Abstract
Recently, there has been a great interest in computer-aided Alzheimer’s Disease (AD) and Mild Cognitive Impairment (MCI) diagnosis. Previous learning based methods defined the diagnosis process as a classification task and directly used the low-level features extracted from neuroimaging data without considering relations among them. However, from a neuroscience point of view, it’s well known that a human brain is a complex system that multiple brain regions are anatomically connected and functionally interact with each other. Therefore, it is natural to hypothesize that the low-level features extracted from neuroimaging data are related to each other in some ways. To this end, in this paper, we first devise a coupled feature representation by utilizing intra-coupled and inter-coupled interaction relationship. Regarding multi-modal data fusion, we propose a novel coupled boosting algorithm that analyzes the pairwise coupled-diversity correlation between modalities. Specifically, we formulate a new weight updating function, which considers both incorrectly and inconsistently classified samples. In our experiments on the ADNI dataset, the proposed method presented the best performance with accuracies of 94.7% and 80.1% for AD vs. Normal Control (NC) and MCI vs. NC classifications, respectively, outperforming the competing methods and the state-of-the-art methods.
1. Introduction
Alzheimer’s Disease (AD) and its early stage, Mild Cognitive Impairment (MCI), are becoming the most prevalent neurodegenerative brain diseases in elderly people world-wide. According to [1], the prevalence of AD will rise dramatically during the next 20 years, and 1 in 85 people will be affected by 2050. To this end, there have been a lot of efforts on investigating the underlying biological or neurological mechanisms and also discovering biomarkers for early diagnosis or prognosis of AD and MCI. Neuroimaging tools such as Magnetic Resonance Imaging (MRI) [2], Positron Emission Tomography (PET) [9], and functional MRI (fMRI) [4] have played the key roles in those works, and different neuroimaging tools can convey different information for diagnosis. Recent studies have shown that information fusion from multiple modalities can thus help enhance the diagnostic performance [5, 8, 16, 19, 22, 26, 25].
Regarding the multi-modal fusion, most of the previous methods first extracted features from each modality (e.g., gray matter tissue volume from MRI, mean signal intensities from PET), trained a typical classifier to model the training examples for each modality independently, and then combined the outputs from classifiers in an ensemble way for a final decision. Here, we should note that, to our best knowledge, those methods assumed the conditional independence among the features. However, since we extract features in a homogeneous way, e.g., statistical information from particular Region Of Interests (ROIs) in a brain, they are naturally related to each other in certain ways. Furthermore, it’s important to combine multi-modal information in a systematic manner.
To this end, in this paper, we design a new framework, in which we consider the feature-level coupled-interaction analysis and modality-level coupled-interaction analysis. Specifically, for the feature-level coupled-interaction, we devise a coupled-feature representation using intra-coupled interaction (correlations between features and their own powers) and inter-coupled interaction (correlations between features and the powers of other features) [19]. For the modality-level coupled-interaction, we propose a novel coupled boosting method that analyzes the pairwise coupled-diversity correlation between modalities. We illustrate the major difference between the previous methods and our new method in Fig. 1.
Figure 1.
The difference of the previous methods and our proposed method working on the AD/MCI diagnosis.
Fig. 2 schematizes the proposed framework, where we adopt two neuroimaging modalities of MRI and PET. Without loss of generality, we denote the MRI as modality A, and the PET as modality B. After the image preprocessing and low-level feature extraction, we find a coupled feature representation [19] by mapping the original feature vectors into the expanded feature vectors via both linear and nonlinear fashion. We then select the label-related features from the expanded feature vectors by means of Least Absolute Shrinkage and Selection Operator (LASSO), which is one of the most widely used feature selection methods in the literature. Finally, the proposed coupled boosting algorithm trains the base learners that analyze the pairwise coupled-diversity correlation between modalities at multiple rounds. Our major contributions can be two folds:
We propose a novel coupled boosting algorithm that makes a full use of the pairwise coupled diversity between multi-modal data (i.e., MRI and PET) to improve the generalization power. Unlike the previous boosting algorithms [10, 11] that usually focused on single-modal data classification, the proposed coupled boosting algorithm introduces the large diversity theory in ensemble learning and thus deals with multi-modal data classification problems, but still maintaining the general steps of AdaBoost.
A coupled feature representation method is employed to analyze the intra-coupled and inter-coupled interaction among features for AD/MCI diagnosis, which can successfully capture the intrinsic linear and nonlinear information.
