Abstract
Background
Killer cell immunoglobulin-like receptors (KIRs) show extensive variation in genetic content and allelic polymorphi sms among different populations.
Materials and methods
We analyzed the distribution of KIR genes in the Tibetan ethnic minority of Lhasa city, the Uyghur and Kazakh ethnic minorities of Urumqi city populations in China. Genotyping of 16 KIR genes was tested in 479 randomly selected individuals using the multiple PCR-SSP method.
Results
A total of 42 KIR genotypes were detected, of which, 29 were predicted to be AB genotypes, 12 were BB genotypes and one was AA genotypes. 27 KIR genotypes were identified in Kazakhs, 30 KIR genotypes were identified in Uyghurs and 20 KIR genotypes were identified in Tibetans. The predominant genotype 1(AA genotypes) occurred most frequently in Tibetans (52.7%, 118/224), Kazakhs (43.2%, 54/125) and Uyghurs (34.9%, 45/130). Not only the four framework genes were present in all individuals, but the pseudogene 2DP1 could also be detected in all Uyghur individuals. Tibetans were different from Kazakh and Uyghur groups in KIR genetic content and KIR allelic variation. Intriguingly, Tibetans (29.5%, 66/224) had lower frequencies of 2DS4-v when compared with Uyghurs (60.8%, 79/130) and Kazakh s (59.2%, 74/125). Uyghurs (25.4%, 33/130) displayed higher frequencies of Bx genotypes with C4Tx (absence of KIR3DS1-2DL5-2DS5-2DS1) than both Kazakhs (11.2%, 14/125) and Tibetans (3.6%, 8/224).
Conclusions
The study showed that profile of KIR genotypes in three ethnic minority populations in China displayed ethnic diversity. It could be valuable for enriching the ethnical information resources for KIR gene, as well as facilitating further research on KIR-related diseases.
Electronic supplementary material
The online version of this article (doi:10.1186/s12967-015-0544-7) contains supplementary material, which is available to authorized users.
Keywords: KIR gene, Genotype, Haplotype, Ethnic minority populations in China
Background
Killer cell immunoglobulin-like receptors (KIRs) are a group of glycoproteins expressed on the surface of Natural killer (NK) cells [1, 2]. NK cells are important components of the innate immune system that take part in various immune responses to different infectious agents [3].
KIR genes are located on human chromosome 19q13.4. Up to now, 16 KIR gene loci have been identified, including two pseudogenes (KIR 2DP1 and 3DP1). Four framework KIR genes (2DL4, 3DL2, 3DL3 and 3DP1) exist in almost all populations [4]. Fourteen functional KIR genes have been identified and confirmed, of which seven are inhibitory (KIR2DL1-3, 2DL5 and 3DL1-3) and six are activating (KIR2DS1-5 and 3DS1), with only KIR2DL4 showing both inhibitory and activating potential [1, 5, 21]. According to the numbers and types, KIR haplotypes are broadly classified into two groups: group A haplotypes have a fixed KIR gene (2DL1, 2DL3-4, 3DL1-3, 2DS4, 2DP1 and 3DP1), in contrast, group B haplotypes have variations in genetic content which is comprised of several genes and alleles that are not found in A haplotypes. Especially, KIR2DS1-3, 2DS5, 2DL2, 2DL5 and 3DS1 are associated only with group B haplotypes, thus B haplotypes generally encode more activating KIR receptors than A haplotypes which encode a single activating receptor (KIR2DS4). Both group A and B are present in all populations, but their frequencies vary considerably [1, 6]. Many individuals carry two copies of A haplotypes (AA genotypes). The remainders carry Bx genotypes, which are consisted of either one copy of A haplotype and one copy of B haplotype (AB genotypes) or two copies of B haplotypes (BB genotypes). Based on the presence and absence of two distinct gene clusters (C4, KIR2DS2-2DL2-2DS3-2DL5; T4, KIR3DS1-2DL5-2DS5-2DS1), Bx genotype carries are divided into four subsets: C4Tx (presence of C4 and absence of T4), CxT4 (absence of C4 and presence of T4), C4T4 (presence of both C4 and T4), and CxTx (absence of both C4 and T4) [1, 7, 8].
