Abstract
Background
Plasmodium vivax is the second most prevalent human malaria parasite in Bangladesh; however, there are no data of its genetic diversity. Several molecular markers are available where Pvcsp, Pvmsp 1 and Pvmsp 3α are most commonly used for P. vivax genotyping studies. The aim of the study was to investigate the population structure of P. vivax in Bangladesh.
Methods
A total of 102 P. vivax-positive blood samples were collected from different malaria-endemic areas in Bangladesh and subsequently analysed for those three genotyping markers. Nested PCR was performed for diagnosis and genotyping analysis followed by PCR–RFLP to detect genetic diversity using Pvcsp, Pvmsp 1 and Pvmsp 3α markers.
Results
Analysis of Pvcsp showed that the VK210 repeat type was highly prevalent (64.7%, 66/102) compared to VK247 (35.3%, 36/102), although the prevalence of VK247 was higher than other Southeast Asian countries. Analysis of these three genes revealed a diverse, circulating population of P. vivax where a total of ten, 56 and 35 distinct genotypes were detected for Pvcsp, Pvmsp 1 and Pvmsp 3α, respectively.
Conclusion
This genotyping observation of P. vivax is the first report from Bangladesh and will provide valuable information for establishing the genotyping methods and circulating genetic variants of these three markers available in Bangladesh.
Keywords: Plasmodium vivax, Genetic diversity, Pvcsp. Pvmsp 1, Pvmsp 3α, Bangladesh
Background
Approximately 3.2 billion people were at risk of malaria globally in 2013 [1]. Among five Plasmodium species causing malaria in humans, Plasmodium falciparum is the most deadly and predominant, followed by Plasmodium vivax, which is less virulent but has a wide geographical distribution. About 8% of estimated cases globally are due to P. vivax, but its incidence outside the African continent is in similar proportion to P. falciparum [1]. Although once thought benign, P. vivax has recently been found to be associated with severe anaemia, respiratory distress, malnutrition [2], and recurrent haemolysis [3]. It has been reported from clinical findings from Thailand and India that vivax malaria during pregnancy causes maternal anaemia and a significant reduction in mean birth weight [4, 5].
Drug resistance is a growing problem. Resistance to sulfadoxine-pyrimethamine (SP) and chloroquine by P. falciparum has led to an increase in morbidity and mortality [6]. Worryingly, SP and chloroquine resistance have also been reported for P. vivax recently [6–8], which calls for measures to restrict the spread of drug resistance. One of the important tools to monitor drug resistance is to study the molecular markers in malaria parasites involved in consecutive transmission [9].
Examining the genetic diversity and population structure of P. vivax parasites provides insights into the transmission dynamics of vivax malaria, which are important to help support and monitor malaria control measures, including the design and evaluation of new drugs and vaccines [10, 11]. Various large-scale studies have been conducted for P. falciparum and the presence and dynamics of different single or multiple polymorphic genes encoding different antigens have been investigated [12–14]. In recent studies, well-characterized polymorphic antigenic regions in both pre-erythrocytic and erythrocytic genes have been widely used to analyse genetic diversity patterns in P. vivax populations [15–17]. For molecular genotyping studies of P. vivax, three polymorphic single copy genes have been in common use: Pvcsp coding for the circumsporozoite protein, which is responsible for binding of sporozoite to liver cells and contains two types of repeat elements (either VK210 or VK247) [15, 18, 19]; Pvmsp 1 (coding for the merozoite surface protein 1), which is involved in the parasite’s invasion to red blood cells and contains 13 inter-allele conserved and highly variable blocks, where variable blocks are: block 2 (F1 region), 6–8 (F2 region) and 10 (F3 region) [18]; and, Pvmsp 3α, which initiates antibody-dependent, cell-mediated inhibition during repeated malaria infection by triggering the binding of antibody to monocytes and consists of an alanine-rich central domain [20].
While P. vivax is the second most malaria-causing parasite in Bangladesh [21], it receives relatively little attention [22]. There is no information available on the circulating strains of P. vivax across endemic areas of Bangladesh. The primary objective of this study was to document the genetic diversity of P. vivax by three well-established markers (Pvcsp, Pvmsp 1 and Pvmsp 3α) in some selected endemic areas of Bangladesh.
Methods
Study sample
A total of 102 P. vivax mono-infected blood samples (day 0) from five malaria-endemic districts in Bangladesh: Bandarban (9), Cox’s Bazar (73), Khagrachari (15), Rangamati (1), and Netrokona (4), were considered for this study. The age range of patients was 2–50 years, with median age of 25.5 years. All the samples were collected from patients with febrile illness at different upazila (sub-district) health complexes (UHC) of the aforementioned districts (Figure 1) from May 2009 to April 2014, but no samples were collected in 2011 (total 35, 39, four, four, and 20 samples were collected in the years 2009, 2010, 2012, 2013, and 2014, respectively). These samples were referred to microscopy for malaria diagnosis. No follow-up data were collected from any of the patients. All samples were positive in microscopy and nested PCR [23] and/or real-time PCR for P. vivax mono-infection [22]. Most of the samples were used for different studies reported elsewhere [22, 24, 25] and the studies were approved by the Research Review Committee and Ethical Review Committee of the International Centre for Diarrhoeal Disease Research Bangladesh (icddr,b). All the patients consented to further use of blood samples.