Figure 2.
An illustration of the proposed framework for AD diagnosis.
The rest of this paper is organized as follows. In Section 2, we briefly introduce the related work for AD/MCI diagnosis with multi-modal data. The MRI and PET image processing and feature extraction are described in Section 3. In Section 4, we propose our new coupled feature representation and coupled boosting algorithm. Experimental results and performance comparisons with competing methods are presented in Section 5. We conclude this paper by summarizing the proposed method and also discussing the obtained results in Section 6.
2. Related Work
Recent studies have shown that fusing the complementary information from multiple modalities helps enhance the AD/MCI diagnostic accuracy [5, 8, 16, 18, 19, 22, 26]. In general, we can divide the previous methods into two categories: feature-concatenation approach and kernel-based approach. The first approach simply concatenates features from different modalities into a long feature vector and then build a classifier to find the relations implicitly. For example, Kohannim et al. concatenated features from modalities into a vector and trained a Support Vector Machine (SVM) classifier using the concatenated feature vectors [8]. Walhovd et al. applied multi-method stepwise logistic regression analyses [18], and Westman et al. exploited a hierarchical modeling of orthogonal partial least squares to latent structures [22]. Meanwhile, the latter approach fuses multi-modal information by means of a kernel technique, in which the original feature vectors are mapped into a higher dimensional space via different types of kernels. For example, Hinrichs et al. [5] and Zhang et al. [26], independently, utilized a multi-kernel SVM to combine information from different modalities.
However, none of these methods considered relational information inherent in the original feature vectors. Since a human brain is a highly complicated system and multiple brain regions interact consistently, we believe that there exist relations among brain regions, and therefore the features extracted from multiple ROIs are also highly correlated to each other. Recently, Suk and Shen used a deep learning method to find the latent high-level features that capture the relations inherent in the low-level features and achieved prominent results [16].
In this paper, we focus on the problems of feature representation and multi-modal data fusion for AD/MCI diagnosis. Specifically, we devise a feature-coupled representation by considering intra-coupled and inter-coupled interactions among features within a modality, and then propose a novel coupled boosting algorithm that systematically combine multi-modal information by building base learners at multiple rounds and finally combine them in an ensemble manner for a final decision.
3. Preprocessing and Feature Extraction
The MRI and PET images are preprocessed to extract ROI-based features. For MR images, we first perform anterior commissure-posterior commissure correction using MIPAV software1, and then re-sample the images to 256×256×256 resolution. The intensity inhomogeneity correction and skull stripping are performed by [14] and [20], respectively. The MR images are then segmented into three different types of tissues, i.e., Gray Matter (GM), White Matter (WM) and CerebroSpinal Fluid (CSF), using FAST [27] in the FMRIB Software Library (FSL) package [23]. After registration using HAMMER [12], we obtain the subject-labeled images based on a template with 93 manually labeled ROIs [7]. For each subject, we use the volumes of GM tissue of the 93 ROIs, which are normalized by the total intracranial volume (which is estimated by the summation of GM, WM and CSF volumes from all ROIs), as features. For PET images, we first align them to their respective MR images through affine transformation, and then compute the average intensity of each ROI as feature. In summary, we have 93 features from MRI and 93 features from PET, and, hereafter, we regard these features as original low-level features.
4. Proposed Method
4.1. Notations
In this paper, we denote matrices as boldface uppercase letters, vectors as boldface lowercase letters, and scalars as normal italic letters, respectively. For a matrix Z = [Zji], its j-th row is denoted as Zj,‥ For the k-th element in a vector zi, we denote it as zi,k. We further denote a transpose operator of a vector or a matrix with a superscript⊤.
Suppose that we have a set of samples , where m and n denote, respectively, the number of training samples and the number of testing samples, and without loss of generality we assume that the samples are sorted in the order of training and testing samples. Here, and denote, respectively, the original feature vector from MRI (modality A) and PET (modality B) of the i-th sample (i = 1, …, m + n), and yi ∈ {− 1, +1} is the ground truth label of the i-th sample. pA and pB denote the dimensionality of MRI and PET feature vectors, respectively2. Then, we can represent the whole original feature samples with matrices and .