Usually, Group B haplotypes encode more activating KIR genes. In individuals with AA genotype, the alleles of KIR2DS4 can encode either functional (KIR2DS4-f) or non-functional (KIR2DS4-v) variants [9, 10]. KIR2DS4-v alleles differ from the KIR2DS4-f alleles in that the former have a 22-bp deletion in exon 5, which leads to a frame shift mutation and produces a premature stop codon, preventing the formation of a functional membrane-bound receptor domain [10]. Therefore, individuals with AA genotypes which harbor KIR2DS4-v alleles will not have a membrane-bound KIR2DS4 protein, but rather a soluble form of KIR2DS4. KIR2DS4-f and 2DS4-v frequencies vary amongst different populations. Increasing attention has been paid to the role of KIR genetic content and allelic variation on infectious diseases such as Hepatitis C and HIV, with some studies dedicating a specific focus on KIR 2DS4 [9–14]. KIR gene diversities have been studied in many different geographical populations as previously reported [1, 9, 15–23]. There are 56 ethnic groups in China [24]. Most of the studies on KIR gene diversity have been reported on Han populations in China [18–21]. A large-scale survey on transfusion-transmitted HIV-1/2 infection among Chinese blood donors conducted by our institute showed that the positive rate for HIV infection was higher in some ethnic minorities (including data accumulated at Urumqi Blood Center, Xinjiang) than Han majority donors [25]. Expanding our understanding of the ethnic intermarriage and possibly random demographic factors could help us to determine the variation of KIR gene frequencies, which might be useful for future research on ethnicity-based diseases. Hence, in this study we chose the main minorities in Urumqi city of Northwest China, the Uyghur (comprised 10,069,346 persons [26], presented a typical mixture of Eastern and Western anthropometric traits [27]) and Kazakh (comprised 1,462,588 persons [26]), and the Tibetan (the major ethnic minority in Tibet, comprised 6,282,000 persons [26]), mostly living in Lhasa city of Southwest China, as the research objects. (Additional file 1: Figure S1).
Methods
Samples and DNA isolation
A total of 479 randomly selected donors were studied from three ethnic minority populations in China, including Tibetan (N = 224) volunteer blood donors in Lhasa (April 2011 to March 2012), Kazakh (N = 125) and Uyghur (N = 130) volunteer blood donors in Urumqi (March to August 2012). Informed consents of all samples used in this study were approved by the ethical Committee of Institute of Blood Transfusion. Genomic DNA was extracted from PBMC by TIANGEN blood DNA kit (Cat: #DP318-03), following the manufacturer’s instructions. The concentration of extracted DNA was adjusted to 50–100 ng/μL, and the O.D. 260/O.D. 280 ratio ranged from 1.6 to 1.9.
KIR PCR-SSP genotyping
KIR genes were typed for the presence or absence of the 14 KIR genes, including KIR2DL1-5, 2DS1-5, 3DL1-3 and the 2 pseudogenes (3DP1 and 2DP1), using multiplex PCR-SSP. In order to ensure accuracy, two different sets of primers for amplification (except 3DP1 and 2DS1) were used while HLA-DRB1 intron amplification primers were added to the third well as a positive control. Primer designs were adopted from those previously reported by Campbell et al. [28]. The primer arrangement was referred in Table 1 PCR master-mix for the multiplex PCR-SSP which contained 3 μL of primer (10 μM) pairs, 1.5 μL of ddH2O, 0.5 μL of DNA, 5 μL of GoTaq green master mix (Promega, Cat: #M7123), for a total of 10 μL. The conditions for PCR cycles were as followed: five cycles at 94°C for 3 min, 94°C for 15 s, 65°C for 15 s, 72°C for 30 s; twenty-one cycles at 94°C for 15 s, 60°C for 15 s, 72°C for 30 s; four cycles at 94°C for 15 s, 55°C for 1 min, 72°C for 2 min; lastly, at 72°C for 7 min and stored at 4°C. PCR products were visualized under UV light after electrophoresis in 3% agarose gel and stained with ethidium bromide, using DL1000 DNA Marker (TIANGEN, Cat: #MD108).
Table 1.
Multiple PCR primers combination pattern
| Mix | Locus (bp) | Locus (bp) | Locus (bp) | Locus (bp) |
|---|---|---|---|---|
| 1 | 2DL1 (330) | 2DL2 (173) | 2DL2 (151) | |
| 2 | 2DS3 (190) | 2DP1 (89) | ||
| 3 | DRB1 (800) | 3DS1 (300) | 3DL1 (191) | |
| 4 | 3DP1 (399/280)a | 2DL5 (214) | 2DL5 (191) | 2DS1 (102) |
| 5 | 2DL3 (800) | 2DL4 (254) | ||
| 6 | 2DL3 (550) | 2DS3 (242) | 3DS1 (180) | |
| 7 | 2DS4 (204) | 3DL1 (186) | 2DL1 (146) | |
| 8 | 2DS4 (179/219)b | 2DS5 (126) | ||
| 9 | 3DL3 (232) | 2DS5 (178) | 3DL2 (130) | |
| 10 | 2DP1 (205) | 2DS2 (175) | ||
| 11 | 2DS2 (240) | |||
| 12 | 2DL4 (288) | 3DL2 (242) | 3DL3 (165) |
aThe length of 3DP1 is 399 bp and the variant 3DP1v is 280 bp.
bThe length of 2DS4-f is 219 bp and the variant 2DS4-v is 197 bp.