Figure 1.
Geographical map of the study areas.
Genotyping PCR
Positive samples were analysed using nested PCR for Pvcsp, Pvmsp 1 (containing variable blocks 2, 6–8 and 10, designated as F1, F2 and F3, respectively) and Pvmsp 3α markers as described previously [9, 20]. All amplification reactions were carried out with some modifications. Briefly, for the nested round of the PCR, reaction volume was 50 µL, where 2.5 µL of 50 times diluted initial PCR product was used as template. The PCR products were analysed by ethidium bromide stained 1.5% (for Pvcsp and Pvmsp 1) and 0.8% (for Pvmsp 3α) agarose gel electrophoresis.
RFLP analysis of Pvcsp, Pvmsp 1 and Pvmsp 3α
Restriction enzymes Alu I, Bst NI, Scr FI, Bbs I, and MboII for Pvcsp; Alu I and Mnl I for F2 region of Pvmsp 1, and Hha I and Alu I for Pvmsp 3α were used, as described previously for restriction fragment length polymorphism analysis of the PCR products (PCR–RFLP) [9, 20]. In brief, 10 μL of the amplified PCR product were digested individually with the restriction enzyme in 20 µL reaction volume at 37°C for 3 h. All the restriction enzymes were obtained from New England Biolabs Inc, USA. Electrophoresis was performed on ethidium bromide stained 1.5% agarose gel in TBE buffer, and was visualized under UV illumination.
Allele detection
The repeat types of Pvcsp were classified based on restriction digestion [9]. Analysis of number of genotypes and size polymorphisms in Pvcsp, Pvmsp 1 F1, F2, F3, and Pvmsp 3α was done by grouping the band sizes of electrophoresis, differing by 25 bp. Thereafter, each 25-bp interval was defined as a distinct genotype [14, 26]. All the data were computed and analysed in Microsoft Excel 2007 (Microsoft Corp, USA) and Chi square (χ2) test was performed to compare genetic diversity of markers with study districts using SPSS 19.0 (IBM Corp, USA). A p value of ≤0.05 was considered significant.
Heterozygosity (HE) and mean multiplicity of infection (MOI)
The expected heterozygosity (HE) was calculated by using the formula , where n = sample size, Pi = allele frequency. The theoretical probability of infection by two parasites with the same allele was calculated as [27]. The combined probability was calculated by multiplying the probabilities P for all marker genes, which indicates that two independent clones share the same genotype for all marker genes, assuming that all the loci sort individually from each other. The mean multiplicity of infection (MOI) was calculated by dividing the total number of clones by the number of PCR-positive samples for each marker gene.
Results
Allelic diversity of Pvcsp
PCR–RFLP analysis of Pvcsp by AluI and BstI of 102 isolates revealed the presence of both VK210 (66/64.7%) and VK247 (36/35.3%) repeat types (Figure 2). Digestion products were less than 150 bp for both digestion. No isolate harboured both VK210 and VK247 repeat type infection. For Pvcsp, ten different allelic variants were detected, equally (five from each) representing the VK210 and VK247 repeat types. Each genotype was labelled alphabetically from A to E (650–775 bp) for both repeat types. Variant B was found abundant (0.250) for VK210, whereas for VK247, the highest frequency (0.125), was found with variant C and D. To increase the genotyping resolution of Pvcsp, the presence and absence of pre- and post-repeat region of VK210 (recognized by Scr FI and Bbs I, respectively) and pre-repeat region of VK247 (recognized by MboII) were analysed by RFLP. A total of 14 different allelic types was found for VK210 repeat type after RFLP (Table 1). Geographical distributions of both repeat types are provided in Figure 3. These allelic variants were found from 109 bands upon PCR; the highest frequency, 0.127, was found for both VK210c and VK210g (Table 1). Among these two variants, VK210c was found almost equally distributed in the study areas while VK210g was found only in Khagrachari and Cox’s Bazar (Figure 3).
Figure 2.