4.2. Coupled Feature Representation
To our best knowledge, the previous work that addressed the AD/MCI diagnosis as a classification problem and used original low-level features without considering their relationships. However, it’s well known that a human brain is a complex system that multiple brain regions are anatomically connected and functionally interact with each other for tasks. That is, it is natural to hypothesize that the original features extracted from MRI and PET in multiple ROIs are related to each other in a way. To this end, in this work, we propose to find such latent relations among features with a coupled feature representation method [19] and use the high-level information for the AD/MCI diagnosis.
In particular, we consider two types of relational information inherent in the original low-level features: ‘intra-coupled interaction’ with correlations between features and their own powers, and ‘inter-coupled interaction’ with correlations between features and the powers of other features. Here, we should note that we find the coupled feature representation for each modality individually. That is, although we describe the feature representation for a modality A, it’s equally applicable to a modality B.
For an original feature vector of the modality A in the i-th sample, we map it to an expanded feature space with the incorporation of linear and nonlinear information by means of a matrix expansion as follows:
where indicates the e-th power of the numerical value and in this case e ∈ {1, 2}.
Utilizing the matrix expansion described above, we first define an intra-coupled interaction, which considers the correlations between the j-th feature and its own powers as follows:
where denotes a Pearson’s correlation coefficient between and , e1 = {1, 2, ⋯, E}, e2 = {1, 2, ⋯, E}, and E is a maximal power.
Besides the intra-coupled interaction, we also define an inter-coupled interaction that captures the correlations between the j-th feature and the powers of other features as follows:
where qj = [1, ⋯, j − 1, j + 1, ⋯, pA]⊤ ∈ ℝpA − 1, and is a Pearson’s correlation coefficient between and . Note that we use both the training and testing samples in inter-coupled interaction estimation for robustness by taking advantage of the information from testing samples, but the testing samples are not further involved in the following steps, i.e., feature selection and classifier learning.
Let , and w = [1/(1!), 1/(2!), …, 1/(E!)]. We integrate the intra-coupled interaction and inter-coupled interaction to obtain the coupled feature representation of the j-th feature for the i-th sample as follows:
where ⊙ and ⊗ denote, respectively, a Hadamard product and a matrix multiplication. Therefore, the final coupled feature representation for the i-th sample can be represented as follows:
| (1) |
We then apply a feature selection method to focus on only the label-related features. Specifically, we use a LASSO method [17] that selects features with a sparsity constraint in an objective function. In the following, we denote the dimension-reduced coupled-feature representation of the modality A for the i-th sample as .
4.3. Coupled Boosting
In a nutshell, our coupled boosting algorithm follows the general steps of AdaBoost [3]: iteration of (1) drawing training samples and (2) learning a base learner and determining the respective weight function.
Let T denotes the total number of iterations, and and are, respectively, the trained base learners3 for modalities of A and B at the t-th iteration. For the base learners at t-th iteration, one for each modality, we have the respective weight distributions, represented by and , where m is the number of training samples in our dataset.
At the t-th iteration, we draw samples according to the weight distributions and use them to train our base learners. After training base learners of and , we then measure the errors over the total m training samples as follows:
| (2) |
| (3) |
where and denote, respectively, the output label of the base learners and for inputs and , and l (a, b) is a loss function that outputs 1 if and only if a ≠ b, and 0 otherwise. Based on these errors, we determine the weight distributions and for the next round.
Therefore, the core problem in our coupled boosting is the way of updating the weight distribution functions. For a binary classification problem with two modalities, there exist four different cases (see Table. 1): (Case 1) Both samples and are correctly classified; (Case 2 and Case 3) One of and is incorrectly classified; (Case 4) Both and are incorrectly classified and their predicted labels and are consistent with each other. Here, it is clear that we need to increase the weights of the samples belonging to Case 2, Case 3, and Case 4 compared to the samples belonging to Case 1 for the next round.
Table 1.
Four different cases in binary classification with two modalities. (O: correct, X: incorrect)
| Case 1 | O | O | O |
| Case 2 | O | X | X |
| Case 3 | X | O | X |
| Case 4 | X | X | O |
First, for the incorrectly classified samples, we apply the following rule (similar to AdaBoost):
| (4) |
| (5) |
where and , and and are the normalizing factors.
Meanwhile, for the inconsistently classified samples (Case 2 & Case 3), we impose the pairwise coupled-diversity to strengthen the generalization power, which has been mathematically validated in [6, 13]. Formally, we denote the pairwise coupled-diversity function as follows:
| (6) |
where δ(·) is a Dirac Delta function.
