007). In non-COPD, 4 SNPs had a p-value <10−5 in the meta-analysis, including a SNP (rs4863687) in the MAML3 gene, the T-allele showing modest association with CMH (p=7.57×10−6, OR=1.48) and with significantly increased MAML3 expression in lung tissue (p=2.59×10−12).
Conclusions
Our data suggest the potential for differential genetic backgrounds of CMH in individuals with and without COPD.
Introduction
Chronic mucus hypersecretion (CMH) can be present in individuals with and without COPD. The prevalence of CMH varies from 3.5% to 12.7% in the general population depending on the population studied and the CMH definition used [1,2]. The prevalence of CMH is much higher In individuals with COPD (30%) and increases with the severity of airflow limitation [3,4]. Some risk factors for COPD and CMH overlap, like smoking, occupational exposures and bacterial infections [5-9].
However, not all heavy smokers have CMH, which may be explained by a genetic contribution to CMH, as evidenced by familial aggregation of mucus overproduction and higher concordance of CMH in monozygotic than in dizygotic twins [10-12]. So far, only two genetic studies on CMH have been published. One study suggested that CTLA4 is associated with chronic bronchitis in individuals with COPD without a direct association with COPD itself [13]. A second study showed that a SNP (rs6577641) in the SATB1 gene was strongly associated with CMH in a heavy smoking population [14].
Since not all individuals with COPD have CMH and conversely not all individuals with CMH have COPD, the question arises whether similar or differential genetic factors are involved in the development of CMH in individuals with and without COPD. Therefore, we performed a genome wide association study on CMH in a group of male individuals with COPD and a group without COPD, from the same heavy smoking general population based cohort (NELSON) [15]. Subsequently, we evaluated our findings on the association with CMH in replication cohorts including individuals with and without COPD, and searched for features of our most significant findings.
Methods
Ethics Statement
The Dutch Ministry of Health and the Medical Ethics Committee of the hospital approved the study protocol for the Dutch centers. Ethics approval and written informed consent was obtained from all participants in the studies. For detailed information, see Supplement.
Identification population
Male Caucasian participants from Groningen and Utrecht were included from the Dutch NELSON study [15], a heavy smoking population based lung cancer screening study. Information on CMH and smoking behavior was collected by questionnaires as published previously [14]. Spirometry was performed according to the European Respiratory Society guidelines, including forced expiratory volume in 1 sec (FEV1) and forced vital capacity (FVC), without using a bronchodilator [16]. COPD was defined as FEV1/FVC < 0.70. To assess whether different genetic factors contribute to the presence of CMH in smoking individuals with and without COPD, we conducted two genome wide association (GWA) studies; one in NELSON-individuals with COPD (NELSON-COPD) and a second in NELSON participants without COPD (NELSON-non-COPD) [15].
Replication populations
Top hits associated with CMH in NELSON-COPD were in silico analyzed in individuals with ≥ 5 pack-years smoking and FEV1/FVC < 0.70 from four independent, Caucasian COPD-cohorts: GenKOLS, COPDGene, ECLIPSE and MESA [17-20]. Subsequently meta-analyses were performed across these replication cohorts, and across NELSON-COPD and these replication cohorts.
Top hits associated with CMH in NELSON-non-COPD, were analyzed in the general population cohort LifeLines by selecting individuals without COPD and ≥ 5 pack-years smoking.
A description of the replication cohorts is given in the supplementary file. Details on the identification and replication cohorts concerning genotyping method, genotyping imputation software, and CMH and COPD definitions are given in Supplementary Table 1.
Functional relevance of identified top SNPs
We assessed whether the top SNPs in individuals with and without COPD were associated with gene expression levels in human lungs. Expression quantitative trait loci (eQTLs) were identified in 1,095 lung tissues from three independent cohorts recruited from Laval University, University of British Columbia, and University of Groningen as described previously [21].
Additionally, we assessed whether CMH was associated with mRNA expression of candidate genes in bronchial biopsies from 77 COPD participants in the Groningen and Leiden Universities study of Corticosteroids in Obstructive Lung Disease study (GLUCOLD) [22,23]. Details on the methods are given in the Supplement.
Statistical analysis
General characteristics of CMH-cases and controls were compared using Student's t- and Mann-Whitney-U tests for continuous variables as appropriate and χ2 tests for dichotomous variables with SPSS 20.0. Quality control (QC) of genotyping, regression- and meta-analyses were performed with PLINK 1.07 [24]. QC was performed in cases and controls according to the following exclusion criteria: SNPs with call rate < 95%, Minor Allele Frequency (MAF) < 0.05, proportion of individuals for which no genotype was called (mind) < 0.95 and Hardy Weinberg equilibrium (HWE) p < 0.0001. Ethnic outliers, duplicates and relatives were removed (based on the top two components from multidimensional scaling). Logistic regression analysis under an additive genetic model with adjustment for center and smoking (ex/current) was used to identify SNPs associated with CMH in NELSON participants in two separate analyses. SNPs were included for replication if there was any nominally significant association between CMH and a SNP (p < 2.0 × 10−4) and analyzed using additional adjustment for gender as the replication cohorts also included females.
Results
Populations
After QC, out of 3,005 NELSON participants, 2,799 remained. Females were excluded as only 48 were present after QC. 2,194 NELSON males with complete information on CMH, spirometry and smoking history were analyzed including 849 with and 1,345 without COPD. The prevalence of CMH in individuals with COPD was 39.8% (n = 338) and in individuals without COPD 25.4% (n = 342). Demographic and clinical characteristics of NELSON participants with COPD and of the four COPD-replication cohorts are presented in Table 1 [17-20].
Table 1.
Characteristics of individuals with and without CMH, in NELSON-COPD and in replication COPD cohorts.
| NELSON | GenKOLS | COPDGene | ECLIPSE | MESA | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| +CMH | −CMH | p | +CMH | −CMH | p | +CMH | −CMH | p | +CMH | −CMH | p | +CMH | −CMH | p | |
| N (%) |
338 (39.9) |
511 (60.1) |
487 (57.1) |
364 (42.7) |
182 (36.6) |
315 (63.4) |
643 (38.1) |
1,045 (61.9) |
50 (21.4) |
184 (78.6) |
|||||
| Age, yrs | 61.5 (5.9) |
61.2 (5.4) |
0.44 | 65.8 (10.0) |
65.2 (10.0) |
0.36 | 63.9 (7.8) |
65.2 (8.3) |
0.09 | 62.9 (7.6) |
64.1 (6.8) |
0.37 | 64.8 (9.4) |
65.6 (9.1) |
0.61 |
| Female, % | 0 | 0 | 0 | 0 | 39.0 | 57.1 | 0.001 | 24.7 | 38.5 | <0.001 | 58.0 | 64.7 | 0.39 | ||
| Pack-years | 38.7 (20- 140) |
38.7 (20- 119) |
0.044 | 33.2 (5- 119) |
31.2 (5- 130) |
0.16 | 47.8 (11- 238) |
47.6 (10- 146) |
0.16 | 45.0 (6- 220) |
45.0 (10- 205) |
0.10 | 47.0 (6- 135) |
40.6 (5- 167) |
0.19 |
| Current smoking, % |
74.8 | 50.2 | <0.001 | 53.5 | 39.7 | <0.001 | 42.9 | 23.5 | <0.001 | 45.1 | 27.0 | <0.001 | 38.0 | 12.5 | <0.001 |
| FEV1, %predicted | 81.8 (19.8) |
86.3 (7.1) |
<0.001 | 48.2 (17.5) |
54.0 (16.8) |
<0.001 | 46.5 (18.1) |
49.9 (18.5) |
0.044 | 46.7 (15.4) |
48.2 (15.7) |
<0.001 | 67.5 (18.6) |
75.4 (17.4) |
0.005 |
| FEV1/FVC, % | 60.1 (8.6) |
62.5 (7.1) |
<0.001 | 49.7 (13.4) |
53.5 (12.2) |
<0.001 | 45.5 (11.9) |
48.6 (13.8) |
0.007 | 44.3 (11.8) |
49.7 (13.3) |
<0.001 | 59.4 (10.5) |
62.6 (7.2) |
0.014 |
CMH = chronic mucus hypersecretion; Mean (standard deviation) shown for normally distributed continuous data and median (range) for non-normally distributed continuous data.
Demographic and clinical characteristics of NELSON participants without COPD and the replication cohort LifeLines are presented in Table 2.
Table 2.
Characteristics of individuals with and without CMH, in NELSON-non-COPD and in the Lifelines cohort.
| NELSON | LifeLines | |||||
|---|---|---|---|---|---|---|
| + CMH | - CMH | p | + CMH | - CMH | p | |
| N, (% ) | 342 (25.4) | 1,006 (74.6) | 130 (5.3) | 2,313 (94.7) | ||
| Age, yrs | 59.6 (5.3) | 59.8 (5.3) | 0.61 | 47.2 (10.7) | 47.4 (9.7) | 0.82 |
| Female, % | 0 | 0 | 46.2 | 53.4 | 0.11 | |
| Pack-years | 38.0 (22-140) | 34.2 (20-133) | 0.029 | 15.5 (5-84) | 13.0 (5-75) | <0.001 |
| Current smoking, % | 70.8 | 45.2 | <0.001 | 60.0 | 43.1 | <0.001 |
| FEV1, %predicted | 105.2 (13.1) | 107.6 (13.4) | 0.62 | 100.5 (14.2) | 103.6 (12.8) | 0.008 |
| FEV1/FVC, % | 78.0 (4.6) | 78.1 (4.5) | 0.003 | 77.1 (4.4) | 78.0 (4.8) | 0.040 |
CMH = chronic mucus hypersecretion; Mean (standard deviation) shown for normally distributed continuous data and median (range) for non-normally distributed continuous data.
In all cohorts, irrespective of COPD status, individuals with CMH had significantly lower lung function and were significantly more often current smokers compared to individuals without CMH.
Genome wide analyses in NELSON participants with COPD
After QC, out of 620,901 SNPs 522,636 remained for GWA analysis in 849 individuals with COPD, 338 with and 511 without CMH. The QQ-plot showed no indication of population stratification (λ = 1.002). The p-values of the GWA study are presented in the Manhattan plot (Figure 1). A total of 78 SNPs were associated with CMH at a p < 2 × 10−4 (Table 3). SNP rs626326 located in an intron in the StAR-related lipid transfer (START) domain containing 13 gene (STARD13) on chromosome 13q13.1 showed the strongest association with CMH (p = 3.99 × 10−6, OR 1.632).
Figure 1.

Quantile-quantile plot (left) and Manhattan plot (right) of GWA results for association of SNPs with CMH in NELSON participants with COPD.
Table 3.
Association of SNPs with CMH in identification analysis (NELSON-COPD) and in replication cohorts and subsequent meta-analysis across identification and replication cohorts.