RFLP gel pictures of three markers. a Alu I digestion of Pvcsp PCR products for VK210 type, digested (lane 2, 4, 6, 9–13), undigested (lane 3), marker [lane 7, 8 (100 bp)]. b Bst NI digestion for VK247 type, digested (lane 3–7, 11, 13–17), undigested (lane 1, 2, 8, 10, 12). c Digestion of Pvcsp PCR product with Bbs I for presence of post repeat sequence, digested (lane 2, 4, 6–9, 12), undigested (lane 3, 5, 10, 11, 13), 100 bp marker (lane 1). d Scr FI digestion of Pvcsp product for pre-repeat site, digested (lane 1, 2, 5, 8, 9, 11, 12), undigested (lane 4, 6, 7, 10, 13), 100-bp marker (lane 1). e, f Restriction digestion pattern of Pvmsp-1 F-2 PCR product using Mnl I and Alu I, respectively, 100-bp marker was used in both gel. g, h Digestion pattern of Pvmsp 3α PCR product using Alu I and Hha I, respectively. Presence of a common 1,000-bp fragment observed for Hha I digestion. 50-bp and 100-bp markers (lane 1 of both gel) were used in Alu I and Hha I digestions, respectively.
Table 1.
Allelic variant frequency of Pvcsp by size, repeat types and insertion of pre- and post-repeats
| Allele | Size | Pre-repeat | Post-repeat | n | Frequency |
|---|---|---|---|---|---|
| VK210a | A (650–675 bp) | Yes | Yes | 1 | 0.010 |
| VK210b | A (650–675 bp) | Yes | No | 4 | 0.039 |
| VK210c | A (650–675 bp) | No | No | 13 | 0.127 |
| VK210d | B (675–700 bp) | Yes | Yes | 3 | 0.029 |
| VK210e | B (675–700 bp) | Yes | No | 10 | 0.098 |
| VK210f | B (675–700 bp) | No | Yes | 4 | 0.039 |
| VK210g | B (675–700 bp) | No | No | 13 | 0.127 |
| VK210h | C (701–725 bp) | Yes | Yes | 2 | 0.020 |
| VK210i | C (701–725 bp) | Yes | No | 5 | 0.049 |
| VK210j | C (701–725 bp) | No | Yes | 3 | 0.029 |
| VK210k | C (701–725 bp) | No | No | 6 | 0.059 |
| VK210l | D (725–750 bp) | No | Yes | 1 | 0.010 |
| VK210m | E (750–775 bp) | Yes | No | 4 | 0.039 |
| VK210n | E (750–775 bp) | No | No | 2 | 0.020 |
| VK247A | A (650–675 bp) | No | ND | 4 | 0.039 |
| VK247B | B (675–700 bp) | No | ND | 5 | 0.049 |
| VK247C | C (701–725 bp) | No | ND | 12 | 0.118 |
| VK247D | D (725–750 bp) | No | ND | 12 | 0.118 |
| VK247E | E (750–775 bp) | No | ND | 3 | 0.029 |
ND not done.
Figure 3.
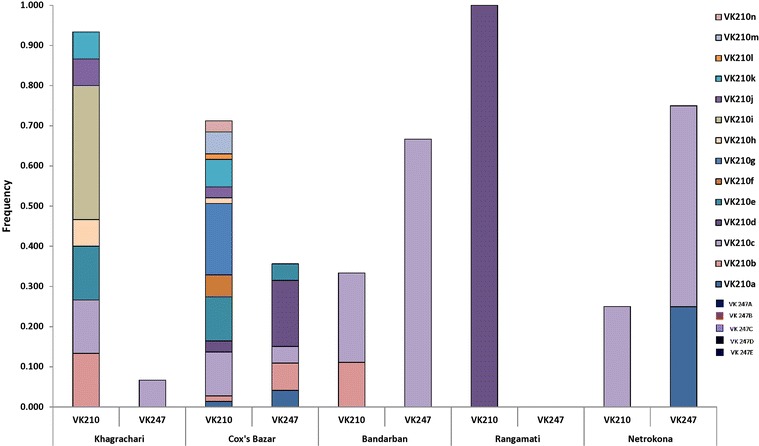
Distribution of Pvcsp repeat types and presence of pre- and post-repeats in Bandarban, Cox’s Bazar, Khagrachari, Rangamati, and Netrokona.
Allelic diversity of Pvmsp 1
A total of 56 distinct genotypes were observed in Pvmsp 1 PCR. The geographical distribution for F1 and F3 region variants with their size variants (labelled alphabetically) is provided in Figure 4. For F1, six size variants (A–F), two (A and B) for F2 and three size variants (A–C) for F3 region were observed. For F1, variant A (350–374 bp) was the dominant variant (38% of the bands observed), variant B (1,200 bp) for F2 (57%) and variant A (250–274 bp) for F3 (62%) was the dominant allelic variant (Table 2; Figure 4). Increased genotype resolution was observed by RFLP for Pvmsp 1 F2 fragment. In total, seven different Alu I patterns and five Mnl I patterns were observed (Table 2). Mixed patterns were also observed in both digestions where the sums of total band size exceeded the band size upon PCR. The highest frequency was found in Cox’s Bazar for both Ba7 (0.324) and Bm3 (0.333) in Alu I and Mnl I digestion, respectively (Table 2). Geographical distributions of all variants of these three regions (F-1, F-2 and F-3) are shown in Table 2 and Figure 4.