By combining the functions of Eq.(4), Eq.(5) and Eq.(6), we define the following weight factors:
| (7) |
| (8) |
where and . Then, our new weight distributions can be estimated as follows:
| (9) |
| (10) |
where and are the normalizing factors.
Algorithm 1.
Pseudo-code of a coupled boosting algorithm
| Input: Data , | |
| and the maximum number of iteration T | |
| Output: Ensemble classifier H(xA, xB) | |
| 1: | |
| 2: | for t = 1, ⋯, T do |
| 3: | Train a base learner with samples drawn with |
| 4: | Train a base learner with samples drawn with |
| 5: | Compute errors and by Eq.(2) and Eq.(3) |
| 6: | if then |
| 7: | |
| 8: | |
| 9: | Update and by Eq.(9) and Eq.(10) |
| 10: | end if |
| 11: | end for |
| 12: | H(xA, xB) defined by Eq.(11) |
After T iterations, we can finally get an ensemble classifier H(xA, xB) that makes a final decision via weighted majority voting on all the base learners as follows:
| (11) |
where sgn(·) is a sign function. We present the pseudo-code of the proposed coupled boosting method in Algorithm. 1.
5. Experimental Results and Analysis
5.1. Experimental Setup
We conducted experiments on the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset4, which has been considered as the benchmark database for performance evaluation of various methods for AD/MCI diagnosis. In our experiments, we used baseline MRI and PET data obtained from 202 subjects of 51 AD patients, 99 MCI patients5, and 52 healthy Normal Controls (NC).
Following the related works [21, 26], we considered two binary classification tasks: AD vs. NC and MCI vs. NC. We employed four different metrics, namely, classification ACCuracy (ACC), SPEcificity (SPE), SENsitivity (SEN), and Area Under Receiver Operating Characteristic (ROC) Curve (AUC) to compare the proposed method with the previous methods. Particularly, ACC is calculated as the number of correctly classified testing samples divided by the total number of testing samples. SPE means the proportion of correctly classified NC samples. SEN means the proportion of correctly classified patient samples. Regarding the four metrics under consideration, the higher the values are, the better the respective method is. Due to a limited number of samples, we used a 10-fold Cross Validation (CV) technique in evaluating the performance, and repeated the 10-fold CV 100 times to reduce the possible bias that could be raised during data partition. For the base learner in our coupled boosting, we used a linear SVM or a Sparse Representation-based Classifier (SRC), which are the most widely used classification methods in many real applications [15, 24, 26]. The model parameters of the base learners were determined by a nested CV on the training samples. Instead of considering the full combination of parameters which is computationally very expensive, we chose the parameters for each base learning separately that helps reduce the computational cost greatly. The sparsity control parameter in LASSO, and and in Eq.(7) and Eq.(8) were also chosen by a nested CV on the training samples. Hereafter, we denote our Coupled Boosting (CB) with SVM or SRC as CB-SVM or CB-SRC, respectively.
5.2. Coupled Feature Representation Evaluation
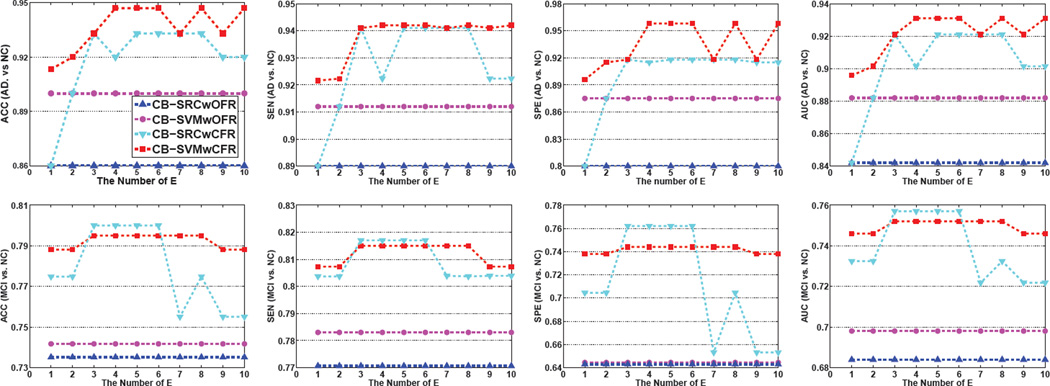
To validate the advantage of the coupled feature representation, we compared the performances of CB-SRC trained on the Original Feature Representations (CB-SRCwOFR) with those of CB-SRC trained on the Coupled Feature Representations (CB-SRCwCFR). We also compared results of CB-SVMwOFR with those of CB-SVMwCFR. In Fig. 3, we presented the performance changes of the competing methods by varying the power expansion value from 1 to 10. For all the methods, we applied LASSO on their respective feature representations.