| CHR | SNP | NELSON-COPD | GenKOLS | COPDGene | ECLIPSE | MESA | Meta-analysis across identification (NELSON-COPD) and replication cohorts (GenKOLS, COPDGene, ECLIPSE and MESA) | Direction of effect | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rank | p | OR | p | OR | p | OR | p | OR | p | OR | rank | p# | OR# | Q | |||
| 1 | rs2810587 | 33 | 9.90E-05 | 1.59 | 3.99E-01 | 1.10 | 3.10E-01 | 0.85 | 2.30E-01 | 0.90 | 6.49E-02 | 0.57 | 77 | 9.88E-01 | 1 | <0.001 | + + - - - |
| 1 | rs17518769 | 28 | 8.94E-05 | 2.03 | 1.49E-01 | 0.73 | 1.00E+00 | 1.00 | 3.00E-01 | 1.15 | 8.11E-02 | 0.55 | 70 | 8.59E-01 | 1.04 | 0.001 | + - 0 + - |
| 1 | rs10753077 | 3 | 1.65E-05 | 1.79 | 4.95E-01 | 1.10 | 8.20E-01 | 1.05 | 6.70E-01 | 1.04 | 7.04E-01 | 1.15 | 14 | 5.44E-03 | 1.2 | 0.020 | + + + 0 + |
| 1 | rs12410049 | 49 | 1.38E-04 | 1.79 | 7.96E-01 | 1.04 | 4.20E-01 | 0.84 | 2.90E-01 | 0.88 | 9.02E-01 | 0.96 | 61 | 6.43E-01 | 1.07 | 0.004 | + 0 - - - |
| 1 | rs2001475 | 50 | 1.38E-04 | 1.79 | 7.96E-01 | 1.04 | 4.20E-01 | 0.84 | 2.90E-01 | 0.88 | 9.28E-01 | 0.97 | 60 | 6.37E-01 | 1.08 | 0.004 | + 0 - - 0 |
| 1 | rs3123695 | 36 | 1.08E-04 | 1.85 | 2.12E-01 | 0.78 | 7.40E-01 | 0.92 | 3.90E-01 | 0.90 | 6.49E-01 | 0.83 | 72 | 8.84E-01 | 1.03 | 0.002 | + - - - - |
| 2 | rs4671197 | 63 | 1.67E-04 | 1.50 | 6.85E-01 | 0.96 | 3.90E-01 | 1.15 | 3.90E-01 | 1.07 | 5.82E-01 | 0.86 | 24 | 2.01E-02 | 1.13 | 0.030 | + 0 + + - |
| 2 | rs216626 | 25 | 7.95E-05 | 1.89 | 2.44E-01 | 1.22 | 8.80E-01 | 1.03 | 2.50E-01 | 1.14 | 1.93E-01 | 0.67 | 13 | 4.94E-03 | 1.23 | 0.016 | + + 0 + - |
| 2 | rs216640 | 59 | 1.55E-04 | 1.86 | 2.55E-01 | 1.21 | 8.40E-01 | 1.04 | 2.70E-01 | 1.13 | 1.84E-01 | 0.67 | 17 | 8.06E-03 | 1.21 | 0.020 | + + 0 + - |
| 2 | rs3821072 | 20 | 6.69E-05 | 1.93 | 2.00E-01 | 1.25 | 7.90E-01 | 1.06 | 3.50E-01 | 1.11 | 1.89E-01 | 0.67 | 15 | 6.25E-03 | 1.22 | 0.013 | + + + + - |
| 2 | rs6760631 | 68 | 1.78E-04 | 0.60 | 4.55E-01 | 0.91 | 5.00E-02 | 1.35 | 5.20E-01 | 1.06 | 4.37E-02 | 0.61 | 43 | 3.84E-01 | 0.88 | <0.001 | - - + + - |
| 3 | rs6442701 | 70 | 1.82E-04 | 0.66 | 7.29E-01 | 0.96 | 3.90E-01 | 0.88 | 9.50E-01 | 1.00 | 1.57E-01 | 1.45 | 32 | 5.92E-02 | 0.91 | 0.010 | - 0 - 0 + |
| 3 | rs6799163 | 73 | 1.90E-04 | 0.66 | 7.11E-01 | 0.96 | 4.70E-01 | 0.90 | 9.30E-01 | 0.99 | 25 | 2.44E-02 | 0.89 | 0.023 | - 0 - 0 x | ||
| 3 | rs492476 | 67 | 1.76E-04 | 0.64 | 1.14E-01 | 1.20 | 1.10E-01 | 1.28 | 7.90E-01 | 0.98 | 4.64E-01 | 1.24 | 73 | 9.28E-01 | 1.01 | 0.001 | - + + - + |
| 3 | rs4420851 | 69 | 1.80E-04 | 0.65 | 1.20E-01 | 1.19 | 1.30E-01 | 1.26 | 6.70E-01 | 0.96 | 4.79E-01 | 1.23 | 78 | 9.95E-01 | 1 | 0.001 | - + + 0 + |
| 3 | rs547906 | 39 | 1.13E-04 | 1.54 | 9.05E-01 | 0.99 | 7.00E-02 | 1.29 | 2.10E-01 | 0.90 | 9.57E-01 | 0.99 | 40 | 3.22E-01 | 1.12 | 0.002 | + 0 + - 0 |
| 3 | rs12632517 | 29 | 9.02E-05 | 1.56 | 9.23E-01 | 1.01 | 1.00E-01 | 1.27 | 5.00E-02 | 0.85 | 9.28E-01 | 0.98 | 45 | 4.12E-01 | 1.11 | <0.001 | + 0 + - 0 |
| 3 | rs4515036 | 40 | 1.16E-04 | 1.55 | 9.76E-01 | 1.00 | 1.00E-01 | 1.27 | 4.00E-02 | 0.85 | 9.28E-01 | 0.98 | 46 | 4.31E-01 | 1.11 | <0.001 | + 0 + - 0 |
| 3 | rs9826025 | 30 | 9.30E-05 | 1.56 | 8.16E-01 | 0.97 | 1.00E-01 | 1.27 | 4.00E-02 | 0.85 | 9.96E-01 | 1.00 | 47 | 4.43E-01 | 1.11 | <0.001 | + 0 + - 0 |
| 3 | rs3856798 | 66 | 1.74E-04 | 0.55 | 1.93E-01 | 1.21 | 5.50E-01 | 1.13 | 7.70E-01 | 1.03 | 2.33E-02 | 2.63 | 63 | 7.45E-01 | 1.09 | <0.001 | - + + 0 + |
| 3 | rs2447616 | 47 | 1.34E-04 | 0.54 | 2.02E-01 | 1.21 | 5.10E-01 | 1.14 | 7.60E-01 | 1.03 | 3.48E-02 | 2.52 | 69 | 8.37E-01 | 1.04 | <0.001 | - + + 0 + |
| 3 | rs9831604 | 55 | 1.47E-04 | 0.55 | 1.73E-01 | 1.22 | 5.10E-01 | 1.14 | 8.40E-01 | 1.02 | 2.30E-02 | 2.62 | 67 | 7.94E-01 | 1.05 | <0.001 | - + + 0 + |
| 3 | rs339668 | 34 | 1.02E-04 | 1.51 | 1.61E-01 | 1.15 | 2.00E-02 | 0.71 | 8.20E-01 | 1.02 | 4.08E-01 | 0.81 | 65 | 7.58E-01 | 1.04 | 0.001 | + + - 0 - |
| 3 | rs12485872 | 27 | 8.24E-05 | 1.85 | 2.15E-01 | 0.84 | 6.70E-01 | 1.09 | 9.00E-01 | 1.01 | 5.27E-01 | 1.30 | 44 | 3.90E-01 | 1.21 | 0.003 | + - + 0 0 |
| 4 | rs4306981 | 12 | 4.40E-05 | 1.57 | 4.84E-02 | 1.25 | 6.70E-01 | 0.94 | 8.90E-01 | 0.99 | 1.32E-01 | 1.52 | 10 | 4.12E-03 | 1.16 | 0.005 | + + - 0 + |
| 5 | rs7732527 | 43 | 1.25E-04 | 1.50 | 4.38E-01 | 1.08 | 8.00E-01 | 1.03 | 9.00E-01 | 1.01 | 7.12E-01 | 0.92 | 26 | 2.46E-02 | 1.12 | 0.033 | + + 0 0 - |
| 5 | rs4867387 | 23 | 6.82E-05 | 1.73 | 4.28E-01 | 1.12 | 7.10E-01 | 0.92 | 6.50E-01 | 1.05 | 4.80E-01 | 1.27 | 16 | 7.70E-03 | 1.2 | 0.037 | + + - + + |
| 5 | rs11111 | 21 | 6.70E-05 | 0.56 | 7.72E-01 | 1.04 | 1.60E-01 | 0.76 | 2.40E-01 | 0.89 | 6.12E-01 | 0.84 | 8 | 2.74E-03 | 0.82 | 0.033 | - 0 - - - |
| 5 | rs10461985 | 71 | 1.82E-04 | 0.52 | 1.87E-01 | 0.78 | 9.80E-01 | 1.00 | 2.00E-02 | 0.74 | 3.70E-01 | 0.69 | 1 | 5.43E-05 | 0.71 | 0.228 | - - 0 - - |
| 5 | rs1501977 | 19 | 6.48E-05 | 0.62 | 1.94E-01 | 1.16 | 1.90E-01 | 0.81 | 6.00E-01 | 1.05 | 4.14E-01 | 0.78 | 39 | 3.13E-01 | 0.88 | 0.001 | - + - + - |
| 5 | rs1229729 | 52 | 1.42E-04 | 0.66 | 4.91E-01 | 1.07 | 2.50E-01 | 1.17 | 1.90E-01 | 1.11 | 9.62E-01 | 1.01 | 71 | 8.80E-01 | 0.98 | 0.001 | - + + + 0 |
| 5 | rs1229708 | 11 | 4.39E-05 | 1.54 | 8.06E-01 | 0.98 | 3.50E-01 | 0.88 | 7.60E-01 | 0.98 | 4.78E-01 | 1.19 | 48 | 4.48E-01 | 1.08 | 0.003 | + 0 - 0 + |
| 5 | rs7736228 | 74 | 1.91E-04 | 0.64 | 5.68E-01 | 0.94 | 1.70E-01 | 0.81 | 2.80E-01 | 0.91 | 7.86E-01 | 1.08 | 5 | 1.94E-03 | 0.85 | 0.100 | - - - - + |
| 5 | rs13178728 | 78 | 1.99E-04 | 1.91 | 8.49E-01 | 1.04 | 4.30E-01 | 1.22 | 9.70E-01 | 1.00 | 2.14E-01 | 1.80 | 21 | 1.59E-02 | 1.23 | 0.037 | 0 0 + 0 + |
| 5 | rs13159558 | 56 | 1.49E-04 | 2.20 | 4.07E-01 | 1.18 | 7.50E-01 | 1.09 | 3.00E-01 | 0.87 | 4.90E-01 | 1.92 | 6 | 2.14E-03 | 1.48 | 0.101 | + + + - + |
| 6 | rs7751774 | 22 | 6.77E-05 | 0.52 | 2.06E-01 | 0.82 | 5.40E-01 | 0.88 | 7.50E-01 | 0.96 | 3.32E-01 | 0.72 | 7 | 2.23E-03 | 0.8 | 0.049 | - - - 0 - |
| 6 | rs1360811 | 14 | 5.80E-05 | 0.51 | 2.83E-01 | 0.84 | 4.10E-01 | 0.85 | 4.40E-01 | 0.92 | 4.82E-01 | 0.79 | 4 | 1.50E-03 | 0.8 | 0.062 | - - - - - |
| 6 | rs9503979 | 15 | 5.80E-05 | 0.51 | 2.88E-01 | 0.85 | 4.10E-01 | 0.84 | 4.10E-01 | 0.91 | 4.83E-01 | 0.79 | 3 | 1.13E-03 | 0.79 | 0.070 | - - - - - |
| 6 | rs6933317 | 31 | 9.44E-05 | 1.49 | 5.91E-01 | 0.95 | 6.90E-01 | 1.06 | 4.80E-01 | 1.06 | 8.54E-01 | 0.96 | 28 | 3.09E-02 | 1.11 | 0.020 | + - + + - |
| 6 | rs6940071 | 13 | 5.66E-05 | 1.52 | 9.38E-01 | 0.99 | 6.80E-01 | 1.06 | 1.30E-01 | 1.13 | 8.05E-01 | 0.94 | 9 | 3.46E-03 | 1.16 | 0.036 | + 0 + + - |
| 6 | rs12527298 | 64 | 1.69E-04 | 0.68 | 8.42E-01 | 0.98 | 7.70E-01 | 0.96 | 4.10E-01 | 0.94 | 9.54E-01 | 0.99 | 19 | 1.34E-02 | 0.89 | 0.067 | - 0 0 - 0 |
| 6 | rs12527846 | 53 | 1.42E-04 | 0.67 | 8.97E-01 | 0.99 | 7.70E-01 | 0.96 | 3.70E-01 | 0.93 | 8.92E-01 | 1.04 | 20 | 1.36E-02 | 0.86 | 0.037 | - 0 0 - 0 |
| 6 | rs12211633 | 76 | 1.95E-04 | 0.64 | 5.54E-01 | 0.94 | 7.20E-01 | 1.06 | 6.30E-01 | 1.04 | 2.18E-01 | 1.48 | 38 | 2.10E-01 | 0.94 | 0.006 | - - + 0 + |
| 6 | rs2682185 | 51 | 1.38E-04 | 2.04 | 7.78E-01 | 1.05 | 9.90E-01 | 1.00 | 4.40E-01 | 1.11 | 4.50E-01 | 0.73 | 27 | 2.69E-02 | 1.21 | 0.028 | + + 0 + - |
| 6 | rs164301 | 8 | 3.82E-05 | 0.64 | 9.34E-01 | 1.01 | 4.20E-01 | 1.12 | 8.70E-01 | 0.99 | 7.29E-01 | 1.09 | 51 | 5.14E-01 | 0.94 | 0.004 | - 0 + 0 + |
| 6 | rs9365242 | 5 | 2.55E-05 | 0.55 | 4.29E-01 | 0.91 | 5.20E-01 | 1.12 | 9.80E-01 | 1.00 | 9.84E-01 | 1.01 | 29 | 4.04E-02 | 0.88 | 0.006 | - - + 0 0 |
| 6 | rs12055716 | 24 | 7.26E-05 | 0.59 | 5.95E-01 | 0.94 | 7.10E-01 | 1.06 | 5.40E-01 | 0.95 | 7.32E-01 | 1.11 | 23 | 1.97E-02 | 0.84 | 0.013 | - - + - + |