Figure 4.
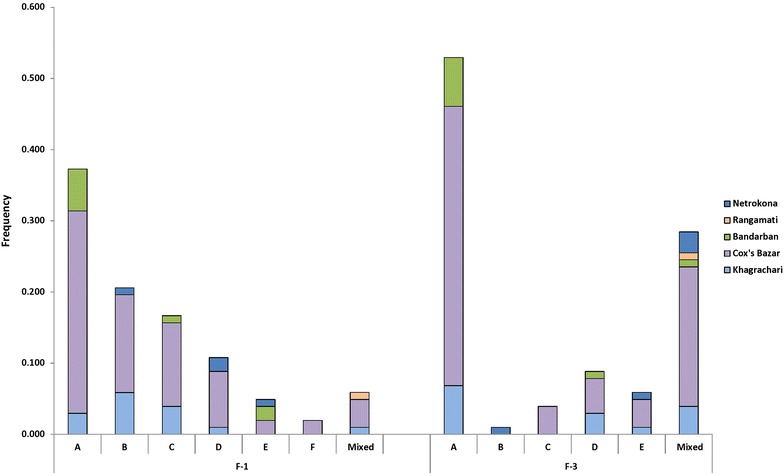
Size fragment distribution of Pvmsp 1 F-1 and F-3 regions in five endemic areas.
Table 2.
Allele frequency of Pvmsp-1 F-2 region with geographical distribution
| Allele | Size | n (frequency) | ||||
|---|---|---|---|---|---|---|
| Khagrachari | Cox’s Bazar | Bandarban | Rangamati | Netrokona | ||
| Aa1 | A | 0 (0) | 1 (0.010) | 0 (0) | 0 (0) | 0 (0) |
| Aa2 | A | 0 (0) | 2 (0.020) | 0 (0) | 0 (0) | 0 (0) |
| Aa3 | A | 1 (0.010) | 5 (0.049) | 0 (0) | 0 (0) | 0 (0) |
| Aa4 | A | 0 (0) | 0 (0) | 1 (0.010) | 0 (0) | 0 (0) |
| Aa5 | A | 0 (0) | 3 (0.029) | 0 (0) | 0 (0) | 1 (0.010) |
| Aa6 | A | 2 (0.020) | 3 (0.029) | 1 (0.010) | 0 (0) | 0 (0) |
| Aa7 | A | 3 (0.029) | 7 (0.069) | 0 (0) | 0 (0) | 1 (0.010) |
| Ba3 | B | 0 (0) | 1 (0.010) | 0 (0) | 0 (0) | 0 (0) |
| Ba5 | B | 0 (0) | 1 (0.010) | 1 (0.010) | 0 (0) | 0 (0) |
| Ba7 | B | 3 (0.029) | 33 (0.324) | 3 (0.029) | 0 (0) | 1 (0.010) |
| Mixed | 4 (0.039) | 5 (0.049) | 0 (0) | 1 (0.010) | 1 (0.010) | |
| Not digested | 1 (0.010) | 5 (0.049) | 3 (0.029) | 0 (0) | 0 (0) | |
| Am1 | A | 0 (0) | 2 (0.020) | 0 (0) | 0 (0) | 0 (0) |
| Am2 | A | 0 (0) | 0 (0) | 1 (0.010) | 0 (0) | 0 (0) |
| Am3 | A | 4 (0.039) | 15 (0.147) | 1 (0.010) | 0 (0) | 2 (0.020) |
| Am4 | A | 1 (0.010) | 3 (0.029) | 0 (0) | 0 (0) | 0 (0) |
| Am5 | A | 0 (0) | 1(0.010) | 0 (0) | 0 (0) | 0 (0) |
| Bm2 | B | 0 (0) | 1 (0.010) | 0 (0) | 0 (0) | 0 (0) |
| Bm3 | B | 3 (0.029) | 34 (0.333) | 5 (0.049) | 0 (0) | 1 (0.010) |
| Bm4 | B | 0 (0) | 1 (0.010) | 1 (0.010) | 0 (0) | 0 (0) |
| Mixed | 4 (0.039) | 5 (0.049) | 1 (0.010) | 1 (0.010) | 1 (0.010) | |
| Not digested | 2 (0.020) | 4 (0.039) | 0 (0) | 0 (0) | 0 (0) | |
Allelic diversity of Pvmsp 3α
A total of 102 isolates were successfully amplified by nested PCR for Pvmsp 3α, which showed distinct size polymorphism with three allelic forms, labelled here as A, B and C. Seventy-five of these were A (1,900 bp), five of B (1,400 bp) and nine of C (1,100 bp). The remaining 13 isolates were mixed genotypes (Table 3). PCR–RFLP analysis by Alu I and Hha I restriction enzymes showed a greater genotype distinction among the isolates. Alu I digestion resulted in 19 different allelic variants for Pvmsp 3α, while Hha I digestion resulted in 16 distinguishable variants found from the study areas (Figure 5). A common clear restriction pattern of 1,000 bp fragment was observed in all samples in Hha I digestion (Figure 2). Thus, different patterns of smaller fragments (100–500 bp) were included for the genotyping analysis. Mixed infection was observed in 13 isolates with more than one PCR product of different sizes in a single sample or when the sum of the restriction fragments sizes exceeded the size of the PCR products.