Figure 3.
The effectiveness of coupled feature representation.
For both AD vs. NC and MCI vs. NC classifications, the proposed CB-SRCwCFR and CB-SVMwCFR outperformed CB-SRCwOFR and CB-SVMwOFR, respectively, in terms of ACC, SEN, SPE, and AUC. Specifically, the proposed method of CB-SRC/CB-SVM improved by 6%/4% (ACC), 3.30%/1.2% (SEN), 11%/9.5% (SPE), and 6%/3.50% (AUC) for AD diagnosis, and by 4.5%/0.69% (ACC), 2.45%/0.77% (SEN), 9.53%/0.6% (SPE), and 4.43%/0.6% (AUC) for MCI diagnosis. That is, the coupled feature representation is helpful to improve the diagnostic accuracy for both AD vs. NC and MCI vs. NC classifications with the base learners of SRC and SVM.
Regarding the expansion parameter E, a small E deteriorates the classification performance while a large E increases the unnecessary computation burden. In the following experiments, we fixed E to 5.
5.3. Feature Selection Evaluation
In order to show the efficacy of feature selection, we compared the performances of CB-SRC or CB-SVM without feature selection (called as CB-SRCwoFS or CB-SVMwoFS) with those of the same methods with feature selection (called as CB-SRCwFS or CB-SVMwFS) in Fig. 4. It is obvious that feature selection helped improve the classification results both for CB-SRC and CB-SVM (see Table. 2).
Figure 4.

The effectiveness of feature selection by LASSO, compared with the methods without feature selection.
Table 2.
Performance improvements (%) by feature selection.
| Methods | ACC | SEN | SPE | AUC | |
|---|---|---|---|---|---|
| AD vs. NC | CB-SRC | 6.00 | 3.30 | 11.00 | 6.00 |
| CB-SVM | 4.00 | 1.20 | 9.50 | 3.50 | |
| MCI vs. NC | CB-SRC | 4.50 | 2.45 | 9.53 | 4.43 |
| CB-SVM | 0.69 | 0.77 | 0.60 | 0.60 | |
5.4. Coupled Boosting Evaluation
If we set the parameters and to 0, then the proposed coupled boosting algorithm becomes the conventional AdaBoost by degenerating the weights for the inconsistently classified samples. In order to show the validity of the newly devised formulation for weight distributions, we compared the proposed method with AdaBoost, for which we also used SRC or SVM as a base learner (called as AdaBoost-SRC or AdaBoost-SVM) (see Fig. 5). Although there exist cases that the proposed method showed lower performance, i.e., −0.7% (SEN) with CB-SVM in AD vs. NC, and −1.3% (SEN) with CB-SRC and −2.6% (SPE) with CB-SVM in MCI vs. NC, overall, our method was statistically superior to AdaBoost with p-values of 1.1e-17 (CB-SRC), 3.7e-05 (CB-SVM) in AD vs. NC, and 0.0061 (CB-SRC), 2.7e-12 (CB-SVM) in MCI vs. NC. Thanks to the pairwise coupled-diversity, the proposed methods of CB-SRCwFS and CB-SVMwFS outperformed both AdaBoost-SRC and AdaBoost-SVM.
Figure 5.

The effectiveness of pairwise coupled-diversity employed in coupled boosting, compared with AdaBoost-SRC and AdaBoost-SVM (not considering the pairwise coupled-diversity).
5.5. Comparison with Previous Methods
We also compared the classification performances of the proposed method with those of the state-of-the-art methods, namely, Wang et al.’s method [21] and Zhang et al.’s method [26]. For a comparison purpose, we also present the performance of four single-modality based methods: (1) SRC with MRI features (SRC-MRI), SRC with PET features (SRC-PET), SVM with MRI features (SVM-MRI), and SVM with PET features (SVM-PET). For SRC-MRI, SRC-PET, SVM-MRI, and SVM-PET, the parameters (e.g., the sparsity regularization parameter in SRC) in the respective models were determined by a nested CV on the training samples. Table. 3 compares the performances of the competing methods. For Wang et al.’s method [21] and Zhang et al.’s method [26], we presented the best performances reported in the respective papers. Our method achieved the best performance for both AD vs. NC (by CB-SVM) and MCI vs. NC (by CB-SRC) classifications. Regarding AUCs, we repeated the 10-fold CV 100 times to reduce the possible bias caused by different data partition, and then obtained the mean AUC value. Our best AUCs were 0.975 (AD vs. NC) and 0.833 (MCI vs. NC), better than results in [21].