| 6 | rs9295312 | 17 | 5.96E-05 | 1.84 | 7.19E-01 | 0.95 | 6.10E-01 | 0.91 | 2.90E-01 | 0.89 | 7.20E-01 | 1.13 | 54 | 5.64E-01 | 1.09 | 0.002 | + - - - + |
| 8 | rs4875186 | 42 | 1.23E-04 | 1.91 | 8.46E-01 | 0.97 | 6.80E-01 | 1.09 | 2.80E-01 | 0.87 | 8.81E-01 | 0.95 | 50 | 4.93E-01 | 1.12 | 0.004 | + 0 + - - |
| 8 | rs7830870 | 16 | 5.81E-05 | 1.67 | 7.27E-01 | 1.04 | 1.00E-01 | 1.32 | 7.40E-01 | 1.03 | 6.98E-01 | 1.14 | 12 | 4.81E-03 | 1.18 | 0.024 | + 0 + 0 + |
| 8 | rs1864773 | 7 | 2.90E-05 | 1.88 | 9.14E-01 | 1.02 | 9.80E-01 | 0.99 | 8.80E-01 | 0.98 | 6.34E-01 | 1.18 | 31 | 4.62E-02 | 1.15 | 0.008 | + 0 0 0 + |
| 8 | rs7840848 | 37 | 1.10E-04 | 1.51 | 6.09E-01 | 1.05 | 5.60E-01 | 1.08 | 5.20E-01 | 0.95 | 4.29E-01 | 0.82 | 35 | 8.90E-02 | 1.09 | 0.008 | + + + - - |
| 8 | rs2289001 | 46 | 1.33E-04 | 1.53 | 8.58E-01 | 1.02 | 6.80E-01 | 1.07 | 3.30E-01 | 0.92 | 2.68E-01 | 1.38 | 37 | 1.27E-01 | 1.08 | 0.005 | + 0 + - + |
| 11 | rs6483640 | 75 | 1.93E-04 | 1.47 | 1.97E-01 | 1.14 | 5.80E-01 | 1.08 | 8.50E-01 | 1.02 | 7.15E-01 | 1.11 | 11 | 4.63E-03 | 1.15 | 0.088 | + + + 0 + |
| 11 | rs2217032 | 54 | 1.43E-04 | 1.51 | 6.22E-01 | 1.05 | 3.00E-01 | 1.15 | 1.20E-01 | 1.13 | 9.30E-01 | 0.98 | 2 | 1.05E-03 | 1.18 | 0.119 | + + + + - |
| 11 | rs2292730 | 48 | 1.36E-04 | 0.67 | 8.59E-01 | 0.98 | 2.50E-01 | 0.85 | 4.60E-01 | 1.06 | 7.80E-02 | 1.61 | 56 | 5.89E-01 | 0.94 | 0.002 | - 0 - + + |
| 11 | rs7935816 | 18 | 6.40E-05 | 0.63 | 1.64E-01 | 1.17 | 9.10E-01 | 0.98 | 1.40E-01 | 1.13 | 5.43E-01 | 0.84 | 59 | 6.36E-01 | 0.94 | <0.001 | - + 0 + - |
| 12 | rs7304675 | 77 | 1.95E-04 | 0.66 | 9.16E-01 | 0.99 | 8.90E-01 | 0.98 | 5.00E-01 | 1.05 | 1.13E-02 | 2.17 | 75 | 9.54E-01 | 0.99 | 0.001 | - 0 0 + + |
| 12 | rs812512 | 35 | 1.07E-04 | 1.51 | 7.33E-01 | 0.97 | 7.90E-01 | 0.96 | 1.00E-02 | 0.81 | 3.94E-01 | 0.79 | 76 | 9.85E-01 | 1 | <0.001 | + - - - - |
| 13 | rs495680 | 6 | 2.78E-05 | 0.63 | 4.08E-02 | 1.24 | 9.60E-01 | 1.01 | 6.00E-01 | 0.96 | 9.63E-01 | 1.01 | 58 | 6.30E-01 | 0.94 | <0.001 | - + 0 0 0 |
| 13 | rs626326 | 1 | 3.99E-06 | 1.63 | 9.16E-02 | 0.84 | 1.00E-01 | 0.79 | 8.60E-01 | 0.99 | 7.54E-01 | 0.93 | 74 | 9.42E-01 | 1.01 | <0.001 | +---- |
| 13 | rs2858808 | 4 | 1.79E-05 | 0.60 | 5.85E-01 | 1.06 | 4.10E-01 | 0.88 | 7.30E-01 | 1.03 | 3.74E-01 | 1.25 | 49 | 4.82E-01 | 0.92 | 0.001 | - + - 0 + |
| 13 | rs523523 | 2 | 1.32E-05 | 0.64 | 3.31E-01 | 1.10 | 1.60E-01 | 1.22 | 8.70E-01 | 0.99 | 8.83E-01 | 1.04 | 64 | 7.49E-01 | 0.96 | <0.001 | - + + 0 0 |
| 13 | rs2697092 | 57 | 1.49E-04 | 1.62 | 3.34E-01 | 1.12 | 3.30E-01 | 0.84 | 3.80E-01 | 1.09 | 9.15E-01 | 1.03 | 18 | 1.13E-02 | 1.16 | 0.029 | + + - + 0 |
| 15 | rs8041061 | 61 | 1.60E-04 | 1.47 | 8.00E-01 | 1.03 | 5.60E-01 | 1.08 | 9.40E-01 | 0.99 | 2.67E-01 | 0.76 | 34 | 6.83E-02 | 1.09 | 0.014 | - 0 - 0 + |
| 15 | rs809736 | 62 | 1.62E-04 | 0.64 | 9.12E-01 | 1.01 | 4.20E-01 | 0.87 | 8.10E-01 | 0.98 | 5.78E-01 | 1.17 | 30 | 4.35E-02 | 0.89 | 0.024 | - 0 - 0 + |
| 18 | rs8088174 | 72 | 1.87E-04 | 1.64 | 3.77E-02 | 0.76 | 8.30E-01 | 0.96 | 4.70E-01 | 0.93 | 8.24E-01 | 1.08 | 68 | 8.32E-01 | 1.03 | 0.001 | + - 0 - + |
| 20 | rs6085660 | 10 | 4.03E-05 | 1.55 | 2.42E-01 | 0.89 | 9.10E-01 | 0.98 | 1.10E-01 | 1.13 | 9.41E-01 | 0.98 | 42 | 3.69E-01 | 1.1 | 0.004 | + - 0 + 0 |
| 20 | rs1500545 | 60 | 1.59E-04 | 1.49 | 2.86E-01 | 0.90 | 9.90E-01 | 1.00 | 2.50E-01 | 1.09 | 6.86E-01 | 0.91 | 33 | 6.50E-02 | 1.1 | 0.010 | + - 0 + - |
| 20 | rs6055258 | 58 | 1.53E-04 | 0.67 | 2.57E-01 | 0.89 | 4.00E-02 | 1.34 | 2.70E-01 | 0.92 | 5.68E-01 | 1.16 | 66 | 7.87E-01 | 0.96 | 0.001 | - - + - + |
| 20 | rs969111 | 45 | 1.27E-04 | 0.67 | 2.76E-01 | 0.90 | 4.00E-02 | 1.34 | 2.60E-01 | 0.92 | 4.90E-01 | 1.19 | 57 | 5.99E-01 | 0.94 | 0.002 | - - + - + |
| 20 | rs1008096 | 44 | 1.26E-04 | 0.67 | 2.41E-01 | 0.89 | 4.00E-02 | 1.34 | 2.70E-01 | 0.92 | 4.85E-01 | 1.20 | 55 | 5.89E-01 | 0.94 | 0.002 | - - + - + |
| 20 | rs6118681 | 38 | 1.12E-04 | 1.51 | 2.46E-01 | 0.89 | 4.20E-01 | 1.13 | 1.40E-01 | 0.89 | 6.16E-01 | 1.14 | 52 | 5.25E-01 | 1.08 | 0.001 | + - + - + |
| 20 | rs6141026 | 9 | 3.98E-05 | 1.69 | 5.32E-01 | 0.93 | 5.60E-01 | 1.11 | 4.30E-01 | 1.08 | 7.41E-01 | 1.10 | 22 | 1.73E-02 | 1.16 | 0.013 | + - + + + |
| 20 | rs6081741 | 65 | 1.71E-04 | 0.63 | 9.73E-01 | 1.00 | 6.00E-01 | 1.08 | 7.80E-01 | 0.98 | 6.74E-01 | 1.14 | 36 | 1.05E-01 | 0.91 | 0.018 | - 0 + 0 + |
| 20 | rs6013773 | 41 | 1.18E-04 | 0.67 | 8.80E-01 | 1.02 | 1.90E-01 | 1.20 | 2.40E-01 | 1.09 | 6.22E-01 | 0.88 | 62 | 6.94E-01 | 0.96 | 0.002 | - 0 + + - |
| 23 | rs5927035 | 32 | 9.52E-05 | 1.78 | 1.76E-01 | 0.85 | 9.10E-01 | 0.99 | 53 | 5.34E-01 | 1.13 | <0.001 | + - x 0 x | ||||
| 23 | rs2879751 | 26 | 8.10E-05 | 1.79 | 9.90E-01 | 1.00 | 41 | 3.24E-01 | 1.33 | 0.003 | + x x 0 x | ||||||
CMH is chronic mucus hypersecretion; OR is odds ratio; Q = p-value for heterogeneity;
p# = fixed p-value if p-value for heterogeneity > 0.005 and random p-value if p-value for heterogeneity < 0.005;
OR# = fixed OR if p-value for heterogeneity > 0.005 and random OR if p-value for heterogeneity < 0.005;
Direction of effect in identification and replication cohorts is presented in the following order: NELSON-COPD, GenKOLS, COPDGene, ECLIPSE and MESA; Direction of effect: - = OR ≤ 0.95, 0 = 0.95 ≤ OR ≤ 1.05, 1 = OR ≥ 1.05, x = not applicable;
An empty box = SNP was not analyzed in the corresponding replication cohort.
When performing replication in males only, i.e. the same gender as in the identification cohort, results were comparable with all SNP effects in the same direction, but with lower significance due to the deletion of 714 females and hence lower power.
Replication of top SNPs in four COPD cohorts
Table 3 shows the results of the 78 SNPs that were analyzed in 3,106 individuals with COPD, including 1,198 with and 1,908 without CMH, participating in 4 different COPD cohorts. Meta-analyses of these 78 SNPs across the replication cohorts showed borderline association to six SNPs with CMH and a similar direction of effect (combined p-values ranging from 1.02 × 10−2 to 9.49 × 10−2).
The strongest association in the meta-analysis across identification and replication cohorts was observed for rs10461985 on chromosome 5p13.2 showing effects in the same direction in NELSON participants with COPD and the replication cohorts (p = 5.43 × 10−5, OR = 0.714, Table 3), except for COPDGene that showed no effect. SNP rs10461985 is located in an intron in the glial cell line-derived neurotrophic factor antisense RNA 1 gene (GDNF-AS1).
Functional relevance of rs10461985 and GDNF
The Affymetrix chip used to investigate mRNA expression in airway wall biopsies of COPD patients did not have probe set for the GDNF-AS1 gene. As the role of GDNF-AS1 as an antisense RNA is to prevent translation of GDNF, we assessed the association of the mRNA expression of this gene and CMH. GDNF mRNA expression was found to be significantly lower in bronchial biopsies of COPD patients with CMH than those without CMH (b = −2.8, p = 0.007).
Genome wide analyses in NELSON participants without COPD
The same 522,636 SNPs were analyzed in 1,348 NELSON participants without, 342 with and 1,006 without CMH. The QQ-plot confirmed that there was no population stratification (λ = 1.009). The p-values of this GWA study are presented in the Manhattan plot (Figure 2). There were 79 SNPs associated with CMH with a p < 2.0 × 10−4 (Table 4).
Figure 2.

Quantile-quantile plot (left) and Manhattan plot (right) of GWA results for association of SNPs with CMH in NELSON participants without COPD.
Table 4.