Table 3.
Size variant observations of Pvmsp 3α
| Area | |||||||
|---|---|---|---|---|---|---|---|
| Khagrachari | Cox’s Bazar | Bandarban | Rangamati | Netrokona | Total | Frequency | |
| A (1,900 bp) | 10 | 56 | 6 | 1 | 2 | 75 | 0.735 |
| B (1,400 bp) | 1 | 2 | 1 | 0 | 1 | 5 | 0.049 |
| C (1,100 bp) | 2 | 5 | 2 | 0 | 0 | 9 | 0.088 |
| Mixed | 2 | 10 | 0 | 0 | 1 | 13 | 0.127 |
Figure 5.
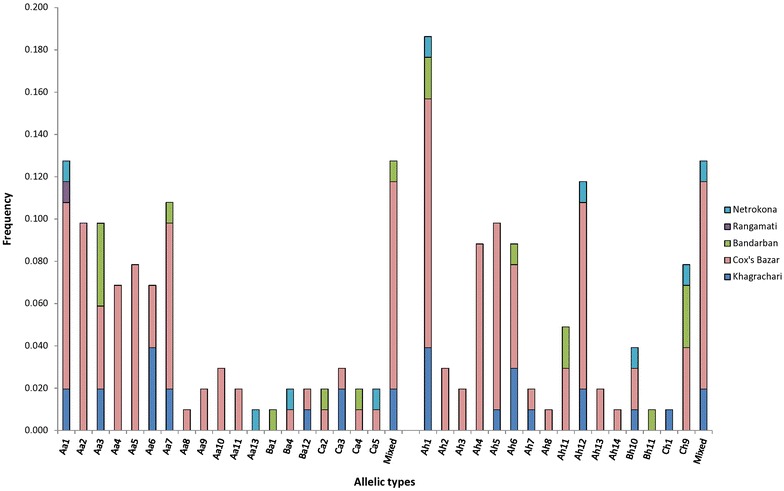
Geographical distribution of Pvmsp 3α marker using Alu I and Hha I restriction enzymes.
The MOI is higher for Pvmsp 1 (1.17) among these three markers. However, expected HE is higher for Pvcsp but low in Pvmsp 3α (Table 4). Also, both MOI and HE were calculated for each study year (Table 5) while low MOI and HE were found only in 2012 for Pvcsp and Pvmsp 3α. Significant differences were observed for Pvcsp (p = 0.013), F1 and F2 variable regions of Pvmsp 1 (p = 0.006 and 0.027, respectively) according to areas but not for others (p = 0.364 and 0.595, respectively for Pvmsp 1 F3 region and Pvmsp 3α).
Table 4.
Multiplicity of infection and expected heterozygosity (HE) of three different P. vivax polymorphic markers (based on PCR)
| Marker | |||
|---|---|---|---|
| Pvcsp | Pvmsp 1 | Pvmsp 3α | |
| Number of PCR positive samples (n) | 102 | 102 | 102 |
| Mean MOI | 1.07 | 1.17 | 1.13 |
| HE | 0.777 | 0.689 | 0.437 |
| 0.230 | 0.318 | 0.567 | |
| Combined probabilitya | 0.041 | ||
MOI mean multiplicity of infection, H E heterozygosity (expected).
aCalculated by multiplying the probabilities ‘P’ for all markers, which defines any of two independent clones share the same genotype.
Table 5.
Multiplicity of infection and expected heterozygosity (HE) values according to years
| Years | Pvcsp | Pvmsp 1 | Pvmsp 3α | |||
|---|---|---|---|---|---|---|
| Mean MOI | HE | Mean MOI | HE | Mean MOI | HE | |
| 2009 | 1.14 | 0.81 | 1.21 | 0.72 | 1.14 | 0.66 |
| 2010 | 1.00 | 0.66 | 1.09 | 0.69 | 1.13 | 0.31 |
| 2012 | 1.00 | 0.00 | 1.33 | 0.72 | 1.00 | 0.00 |
| 2013 | 1.00 | 0.83 | 1.25 | 0.83 | 1.00 | 0.92 |
Discussion
In this first report of genetic diversity of P. vivax in Bangladesh, a highly diverse P. vivax population was documented based on three markers Pvcsp, Pvmsp 1and Pvmsp 3α in malaria-endemic regions of the country.