Table 3.
Performance comparison with the state-of-the-art methods. The best results are marked in boldface.
| Methods | AD vs. NC | MCI vs. NC | ||||||
|---|---|---|---|---|---|---|---|---|
| ACC | SEN | SPE | AUC | ACC | SEN | SPE | AUC | |
| SRC-MRI | 0.837 | 0.783 | 0.891 | 0.817 | 0.714 | 0.794 | 0.554 | 0.741 |
| SRC-PET | 0.847 | 0.803 | 0.891 | 0.827 | 0.721 | 0.800 | 0.569 | 0.748 |
| SVM-MRI | 0.860 | 0.890 | 0.800 | 0.842 | 0.723 | 0.782 | 0.600 | 0.688 |
| SVM-PET | 0.833 | 0.856 | 0.783 | 0.802 | 0.735 | 0.771 | 0.643 | 0.684 |
| Zhang et al. [26] | 0.932 | 0.930 | 0.933 | N.A. | 0.764 | 0.818 | 0.633 | N.A. |
| Wang et al. [21] | 0.933 | 0.900 | 0.966 | 0.970 | 0.789 | 0.856 | 0.663 | 0.807 |
| CB-SVM | 0.947 | 0.942 | 0.969 | 0.931 | 0.795 | 0.815 | 0.744 | 0.752 |
| CB-SRC | 0.933 | 0.941 | 0.918 | 0.921 | 0.801 | 0.817 | 0.762 | 0.757 |
It is noteworthy that (1) coupled features assigned the lower weight for the higher power features, thus preventing the possible over-fitting caused by complex high power representations and (2) boosting-like algorithm is a well-known classifier which often does not overfit data by increasing the ensemble margin according to several empirical and theoretical studies. In this regard, we can say that our experimental results were not suffered from overfitting.
6. Conclusion
Recently, by addressing the AD/MCI diagnosis process as a classification problem, most of the previous methods assume the conditional independence among the low-level features extracted from neuroimaging data. However, since a human brain is a complex system in which different regions interact for cognitive tasks, it is obvious that the features are naturally correlated to each other. Furthermore, there also exists relational information between different imaging modalities such as MRI and PET.
In this paper, we devised a coupled feature representation with intra-coupled and inter-coupled interaction relationship by means of a matrix expansion. Regarding multimodal data fusion, we proposed a novel coupled boosting algorithm that analyzes the pairwise coupled-diversity correlation between modalities. Specifically, we formulated a method of updating weight distribution functions, which jointly considered both incorrectly and inconsistently classified samples. From our experiments on the publicly available ADNI dataset, we validated the effectiveness of the proposed method both on the AD vs. NC and the MCI vs. NC diagnosis by comparing with the competing methods and the state-of-the-art methods.
Acknowledgements
The work was support by NSFC (61035003, 61175042, 61321491, 61305068), Jiangsu 973 (BK2011005), Jiangsu NSF (BK20130581), the Program for New Century Excellent Talents (NCET-10-0476) and Jiangsu Clinical Medicine Special Program (BL2013033). The work was also supported by the grants from National Institute of Health (EB006733, EB008374, EB009634, AG041721, MH100217, AG042599).
Footnotes
In this work, pA = 93 and pB = 93.
For a base learner, in this work, we use an SVM or a Sparse Representation Classifier (SRC).
Including 43 MCI converters and 56 MCI non-converters.