Association of SNPs with CMH in identification analysis (NELSON-non-COPD) and in replication in LifeLines and subsequent meta-analysis across NELSON-non-COPD and LifeLines.
| CHR | SNP | BP | minor allele | NELSON-non-COPD | LifeLines | META-ANALYSIS across NELSON-non-COPD and LifeLines | Closest gene(s) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF | rank | P | OR | P | OR | rank | P# | OR# | Q | |||||
| 1 | rs2817896 | 22988636 | G | 0.26 | 59 | 1.16E-04 | 1.47 | 1.09E-01 | 1.26 | 8 | 4.66E-05 | 1.40 | 0.362 | EPHB2* |
| 1 | rs893961 | 22990760 | G | 0.25 | 66 | 1.81E-04 | 1.46 | 8.86E-02 | 1.28 | 9 | 5.30E-05 | 1.39 | 0.445 | EPHB2* |
| 1 | rs11208807 | 66407509 | A | 0.31 | 57 | 1.50E-04 | 1.43 | 2.55E-01 | 1.17 | 23 | 1.65E-04 | 1.34 | 0.228 | PDE4B* |
| 1 | rs2208370 | 170221954 | A | 0.39 | 53 | 1.98E-04 | 1.42 | 7.22E-01 | 1.07 | 35 | 5.51E-04 | 1.33 | 0.154 | DNM3* |
| 1 | rs3845529 | 214203243 | C | 0.42 | 73 | 1.96E-04 | 0.7 | 4.98E-03 | 0.67 | 1 | 3.25E-06 | 0.69 | 0.780 | USH2A* |
| 1 | rs629199 | 232830726 | A | 0.19 | 65 | 1.24E-04 | 1.54 | 3.64E-01 | 1.25 | 17 | 1.10E-04 | 1.48 | 0.445 | IRF2BP2 & PP2672 |
| 1 | rs12028329 | 245477414 | G | 0.25 | 46 | 2.20E-05 | 1.55 | 6.74E-01 | 1.07 | 21 | 1.47E-04 | 1.39 | 0.052 | LOC441931 & VN1R5 |
| 2 | rs1476151 | 125744258 | G | 0.46 | 19 | 1.08E-04 | 1.43 | 5.37E-01 | 0.91 | 62 | 2.98E-03 | 1.26 | 0.010 | CNTP5 & LOC150554 |
| 2 | rs13028050 | 125844903 | A | 0.42 | 29 | 1.25E-04 | 0.7 | 7.36E-01 | 1.05 | 61 | 2.71E-03 | 0.79 | 0.016 | CNTP5 & LOC150554 |
| 3 | rs17776719 | 11615481 | G | 0.13 | 42 | 6.72E-05 | 1.64 | 5.58E-01 | 0.84 | 34 | 5.49E-04 | 1.49 | 0.038 | VGLL4* |
| 3 | rs2956507 | 13682301 | A | 0.35 | 21 | 6.61E-05 | 0.68 | 7.82E-01 | 1.04 | 56 | 2.06E-03 | 0.78 | 0.011 | FBLN2 & WNT7A |
| 3 | rs6792244 | 13692200 | A | 0.42 | 28 | 5.77E-05 | 0.68 | 6.74E-01 | 1.07 | 49 | 1.28E-03 | 0.77 | 0.014 | FBLN2 & WNT7A |
| 3 | rs6775581 | 13695098 | G | 0.42 | 16 | 1.22E-05 | 0.66 | 6.80E-01 | 1.07 | 30 | 4.24E-04 | 0.75 | 0.009 | FBLN2 & WNT7A |
| 3 | rs6781368 | 13701841 | G | 0.43 | 14 | 2.02E-05 | 0.67 | 8.42E-01 | 1.03 | 42 | 8.12E-04 | 0.77 | 0.008 | FBLN2 & WNT7A |
| 3 | rs6794344 | 13701889 | A | 0.46 | 24 | 8.84E-05 | 0.7 | 7.82E-01 | 1.04 | 59 | 2.51E-03 | 0.80 | 0.012 | FBLN2 & WNT7A |
| 3 | rs6795216 | 13705683 | C | 0.46 | 41 | 1.06E-04 | 0.7 | 9.03E-01 | 1.02 | 47 | 1.13E-03 | 0.77 | 0.035 | FBLN2 & WNT7A |
| 3 | rs2974399 | 13740911 | A | 0.45 | 30 | 2.89E-05 | 0.68 | 7.99E-01 | 1.04 | 33 | 5.38E-04 | 0.76 | 0.018 | FBLN2 & WNT7A |
| 3 | rs6768597 | 20394587 | G | 0.3 | 50 | 7.05E-05 | 0.66 | 3.17E-01 | 0.87 | 20 | 1.44E-04 | 0.73 | 0.125 | SGOL1 & VENTXP7 |
| 3 | rs9682418 | 72180217 | G | 0.27 | 70 | 9.15E-05 | 1.48 | 4.91E-02 | 1.32 | 5 | 1.52E-05 | 1.43 | 0.494 | PROK2 & CCDC137P |
| 3 | rs11714053 | 133332100 | A | 0.17 | 37 | 3.49E-05 | 1.61 | 5.06E-01 | 0.84 | 28 | 3.74E-04 | 1.46 | 0.026 | CPNE4 & LOC729674 |
| 3 | rs1403428 | 149752754 | A | 0.22 | 52 | 5.96E-05 | 1.55 | 3.27E-01 | 1.16 | 19 | 1.18E-04 | 1.41 | 0.133 | LOC344741 & RPL38P1 |
| 3 | rs9825199 | 196385873 | A | 0.06 | 17 | 4.83E-05 | 2.02 | 4.88E-01 | 0.81 | 50 | 1.38E-03 | 1.62 | 0.009 | C3orf21* |
| 3 | rs3796160 | 196387903 | A | 0.06 | 22 | 6.76E-05 | 2 | 5.17E-01 | 0.82 | 52 | 1.74E-03 | 1.60 | 0.011 | C3orf21* |
| 4 | rs17447715 | 80821889 | A | 0.19 | 58 | 1.94E-04 | 0.62 | 1.52E-01 | 0.78 | 18 | 1.16E-04 | 0.67 | 0.295 | OR7E94P & GDEP |
| 4 | rs6858670 | 137477830 | G | 0.47 | 32 | 1.29E-04 | 1.42 | 9.08E-01 | 0.99 | 57 | 2.13E-03 | 1.26 | 0.022 | LOC100132574 & LOC646316 |
| 4 | rs7688325 | 137479502 | A | 0.47 | 35 | 1.65E-04 | 1.41 | 8.99E-01 | 0.98 | 60 | 2.54E-03 | 1.25 | 0.024 | LOC100132574 & LOC646316 |
| 4 | rs4863687 | 140897731 | A | 0.28 | 72 | 1.89E-04 | 1.45 | 1.22E-02 | 1.57 | 3 | 7.57E-06 | 1.48 | 0.688 | MAML3* |
| 4 | rs6552407 | 181166606 | A | 0.25 | 1 | 2.38E-05 | 1.55 | 7.85E-02 | 0.76 | 73 | 8.04E-01 | 1.09 | 0.000 | LOC391719&hCG_2025798 |
| 5 | rs1816237 | 33076569 | G | 0.11 | 49 | 1.27E-04 | 0.53 | 8.00E-01 | 0.93 | 32 | 5.09E-04 | 0.61 | 0.102 | LOC340113 & LOC728553 |
| 5 | rs4836527 | 122670280 | A | 0.4 | 33 | 1.45E-04 | 1.41 | 5.38E-01 | 0.9 | 54 | 1.96E-03 | 1.28 | 0.022 | PRDM6 & CEP120 |
| 5 | rs13183447 | 172004970 | A | 0.39 | 4 | 9.28E-06 | 0.65 | 3.04E-01 | 1.17 | 70 | 6.13E-01 | 0.86 | 0.001 | SH3PXD2B & LOC100130394 |
| 5 | rs262020 | 177896923 | A | 0.39 | 54 | 5.78E-05 | 0.68 | 8.99E-01 | 0.97 | 24 | 1.68E-04 | 0.71 | 0.154 | COL23A1* |
| 6 | rs7770889 | 96965174 | A | 0.37 | 60 | 9.92E-05 | 1.45 | 3.65E-01 | 1.19 | 13 | 9.81E-05 | 1.40 | 0.368 | FUT9 & KIAA0776 |
| 6 | rs9486181 | 96974853 | G | 0.36 | 63 | 1.30E-04 | 1.45 | 2.82E-01 | 1.22 | 14 | 1.03E-04 | 1.40 | 0.410 | FUT9 & KIAA0776 |
| 6 | rs4425602 | 97000627 | G | 0.36 | 61 | 1.30E-04 | 1.45 | 2.93E-01 | 1.21 | 16 | 1.08E-04 | 1.39 | 0.396 | FUT9 & KIAA0776 |
| 6 | rs3860243 | 97012024 | A | 0.36 | 62 | 1.21E-04 | 1.45 | 2.79E-01 | 1.22 | 12 | 9.32E-05 | 1.40 | 0.402 | FUT9 & KIAA0776 |
| 6 | rs12207471 | 97070503 | A | 0.36 | 47 | 1.30E-04 | 1.45 | 9.17E-01 | 1.02 | 43 | 8.20E-04 | 1.32 | 0.064 | FUT9 & KIAA0776 |
| 6 | rs9398148 | 97170276 | G | 0.34 | 64 | 1.39E-04 | 1.45 | 2.97E-01 | 1.23 | 15 | 1.05E-04 | 1.40 | 0.442 | FHL5* |
| 6 | rs9375195 | 98669441 | G | 0.48 | 40 | 1.35E-04 | 1.42 | 9.58E-01 | 1.01 | 53 | 1.78E-03 | 1.26 | 0.029 | C6orf167 & LOC100129158 |
| 6 | rs2151522 | 127251786 | A | 0.39 | 55 | 1.45E-04 | 1.43 | 2.21E-01 | 1.17 | 22 | 1.57E-04 | 1.33 | 0.196 | LOC442257 & RSPO3 |
| 7 | rs10499977 | 108947923 | A | 0.33 | 31 | 4.81E-05 | 1.48 | 6.02E-01 | 0.91 | 41 | 7.41E-04 | 1.34 | 0.020 | LOC646614 & LOC100128056 |
| 7 | rs12538214 | 154969302 | A | 0.25 | 48 | 1.75E-04 | 1.48 | 5.29E-01 | 1.1 | 40 | 6.48E-04 | 1.34 | 0.092 | EN2 & CNPY1 |
| 8 | rs7007974 | 8839477 | G | 0.1 | 56 | 1.48E-04 | 1.69 | 2.75E-01 | 1.24 | 25 | 1.82E-04 | 1.53 | 0.208 | MRPS18CP2 & LOC645960 |
| 8 | rs13265648 | 73208111 | A | 0.49 | 2 | 1.38E-04 | 0.7 | 8.67E-02 | 1.25 | 72 | 7.98E-01 | 0.93 | 0.000 | TRPA1 & LOC392232 |
| 8 | rs16886291 | 115780612 | A | 0.12 | 44 | 1.90E-04 | 0.55 | 6.96E-01 | 0.92 | 51 | 1.46E-03 | 0.67 | 0.047 | hCG_1644355 & TRPS1 |
| 9 | rs10119913 | 29254328 | C | 0.3 | 3 | 1.61E-04 | 0.68 | 5.54E-02 | 1.5 | 74 | 9.74E-01 | 0.99 | 0.001 | LINGO2 & LOC286239 |
| 10 | rs10827563 | 36255556 | G | 0.48 | 38 | 1.04E-04 | 1.43 | 5.15E-01 | 0.88 | 48 | 1.14E-03 | 1.31 | 0.027 | LOC439954 & PBEF2 |
| 10 | rs2696310 | 36262016 | G | 0.44 | 7 | 1.55E-05 | 1.5 | 6.65E-01 | 0.95 | 68 | 4.27E-01 | 1.20 | 0.004 | LOC439954 & PBEF2 |