Statistical analysis
Genotypic frequency = n/N (n represented numbers of positive genotypes and N was the total number of individuals tested). The carrier frequencies (CF) of the KIR genes were determined by dividing the number of positive typing reactions by the total number of typed individuals. The estimated gene frequencies were calculated using the formula GF = 1 − (1 − CF)1/2. The frequencies of A and B haplotypes were calculated using the following formula: group A = 2nAA + nAB/2N and group-B = 2nBB + nAB/2N, where nAA, nAB, and nBB were the numbers of AA, AB and BB genotypes. Differences in the carrier frequencies between the studied population and other populations previously published were assessed by the standard Chi square test (x2) using statistical software PEMS3.1 Medicine, and p < 0.05 was considered to be statistically significant. Subsets of KIR genetic content profiles and their frequencies in populations were carried out using statistical software Graph Pad Prism5. Principal components analysis (PCA) of 22 populations [15] was performed using an R package: psych software. Genetic distances were calculated by Nei’s method using phylip [29]. On the basis of the Nei’s genetic distance, a dendrogram was constructed by the neighbor-joining method [30] and visualized with MEGA software in order to compare the frequencies of KIR genes in 22 populations [15], using bootstrap values are calculated from 100 trees.
Results
KIR genotypes and haplotypes
A total of 479 individuals, 42 genotypes were found to carry a different number and combination of 16 KIR genes (Figure 1), of which, 29 were predicted to be AB genotypes, 12 were predicted to be BB genotypes and one was AA genotype. 27 KIR genotypes were identified in Kazakhs, 30 KIR genotypes were identified in Uyghurs and 20 KIR genotypes were identified in Tibetans. The predominant genotype 1 (AA genotypes) occurred most frequently in Tibetans (52.7%, 118/224), Kazakhs (43.2%, 54/125) and Uyghurs (34.9%, 45/130). Variations in genotypes and haplotypes were significant between the Tibetan and Kazakh/Uyghur groups (Table 2): 3 displayed significant statistical difference between Tibetan and Kazakh populations, 6 displayed significant statistical difference between Tibetan and Uyghur populations, and 2 displayed significant statistical difference between Uyghur and Kazakh populations.
Figure 1.

KIR gene content diversity of three Chinese populations, within 479 unrelated individuals, 42 genotypes that differed by the presence (shaded box) and absence (white box) of 16 KIR genes were detected. The frequency of each genotype is presented in percentage frequency (%F) and defined as the number of individuals carrying the genotype (n) divided by the number of individuals studied (N) in the provided population.
Table 2.
Comparison of genotypes, haplotypes and linkage groups in Tibetan, Kazakh and Uyghur populations
| Types | Tibetan | Kazakh | Uyghur | P values | ||
|---|---|---|---|---|---|---|
| N = 224 | N = 125 | N = 130 | Tibetan vs Kazakh | Tibetan vs Uyghur | Kazakh vs Uyghur | |
| %F(n) | %F(n) | %F(n) | ||||
| AA genotypes | 52.7 (118) | 43.2 (54) | 34.9 (45) | 0.0010 | ||
| BB genotypes | 0.9 (2) | 4.0 (5) | 9.0 (12) | 0.0471 | 0.0001 | |
| AB genotypes | 46.4 (104) | 52.8 (66) | 56.1 (73) | |||
| C4Tx genotypes | 3.6 (8) | 11.2 (14) | 25.4 (33) | 0.0049 | <0.0001 | 0.0035 |
| C4T4 genotypes | 4.9 (11) | 4.8 (6) | 1.6 (2) | |||
| CxT4 genotypes | 24.1 (54) | 21.6 (27) | 20.0 (26) | |||
| CxTx genotypes | 14.7 (33) | 15.2 (19) | 16.2 (21) | |||
| A haplogroups | 75.9 (170) | 69.6 (87) | 63.0 (82) | 0.0103 | ||
| B haplogroups | 24.1 (54) | 30.4 (38) | 36.9 (48) | 0.0103 | ||
| C4 gene-cluster | 8.5 (19) | 16.0 (20) | 26.9 (35) | 0.0326 | <0.0001 | 0.0340 |
| T4 gene-cluster | 29.0 (65) | 27.2 (34) | 21.5 (28) | |||
The haplotype A and B were determined by using the following formula: group A = 2nAA + nAB/2N and group B = 2nBB + nAB/2N, where nAA, nAB, and nBB are the numbers of AA, AB, and BB genotypes, N = total number of population. The p values are given only for those pairwise comparisons indicating significant (P < 0.05) differences.
