Abstract
Objective
Combined antiretroviral treatment (cART) has changed the clinical presentation of HIV-associated neurocognitive disorders (HAND) to that of the milder forms of the disease. Asymptomatic neurocognitive impairment (ANI) is now more prevalent and is associated with increased morbidity and mortality risk in HIV-1–infected people. HIV-1 envelope (env) genetic heterogeneity has been detected within the central nervous system (CNS) of individuals with ANI. Changes within env determine co-receptor use, cellular tropism, and neuropathogenesis. We hypothesize that compartmental changes are associated with HIV-1 env C2V4 during ANI and sought to analyze paired HIV-1 env sequences from plasma and cerebrospinal fluid (CSF) of a female subject undergoing long-term cART.
Methods
Paired plasma and CSF samples were collected at 12-month intervals and HIV-1 env C2V4 was cloned and sequenced.
Results
Phylogenetic analysis of paired samples consistently showed genetic variants unique to the CSF. Phenotypic prediction showed CCR5 (R5) variants for all CSF-derived sequences and showed minor X4 variants (or dual-tropic) in the plasma at later time points. Viral compartmentalization was evident throughout the study, suggesting that the occurrence of distinctive env strains may contribute to the neuropathogenesis of HAND.
Conclusions
Our study provides new insights about the genetic characteristics within the C2V4 of HIV-1 env that persist after long-term cART and during the course of persistent ANI.
Keywords: HIV, HAND, envelope, evolution, asymptomatic neurocognitive impairment, plasma, cerebrospinal fluid
Introduction
HIV+ subjects may develop neurocognitive changes within the early stages of infection [1]. HIV-1–associated neurocognitive disorders (HAND) is a collective term for the neurological morbidities and neurocognitive impairment (NI) that develop in approximately 40 to 56% of HIV-1 cases [2–4]. The clinical categories of HAND are based on the disease severity: asymptomatic neurocognitive impairment (ANI), mild cognitive motor disorder (MCMD), and HIV-1–associated dementia (HAD) [5]. ANI, MCMD, and HAD have an estimated prevalence of 33%, 12%, and 2%, respectively [3]. Since the advent of combined antiretroviral treatment (cART), the clinical features of HAND have shifted from those associated with the severe forms of the disease to those associated with its milder forms [4]. Consequently, the incidence of HIV+ cases presenting severe HAND has declined, resulting in a greater prevalence of cases with the asymptomatic form [3, 4]. HIV+ subjects with neurological comorbidities are at greater risk of mortality than those that are neurologically intact [2]. HIV+ subjects with ANI have a greater likelihood of symptomatic neurocognitive decline, a prognosis that seems to be overrepresented in females and in subjects with low nadir CD4+ levels [6].
In the plasma of recently infected individuals, HIV-1 genetic diversity is low [7], and the invading viral population is exposed to selective pressures that affect viral fitness, thereby allowing unique viral genomes to prevail [8, 9]. In addition, the diversification of viral isolates is influenced by other means, such as error-prone reverse transcriptase activity [10], extensive viral recombination [11, 12], and viral escape from adaptive immunity [13, 14]. In the CNS, viral diversity is constrained as well by the receptor and co-receptor expression phenotypes of the immune cells in the brain [15–17]. HIV-1 env evolution in the CNS occurs independently from the periphery [18, 19]. HIV-1 env variants from the brains or cerebrospinal fluid (CSF) of HIV+ subjects with or without overt NI are genetically compartmentalized [20–23]. Functional studies of full-length HIV-1 env obtained from the brain tissue of HIV+ subjects with NI demonstrate enhanced CD4-gp120 interactions and a lower CD4 dependence [24–26]. These observations highlight the functional relevance of genetic compartmentalization in providing phenotypic advantages for infectivity. While V3 loop variation is considered the major determinant for modulating cellular tropism and co-receptor usage [27–29], interactions of V3 with other regions are known to influence these mechanisms as well [30, 31].
The neurotropic and neurovirulent capacity of HIV-1 envs are characterized by M-tropism [24, 26, 28, 32]. Indeed, envs recovered from the brains of demented patients are predominantly CCR5 and M-tropic in vitro [33]. The relationship between the characteristics which specify for CCR5 usage compared to the determinants of M-tropism are highly complex and are not limited to the V3 loop [24, 34–37]. One mechanism by which HIV-1 env M-tropism is enhanced, is the deletion of a single potential N-linked glycosylation (PNLG) site at position 386 of the V4 region [38]. In highly M-tropic variants, Env has an enhanced ability to interact CD4 at low density and the resulting CD4-gp120 bound complex simultaneously adopts a conformation that facilitates the interaction with the N-terminus of the CCR5 co-receptor [39]. In contrast, the fusogenic mechanism of CXCR4-restricted Env in macrophages requires a critical isoleucine residue at position 326 of the V3 region coupled with additional determinants mapped to the V1/V2 and V5 regions [28]. Taken together, these findings highlight the importance of studying genetic variability within HIV-1 env as an additional risk factor of NI. In our study, we used an in silico approach to analyze the C2V4 regions in a total of 60 env sequences obtained from cell-free CSF and plasma samples from a single donor across multiple time points with chronic and stable ANI.
Materials and Methods
Ethics statement
This study was approved by the Institutional Review Boards of Ponce Health Sciences University and the University of Puerto Rico, Medical Sciences Campus. The samples were devoid of any personal identification.
Samples
Paired cell-free plasma and cerebrospinal fluid (CSF) samples were collected once a year during the evaluation period of the subject within the Specialized Neurosciences Research Program (SNRP) cohort at University of Puerto Rico, Medical Sciences Campus [40, 41]. The inclusion criteria, exclusion criteria, and evaluation have been previously described [42]. The samples analyzed correspond to evaluations 3, 5, and 7 out of a total of 9 evaluations that were made over a 4.5-year period. The subject had been diagnosed with asymptomatic neurocognitive impairment (ANI) [5] using a modified AAN guideline, since the samples were obtained and the evaluation performed before the HAND diagnosis [41]. The ANI diagnosis remained unchanged for more than 2.5 years. Plasma and CSF viral loads were determined with the use of an ultra-sensitive RNA Roche Amplicor at an ACTG-certified laboratory, using a detection range of 50 to 75000 copies of RNA/mL. Cognitive status was determined using the American Academy of Neurology HIV-associated dementia guideline that had been modified (1991, 1996, AAN criteria, American Academy of Neurology AIDS Task Force, m-AAN criteria) to include an asymptomatic stage (described previously) [42]. Information related to cART therapy and CD4+ counts was provided by the SNRP group.