| 10 | rs2767073 | 36269018 | A | 0.44 | 8 | 4.75E-06 | 1.54 | 5.86E-01 | 0.92 | 26 | 2.21E-04 | 1.35 | 0.006 | LOC439954 & PBEF2 |
| 10 | rs1571136 | 36270927 | G | 0.44 | 18 | 1.57E-05 | 1.5 | 6.14E-01 | 0.92 | 31 | 4.56E-04 | 1.33 | 0.010 | LOC439954 & PBEF2 |
| 10 | rs2804852 | 36277541 | A | 0.42 | 39 | 8.39E-05 | 1.44 | 6.53E-01 | 0.92 | 45 | 1.01E-03 | 1.31 | 0.028 | LOC439954 & PBEF2 |
| 11 | rs2071461 | 11330536 | G | 0.24 | 26 | 3.86E-05 | 1.52 | 3.12E-01 | 0.78 | 37 | 6.06E-04 | 1.38 | 0.013 | GALNTL4* |
| 11 | rs3903687 | 35288218 | G | 0.37 | 10 | 1.40E-04 | 1.43 | 4.90E-01 | 0.91 | 67 | 6.03E-03 | 1.24 | 0.006 | SLC1A2 |
| 11 | rs474158 | 105342254 | A | 0.07 | 36 | 3.28E-06 | 2.17 | 7.05E-01 | 1.1 | 7 | 4.35E-05 | 1.76 | 0.024 | GRIA4* |
| 11 | rs2288403 | 129243199 | G | 0.17 | 71 | 1.63E-04 | 0.6 | 6.27E-02 | 0.69 | 6 | 3.00E-05 | 0.63 | 0.604 | NFRKB* |
| 12 | rs10459134 | 5750112 | A | 0.18 | 13 | 1.47E-04 | 1.55 | 5.12E-01 | 0.89 | 65 | 5.21E-03 | 1.31 | 0.008 | TMEM16B* |
| 12 | rs7959932 | 23931073 | G | 0.32 | 9 | 2.74E-05 | 1.49 | 2.08E-01 | 0.74 | 39 | 6.34E-04 | 1.35 | 0.006 | SOX5* |
| 12 | rs7308636 | 23942557 | A | 0.31 | 15 | 3.27E-05 | 1.48 | 2.34E-01 | 0.75 | 38 | 6.25E-04 | 1.35 | 0.008 | SOX5* |
| 12 | rs1690139 | 74558944 | G | 0.11 | 74 | 1.76E-04 | 1.67 | 1.11E-02 | 1.69 | 2 | 5.91E-06 | 1.67 | 0.951 | LOC100130336 & LOC100131830 |
| 13 | rs9300394 | 86801456 | A | 0.29 | 27 | 1.52E-04 | 0.67 | 6.11E-01 | 1.09 | 64 | 3.67E-03 | 0.77 | 0.013 | LOC100130117 & hCG_1795283 |
| 13 | rs4514531 | 86805556 | G | 0.29 | 23 | 7.12E-05 | 0.66 | 6.32E-01 | 1.08 | 55 | 1.99E-03 | 0.76 | 0.011 | LOC100130117 & hCG_1795283 |
| 13 | rs944899 | 111798962 | A | 0.46 | 69 | 5.76E-05 | 1.46 | 4.05E-02 | 1.3 | 4 | 8.40E-06 | 1.40 | 0.476 | SOX1 |
| 15 | rs12594495 | 20499445 | G | 0.26 | 6 | 3.44E-05 | 0.62 | 5.49E-01 | 1.09 | 69 | 4.71E-01 | 0.82 | 0.002 | CYFIP1* |
| 15 | rs8042800 | 57638092 | A | 0.3 | 5 | 1.36E-04 | 0.67 | 2.60E-01 | 1.17 | 71 | 6.39E-01 | 0.88 | 0.001 | FAMS1A & GCNT3 |
| 15 | rs3784350 | 66429101 | A | 0.37 | 11 | 7.25E-05 | 0.68 | 6.38E-01 | 1.07 | 63 | 3.47E-03 | 0.79 | 0.006 | ITGA11* |
| 15 | rs1348533 | 84527598 | A | 0.2 | 12 | 1.67E-04 | 0.63 | 4.36E-01 | 1.17 | 66 | 5.73E-03 | 0.75 | 0.008 | AGBL1 |
| 15 | rs8043332 | 96890829 | A | 0.3 | 20 | 1.85E-05 | 1.51 | 3.68E-01 | 0.82 | 29 | 3.84E-04 | 1.36 | 0.011 | FAM169B & IGF1R |
| 16 | rs1978316 | 6277315 | A | 0.19 | 67 | 1.44E-04 | 1.53 | 1.85E-01 | 1.29 | 11 | 7.70E-05 | 1.46 | 0.448 | A2BP1* |
| 16 | rs1344471 | 6278829 | A | 0.19 | 68 | 1.36E-04 | 1.53 | 1.84E-01 | 1.29 | 10 | 7.31E-05 | 1.47 | 0.449 | A2BP1* |
| 16 | rs12443545 | 82156133 | A | 0.19 | 45 | 1.31E-04 | 0.62 | 5.94E-01 | 1.18 | 44 | 8.58E-04 | 0.68 | 0.051 | CDH13* |
| 16 | rs12918351 | 82156354 | G | 0.2 | 43 | 1.30E-04 | 0.62 | 9.35E-01 | 0.98 | 46 | 1.12E-03 | 0.71 | 0.044 | CDH13* |
| 17 | rs1508960 | 49024530 | G | 0.3 | 25 | 8.74E-05 | 1.45 | 7.06E-01 | 0.95 | 58 | 2.36E-03 | 1.27 | 0.012 | LOC645163 & LOC645173 |
| 20 | rs6042209 | 1354212 | A | 0.18 | 34 | 3.64E-05 | 1.59 | 9.79E-01 | 1 | 36 | 5.69E-04 | 1.38 | 0.023 | FKBP1A & NSFL1C |
| 21 | rs2032257 | 26696741 | A | 0.39 | 51 | 1.30E-04 | 0.69 | 3.58E-01 | 0.88 | 27 | 2.78E-04 | 0.75 | 0.131 | APP & CYYR1 |
CMH is chronic mucus hypersecretion; OR is odds ratio; Q = p-value for heterogeneity;
p# = fixed p-value if p-value for heterogeneity > 0.005 and random p-value if p-value for heterogeneity < 0.005;
OR# = fixed OR if p-value for heterogeneity > 0.005 and random OR if p-value for heterogeneity < 0.005;
Direction of effect in identification and replication cohorts is presented in the following order: NELSON-non-COPD, LifeLines; Direction of effect: - = OR ≤ 0.95, 0 = 0.95 ≤ OR ≤ 1.05, 1 = OR ≥ 1.05, x = not applicable
Replication of top SNPs in the general population based LifeLines cohort
Genotypes from 74 of the 79 SNPs with a p < 2.0 × 10−4 were available from the general population based LifeLines cohort, including 130 individuals with CMH and 2,313 without CMH. Ten SNPs showed some association with CMH in LifeLines (p < 10−1) and among these, 7 SNPs had effects in the same direction in the NELSON participants without COPD and in LifeLines (Table 4). In the meta-analysis across this NELSON population and LifeLines 4 SNPs were associated with CMH with a p < 10−5:
rs3845529 on chromosome 1q41; p = 3.25 × 10−6 (OR = 0.693), located in an intron in the Usher syndrome 2A gene (USH2A);
rs1690139 on chromosome 12q; p = 5.91 × 10−6 (OR = 1.673), located in a gene desert between LOC100130336 and LOC100131830;
rs4863687 on chromosome 4q28; p = 7.57 × 10−6 (OR = 1.476), located in an intron in the mastermind-like 3 gene (MAML3);
rs944899 on chromosome 13q34; p = 8.40 × 10−6 (OR = 1.399), located near (< 25 kb) the SRY (sex determining region Y)-box 1 gene (SOX1).
Functional relevance of identified top SNPs associated with CMH in individuals without COPD
The rs3845529 genotypes showed no significant eQTL effect on USHA2 mRNA expression levels and rs944899 genotypes not on SOX1 mRNA expression levels in lung tissue (p ≈ 7 × 10−1). In contrast, a strong effect of rs4863687 genotypes (CC = 622, TC = 408, TT = 66) on MAML3 mRNA expression levels was shown; the CMH associated risk allele T was significantly associated with higher expression of MAML3 (p = 2.59 × 10−12) (Affymetrix ID: 100146901-TGI-at, Ensemble ID: NM-018717) (Figure 3).
Figure 3.
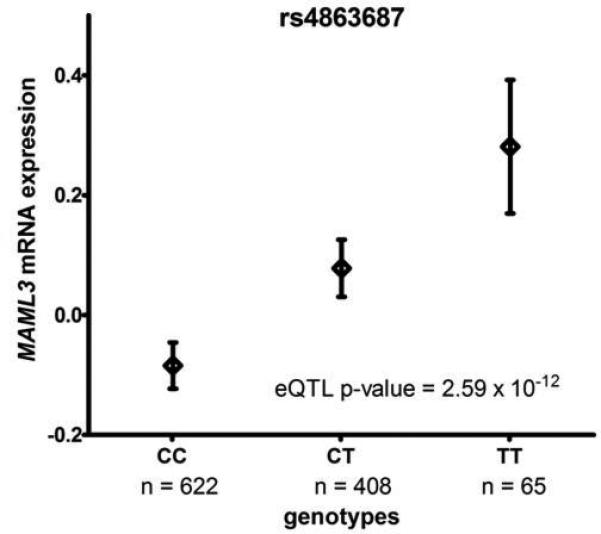
Boxplots of lung gene expression levels for MAML3 according to genotype groups for SNP rs4868687 in 1,095 individuals.
Gene expression profiles of genes close to rs1690139 were not present on the Affymetrix array for the eQTL-analyses.
Overlap of top SNPs associated with CMH in COPD and non-COPD
Comparison of top SNPs in the GWA study in NELSON participants with COPD (5,146 SNPs, p < 10−2) and in the GWA study in NELSON participants without COPD (5,186 SNPs, p < 10−2) showed 60 overlapping SNPs (Table 5). When only SNPs with a p-value < 10−3 were considered, only one overlapping SNP was observed: rs4306981, located close to (64kb) the progestin and adipoQ receptor family member III gene (PAQR3) on chromosome 4q21.21 (p = 4.40 × 10−5 in individuals with COPD and 5.73 × 10−4 in those without COPD) with effects in the same direction in both analyses (OR = 1.57 and OR = 1.40, respectively). Follow up of this SNP in COPD cohorts did not confirm this association (meta-analysis across NELSON and replication cohorts p = 4.12 × 10−3).
Table 5.