Both VK210 and VK247 repeat types of Pvcsp were found in field isolates, where VK210 has higher prevalence. Previously, both the types were confirmed in a mosquito population from endemic areas of Bangladesh by CSP-ELISA where VK210 repeat types were also reported with higher prevalence [28, 29]. The prevalence of VK210 has been reported from other studies in Southeast Asia [9, 30, 31]. However, the VK247 repeat type is comparatively higher in Southeast Asia than other countries [18, 30–32]. Among the study areas, VK247 was observed highest in Netrokona (75%) followed by Bandarban (66.7%) of total P. vivax population in the respective areas, and none in Rangamati. The differences in frequency of VK247 type may be due to vector species distribution and their increased susceptibility to infection by VK247 repeat type and/or host immune pressure to certain Pvcsp repeat type [30, 32].
In this study, three different variable fragments (F1, F2 and F3) were analysed for Pvmsp 1 marker and a total of 56 distinct variants were distinguished through size polymorphism and PCR–RFLP. This indicates extensive polymorphisms in the Pvmsp 1 gene. In Thailand and India, less polymorphism has been reported [9, 30]. PCR–RFLP analysis showed high polymorphism in F2 fragments and out of a total of 102 PCR samples, seven different Alu I patterns and five different Mnl I patterns were observed, which is greater than other studies (Thailand, India, Pakistan) [9, 18, 30]. These variations indicate high genetic diversity in all the study areas and suggest that the Pvmsp 1 gene is under selective pressure for the parasite’s survival and transmission [18].The Pvmsp 3α gene is a reliable molecular marker for genotyping study of P. vivax. Three different allelic variants were observed, of which size variant A (1,900 bp) was predominant as reported in other studies [30–33], but not in a study in Thailand [19]. Diverse RFLP patterns were observed for both Alu I and Hha I enzymes, with 19 and 16 distinct variants respectively, which is fewer than reported elsewhere [31, 32], but almost the same as another study from India [30]. While the findings are not directly comparable, with a dissimilar sampling strategy in use in this study, these values are strikingly higher in respect of the low endemicity of vivax malaria in certain regions, such as Thailand. Biological features of P. vivax, such as earlier gametocytogenesis, ie., production of gametocytes in the presymptomatic period before the drug treatment is initiated, and relapse could be the reason behind this extensive polymorphism. Earlier gametocytogenesis and relapses might allow for more efficient transmission to the vector mosquitoes [15].
In this study, extensive genetic diversity of all three markers in P. vivax populations was observed in Bangladesh. Diverse anopheline fauna and their susceptibility to infections by different parasite types [28, 34] can be vital reasons for this diversity. Most of the study areas are populated with different ethnic groups and there may be the presence of different host immune responses to the parasite, which can support this diversity. Also, migration of people from one country to another may carry different parasite variants that increase diversity to the gene pool [32], which is common on the Bangladesh-Myanmar-India border areas.
Conclusion
High genetic diversity based on Pvcsp, Pvmsp1 and Pvmsp3α for P. vivax in clinical isolates was observed in Bangladesh. Establishment of genotyping methods for these three polymorphic markers and the knowledge from this study will provide valuable support for future genotyping study of recurrent infection, which will help in drug efficacy and drug resistance observation.
Authors’ contributions
MSA, WAK and RH conceived and designed the study. MGK, RE and ANM performed laboratory assays. MGK, RE, ANM, and MSA analysed the data. MGK, RE and MSA drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This study was funded by core donors which provide unrestricted support to icddr,b for its operations and research. Current donors providing unrestricted support include: Australian Agency for International Development (AusAID), Government of the People’s Republic of Bangladesh; Canadian International Development Agency (CIDA), Swedish International Development Cooperation Agency (Sida), and the Department for International Development, UK (DFID). We gratefully acknowledge these donors for their support and commitment to icddr,b’s research efforts. We also appreciate the valuable contribution of H M Al-Amin, Khalid Eakbal Anik, Md Imtiaz Khalil, Nusrat Jahan, Maisha Khair, Khaja Md Mohiuddin, Sultan Mahmud, and Shariar Mustafa in this study. We thank Jocalyn Clark for her comments on earlier versions of this manuscript.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Contributor Information
Mohammad Golam Kibria, Email: golam.kibria@icddrb.org.
Rubayet Elahi, Email: rubayet@vt.edu.
Abu Naser Mohon, Email: manmohon@ucalgary.ca.
Wasif A Khan, Email: wakhan@icddrb.org.
Rashidul Haque, Email: rhaque@icddrb.org.
Mohammad Shafiul Alam, Email: shafiul@icddrb.org.