Viral RNA extraction, envelope amplification, and cloning
The samples were thawed on ice and centrifuged, followed by total viral RNA extraction using QIAamp® Ultrasense® Virus Kit from QIAGEN®, according to the manufacturer’s instruction. A 525 bp fragment containing the HIV-1 env C2-V4 variable region was amplified by RT-PCR and nested PCR using primers against the conserved regions C2 and C4 (forward primer: 5′-CTGTTAAATGGCAGTCTAGC-3′; reverse primer: 5′-TGATGGGAGGGGTATACATT-3′ [43]. The PCR-amplified product was verified by agarose gel electrophoresis, gel-purified, and cloned into cloning vector pCR2.1 (Invitrogen). Eight to 10 clones of each sample were sent to MCLAB (South San Francisco, CA) for sequencing.
Sequence alignment and phylogenetic analyses
All the sequence files were first manually verified and edited as necessary using the software Chromas Lite 2.0 (Technelysium Pty Ltd, Australia). Edited sequences were aligned using BioEdit version 7.0.5.2 [44] and Clustal W [45]. The Clustal W program (runs within BioEdit) was set to perform multiple sequence alignments using the default penalties. The sequences were then subjected to phylogenetic analysis, including tree construction, and divergence and diversity calculations using MEGA 6 [46]. Finally, sequences were translated to amino acids using BioEdit. Envelope gene sequences from Clade B HIV-1 HXB2, BaL, 89.6 and JR-CSF were used as reference sequences for CCR5-, CXCR4-, and dual- tropic strains, respectively. The evolutionary history was inferred using the neighbor-joining method [47], whereas the evolutionary distances were computed using the Jukes–Cantor method [48] and are in the units of the number of base substitutions per site. The cumulative number of synonymous and non-synonymous substitutions was estimated using the Synonymous/Non-synonymous Analysis Program (SNAP; http://hiv-web.lanl.gov), which calculates rates of nucleotide substitutions from a set of codon-aligned nucleotide sequences (based on the method of Nei and Gojobori) [49–51].
Prediction of co-receptor usage and cellular tropism
We used previously validated tools for the prediction of co-receptor use and cellular tropism based on the V3 genotype [52]. These methods are based on the principle of the net V3 loop charge in combination with the presence of basic amino acid residues (i.e., lysine and aspartate) at position 11/25 in clade B subtypes. V3 sequences with a net charge ≥ +5 are associated with the X4 and R5/X4 phenotype, whereas a net charge of < +5 predicts an R5 phenotype [53, 54]. The genotypic algorithm position-specific scoring matrix (PSSM) (http://indra.mullins.microbiol.washington.edu/webpssm/) allows the prediction of co-receptor usage with a 97% specificity and 68% sensitivity [52, 55, 56]. We also supplemented co-receptor prediction analysis by using the geno2pheno tool (http://coreceptor.bioinf.mpi-inf.mpg.de/) [57].
Potential N-linked glycosylation sites (PNLGs)
The N-GlycoSite tool (http://www.hiv.lanl.gov) was used to highlight PNLGs from motifs with the amino acid context of N-X-S/T, in which an arginine (N) in the 1st position is followed by any amino acid (X, with the exception of proline) and a threonine or serine at the 3rd position [58].
Nucleotide sequences
Sequences in this report are available from GenBank (accession numbers: KM247301 to KM247359).
Results
Clinical findings and description of the study subject
We analyzed the HIV env C2V4 region obtained from paired longitudinal plasma and CSF from a single HIV-infected woman with an ANI diagnosis. She was infected by heterosexual contact, had a negative toxicology report, and was negative for the hepatitis C virus. Table 1 summarizes the clinical data of the subject. The subject was evaluated in the SNRP cohort for 4.5 years, with evaluations every 6 months, for a total of 9 evaluations [40, 42]. The data presented correspond to evaluations 3, 5, and 7. Initially, she was undergoing a cART regimen consisting of AZT-3TC (Combivir) and efavirenz (Sustiva) by evaluation 3 and 5 and was subsequently modified to receive AZT-3TC (Combivir) and Nelfinavir (Viracept) by visit 7 (Table 1). During all visits, the ANI diagnosis remained consistent. A decrease in the performance of her neuropsychological compound score was observed in evaluation 3, going from −0.5 to −0.8. A continuous decline in her CD4+ cell count was also observed, decreasing from 408 cells/μL (initial evaluation) to 58 cells/μL, being lowest at evaluation 7. With time, the CD4+ cell count increased slightly to 80 cells/μL (data not shown).
Table 1. Clinical history of study subject.
Blood and CSF samples were collected in three consecutive visits one year apart. Blood samples were used to determine CD4+ T-cell counts and viral load and the CSF sample was used for viral load. The neurological status of the subject was examined in each visit by a battery of neurological tests.
| Evaluation | Age | CD4+ T-cell count | Antiretrovirals | Plasma viral load (RNA copies/mL) | CSF viral load (RNA copies/mL) | Clinical (Neurological)/BDI* |
|---|---|---|---|---|---|---|
| 3 | 41 | 183 | AZT-3TC (Combivir); Efavirenz (Sustiva) | 138,255 | 4,345 | Asymptomatic/2 |
| 5 | 42 | 235 | AZT-3TC (Combivir); Efavirenz (Sustiva) | 1,580 | < 50 | Asymptomatic/0 |
| 7 | 43 | 58 | AZT-3TC (Combivir); Nelfinavir (Viracept) | 116,934 | < 50 | Asymptomatic/0 |
Cognitive impairment was defined according to the American Academy of Neurology (AAN) HIV criteria modified to identify asymptomatic cognitive impairment as described in the Materials and Methods.
BDI= Beck’s depression index.
CSF variants are unique within the C2V4 region of env
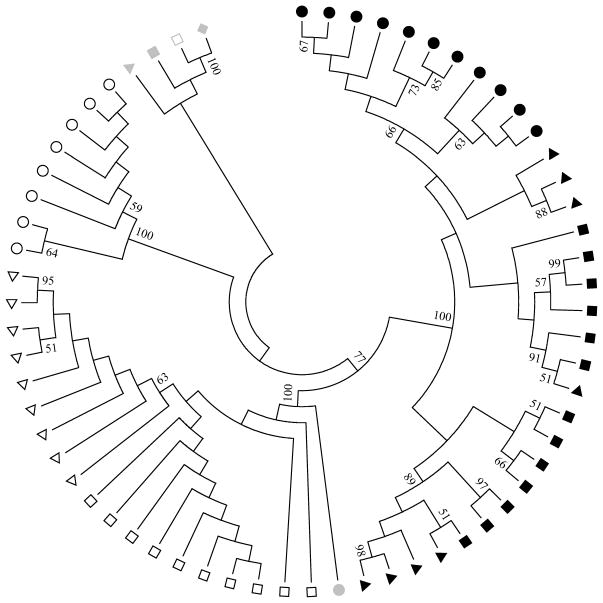
We examined the HIV-1 C2V4 region from both plasma- and CSF-derived sequences as surrogate sources of peripheral and CNS viral isolates, respectively, to assess whether viral compartmentalization was evident. A consensus bootstrap tree was inferred by using 1000 replicates (Figure 1). Neighbor-joining phylogenetic reconstruction revealed compartmentalization between the CSF and plasma sequences. The nucleotide composition within the C2V4 region revealed the occurrence of unique CNS env subpopulations by visit 7 (Figure 1, open circles). CSF sequences from visits 3 and 5 were phylogenetically closer to HIV-1 BaL (Figure 1, grey circle). Although we did not detect intermixed sequences between compartments, we found that in the plasma compartment, C2V4 sequences were intermixed in visits 3 and 5 (Figure 1).