Comparison of SNPs associated with CMH and p-value < 10−2 present in NELSON-COPD and NELSON-non-COPD
| CHR | SNP | BP | minor allele | NELSON-COPD | NELSON-non-COPD | Direction of effect | in or close to gene(s) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF | rank | P | OR | MAF | rank | P | OR | ||||||
| 1 | rs6677529 | 160530378 | A | 0.19 | 48 | 7.24E-03 | 1.42 | 0.17 | 10 | 1.03E-03 | 1.45 | + + | NOS1AP* |
| 3 | rs12632852 | 11593682 | G | 0.40 | 2 | 3.20E-04 | 0.67 | 0.39 | 52 | 8.70E-03 | 1.28 | - + | VGLL4* |
| 3 | rs2574704 | 11630381 | G | 0.29 | 26 | 3.94E-03 | 0.72 | 0.29 | 4 | 5.25E-04 | 1.40 | - + | VGLL4* |
| 3 | rs2574720 | 11635412 | C | 0.26 | 7 | 1.08E-03 | 0.68 | 0.26 | 3 | 3.97E-04 | 1.43 | - + | VGLL4* |
| 3 | rs2616551 | 11642123 | G | 0.18 | 54 | 7.91E-03 | 0.69 | 0.18 | 2 | 3.57E-04 | 1.50 | - + | VGLL4* |
| 3 | rs12374151 | 16605508 | A | 0.12 | 18 | 2.83E-03 | 0.61 | 0.13 | 48 | 7.25E-03 | 1.43 | - + | DAZL* |
| 3 | rs9852824 | 24397993 | A | 0.46 | 50 | 7.51E-03 | 1.32 | 0.46 | 60 | 9.90E-03 | 0.79 | + - | THRB* |
| 3 | rs3796150 | 66584924 | A | 0.20 | 55 | 8.54E-03 | 0.70 | 0.17 | 32 | 4.73E-03 | 0.70 | - - | LRIG1* |
| 3 | rs7648171 | 106704936 | G | 0.20 | 41 | 6.16E-03 | 0.70 | 0.21 | 36 | 6.03E-03 | 0.73 | - - | ALCAM* |
| 4 | rs4306981 | 80143145 | G | 0.31 | 1 | 4.40E-05 | 1.57 | 0.30 | 6 | 5.73E-04 | 1.40 | + + | PAQR3 & ARD1B |
| 4 | rs10518211 | 80156089 | G | 0.48 | 21 | 3.50E-03 | 1.35 | 0.48 | 20 | 1.93E-03 | 1.33 | + + | PAQR3 & ARD1B |
| 4 | rs4834752 | 120275247 | A | 0.42 | 12 | 1.97E-03 | 0.72 | 0.44 | 15 | 1.30E-03 | 1.34 | - + | MYOZ2* |
| 4 | rs1017710 | 180937258 | A | 0.07 | 5 | 9.14E-04 | 1.97 | 0.07 | 37 | 6.23E-03 | 0.58 | + - | LOC391719 & hCG_2025798 |
| 4 | rs17068194 | 180952052 | A | 0.07 | 6 | 9.14E-04 | 1.97 | 0.07 | 41 | 6.71E-03 | 0.58 | + - | LOC391719 & hCG_2025798 |
| 5 | rs365294 | 3476838 | A | 0.38 | 45 | 6.74E-03 | 1.34 | 0.37 | 8 | 7.47E-04 | 1.38 | + + | LOC100132531 & IRX1 |
| 5 | rs1995385 | 73415681 | G | 0.23 | 4 | 6.71E-04 | 0.65 | 0.23 | 58 | 9.39E-03 | 1.32 | - + | RGNEF & ENC1 |
| 5 | rs718164 | 73417137 | G | 0.23 | 3 | 5.37E-04 | 0.64 | 0.23 | 57 | 9.37E-03 | 1.32 | - + | RGNEF & ENC2 |
| 5 | rs11738681 | 176694141 | G | 0.33 | 43 | 6.35E-03 | 0.74 | 0.32 | 43 | 6.79E-03 | 0.76 | - - | LMAN2* |
| 5 | rs11949401 | 176698595 | G | 0.33 | 36 | 5.26E-03 | 0.73 | 0.31 | 53 | 8.76E-03 | 0.76 | - - | LMAN2* |
| 5 | rs9313758 | 176705697 | C | 0.33 | 44 | 6.35E-03 | 0.74 | 0.31 | 42 | 6.76E-03 | 0.76 | - - | LMAN2* |
| 5 | rs4532376 | 176707009 | A | 0.33 | 33 | 4.86E-03 | 0.73 | 0.31 | 33 | 5.13E-03 | 0.75 | - - | LMAN2* |
| 5 | rs4131289 | 176713151 | A | 0.33 | 40 | 5.88E-03 | 0.74 | 0.31 | 29 | 4.15E-03 | 0.74 | - - | LMAN2 & RGS14 |
| 6 | rs10457138 | 106460454 | G | 0.27 | 15 | 2.47E-03 | 0.70 | 0.26 | 17 | 1.66E-03 | 1.37 | - + | LOC100130683 & PRDM1 |
| 7 | rs40463 | 40915342 | A | 0.12 | 24 | 3.65E-03 | 1.55 | 0.13 | 51 | 8.30E-03 | 0.68 | + - | C7orf10 & INHBA |
| 7 | rs4729686 | 100747270 | A | 0.07 | 13 | 2.18E-03 | 0.50 | 0.07 | 22 | 2.76E-03 | 1.67 | - + | RABL5* |
| 7 | rs2905286 | 112081312 | G | 0.48 | 57 | 9.04E-03 | 0.76 | 0.48 | 39 | 6.56E-03 | 0.78 | - - | NPM1P14 & LOC100128875 |
| 8 | rs2055516 | 769714 | C | 0.25 | 11 | 1.85E-03 | 1.46 | 0.25 | 14 | 1.27E-03 | 1.40 | + + | C8orf68* |
| 8 | rs10105558 | 783149 | A | 0.25 | 27 | 4.04E-03 | 1.42 | 0.25 | 28 | 3.65E-03 | 1.35 | + + | C8orf68* |
| 8 | rs13282923 | 4473969 | G | 0.29 | 29 | 4.10E-03 | 1.38 | 0.29 | 18 | 1.82E-03 | 0.72 | + - | CSMD1* |
| 8 | rs13273819 | 135514435 | A | 0.23 | 35 | 5.25E-03 | 1.39 | 0.23 | 54 | 9.15E-03 | 1.32 | + + | LOC100129104 & ZFAT |
| 9 | rs530582 | 134354849 | G | 0.15 | 17 | 2.76E-03 | 0.64 | 0.17 | 7 | 6.63E-04 | 1.49 | - + | RP11-738I14.8* |
| 10 | rs10903396 | 1208030 | G | 0.46 | 28 | 4.06E-03 | 0.74 | 0.46 | 38 | 6.26E-03 | 0.78 | - - | C10orf139 & LOC100130729 |
| 10 | rs10905113 | 7246430 | G | 0.44 | 8 | 1.14E-03 | 1.41 | 0.44 | 50 | 8.12E-03 | 0.79 | + - | SFMBT2* |
| 10 | rs17601717 | 52831431 | G | 0.23 | 39 | 5.38E-03 | 0.71 | 0.25 | 40 | 6.57E-03 | 1.32 | - + | PRKG1* |
| 10 | rs7902476 | 72693742 | A | 0.11 | 25 | 3.70E-03 | 0.60 | 0.12 | 26 | 3.37E-03 | 0.64 | - - | UNC5B* |
| 11 | rs2273688 | 35295319 | A | 0.27 | 31 | 4.49E-03 | 0.71 | 0.28 | 16 | 1.56E-03 | 1.40 | - + | SLC1A2* |
| 11 | rs10768129 | 35319065 | A | 0.27 | 47 | 7.02E-03 | 0.72 | 0.28 | 13 | 1.21E-03 | 1.40 | - + | SLC1A2* |
| 11 | rs7127824 | 35330427 | A | 0.27 | 22 | 3.64E-03 | 0.70 | 0.28 | 11 | 1.14E-03 | 1.40 | - + | SLC1A2* |
| 11 | rs7130967 | 35330584 | A | 0.27 | 23 | 3.64E-03 | 0.70 | 0.28 | 12 | 1.14E-03 | 1.40 | - + | SLC1A2* |
| 11 | rs927352 | 35334090 | A | 0.30 | 58 | 9.36E-03 | 0.73 | 0.31 | 19 | 1.90E-03 | 1.36 | - + | SLC1A2* |
| 11 | rs11033910 | 37021958 | G | 0.28 | 53 | 7.82E-03 | 0.73 | 0.29 | 56 | 9.32E-03 | 1.30 | - + | C11orf74 & LOC100129825 |
| 11 | rs12417575 | 85832165 | G | 0.28 | 37 | 5.31E-03 | 0.72 | 0.27 | 59 | 9.85E-03 | 0.76 | - - | ME3* |
| 11 | rs689051 | 124797700 | A | 0.16 | 10 | 1.43E-03 | 1.58 | 0.15 | 30 | 4.40E-03 | 0.67 | + - | PKNOX2* |
| 12 | rs17179798 | 5184769 | A | 0.24 | 52 | 7.73E-03 | 1.38 | 0.23 | 27 | 3.51E-03 | 1.37 | + + | KCNA5 & LOC387826 |
| 12 | rs1894307 | 11896987 | A | 0.15 | 34 | 4.90E-03 | 1.49 | 0.14 | 9 | 9.39E-04 | 1.50 | + + | ETV6* |
| 12 | rs2255953 | 11902003 | G | 0.23 | 59 | 9.78E-03 | 1.38 | 0.21 | 5 | 5.34E-04 | 1.45 | + + | ETV6* |
| 12 | rs2855708 | 11904839 | G | 0.28 | 30 | 4.10E-03 | 1.40 | 0.27 | 34 | 5.40E-03 | 1.31 | + + | ETV6* |
| 12 | rs1820545 | 39096860 | G | 0.41 | 38 | 5.32E-03 | 0.75 | 0.42 | 31 | 4.47E-03 | 1.29 | - + | LRRK2 & MUC19 |
| 12 | rs7306163 | 39111184 | C | 0.41 | 42 | 6.21E-03 | 0.75 | 0.42 | 35 | 5.50E-03 | 1.28 | - + | MUC19* |
| 14 | rs8009673 | 31412453 | A | 0.14 | 46 | 7.00E-03 | 1.50 | 0.13 | 21 | 2.23E-03 | 1.49 | + + | NUBPL & C14orf128 |
| 14 | rs7155416 | 76021126 | A | 0.12 | 51 | 7.72E-03 | 1.51 | 0.14 | 23 | 3.02E-03 | 1.46 | + + | ESRRB* |
| 14 | rs9323838 | 88789353 | G | 0.37 | 56 | 8.68E-03 | 1.33 | 0.38 | 49 | 7.94E-03 | 0.78 | + - | FOXN3* |
| 15 | rs1531636 | 92404552 | A | 0.36 | 14 | 2.36E-03 | 1.40 | 0.34 | 44 | 7.05E-03 | 1.28 | + + | LOC283682 & LOC100129642 |
| 16 | rs7202333 | 67438996 | G | 0.39 | 32 | 4.76E-03 | 0.73 | 0.37 | 47 | 7.24E-03 | 0.77 | - - | TMCO7* |
| 16 | rs7184633 | 81379514 | A | 0.40 | 19 | 2.93E-03 | 0.73 | 0.40 | 1 | 2.67E-04 | 0.71 | - - | CDH13* |
| 19 | rs10411733 | 62482800 | A | 0.47 | 16 | 2.60E-03 | 0.73 | 0.46 | 25 | 3.29E-03 | 1.31 | - + | ZNF460* |
| 20 | rs2224326 | 19689491 | A | 0.23 | 9 | 1.31E-03 | 0.66 | 0.24 | 46 | 7.15E-03 | 1.31 | - + | LOC100130408* |
| 20 | rs4811610 | 53652782 | G | 0.29 | 60 | 9.92E-03 | 1.33 | 0.31 | 45 | 7.11E-03 | 0.76 | + - | RPL12P4 &CBLN4 |
| 22 | rs2073760 | 17886456 | A | 0.40 | 49 | 7.33E-03 | 1.32 | 0.40 | 24 | 3.20E-03 | 0.76 | + - | CDC45L* |
| 22 | rs467768 | 28291986 | A | 0.14 | 20 | 3.43E-03 | 0.64 | 0.15 | 55 | 9.29E-03 | 0.70 | - - | NIPSNAP1* |
corresponding SNP is present in an intron in this gene
Discussion
In the current study we performed two separate GWA studies on smoking induced CMH, one in individuals with COPD and another in individuals without COPD. We did not find genome wide significance for CMH in either individuals with COPD and without COPD. However, we found suggestive evidence of association of some genes with CMH and differential mRNA expression for some of these genes. Different genes were associated with CMH in smokers with and without COPD. We found one overlapping SNP associated with CMH in NELSON participants with and without COPD with a p-value < 10−3, yet this was not replicated in the validation cohorts. Together our data raise the possibility that the pathogenetic development of CMH is differentially regulated in individuals with and without COPD.
In the analysis of CMH performed in individuals with COPD, we found one SNP, rs10461985, in GDNF-AS1 which has a lower p-value in the replication cohorts compared with the identification analysis (p = 5.43 × 10−5 and p = 1.82 × 10−4 respectively), the SNP showing the same direction of effect in all cohorts except one separately. Antisense RNAs are transcribed to prevent translation of a complementary mRNA by base pairing to it and blocking translation [25]. In this way GDNF-AS1 prevents expression of GDNF. As GDNF expression was significantly lower in bronchial biopsies of COPD patients with CMH than without CMH, this is suggestive for the hypothesis that expression of GDNF-AS1 attenuates CMH. Unfortunately, we were not able to perform a relevant study to assess the expression of GDNF-AS1 in bronchial biopsies of COPD-patients with and without CMH, since GDNF-AS1 was not present on the Affymetrix chip used to investigate mRNA expression in COPD patients (GLUCOLD). GDNF is a neurotrophic factor that can induce plasticity in sensory neurons innervating the respiratory tract and is involved in lung development [26-28]. These data suggest that GDNF is a biologically plausible candidate gene for both COPD and CMH. However, the gene has not been identified in previous GWA studies of lung function or COPD, making it more likely that it is a gene related to CMH in those who have COPD or a gene that interacts with genes associated with COPD. We did not have sufficient power to further investigate the latter possibility.
The SNP rs4863687 which is located in the MAML3 gene on chromosome 4, a transcriptional co-activator for Notch signaling, was associated with CMH in individuals without COPD. It has been suggested that MAM interacts functionally with different transcription factors, including β-catenin and NF-κB both associated with lung inflammation [29]. We found a strong effect of rs4863687 genotype on MAML3 mRNA expression levels; the risk allele T was significantly associated with higher expression of MAML3. These data suggest that MAML3 affects risk for CMH by influencing inflammation. Additionally, it was shown in mice that coordinated cooperation between Wnt signaling and Notch signaling in intestinal epithelium is necessary for the maintenance of proliferative cells and that disruption of the Notch signaling pathway induces goblet cell conversion of crypt proliferative cells [30]. It is conceivable that the role of the Notch signaling pathway is also important in the airway epithelium and that MAML3 may play a role in goblet cell hyperplasia and consequently CMH.
Rs944899, associated with CMH in individuals without COPD, is located close to the SOX1 gene that belongs to a family of transcription factors involved in many tissues and developmental processes. SOX proteins have unique functions in different cell types, and different functions within the same cell type. The specificity of these functions is regulated by protein-protein interactions [31]. SOX proteins also regulate the Wnt signaling pathway, required for the specification and differentiation of lung epithelial cells, by interacting with β-catenin [31]. Since SOX and MAML3 are both associated with β-catenin it is conceivable that there is a link between these genes and CMH.