References
- 1.WHO . World Malaria Report 2014. Geneva: World Health Organization; 2014. [Google Scholar]
- 2.Douglas NM, Pontororing GJ, Lampah DA, Yeo TW, Kenangalem E, Poespoprodjo JR, et al. Mortality attributable to Plasmodium vivax malaria: a clinical audit from Papua, Indonesia. BMC Med. 2014;12:217. doi: 10.1186/s12916-014-0217-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins WE, Jeffery GM, Roberts JM. A retrospective examination of anemia during infection of humans with Plasmodium vivax. Am J Trop Med Hyg. 2003;68:410–412. [PubMed] [Google Scholar]
- 4.Nosten F, Vincenti M, Simpson J, Yei P, Thwai KL, de Vries A, et al. The effects of mefloquine treatment in pregnancy. Clin Infect Dis. 1999;28:808–815. doi: 10.1086/515183. [DOI] [PubMed] [Google Scholar]
- 5.Singh N, Shukla M, Sharma V. Epidemiology of malaria in pregnancy in central India. Bull World Health Organ. 1999;77:567–572. [PMC free article] [PubMed] [Google Scholar]
- 6.Trape J-F. The public health impact of chloroquine resistance in Africa. Am J Trop Med Hyg. 2001;64:12–17. doi: 10.4269/ajtmh.2001.64.12. [DOI] [PubMed] [Google Scholar]
- 7.Garg M, Gopinathan N, Bodhe P, Kshirsagar NA. Vivax malaria resistant to chloroquine: case reports from Bombay. Trans R Soc Trop Med Hyg. 1995;89:656–657. doi: 10.1016/0035-9203(95)90432-8. [DOI] [PubMed] [Google Scholar]
- 8.Goncalves LA, Cravo P, Ferreira MU. Emerging Plasmodium vivax resistance to chloroquine in South America: an overview. Mem Inst Oswaldo Cruz. 2014;109:534–539. doi: 10.1590/0074-0276130579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imwong M, Pukrittayakamee S, Gruner AC, Renia L, Letourneur F, Looareesuwan S, et al. Practical PCR genotyping protocols for Plasmodium vivax using Pvcs and Pvmsp1. Malar J. 2005;4:20. doi: 10.1186/1475-2875-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Read A, Day K. The genetic structure of malaria parasite populations. Parasitol Today. 1992;8:239–242. doi: 10.1016/0169-4758(92)90125-L. [DOI] [PubMed] [Google Scholar]
- 11.Zhong D, Bonizzoni M, Zhou G, Wang G, Chen B, Vardo-Zalik A, et al. Genetic diversity of Plasmodium vivax malaria in China and Myanmar. Infect Genet Evol. 2011;11:1419–1425. doi: 10.1016/j.meegid.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-abd NM, Mahdy MA, Al-Mekhlafi AM, Snounou G, Abdul-Majid NB, Al-Mekhlafi HM, et al. The suitability of P. falciparum merozoite surface proteins 1 and 2 as genetic markers for in vivo drug trials in Yemen. PLoS One. 2013;8:e67853. doi: 10.1371/journal.pone.0067853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atroosh WM, Al-Mekhlafi HM, Mahdy MA, Saif-Ali R, Al-Mekhlafi AM, Surin J. Genetic diversity of Plasmodium falciparum isolates from Pahang, Malaysia based on MSP-1 and MSP-2 genes. Parasit Vectors. 2011;4:233. doi: 10.1186/1756-3305-4-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mwingira F, Nkwengulila G, Schoepflin S, Sumari D, Beck H-P, Snounou G, et al. Plasmodium falciparum msp1, msp2 and glurp allele frequency and diversity in sub-Saharan Africa. Malar J. 2011;10:79. doi: 10.1186/1475-2875-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui L, Escalante AA, Imwong M, Snounou G. The genetic diversity of Plasmodium vivax populations. Trends Parasitol. 2003;19:220–226. doi: 10.1016/S1471-4922(03)00085-0. [DOI] [PubMed] [Google Scholar]
- 16.Patil A, Orjuela-Sánchez P, da Silva-Nunes M, Ferreira MU. Evolutionary dynamics of the immunodominant repeats of the Plasmodium vivax malaria-vaccine candidate circumsporozoite protein (CSP) Infect Genet Evol. 2010;10:298–303. doi: 10.1016/j.meegid.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wickramarachchi T, Premaratne PH, Dias S, Handunnetti SM, Udagama-Randeniya PV. Genetic complexity of Plasmodium vivax infections in Sri Lanka, as reflected at the merozoite-surface-protein-3alpha locus. Ann Trop Med Parasitol. 2010;104:95–108. doi: 10.1179/136485910X12607012374190. [DOI] [PubMed] [Google Scholar]
- 18.Raza A, Ghanchi NK, Thaver AM, Jafri S, Beg MA. Genetic diversity of Plasmodium vivax clinical isolates from southern Pakistan using pvcsp and pvmsp1 genetic markers. Malar J. 2013;12:16. doi: 10.1186/1475-2875-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rungsihirunrat K, Chaijaroenkul W, Siripoon N, Seugorn A, Na-Bangchang K. Genotyping of polymorphic marker (MSP3alpha and MSP3beta) genes of Plasmodium vivax field isolates from malaria endemic of Thailand. Trop Med Int Health. 2011;16:794–801. doi: 10.1111/j.1365-3156.2011.02771.x. [DOI] [PubMed] [Google Scholar]