Figure 1. HIV-1 env sequences from the CSF and the plasma of a female subject with persistent asymptomatic neurocognitive impairment are genetically distinct within the C2V4 region despite ongoing long-term cART regimen.
The evolutionary history was inferred using the neighbor-joining method [47]. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed [65]. Branches corresponding to partitions reproduced in fewer than 50% bootstrap replicates are collapsed. The evolutionary distances were computed using the maximum composite likelihood method [66] and are in the units of the number of base substitutions per site. The analysis involved 64 nucleotide sequences. The codon positions included were 1st+2nd+3rd+non-coding. All positions containing gaps and missing data were eliminated. There were a total of 355 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 [46]. Filled black symbols represent plasma-derived sequences, whereas open symbols represent CSF-derived sequences (visit 3, squares; visit 5, triangles; visit 7, circles). Grey symbols represent env sequences from the following HIV-1 strains: JR-CSF X4R5 (diamond), JR-CSF R5 (open square), HXB2 X4 (triangle), 89.6 X4R5 (square), and BaL R5 (circle).
To further assess nucleotide differences (per evaluation and compartment), we used the nucleotide alignments to calculate: 1) the mean pairwise nucleotide diversity in sequences from plasma and CSF and 2) nucleotide divergence, as a measure of the genetic distance of the sequences between compartments [59, 60]. As expected, we found that genetic diversity in the envs from plasma was higher than it was for CSF envs, and was so at all visits. Nucleotide diversity for plasma sequences during visits 3, 5, and 7 were, respectively, 3.75% (± 0.96%), 3.24% (± 0.90%), and 0.83% (± 0.31%). The mean diversity in the C2V4 of CSF envs was low and accounted for 0.28% (± 0.12%), 0.58% (± 0.24%), and 0.41% (± 0.17%) at, respectively, visits 3, 5, and 7. The divergence between plasma and CSF sequences was 12.6% ± 1.8% and 12.4% ± 1.4% for evaluations 3 and 5 respectively. Sequence divergence was the highest at evaluation 7, 15.8% ± 1.8%. The results of the genetic relationship are in accordance with previous reports stating that the V3 nucleotide diversity of the viral CNS population is lower than that of the plasma population in autologous samples [14, 61].
The inferred amino acid sequences of HIV-1 env isolates were aligned by region using BioEdit 7.0.9. Changes in amino acid composition within the V3 and V4 regions are highlighted in red (Table 2) compared to Clade B consensus sequence (HIV-1 BaL). An analysis of the V3 region from inferred amino acid composition did not reveal any conserved motifs for the plasma or CSF variants. However, we detected a conserved amino acid motif, GGEFFXXNXXXLFNSTW, within the V4 region of all the sequences (Table 3).
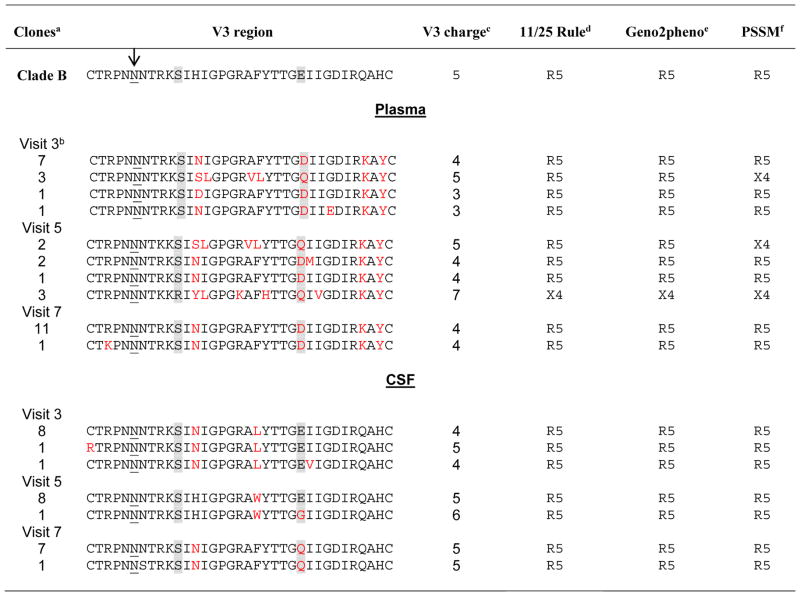
Table 2. V3 amino acid sequences and co-receptor usage prediction by HIV-1 virus clones isolated from plasma and CSF.
Co-receptor usage was determined using various computational methods. V3 loop sequences derived from clones isolated from plasma and CSF are presented. Changes in the amino acid sequence as compared to the V3 loop of the Clade B consensus sequence are highlighted in red. The arrow marks the PNLG site at position 301.
|
The number indicates the total number of clones with the same env V3 sequence.
One clone from visit 3 had a significant deletion of 31 amino acids within the V3 region (not shown).
V3 charge was determined by subtracting the total number of negatively charged amino acids (D+E) from the total number of positively charged amino acids (H+K+R). Higher positive charge has been correlated with the likelihood of CXCR4 use.
Sequences with positively charged amino acids at positions 11 and/or 25 within the V3 loop were classified as having an 11/25 genotype (believed as X4 or X4/R5 strain). Positions 11 and 25 are highlighted in grey.
Geno2pheno, env V3 sequence was aligned to the reference strain HXB2 and predicted as to whether the corresponding virus is capable of using CXCR4 as a coreceptor (R5/X4 or X4 variants) or not (R5 variants).