There are limitations to the study. In this study we did not have post-bronchodilator spirometry therefore therefore some individuals without COPD may have been set in the COPD group. The power of each identification analysis (338 cases and 511 controls in COPD and 342 cases and 1,006 controls in non-COPD) is rather limited, possibly explaining the lack of genome-wide significant findings. Moreover, also some replication cohorts were underpowered and CMH is rather a rough estimate. However, we found suggestive evidence for a genetic contribution to CMH in the full population without stratification for COPD, thus suggesting that power would be more of a problem than the definition of CMH [14]. When we analyzed whether our previously reported gene SATB1 was associated with CMH in individuals with and without COPD, we also found that the significance was considerably reduced, p-values of rs6577641 being 2.52 10−2 and 5.69 10−2 respectively.
In summary, we found no significant overlap in genes associated with CMH in individuals with COPD and in individuals without COPD. In COPD lower GDNF mRNA expression in bronchial biopsies was significantly associated with CMH, possibly by the altered action of GDNF-AS1, our top gene. Furthermore, in individuals without COPD, a top SNP in MAML3 that nominally replicated in the non-COPD cohort was an eQTL in lung tissue. Our results suggest genetic heterogeneity of CMH in individuals with and without COPD and indicate that it is worthwhile to repeat this study in much larger cohorts.
Supplementary Material
Acknowledgements
The authors would like to thank the staff at the Respiratory Health Network Tissue Bank of the FRQS for their valuable assistance.
The authors would like to thank the COPDGene® Investigators - Core Units
Administrative Core: James Crapo, MD (PI), Edwin Silverman, MD, PhD (PI), Barry Make, MD, Elizabeth A. Regan, MD, PhD; Sara Penchev; Rochelle Lantz; Sandra Melanson, MSW, LCSW; Lori Stepp
Genetic Analysis Core: Terri Beaty, PhD, Nan Laird, PhD, Christoph Lange, PhD, Michael Cho, MD, Stephanie Santorico, PhD, John Hokanson, MPH, PhD, Dawn DeMeo, MD, MPH, Nadia Hansel, MD, MPH, Craig Hersh, MD, MPH, Peter Castaldi, MD, MSc, Merry-Lynn McDonald, PhD, Jin Zhou, PhD, Manuel Mattheisen, MD, Emily Wan, MD, Megan Hardin, MD, Jacqueline Hetmanski, MS, Margaret Parker, MS, Tanda Murray, MS
Imaging Core: David Lynch, MB, Joyce Schroeder, MD, John Newell, Jr., MD, John Reilly, MD, Harvey Coxson, PhD, Philip Judy, PhD, Eric Hoffman, PhD, George Washko, MD, Raul San Jose Estepar, PhD, James Ross, MSc, Mustafa Al Qaisi, MD, Jordan Zach, Alex Kluiber, Jered Sieren, Tanya Mann, Deanna Richert, Alexander McKenzie, Jaleh Akhavan, Douglas Stinson PFT QA Core, National Jewish Health: Robert Jensen, PhD
Biological Repository, Johns Hopkins University, Baltimore, MD: Homayoon Farzadegan, PhD, Stacey Meyerer, Shivam Chandan, Samantha Bragan
Data Coordinating Center and Biostatistics, National Jewish Health, Denver, CO: Douglas Everett, PhD, Andre Williams, PhD, Carla Wilson, MS, Anna Forssen, MS, Amber Powell, Joe Piccoli
Epidemiology Core, University of Colorado School of Public Health, Denver, CO: John Hokanson, MPH, PhD, Marci Sontag, PhD, Jennifer Black-Shinn, MPH, Gregory Kinney, MPH, PhDc, Sharon Lutz, MPH, PhD
COPDGene® Investigators: Clinical Centers
Ann Arbor VA: Jeffrey Curtis, MD, Ella Kazerooni, MD
Baylor College of Medicine, Houston, TX: Nicola Hanania, MD, MS, Philip Alapat, MD, Venkata Bandi, MD, Kalpalatha Guntupalli, MD, Elizabeth Guy, MD, Antara Mallampalli, MD, Charles Trinh, MD, Mustafa Atik, MD, Hasan Al-Azzawi, MD, Marc Willis, DO, Susan Pinero, MD, Linda Fahr, MD, Arun Nachiappan, MD, Collin Bray, MD, L. Alexander Frigini, MD, Carlos Farinas, MD, David Katz, MD, Jose Freytes, MD, Anne Marie Marciel, MD
Brigham and Women's Hospital, Boston, MA: Dawn DeMeo, MD, MPH, Craig Hersh, MD, MPH, George Washko, MD, Francine Jacobson, MD, MPH, Hiroto Hatabu, MD, PhD, Peter Clarke, MD, Ritu Gill, MD, Andetta Hunsaker, MD, Beatrice Trotman-Dickenson, MBBS, Rachna Madan, MD
Columbia University, New York, NY: R. Graham Barr, MD, DrPH, Byron Thomashow, MD, John Austin, MD, Belinda D'Souza, MD
Duke University Medical Center, Durham, NC: Neil MacIntyre, Jr., MD, Lacey Washington, MD, H Page McAdams, MD
Reliant Medical Group, Worcester, MA: Richard Rosiello, MD, Timothy Bresnahan, MD, Joseph Bradley, MD, Sharon Kuong, MD, Steven Meller, MD, Suzanne Roland, MD
Health Partners Research Foundation, Minneapolis, MN: Charlene McEvoy, MD, MPH, Joseph Tashjian, MD
Johns Hopkins University, Baltimore, MD: Robert Wise, MD, Nadia Hansel, MD, MPH, Robert Brown, MD, Gregory Diette, MD, Karen Horton, MD
Los Angeles Biomedical Research Institute at Harbor UCLA Medical Center, Torrance, CA: Richard Casaburi, MD, PhD, Janos Porszasz, MD, PhD, Hans Fischer, MD, Matt Budoff, MD Michael E. DeBakey VAMC, Houston, TX: Amir Sharafkhaneh, MD, Charles Trinh, MD, Hirani Kamal, MD, Roham Darvishi, MD, Marc Willis, DO, Susan Pinero, MD, Linda Fahr, MD, Arun Nachiappan, MD, Collin Bray, MD, L. Alexander Frigini, MD, Carlos Farinas, MD, David Katz, MD, Jose Freytes, MD, Anne Marie Marciel, MD
Minneapolis VA: Dennis Niewoehner, MD, Quentin Anderson, MD, Kathryn Rice, MD, Audrey Caine, MD
Morehouse School of Medicine, Atlanta, GA: Marilyn Foreman, MD, MS, Gloria Westney, MD, MS, Eugene Berkowitz, MD, PhD
National Jewish Health, Denver, CO: Russell Bowler, MD, PhD, David Lynch, MB, Joyce Schroeder, MD, Valerie Hale, MD, John Armstrong, II, MD, Debra Dyer, MD, Jonathan Chung, MD, Christian Cox, MD
Temple University, Philadelphia, PA: Gerard Criner, MD, Victor Kim, MD, Nathaniel Marchetti, DO, Aditi Satti, MD, A. James Mamary, MD, Robert Steiner, MD, Chandra Dass, MD, Libby Cone, MD
University of Alabama, Birmingham, AL: William Bailey, MD, Mark Dransfield, MD, Michael Wells, MD, Surya Bhatt, MD, Hrudaya Nath, MD, Satinder Singh, MD
University of California, San Diego, CA: Joe Ramsdell, MD, Paul Friedman, MD
University of Iowa, Iowa City, IA: Alejandro Cornellas, MD, John Newell, Jr., MD, Edwin JR van Beek, MD, PhD
University of Michigan, Ann Arbor, MI: Fernando Martinez, MD, MeiLan Han, MD, Ella Kazerooni, MD
University of Minnesota, Minneapolis, MN: Christine Wendt, MD, Tadashi Allen, MD
University of Pittsburgh, Pittsburgh, PA: Frank Sciurba, MD, Joel Weissfeld, MD, MPH, Carl Fuhrman, MD, Jessica Bon, MD, Danielle Hooper, MD
University of Texas Health Science Center at San Antonio, San Antonio, TX: Antonio Anzueto, MD, Sandra Adams, MD, Carlos Orozco, MD, Mario Ruiz, MD, Amy Mumbower, MD, Ariel Kruger, MD, Carlos Restrepo, MD, Michael Lane, MD
Principal investigators and centers participating in ECLIPSE include: Bulgaria: Y. Ivanov, Pleven; K. Kostov, Sofia. Canada: J. Bourbeau, Montreal; M. Fitzgerald, Vancouver; P. Hernández, Halifax; K. Killian, Hamilton; R. Levy, Vancouver; F. Maltais, Montreal; D. O'Donnell, Kingston. Czech Republic: J. Krepelka, Praha. Denmark: J. Vestbo, Hvidovre. The Netherlands: E. Wouters, Horn. New Zealand: D. Quinn, Wellington. Norway: P. Bakke, Bergen, Slovenia: M. Kosnik, Golnik. Spain: A. Agusti, Jaume Sauleda, Palma de Mallorca. Ukraine: Y. Feschenko, Kiev; V. Gavrisyuk, Kiev; L. Yashina, Kiev. UK: L. Yashina, W. MacNee, Edinburgh; D. Singh, Manchester; J. Wedzicha, London. USA: A. Anzueto, San Antonio, TX; S. Braman, Providence. RI; R. Casaburi, Torrance CA; B. Celli, Boston, MA; G. Giessel, Richmond, VA; M. Gotfried, Phoenix, AZ; G. Greenwald, Rancho Mirage, CA; N. Hanania, Houston, TX; D. Mahler, Lebanon, NH; B. Make, Denver, CO; S. Rennard, Omaha, NE; C. Rochester, New Haven, CT; P. Scanlon, Rochester, MN; D. Schuller, Omaha, NE; F. Sciurba, Pittsburgh, PA; A. Sharafkhaneh, Houston, TX; T. Siler, St Charles, MO; E. Silverman, Boston, MA; A. Wanner, Miami, FL; R. Wise, Baltimore, MD; R. ZuWallack, Hartford, CT.
Steering Committee: H. Coxson (Canada), C. Crim (GlaxoSmithKline, USA), L. Edwards (GlaxoSmithKline, USA), D. Lomas (UK), W. MacNee (UK), E. Silverman (USA), R. Tal Singer (Co-chair, GlaxoSmithKline, USA), J. Vestbo (Co-chair, Denmark), J. Yates (GlaxoSmithKline, USA).
Scientific Committee: A. Agusti (Spain), P. Calverley (UK), B. Celli (USA), C. Crim (GlaxoSmithKline, USA), B. Miller (GlaxoSmithKline, USA), W. MacNee (Chair, UK), S. Rennard (USA), R. Tal-Singer (GlaxoSmithKline, USA), E. Wouters (The Netherlands), J. Yates (GlaxoSmithKline, USA).
Financial support for the study
The COPACETIC study was funded by EU FP7 grant (201379).
The NELSON study was supported by 'Zorg Onderzoek Nederland-Medische Wetenschappen (ZONMW)',' KWF Kankerbestrijiding', 'Stichting Centraal Fonds Reserves van Voormalig Vrijwillige Ziekenfondsverzekeringen (RvvZ).
The LifeLines cohort study was sponsored by the Dutch ministry of Health, Welfare and Sport, the ministry of Economic Affairs, Agriculture and Innovation, the province of Groningen, the European Union (regional development fund), the Northern Netherlands Provinces (SNN), the Netherlands Organisation for Scientific Research (NWO), University Medical Center Groningen (UMCG), University of Groningen, de Nierstichting (the Dutch Kidney Foundation), and the Diabetes Fonds (the Diabetic Foundation).
The COPDGene study was funded by NIH grants R01 HL089856 and R01 HL089897 and by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, and Sunovion.
The ECLIPSE study was funded by GlaxoSmithKline.
Data sampling for the GenKOLS study was funded by GlaxoSmithKline.
The MESA Lung/SHARe Study was funded by NIH grant RC1HL100543. MESA and the MESA SHARe project are conducted and supported by contracts N01-HC-95159 through N01-HC-95169 and RR-024156 from the National Heart, Lung, and Blood Institute (NHLBI). MESA Air is conducted and supported by the United States Environmental Protection Agency (EPA) in collaboration with MESA Air investigators, with support provided by grant RD83169701. Funding for MESA SHARe genotyping was provided by NHLBI Contract N02-HL-6-4278. MESA Family is conducted and supported in collaboration with MESA investigators; support is provided by grants and contracts R01HL071051, R01HL071205, R01HL071250, R01HL071251, R01HL071252, R01HL071258, R01HL071259, M01-RR00425, UL1RR033176, and UL1TR000124. The MESA Lung and MESA COPD Studies are funded by NIH grants R01HL077612 and R01HL093081. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org.