References
- 1.Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer’s disease. Alzheimer’s & dementia. 2007;3(3):186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- 2.Fan Y, Rao H, Hurt H, Giannetta J, Korczykowski M, Shera D, Avants BB, Gee JC, Wang J, Shen D. Multivariate examination of brain abnormality using both structural and functional {MRI} NeuroImage. 2007;36(4):1189–1199. doi: 10.1016/j.neuroimage.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Freund Y, Schapire RE. A decision-theoretic generalization of on-line learning and an application to boosting. Proceedings of the Second European Conference on Computational Learning Theory. 1995:23–37. [Google Scholar]
- 4.Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. PNAS. 2004;101(13):4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinrichs C, Singh V, Xu G, Johnson SC. Predictive markers for AD in a multi-modality framework: An analysis of MCI progression in the ADNI population. NeuroImage. 2011;55(2):574–589. doi: 10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho TK. The random subspace method for constructing decision forests. IEEE TPAMI. 1998;20(8):832–844. [Google Scholar]
- 7.Kakani N, MacDonald D, Holmes C, Evans A. Human brain mapping conference; Human Brain Mapping Conference; 1998. [Google Scholar]
- 8.Kohannim O, et al. Boosting power for clinical trials using classifiers based on multiple biomarkers. Neurobiology of Aging. 2010;31(8):1429–1442. doi: 10.1016/j.neurobiolaging.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nordberg A, Rinne JO, Kadir A, Långström B. The use of PET in Alzheimer disease. Nature Reviews Neurology. 2010;6(2):78–87. doi: 10.1038/nrneurol.2009.217. [DOI] [PubMed] [Google Scholar]
- 10.Saberian MJ, Masnadi-Shirazi H, Vasconcelos N. CVPR. IEEE; 2011. Taylorboost: First and second-order boosting algorithms with explicit margin control; pp. 2929–2934. [Google Scholar]
- 11.Saberian MJ, Vasconcelos N. CVPR. IEEE; 2012. Boosting algorithms for simultaneous feature extraction and selection; pp. 2448–2455. [Google Scholar]
- 12.Shen D, Davatzikos C. HAMMER: Hierarchical attribute matching mechanism for elastic registration. IEEE TMI. 2002;21(11):1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y, Gao Y, Yang Y, Zhang Y, Wang D. Multi-modal sparse representation-based classification for lung needle biopsy images. IEEE TBME. 2013;60(10):2675–2685. doi: 10.1109/TBME.2013.2262099. [DOI] [PubMed] [Google Scholar]
- 14.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE TMI. 1998;17(1):87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 15.Suk H-I, Lee S-W. A novel Bayesian framework for discriminative feature extraction in brain-computer interfaces. IEEE TPAMI. 2013;35(2):286–299. doi: 10.1109/TPAMI.2012.69. [DOI] [PubMed] [Google Scholar]
- 16.Suk H-I, Lee S-W, Shen D. Latent feature representation with stacked auto-encoder for AD/MCI diagnosis. Brain Structure and Function. 2013:1–19. doi: 10.1007/s00429-013-0687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society (Series B) 1996;58:267–288. [Google Scholar]
- 18.Walhovd K, et al. Combining MR imaging, positron-emission tomography, and CSF biomarkers in the diagnosis and prognosis of Alzheimer disease. American Journal of Neuroradiology. 2010;31:347–354. doi: 10.3174/ajnr.A1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, She Z, Cao L. Coupled attribute analysis on numerical data. IJCAI. 2013 [Google Scholar]
- 20.Wang Y, et al. Knowledge-guided robust MRI brain extraction for diverse large-scale neuroimaging studies on humans and non-human primates. PLoS One. 2013 doi: 10.1371/journal.pone.0077810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Liu M, Guo L, Shen D. ISBI. IEEE; 2013. Kernel-based multi-task joint sparse classification for Alzheimer’s disease; pp. 1364–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westman E, Muehlboeck J-S, Simmons A. Combining MRI and CSF measures for classification of Alzheimer’s disease and prediction of mild cognitive impairment conversion. NeuroImage. 2012;62(1):229–238. doi: 10.1016/j.neuroimage.2012.04.056. [DOI] [PubMed] [Google Scholar]
- 23.Woolrich MW, et al. Bayesian analysis of neuroimaging data in fsl. NeuroImage. 2009;45(1):S173–S186. doi: 10.1016/j.neuroimage.2008.10.055. [DOI] [PubMed] [Google Scholar]
- 24.Wright J, Yang AY, Ganesh A, Sastry SS, Ma Y. Robust face recognition via sparse representation. IEEE TPAMI. 2009 Feb;31(2):210–227. doi: 10.1109/TPAMI.2008.79. [DOI] [PubMed] [Google Scholar]
- 25.Zhang D, Shen D. Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease. NeuroImage. 2012;59(2):895–907. doi: 10.1016/j.neuroimage.2011.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D, Wang Y, Zhou L, Yuan H, Shen D. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. NeuroImage. 2011;55(3):856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE TMI. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]