PSSM, Position-specific scoring matrix (PSSMX4R5 and PSSM SINSI)
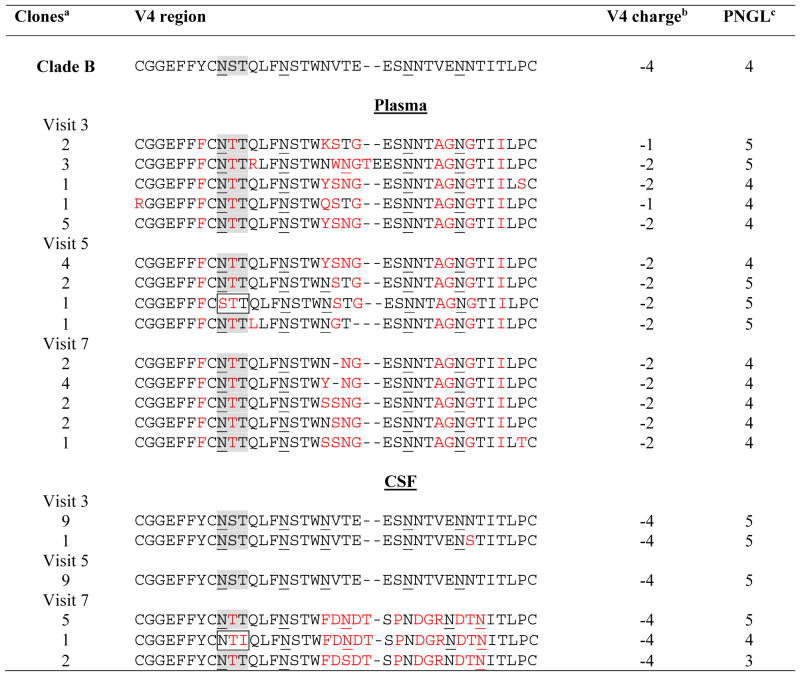
Table 3. V4 amino acid sequences by HIV-1 virus clones isolated from plasma and CSF.
V4 loop sequences derived from clones isolated from plasma and CSF are presented. Changes in the amino acid sequence as compared to the V4 loop of the Clade B consensus sequence are highlighted in red.
|
The number indicates the total number of clones with the same env V4 sequence.
V4 net charge was determined by subtracting the total number of negatively charged amino acids (D+E) from the total number of positively charged amino acids (H+K+R) [28].
PNLG: Potential N-Linked Glycosylation Site; PNLG in position 386 (gray). Loss of PNLG at this site is described to confer higher macrophage tropism and is associated with severe HIV-associated dementia as described by Dunfee et al. 2007.
All envelopes amplified from CSF were predicted to be R5
The amino acid sequence of the V3 loop of HIV-1 env has a major role in determining co-receptor use and cellular tropism. Major co-receptor (R5 or X4) phenotypes can reasonably be predicted by observation of a 35 amino acid segment of the envelope protein extending from amino acids 296 to 330. We explored the relationship using validated genotype inference methods, which predict viral phenotypes and co-receptor usage based on V3 sequences [54, 56]. The deduced amino acid sequences of the envelope V3 region were then used to predict the co-receptor usage and syncytium-inducing properties of all sequences. The data in Table 2 revealed that the presence of env sequences found in CSF were R5-utilizing isolates, whereas plasma-derived envelopes were either an R5 or dual predicted phenotype for the first 2 visits. Plasma clones amplified during the last visit exhibited a predicted R5 phenotype (Table 2).
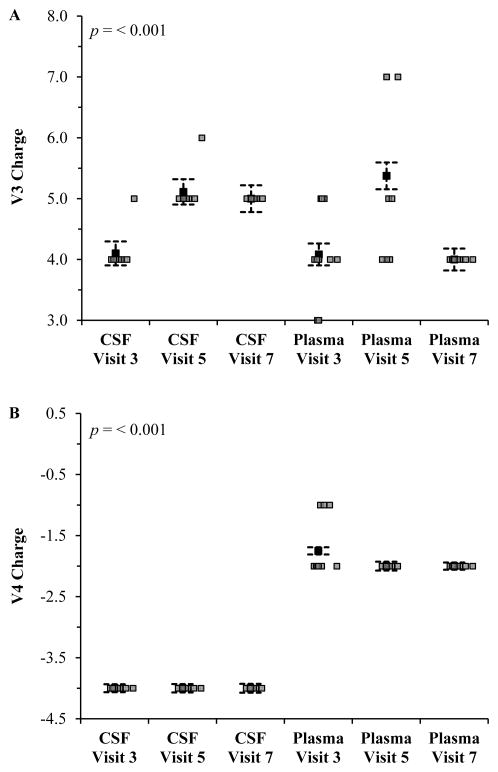
In general, the exclusive CCR5 co-receptor is favored for V3 loops having an overall net positive charge lower than +5, while CXCR4 and dual use are more common among viruses having a highly positive V3 loop greater than or equal to +5 (based on collective charges of amino acid side chains in this variable domain) [53, 54]. Multiple comparisons of the net charges of V3 and V4 regions revealed significant differences between plasma and CSF compartments (Figure 2). However, within each compartment, V3 charge in CSF was significantly different when comparing evaluation 3 to evaluations 5 and 7 (Figure 2A). Whereas for the V4 region, statistical differences were found only between plasma isolates from visits 3 and 7 (Figure 2B). The mean net charge within V4 was −4, and no statistical significances were observed in the CSF variants
Figure 2. The V3 and V4 net charges significantly differed between CSF and plasma isolates during the course of asymptomatic disease and cART.
The net charges of the gp120 regions V3 and V4 were calculated by subtracting the number of negatively charged amino acids from the number of positively charged amino acids. Error bars indicate the mean standard error. Statistical comparisons were done using single factor analysis of variance (ANOVA) followed by Tukey’s test (post hoc). Values of p<0.05 were considered to be statistically significant.
The pattern of potential N-linked glycosylation sites differed in plasma and CSF isolates
The inferred amino acid compositions in V3 and V4 of both CSF and plasma isolates were analyzed using the N-GlycoSite tool (http://www.hiv.lanl.gov). The data analysis showed a conserved PNLG site at position 301 of the V3 region in all isolates from both plasma and CSF (Table 2). In the V4 region, 1 sequence from plasma (visit 5, clone 3; net charge = −2) and 1 sequence from CSF (visit 7, clone 3; net charge = −4) had lost the PNLG at position 386 (Table 3). A conserved motif of the first 18 amino acids (CGGEFFYCNXXQLFNSTW) in V4 was present in all CSF clones. In all plasma clones, the motif CGGEFFFCNTTQLFNSTW had a conserved phenylalanine (F) residue which was not present in CSF plasma isolates (Table 3). The amino acid composition within the V4 region CSF isolates from visit 3 and that from visit 5 were similar to that of HIV-1 BaL, which is consistent with the phylogenetic grouping seen in Figure 1. However, CSF sequences from visit 7 and sequences from plasma had distinct glycosylation patterns in the V4 region (Table 3). We found that 6 of 8 isolates from visit 7 had a unique motif, FDNDTSPNDGRNDTN, and that 3 clones had a loss a PNLG site (Table 2).