The lung eQTL study at Laval University was supported by the Chaire de pneumologie de la Fondation JD Bégin de l'Université Laval, the Fondation de l'Institut universitaire de cardiologie et de pneumologie de Québec, the Respiratory Health Network of the FRQS, the Canadian Institutes of Health Research (MOP - 123369), and the Cancer Research Society and Read for the Cure. Y. Bossé is the recipient of a Junior 2 Research Scholar award from the Fonds de recherche Québec – Santé (FRQS).
Footnotes
Take home message:
Genetic determinants of chronic mucus hypersecretion may differ by COPD status.
Extended Banner/ Group Author
Group Author: the LifeLines Cohort study
LifeLines Cohort Study: BZ Alizadeh1, RA de Boer2, HM Boezen1, M Bruinenberg3, L Franke4, P van der Harst2, HL Hillege1,2, MM van der Klauw5, G Navis6, J Ormel7, DS Postma8, JGM Rosmalen7, JP Slaets9, H Snieder1, RP Stolk1, BHR Wolffenbuttel5, C Wijmenga4
1University of Groningen, University Medical Center Groningen, Department of Epidemiology, Groningen, the Netherlands;
2University of Groningen, University Medical Center Groningen, Department of Cardiology, Groningen, the Netherlands;
3University of Groningen, University Medical Center Groningen, the LifeLines Cohort Study, Groningen, the Netherlands;
4University of Groningen, University Medical Center Groningen, Department of Genetics, Groningen, the Netherlands;
5University of Groningen, University Medical Center Groningen, Department of Endocrinology, Groningen, the Netherlands;
6University of Groningen, University Medical Center Groningen, Department of Internal Medicine, Division of Nephrology, Groningen, the Netherlands;
7University of Groningen, University Medical Center Groningen, Interdisciplinary Center of Psychopathology of Emotion Regulation (ICPE), Department of Psychiatry, Groningen, the Netherlands;
8University of Groningen, University Medical Center Groningen, Department of Pulmonology, Groningen, the Netherlands
References
- 1.de Oca MM, Halbert RJ, Lopez MV, Perez-Padilla R, Talamo C, Moreno D, Muino A, Jardim JR, Valdivia G, Pertuze J, Menezes AM. The chronic bronchitis phenotype in subjects with and without COPD: the PLATINO study. Eur Respir J. 2012;40:28–36. doi: 10.1183/09031936.00141611. [DOI] [PubMed] [Google Scholar]
- 2.Kim V, Criner GJ. Chronic bronchitis and chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2013;187:228–237. doi: 10.1164/rccm.201210-1843CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vestbo J. Epidemiological studies in mucus hypersecretion. Novartis Found Symp. 2002;248:3–12. [PubMed] [Google Scholar]
- 4.Agusti A, Calverley PM, Celli B, Coxson HO, Edwards LD, Lomas DA, MacNee W, Miller BE, Rennard S, Silverman EK, Tal-Singer R, Wouters E, Yates JC, Vestbo J. Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) investigators. Characterisation of COPD heterogeneity in the ECLIPSE cohort. Respir Res. 2010;11:122–9921-11-122. [Google Scholar]
- 5.Dijkstra AE, de Jong K, Boezen HM, Kromhout H, Vermeulen R, Groen HJ, Postma DS, Vonk JM. Risk factors for chronic mucus hypersecretion in individuals with and without COPD: influence of smoking and job exposure on CMH. Occup Environ Med. 2014 doi: 10.1136/oemed-2013-101654. [DOI] [PubMed] [Google Scholar]
- 6.Lange P, Parner J, Prescott E, Vestbo J. Chronic bronchitis in an elderly population. Age Ageing. 2003;32:636–642. doi: 10.1093/ageing/afg108. [DOI] [PubMed] [Google Scholar]
- 7.Patel IS, Seemungal TA, Wilks M, Lloyd-Owen SJ, Donaldson GC, Wedzicha JA. Relationship between bacterial colonisation and the frequency, character, and severity of COPD exacerbations. Thorax. 2002;57:759–764. doi: 10.1136/thorax.57.9.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vestbo J. Chronic bronchitis: should it worry us? Chron Respir Dis. 2004;1:173–176. doi: 10.1191/1479972304cd022rs. [DOI] [PubMed] [Google Scholar]
- 9.Forey BA, Thornton AJ, Lee PN. Systematic review with meta-analysis of the epidemiological evidence relating smoking to COPD, chronic bronchitis and emphysema. BMC Pulm Med. 2011;11:36–2466-11-36. doi: 10.1186/1471-2466-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viegi G, Carrozzi L, Di Pede F, Baldacci S, Pedreschi M, Modena P, Paoletti P. Risk factors for chronic obstructive pulmonary disease in a north Italian rural area. Eur J Epidemiol. 1994;10:725–731. doi: 10.1007/BF01719289. [DOI] [PubMed] [Google Scholar]
- 11.Silverman EK, Chapman HA, Drazen JM, Weiss ST, Rosner B, Campbell EJ, O'DONNELL WJ, Reilly JJ, Ginns L, Mentzer S, Wain J, Speizer FE. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease. Risk to relatives for airflow obstruction and chronic bronchitis. Am J Respir Crit Care Med. 1998;157:1770–1778. doi: 10.1164/ajrccm.157.6.9706014. [DOI] [PubMed] [Google Scholar]
- 12.Hallberg J, Dominicus A, Eriksson UK, Gerhardsson de Verdier M, Pedersen NL, Dahlback M, Nihlen U, Higenbottam T, Svartengren M. Interaction between smoking and genetic factors in the development of chronic bronchitis. Am J Respir Crit Care Med. 2008;177:486–490. doi: 10.1164/rccm.200704-565OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu G, Agusti A, Gulsvik A, Bakke P, Coxson H, Lomas DA, Silverman EK, Pillai SG. CTLA4 gene polymorphisms are associated with chronic bronchitis. Eur Respir J. 2009 doi: 10.1183/09031936.00141808. [DOI] [PubMed] [Google Scholar]
- 14.Dijkstra AE, Smolonska J, van den Berge M, Wijmenga C, Zanen P, Luinge MA, Platteel M, Lammers JW, Dahlback M, Tosh K, Hiemstra PS, Sterk PJ, Spira A, Vestbo J, Nordestgaard BG, Benn M, Nielsen SF, Dahl M, Verschuren WM, Picavet HS, Smit HA, Owsijewitsch M, Kauczor HU, de Koning HJ, Nizankowska-Mogilnicka E, Mejza F, Nastalek P, van Diemen CC, Cho MH, Silverman EK, Crapo JD, Beaty TH, Lomas DA, Bakke P, Gulsvik A, Bosse Y, Obeidat MA, Loth DW, Lahousse L, Rivadeneira F, Uitterlinden AG, Hofman A, Stricker BH, Brusselle GG, van Duijn CM, Brouwer U, Koppelman GH, Vonk JM, Nawijn MC, Groen HJ, Timens W, Boezen HM, Postma DS. LifeLines Cohort study. Susceptibility to chronic mucus hypersecretion, a genome wide association study. PLoS One. 2014;9:e91621. doi: 10.1371/journal.pone.0091621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Klaveren RJ, Oudkerk M, Prokop M, Scholten ET, Nackaerts K, Vernhout R, van Iersel CA, van den Bergh KA, van 't Westeinde S, van der Aalst C, Thunnissen E, Xu DM, Wang Y, Zhao Y, Gietema HA, de Hoop BJ, Groen HJ, de Bock GH, van Ooijen P, Weenink C, Verschakelen J, Lammers JW, Timens W, Willebrand D, Vink A, Mali W, de Koning HJ. Management of lung nodules detected by volume CT scanning. N Engl J Med. 2009;361:2221–2229. doi: 10.1056/NEJMoa0906085. [DOI] [PubMed] [Google Scholar]
- 16.Quanjer PH, Tammeling GJ, Cotes JE, Pedersen OF, Peslin R, Yernault JC. Lung volumes and forced ventilatory flows. Report Working Party Standardization of Lung Function Tests, European Community for Steel and Coal. Official Statement of the European Respiratory Society. Eur Respir J Suppl. 1993;16:5–40. [PubMed] [Google Scholar]
- 17.Cho MH, Castaldi PJ, Wan ES, Siedlinski M, Hersh CP, Demeo DL, Himes BE, Sylvia JS, Klanderman BJ, Ziniti JP, Lange C, Litonjua AA, Sparrow D, Regan EA, Make BJ, Hokanson JE, Murray T, Hetmanski JB, Pillai SG, Kong X, Anderson WH, Tal-Singer R, Lomas DA, Coxson HO, Edwards LD, Macnee W, Vestbo J, Yates JC, Agusti A, Calverley PM, Celli B, Crim C, Rennard S, Wouters E, Bakke P, Gulsvik A, Crapo JD, Beaty TH, Silverman EK. on behalf of the ICGN, ECLIPSE, and COPDGene Investigators. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, Crapo JD. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vestbo J, Anderson W, Coxson HO, Crim C, Dawber F, Edwards L, Hagan G, Knobil K, Lomas DA, MacNee W, Silverman EK, Tal-Singer R. ECLIPSE investigators. Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE). Eur Respir J. 2008;31:869–873. doi: 10.1183/09031936.00111707. [DOI] [PubMed] [Google Scholar]
- 20.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, Greenland P, Jacob DR, Jr, Kronmal R, Liu K, Nelson JC, O'Leary D, Saad MF, Shea S, Szklo M, Tracy RP. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 21.Hao K, Bosse Y, Nickle DC, Pare PD, Postma DS, Laviolette M, Sandford A, Hackett TL, Daley D, Hogg JC, Elliott WM, Couture C, Lamontagne M, Brandsma CA, van den Berge M, Koppelman G, Reicin AS, Nicholson DW, Malkov V, Derry JM, Suver C, Tsou JA, Kulkarni A, Zhang C, Vessey R, Opiteck GJ, Curtis SP, Timens W, Sin DD. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet. 2012;8:e1003029. doi: 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lapperre TS, Snoeck-Stroband JB, Gosman MM, Jansen DF, van Schadewijk A, Thiadens HA, Vonk JM, Boezen HM, Ten Hacken NH, Sont JK, Rabe KF, Kerstjens HA, Hiemstra PS, Timens W, Postma DS, Sterk PJ, Groningen Leiden Universities Corticosteroids in Obstructive Lung Disease Study Group Effect of fluticasone with and without salmeterol on pulmonary outcomes in chronic obstructive pulmonary disease: a randomized trial. Ann Intern Med. 2009;151:517–527. doi: 10.7326/0003-4819-151-8-200910200-00004. [DOI] [PubMed] [Google Scholar]
- 23.van den Berge M, Steiling K, Timens W, Hiemstra PS, Sterk PJ, Heijink IH, Liu G, Alekseyev YO, Lenburg ME, Spira A, Postma DS. Airway gene expression in COPD is dynamic with inhaled corticosteroid treatment and reflects biological pathways associated with disease activity. Thorax. 2014;69:14–23. doi: 10.1136/thoraxjnl-2012-202878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss B, Davidkova G, Zhou LW. Antisense RNA gene therapy for studying and modulating biological processes. Cell Mol Life Sci. 1999;55:334–358. doi: 10.1007/s000180050296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tollet J, Everett AW, Sparrow MP. Development of neural tissue and airway smooth muscle in fetal mouse lung explants: a role for glial-derived neurotrophic factor in lung innervation. Am J Respir Cell Mol Biol. 2002;26:420–429. doi: 10.1165/ajrcmb.26.4.4713. [DOI] [PubMed] [Google Scholar]
- 27.Karihaloo A, Nickel C, Cantley LG. Signals which build a tubule. Nephron Exp Nephrol. 2005;100:e40–5. doi: 10.1159/000084111. [DOI] [PubMed] [Google Scholar]
- 28.Lieu TM, Myers AC, Meeker S, Undem BJ. TRPV1 induction in airway vagal low-threshold mechanosensory neurons by allergen challenge and neurotrophic factors. Am J Physiol Lung Cell Mol Physiol. 2012;302:L941–8. doi: 10.1152/ajplung.00366.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oyama T, Harigaya K, Sasaki N, Okamura Y, Kokubo H, Saga Y, Hozumi K, Suganami A, Tamura Y, Nagase T, Koga H, Nishimura M, Sakamoto R, Sato M, Yoshida N, Kitagawa M. Mastermind-like 1 (MamL1) and mastermind-like 3 (MamL3) are essential for Notch signaling in vivo. Development. 2011;138:5235–5246. doi: 10.1242/dev.062802. [DOI] [PubMed] [Google Scholar]
- 30.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 31.Bernard P, Harley VR. Acquisition of SOX transcription factor specificity through protein-protein interaction, modulation of Wnt signalling and post-translational modification. Int J Biochem Cell Biol. 2010;42:400–410. doi: 10.1016/j.biocel.2009.10.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