- 20.Bruce MC, Galinski MR, Barnwell JW, Snounou G, Day KP. Polymorphism at the merozoite surface protein-3alpha locus of Plasmodium vivax: global and local diversity. Am J Trop Med Hyg. 1999;61:518–525. doi: 10.4269/ajtmh.1999.61.518. [DOI] [PubMed] [Google Scholar]
- 21.Haque U, Overgaard HJ, Clements AC, Norris DE, Islam N, Karim J, et al. Malaria burden and control in Bangladesh and prospects for elimination: an epidemiological and economic assessment. Lancet Glob Health. 2014;2:e98–e105. doi: 10.1016/S2214-109X(13)70176-1. [DOI] [PubMed] [Google Scholar]
- 22.Alam MS, Mohon AN, Mustafa S, Khan WA, Islam N, Karim MJ, et al. Real-time PCR assay and rapid diagnostic tests for the diagnosis of clinically suspected malaria patients in Bangladesh. Malar J. 2011;10:175. doi: 10.1186/1475-2875-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-B. [DOI] [PubMed] [Google Scholar]
- 24.Elahi R, Mohon AN, Khan WA, Haque R, Alam MS. Performance of a HRP-2/pLDH based rapid diagnostic test at the Bangladesh–India–Myanmar border areas for diagnosis of clinical malaria. Malar J. 2013;12:378. doi: 10.1186/1475-2875-12-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohon AN, Elahi R, Podder MP, Mohiuddin K, Hossain MS, Khan WA, et al. Evaluation of the OnSite (Pf/Pan) rapid diagnostic test for diagnosis of clinical malaria. Malar J. 2012;11:415. doi: 10.1186/1475-2875-11-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghanchi NK, Martensson A, Ursing J, Jafri S, Bereczky S, Hussain R, et al. Genetic diversity among Plasmodium falciparum field isolates in Pakistan measured with PCR genotyping of the merozoite surface protein 1 and 2. Malar J. 2010;9:1. doi: 10.1186/1475-2875-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gatton ML, Cheng Q. Can estimates of antimalarial efficacy from field studies be improved? Trends Parasitol. 2008;24:68–73. doi: 10.1016/j.pt.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alam MS, Chakma S, Khan WA, Glass GE, Mohon AN, Elahi R, et al. Diversity of anopheline species and their Plasmodium infection status in rural Bandarban, Bangladesh. Parasit Vectors. 2012;5:150. doi: 10.1186/1756-3305-5-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alam MS, Khan M, Chaudhury N, Deloer S, Nazib F, Bangali AM, et al. Prevalence of anopheline species and their Plasmodium infection status in epidemic-prone border areas of Bangladesh. Malar J. 2010;9:15. doi: 10.1186/1475-2875-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J-R, Imwong M, Nandy A, Chotivanich K, Nontprasert A, Tonomsing N, et al. Genetic diversity of Plasmodium vivax in Kolkata, India. Malar J. 2006;5:71. doi: 10.1186/1475-2875-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zakeri S, Raeisi A, Afsharpad M, Kakar Q, Ghasemi F, Atta H, et al. Molecular characterization of Plasmodium vivax clinical isolates in Pakistan and Iran using pvmsp-1, pvmsp-3alpha and pvcsp genes as molecular markers. Parasitol Int. 2010;59:15–21. doi: 10.1016/j.parint.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Zakeri S, Safi N, Afsharpad M, Butt W, Ghasemi F, Mehrizi AA, et al. Genetic structure of Plasmodium vivax isolates from two malaria endemic areas in Afghanistan. Acta Trop. 2010;113:12–19. doi: 10.1016/j.actatropica.2009.08.025. [DOI] [PubMed] [Google Scholar]
- 33.Adhikari M, Ranjitkar S, Schousboe ML, Alifrangis M, Imwong M, Bhatta DR, et al. Genetic diversity of Plasmodium vivax merozoite surface protein-3alpha (Pvmsp-3alpha) gene in Jhapa District of Nepal. Southeast Asian J Trop Med Public Health. 2012;43:280–286. [PubMed] [Google Scholar]
- 34.Gonzalez-Ceron L, Rodriguez MH, Nettel JC, Villarreal C, Kain KC, Hernandez JE. Differential susceptibilities of Anopheles albimanus and Anopheles pseudopunctipennis to infections with coindigenous Plasmodium vivax variants VK210 and VK247 in southern Mexico. Infect Immun. 1999;67:410–412. doi: 10.1128/iai.67.1.410-412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]