Discussion
We successfully amplified and characterized the C2V4 region of HIV-1 env isolates in matched CSF and plasma samples (collected at 12-month intervals for 3 years) from a female subject diagnosed with ANI [5, 42]. Using the sequence data from matched samples, we conducted phylogenetic analyses that revealed compartmentalization across all sampling times. We expected to find lower genetic diversity in the CSF compared to plasma diversity within sequences of envs at all of the patient’s visits [14, 61]. The results were in accordance with those of previous reports wherein V3 nucleotide diversity of the viral CNS population is lower than that of the plasma population in autologous samples [14, 61]. To our knowledge, this is the first report that describes the mean viral diversity in matched plasma and CSF sequence from a Hispanic female subject with ANI who is undergoing long-term cART.
The levels of CSF viral compartmentalization vary considerably throughout the full course of disease, being high in those who are chronically infected with HAD and low to undetectable in subjects with early and acute HIV-1 infections [19]. Sequence analysis of the plasma and CSF from our study patient indicates that longitudinal compartmentalization in the C2V4 region persists without the development of overt neurocognitive symptoms. Failure to detect intermingled sequences between CSF and plasma suggests the existence of reduced viral trafficking and, in addition, that there might be some restriction on the ability of the virus to migrate freely between the 2 compartments. One possible explanation for the marked plasma/CSF env compartmentalization is the anatomical restriction which in turn provides 2 distinct settings for genetic adaptation within the same host. The brain, considered an immune-privileged site, is anatomically isolated by the blood–brain barrier, is devoid of lymphatic tissues, and is composed of the localized CNS cell population, i.e., astrocytes, microglia, and neurons. Emergence of novel CNS variants may result from both reduced immune pressure [61] and low expression levels of the HIV-1 co-receptors in the CNS [15, 16].
The complexity of virus–cell interaction cellular entry arises from interactions between CD4/gp120 and CCR5 and/or CXCR4 co-receptors which are distinct depending on the tissues infected [28, 39, 62]. Inferring co-receptor usage with bioinformatics tools remains an important tool for assessing the efficacy of pharmacological therapies, such as the use of the CCR5 antagonist, maraviroc, or the CXCR4 antagonist, AMD3100. Our results show that HIV-1 envs vary according to the compartment sampled and the time of sampling. In general, exclusive CCR5 co-receptor usage is favored for V3 loops with an overall net positive charge of less than +5, while CXCR4 and dual usage is more common among viruses with a highly positive V3 loop that ≥ +5 [53, 54]. During the first 2 visits, plasma and CSF envs from our ANI subject carried an equal overall V3 loop charge. At the last evaluation, the CSF isolates carried a higher overall V3 charge than did those found in plasma. We found that CSF-derived isolates (relative to those derived from blood) were enriched with predicted R5 variants. We believe that the use of co-receptor prediction is informative since neurotropic and neurovirulent HIV-1 strains isolated from CNS are reported to enter into macrophages and microglia via CCR5 [24].
We also evaluated whether the amino acid composition in V4 would have value in correlating with Env phenotype. For plasma isolates sampled in evaluation 3, an overall charge of −1 was observed for 3 out of 12 clones (Table 3). In addition, 9 out of 12 clones from evaluation 3, and all isolates from evaluations 5 and 7, carried a net −2 charge within V4. Statistical analyses by group indicated that the overall V4 net charge differed between CSF and plasma groups (p value <0.05). In addition, the V4 region of all CSF sequences (as opposed to the consensus clade B sequence) retained a net charge of −4 [28]. Together, these observations highlight that an overall charge of −4 in the V4 region may be an important genetic feature for the fitness of CSF envs. However, the clinical relevance of V4 loops during the course of HAND remains unclear and requires further study if we are to determine whether it is a useful region associated with HIV-1 neuropathology [38]. Future studies should aim to evaluate the usefulness of the V4 region in terms of its association with the decline of neurocognitive status.
Minor changes within the variable regions of HIV-1 env can provide the virus with several novel characteristics [63]. Among the observed biological changes associated with amino acid sequences are the loss or gain of PNLGs, such as in the case with HIV-1 env [38, 58]. This has consequences in immune evasion, sensitivity to entry inhibitors, the modulation of co-receptor preference and cellular tropism, and altered CD4/CCR5 dependence for cellular entry [13, 25, 28, 32, 38, 58]. We found that 2 clones (plasma and CSF, evaluation 5) lost a PNLG site at position 386 of V4. The importance of identifying PNLGs in association with neurological status was further noted in study a study by Dunfee, et al. [38], in which study an env isolated from the brain of a patient with HIV-1–associated dementia demonstrated an enhanced fusogenic capacity into macrophages in vitro.
All CSF clones had a conserved 18 amino acids motif within V4 (CGGEFFYCNXXQLFNSTW), and isolates from visits 3 and 5 were similar to those of HIV-1 BaL, which is consistent with the monophylogenetic grouping seen in Figure 1. All plasma isolates had a common phenylalanine (F) residue at position 384 of V4, which residue was not present in the CSF isolates. Other groups have described a conserved proline residue within V1 in X4-using M-tropic strains from HIV+ brains; this residue was associated with compartmentalization [28]. Despite the presence of a proline residue in X4-using M-tropic strains, there was no functional advantage characterized [28]. CSF sequences from evaluation 7 showed that the 8 clones had the motif FDNDTSPNDGRNDTN (position 395 to 413) and the unique occurrence of 1 proline and 4 aspartate residues. In our case, we would expect to see a functional impact on these envs from visit 7 since a variation in the glycosylation patterns in the V4 region (3 out of 8 clones) was identified within this motif.
While our study presents some interesting viral evolution data from a Hispanic woman with mild neurocognitive impairment (ANI), it had several limitations. First, we analyzed the C2V4 region of HIV-1 env (thus hindering our analysis of other regions such as V1/V2 and V5) as well as limiting our interpretation of the role of cART in shaping these compartments. However, we would expect to see consistent evolution within other variable regions throughout the course of disease, as previously reported to occur in chronically infected subjects [19]. Second, considering that the available inference bioinformatics tools for co-receptor prediction are not 100% specific, we chose to select those with a greater than 85% concordance of prediction with clade B consensus [64] for our co-receptor analyses. We found that CSF variants were genetically unique within their C2V4 region during the setting of long-term suppressive cART and persistent ANI. In sum, probing the functional phenotypes of CSF-derived HIV-1 env variants will allow for the identification of putative mutations present in the subjects with ANI and for the identification as well of potential for neurotoxic properties that are associated with the C2V4 region.
Acknowledgments
Grant numbers: G12-MD007579, P20RR11126, R01-MH08316-01, U54 NS3011, and GM082406.
The project was supported by the following grants from the National Institutes of Health: NIMHD G12-MD007579, P20RR11126, R01-MH08316-01, U54 NS3011, and NIGMS RISE GM082406. We would like to thank the personnel of the Clinical Research Center for providing the samples and clinical data, the Molecular and Genomics Core (G12-MD007579), the Technological Resources for Core Laboratories (MD007587) for providing technical support, and Mr. Bob Ritchie for providing editing services.
References
- 1.Moore DJ, Letendre SL, Morris S, Umlauf A, Deutsch R, Smith DM, Little S, Rooney A, Franklin DR, Gouaux B, Leblanc S, Rosario D, Fennema-Notestine C, Heaton RK, Ellis RJ, Atkinson JH, Grant I. Neurocognitive functioning in acute or early HIV infection. Journal of neurovirology. 2011;17:50–7. doi: 10.1007/s13365-010-0009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vivithanaporn P, Heo G, Gamble J, Krentz HB, Hoke A, Gill MJ, Power C. Neurologic disease burden in treated HIV/AIDS predicts survival: a population-based study. Neurology. 2010;75:1150–8. doi: 10.1212/WNL.0b013e3181f4d5bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heaton RK, Clifford DB, Franklin DR, Jr, Woods SP, Ake C, Vaida F, Ellis RJ, Letendre SL, Marcotte TD, Atkinson JH, Rivera-Mindt M, Vigil OR, Taylor MJ, Collier AC, Marra CM, Gelman BB, McArthur JC, Morgello S, Simpson DM, McCutchan JA, Abramson I, Gamst A, Fennema-Notestine C, Jernigan TL, Wong J, Grant I, Group C. HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology. 2010;75:2087–96. doi: 10.1212/WNL.0b013e318200d727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heaton RKFD, Ellis RJ, McCutchan JA, Letendre SL, LeBlanc S, Corkran SH, Duarte NA, Clifford DB, Woods SP, Collier AC, Marra CM, Morgello S, Rivera Mindt M, Taylor MJ, Marcotte TD, Hampton Atkinson J, Wolfson T, Gelman BB, McArthur JC, Simpson DM, Abramson I, Gamst A, Fennema-Notestine C, Jernigan TL, Wong J, Grant I the CHARTER and HNRC Groups. HIV-associated neurocognitive disorders before and during the era of combination antiretroviral therapy: differences in rates, nature, and predictors. Journal of neurovirology. 2011;17:3–16. doi: 10.1007/s13365-010-0006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antinori A, AG, Becker JT, Brew BJ, Byrd DA, Cherner M, Clifford DB, Epstein LG, Goodkin K, Gisslen M, Grant I, Heaton RK, Joseph J, Marder K, Marra CM, McArthur JC, Nunn M, Price RW, Pulliam L, Robertson KR, Sacktor N, Valcour V, Wojna VE. Updated Research Nosology for HIV Associated Neurocgonitive Disorders. Neurology. 2007;69:1780–1799. doi: 10.1212/01.WNL.0000287431.88658.8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grant I, Franklin DR, Jr, Deutsch R, Woods SP, Vaida F, Ellis RJ, Letendre SL, Marcotte TD, Atkinson JH, Collier AC, Marra CM, Clifford DB, Gelman BB, McArthur JC, Morgello S, Simpson DM, McCutchan JA, Abramson I, Gamst A, Fennema-Notestine C, Smith DM, Heaton RK, Group C. Asymptomatic HIV-associated neurocognitive impairment increases risk for symptomatic decline. Neurology. 2014;82:2055–62. doi: 10.1212/WNL.0000000000000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kearney M, Maldarelli F, Shao W, Margolick JB, Daar ES, Mellors JW, Rao V, Coffin JM, Palmer S. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. Journal of virology. 2009;83:2715–27. doi: 10.1128/JVI.01960-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biesinger T, Kimata JT. HIV-1 Transmission, Replication Fitness and Disease Progression. Virology: research and treatment. 2008;2008:49–63. [PMC free article] [PubMed] [Google Scholar]
- 9.Goodenow MM, Rose SL, Tuttle DL, Sleasman JW. HIV-1 fitness and macrophages. Journal of leukocyte biology. 2003;74:657–66. doi: 10.1189/jlb.0403186. [DOI] [PubMed] [Google Scholar]
- 10.Roberts JD, Bebenek K, Kunkel TA. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–3. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 11.Charpentier C, Nora T, Tenaillon O, Clavel F, Hance AJ. Extensive recombination among human immunodeficiency virus type 1 quasispecies makes an important contribution to viral diversity in individual patients. J Virol. 2006;80:2472–82. doi: 10.1128/JVI.80.5.2472-2482.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown RJ, Peters PJ, Caron C, Gonzalez-Perez MP, Stones L, Ankghuambom C, Pondei K, McClure CP, Alemnji G, Taylor S, Sharp PM, Clapham PR, Ball JK. Intercompartmental recombination of HIV-1 contributes to env intrahost diversity and modulates viral tropism and sensitivity to entry inhibitors. J Virol. 2011;85:6024–37. doi: 10.1128/JVI.00131-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arendrup M, Nielsen C, Hansen JE, Pedersen C, Mathiesen L, Nielsen JO. Autologous HIV-1 neutralizing antibodies: emergence of neutralization-resistant escape virus and subsequent development of escape virus neutralizing antibodies. Journal of acquired immune deficiency syndromes. 1992;5:303–7. [PubMed] [Google Scholar]
- 14.Van Marle G, Rourke SB, Zhang K, Silva C, Ethier J, Gill MJ, Power C. HIV dementia patients exhibit reduced viral neutralization and increased envelope sequence diversity in blood and brain. Aids. 2002;16:1905–14. doi: 10.1097/00002030-200209270-00007. [DOI] [PubMed] [Google Scholar]
- 15.Lavi E, Strizki JM, Ulrich AM, Zhang W, Fu L, Wang Q, O’Connor M, Hoxie JA, Gonzalez-Scarano F. CXCR-4 (Fusin), a co-receptor for the type 1 human immunodeficiency virus (HIV-1), is expressed in the human brain in a variety of cell types, including microglia and neurons. The American journal of pathology. 1997;151:1035–42. [PMC free article] [PubMed] [Google Scholar]
- 16.Flynn G, Maru S, Loughlin J, Romero IA, Male D. Regulation of chemokine receptor expression in human microglia and astrocytes. Journal of neuroimmunology. 2003;136:84–93. doi: 10.1016/s0165-5728(03)00009-2. [DOI] [PubMed] [Google Scholar]
- 17.van der Meer P, Ulrich AM, Gonzalez-Scarano F, Lavi E. Immunohistochemical analysis of CCR2, CCR3, CCR5, and CXCR4 in the human brain: potential mechanisms for HIV dementia. Experimental and molecular pathology. 2000;69:192–201. doi: 10.1006/exmp.2000.2336. [DOI] [PubMed] [Google Scholar]
- 18.Harrington PR, Nelson JA, Kitrinos KM, Swanstrom R. Independent evolution of human immunodeficiency virus type 1 env V1/V2 and V4/V5 hypervariable regions during chronic infection. J Virol. 2007;81:5413–7. doi: 10.1128/JVI.02554-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrington PR, Schnell G, Letendre SL, Ritola K, Robertson K, Hall C, Burch CL, Jabara CB, Moore DT, Ellis RJ, Price RW, Swanstrom R. Cross-sectional characterization of HIV-1 env compartmentalization in cerebrospinal fluid over the full disease course. Aids. 2009;23:907–15. doi: 10.1097/QAD.0b013e3283299129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritola K, Robertson K, Fiscus SA, Hall C, Swanstrom R. Increased human immunodeficiency virus type 1 (HIV-1) env compartmentalization in the presence of HIV-1-associated dementia. J Virol. 2005;79:10830–4. doi: 10.1128/JVI.79.16.10830-10834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnell G, Joseph S, Spudich S, Price RW, Swanstrom R. HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS pathogens. 2011;7:e1002286. doi: 10.1371/journal.ppat.1002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cunningham AL, Naif H, Saksena N, Lynch G, Chang J, Li S, Jozwiak R, Alali M, Wang B, Fear W, Sloane A, Pemberton L, Brew B. HIV infection of macrophages and pathogenesis of AIDS dementia complex: interaction of the host cell and viral genotype. Journal of leukocyte biology. 1997;62:117–25. doi: 10.1002/jlb.62.1.117. [DOI] [PubMed] [Google Scholar]
- 23.Schnell G, Price RW, Swanstrom R, Spudich S. Compartmentalization and clonal amplification of HIV-1 variants in the cerebrospinal fluid during primary infection. J Virol. 2010;84:2395–407. doi: 10.1128/JVI.01863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorry PR, Taylor J, Holm GH, Mehle A, Morgan T, Cayabyab M, Farzan M, Wang H, Bell JE, Kunstman K, Moore JP, Wolinsky SM, Gabuzda D. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. Journal of virology. 2002;76:6277–92. doi: 10.1128/JVI.76.12.6277-6292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas ER, Dunfee RL, Stanton J, Bogdan D, Taylor J, Kunstman K, Bell JE, Wolinsky SM, Gabuzda D. Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology. 2007;360:105–19. doi: 10.1016/j.virol.2006.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorry PR, Bristol G, Zack JA, Ritola K, Swanstrom R, Birch CJ, Bell JE, Bannert N, Crawford K, Wang H, Schols D, De Clercq E, Kunstman K, Wolinsky SM, Gabuzda D. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J Virol. 2001;75:10073–89. doi: 10.1128/JVI.75.21.10073-10089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Q, Trent JO, Tomaras GD, Wang Z, Murray JL, Conolly SM, Navenot JM, Barry AP, Greenberg ML, Peiper SC. Identification of ENV determinants in V3 that influence the molecular anatomy of CCR5 utilization. Journal of molecular biology. 2000;302:359–75. doi: 10.1006/jmbi.2000.4076. [DOI] [PubMed] [Google Scholar]
- 28.Ghaffari G, Tuttle DL, Briggs D, Burkhardt BR, Bhatt D, Andiman WA, Sleasman JW, Goodenow MM. Complex determinants in human immunodeficiency virus type 1 envelope gp120 mediate CXCR4-dependent infection of macrophages. J Virol. 2005;79:13250–61. doi: 10.1128/JVI.79.21.13250-13261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smit TK, Wang B, Ng T, Osborne R, Brew B, Saksena NK. Varied tropism of HIV-1 isolates derived from different regions of adult brain cortex discriminate between patients with and without AIDS dementia complex (ADC): evidence for neurotropic HIV variants. Virology. 2001;279:509–26. doi: 10.1006/viro.2000.0681. [DOI] [PubMed] [Google Scholar]
- 30.Freed EO, Martin MA. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J Virol. 1994;68:2503–12. doi: 10.1128/jvi.68.4.2503-2512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Del Prete GQ, Leslie GJ, Haggarty B, Jordan AP, Romano J, Hoxie JA. Distinct molecular pathways to X4 tropism for a V3-truncated human immunodeficiency virus type 1 lead to differential coreceptor interactions and sensitivity to a CXCR4 antagonist. J Virol. 2010;84:8777–89. doi: 10.1128/JVI.00333-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunfee R, Thomas ER, Gorry PR, Wang J, Ancuta P, Gabuzda D. Mechanisms of HIV-1 neurotropism. Current HIV research. 2006;4:267–78. doi: 10.2174/157016206777709500. [DOI] [PubMed] [Google Scholar]
- 33.Peters PJ, Bhattacharya J, Hibbitts S, Dittmar MT, Simmons G, Bell J, Simmonds P, Clapham PR. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J Virol. 2004;78:6915–26. doi: 10.1128/JVI.78.13.6915-6926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sterjovski J, Roche M, Churchill MJ, Ellett A, Farrugia W, Gray LR, Cowley D, Poumbourios P, Lee B, Wesselingh SL, Cunningham AL, Ramsland PA, Gorry PR. An altered and more efficient mechanism of CCR5 engagement contributes to macrophage tropism of CCR5-using HIV-1 envelopes. Virology. 2010;404:269–78. doi: 10.1016/j.virol.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sterjovski J, Churchill MJ, Ellett A, Gray LR, Roche MJ, Dunfee RL, Purcell DF, Saksena N, Wang B, Sonza S, Wesselingh SL, Karlsson I, Fenyo EM, Gabuzda D, Cunningham AL, Gorry PR. Asn 362 in gp120 contributes to enhanced fusogenicity by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with AIDS. Retrovirology. 2007;4:89. doi: 10.1186/1742-4690-4-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray L, Sterjovski J, Churchill M, Ellery P, Nasr N, Lewin SR, Crowe SM, Wesselingh SL, Cunningham AL, Gorry PR. Uncoupling coreceptor usage of human immunodeficiency virus type 1 (HIV-1) from macrophage tropism reveals biological properties of CCR5-restricted HIV-1 isolates from patients with acquired immunodeficiency syndrome. Virology. 2005;337:384–98. doi: 10.1016/j.virol.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 37.Rossi F, Querido B, Nimmagadda M, Cocklin S, Navas-Martin S, Martin-Garcia J. The V1-V3 region of a brain-derived HIV-1 envelope glycoprotein determines macrophage tropism, low CD4 dependence, increased fusogenicity and altered sensitivity to entry inhibitors. Retrovirology. 2008;5:89. doi: 10.1186/1742-4690-5-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunfee RL, Thomas ER, Wang J, Kunstman K, Wolinsky SM, Gabuzda D. Loss of the N-linked glycosylation site at position 386 in the HIV envelope V4 region enhances macrophage tropism and is associated with dementia. Virology. 2007;367:222–34. doi: 10.1016/j.virol.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salimi H, Roche M, Webb N, Gray LR, Chikere K, Sterjovski J, Ellett A, Wesselingh SL, Ramsland PA, Lee B, Churchill MJ, Gorry PR. Macrophage-tropic HIV-1 variants from brain demonstrate alterations in the way gp120 engages both CD4 and CCR5. Journal of leukocyte biology. 2013;93:113–26. doi: 10.1189/jlb.0612308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nieves DM, Plaud M, Wojna V, Skolasky R, Melendez LM. Characterization of peripheral blood human immunodeficiency virus isolates from Hispanic women with cognitive impairment. Journal of neurovirology. 2007;13:315–27. doi: 10.1080/13550280701361508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wojna V, Skolasky RL, McArthur JC, Maldonado E, Hechavarria R, Mayo R, Selnes O, Ginebra T, de la Torre T, Garcia H, Kraiselburd E, Melendez-Guerrero LM, Zorrilla CD, Nath A. Spanish validation of the HIV dementia scale in women. AIDS patient care and STDs. 2007;21:930–41. doi: 10.1089/apc.2006.0180. [DOI] [PubMed] [Google Scholar]
- 42.Wojna V, Skolasky RL, Hechavarria R, Mayo R, Selnes O, McArthur JC, Melendez LM, Maldonado E, Zorrilla CD, Garcia H, Kraiselburd E, Nath A. Prevalence of human immunodeficiency virus-associated cognitive impairment in a group of Hispanic women at risk for neurological impairment. Journal of neurovirology. 2006;12:356–64. doi: 10.1080/13550280600964576. [DOI] [PubMed] [Google Scholar]
- 43.Lopez CA, Vazquez M, Hill MD, del Colon MC, Porrata-Doria T, Johnston IC, Lorenzo E. Characterization of HIV-1 RNA forms in the plasma of patients undergoing successful HAART. Archives of virology. 2010;155:895–903. doi: 10.1007/s00705-010-0659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hall TA. BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 45.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic acids research. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular biology and evolution. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 48.Jukes TH, Cantor CR. Mammalian Protein Metabolism. Academic Press; New York: 1969. Evolution of protein molecules; p. 21. [Google Scholar]
- 49.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Molecular biology and evolution. 1986;3:418–26. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 50.Ota T, Nei M. Variance and covariances of the numbers of synonymous and nonsynonymous substitutions per site. Molecular biology and evolution. 1994;11:613–9. doi: 10.1093/oxfordjournals.molbev.a040140. [DOI] [PubMed] [Google Scholar]
- 51.Ganeshan S, Dickover RE, Korber BT, Bryson YJ, Wolinsky SM. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. Journal of virology. 1997;71:663–77. doi: 10.1128/jvi.71.1.663-677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jensen MA, Li FS, van ‘t Wout AB, Nickle DC, Shriner D, He HX, McLaughlin S, Shankarappa R, Margolick JB, Mullins JI. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. Journal of virology. 2003;77:13376–88. doi: 10.1128/JVI.77.24.13376-13388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gray L, Churchill MJ, Sterjovski J, Witlox K, Learmont JC, Sullivan JS, Wesselingh SL, Gabuzda D, Cunningham AL, McPhee DA, Gorry PR. Phenotype and envelope gene diversity of nef-deleted HIV-1 isolated from long-term survivors infected from a single source. Virology journal. 2007;4:75. doi: 10.1186/1743-422X-4-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delobel P, Nugeyre MT, Cazabat M, Pasquier C, Marchou B, Massip P, Barre-Sinoussi F, Israel N, Izopet J. Population-based sequencing of the V3 region of env for predicting the coreceptor usage of human immunodeficiency virus type 1 quasispecies. Journal of clinical microbiology. 2007;45:1572–80. doi: 10.1128/JCM.02090-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raymond S, Delobel P, Mavigner M, Cazabat M, Souyris C, Encinas S, Sandres-Saune K, Pasquier C, Marchou B, Massip P, Izopet J. Genotypic prediction of human immunodeficiency virus type 1 CRF02-AG tropism. Journal of clinical microbiology. 2009;47:2292–4. doi: 10.1128/JCM.02439-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raymond S, Delobel P, Mavigner M, Cazabat M, Souyris C, Sandres-Saune K, Cuzin L, Marchou B, Massip P, Izopet J. Correlation between genotypic predictions based on V3 sequences and phenotypic determination of HIV-1 tropism. Aids. 2008;22:F11–6. doi: 10.1097/QAD.0b013e32830ebcd4. [DOI] [PubMed] [Google Scholar]
- 57.Lengauer T, Sander O, Sierra S, Thielen A, Kaiser R. Bioinformatics prediction of HIV coreceptor usage. Nature biotechnology. 2007;25:1407–10. doi: 10.1038/nbt1371. [DOI] [PubMed] [Google Scholar]
- 58.Zhang M, Gaschen B, Blay W, Foley B, Haigwood N, Kuiken C, Korber B. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004;14:1229–46. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- 59.Kearney MF, Spindler J, Shao W, Yu S, Anderson EM, O’Shea A, Rehm C, Poethke C, Kovacs N, Mellors JW, Coffin JM, Maldarelli F. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS pathogens. 2014;10:e1004010. doi: 10.1371/journal.ppat.1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pillai SK, Pond SL, Liu Y, Good BM, Strain MC, Ellis RJ, Letendre S, Smith DM, Gunthard HF, Grant I, Marcotte TD, McCutchan JA, Richman DD, Wong JK. Genetic attributes of cerebrospinal fluid-derived HIV-1 env. Brain: a journal of neurology. 2006;129:1872–83. doi: 10.1093/brain/awl136. [DOI] [PubMed] [Google Scholar]
- 62.Gray L, Roche M, Churchill MJ, Sterjovski J, Ellett A, Poumbourios P, Sherieff S, Wang B, Saksena N, Purcell DF, Wesselingh S, Cunningham AL, Brew BJ, Gabuzda D, Gorry PR. Tissue-specific sequence alterations in the human immunodeficiency virus type 1 envelope favoring CCR5 usage contribute to persistence of dual-tropic virus in the brain. J Virol. 2009;83:5430–41. doi: 10.1128/JVI.02648-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dunfee RL, Thomas ER, Gorry PR, Wang J, Taylor J, Kunstman K, Wolinsky SM, Gabuzda D. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15160–5. doi: 10.1073/pnas.0605513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garrido C, Roulet V, Chueca N, Poveda E, Aguilera A, Skrabal K, Zahonero N, Carlos S, Garcia F, Faudon JL, Soriano V, de Mendoza C. Evaluation of eight different bioinformatics tools to predict viral tropism in different human immunodeficiency virus type 1 subtypes. Journal of clinical microbiology. 2008;46:887–91. doi: 10.1128/JCM.01611-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Felsenstein J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 66.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11030–5. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]