KIR gene frequency
16 KIR genes were detected in Tibetans, Kazakhs and Uyghurs. The carrier frequency (CF) was listed in Table 3. Four framework genes (2DL4, 3DL2, 3DL3 and pseudogene 3DP1) were present in all individuals. KIR2DL1, 2DL3, 3DL1 and 2DP1 were detected in high frequencies (>90%). Among them, pseudogene 2DP1 can be detected in all individuals of Uyghur. KIR2DL2, 2DL5, 2DS1, 2DS2, 2DS3, 2DS5 and 3DS1 were in low frequencies. In the Uyghurs, the frequencies of KIR2DS4-f and KIR2DS4-v were found to be 62.3% (81/130) and 60.8% (79/130); in the Kazakhs, the frequencies of KIR2DS4-f and KIR2DS4-v were found to be 64.8% (81/125) and 59.2% (74/125); in the Tibetans, the frequencies of KIR2DS4-f and 2DS4-v were found to be 86.6% (194/224) and 29.5% (66/224). Four KIR genes displayed significant statistical difference between the Tibetan and Kazakh populations, eight KIR genes showed significant statistical difference between the Tibetan and Uyghur populations, while no difference was observed between the Kazakh and Uyghur populations (Table 3).
Table 3.
Comparison of carrier frequency of KIR genes in three ethnic minority populations in China
| KIR | Tibetan | Kazakh | Uyghur | P values | ||
|---|---|---|---|---|---|---|
| N = 224 | N = 125 | N = 130 | Tibetan vs Kazakh | Tibetan vs Uyghur | Kazak vs Uyghur | |
| %CF(n) | %CF(n) | %CF(n) | ||||
| A haplotype associated | ||||||
| 2DL1 | 99.1 (222) | 98.4 (123) | 98.5 (128) | |||
| 2DL3 | 98.7 (221) | 95.2 (119) | 92.3 (120) | 0.0056 | ||
| 3DL1 | 97.3 (218) | 95.2 (119) | 91.5 (119) | 0.0028 | ||
| 2DS4-f | 86.6 (194) | 64.8 (81) | 62.3 (81) | <0.0001 | <0.0001 | |
| 2DS4-v | 29.5 (66) | 59.2 (74) | 60.8 (79) | <0.0001 | <0.0001 | |
| B haplotype associated | ||||||
| 2DL2 | 24.6 (35) | 33.6 (42) | 43.8 (57) | 0.0003 | ||
| 2DL5 | 38.4 (86) | 47.2 (59) | 59.2 (77) | 0.0002 | ||
| 2DS1 | 36.2 (81) | 39.2 (49) | 40.8 (53) | |||
| 2DS2 | 25.0 (56) | 36.8 (46) | 46.9 (61) | 0.0277 | <0.0001 | |
| 2DS3 | 12.5 (28) | 23.2 (29) | 33.9 (44) | 0.0146 | <0.0001 | |
| 2DS5 | 31.3 (70) | 33.6 (42) | 33.1 (43) | |||
| 3DS1 | 35.3 (79) | 36.0 (45) | 39.2 (51) | |||
| Framework/pseudogenes | ||||||
| 2DL4 | 100 (224) | 100 (125) | 100 (130) | |||
| 3DL2 | 100 (224) | 100 (125) | 100 (130) | |||
| 3DL3 | 100 (224) | 100 (125) | 100 (130) | |||
| 2DP1 | 99.1 (222) | 98.4 (123) | 100 (130) | |||
| 3DP1 | 100 (224) | 100 (125) | 100 (130) | |||
Frequency (%CF) of carriers of each gene is expressed as a percentage and defined as the number of donors carrying the gene (n) divided by the number of donors studied (N) in the given populations. The p values are given only for those pairwise comparisons indicating significant (P < 0.05) difference.
Genetic relationships between populations
The PCA was generated depending on the basis of KIR gene frequencies of the three groups with other previously studied populations (as shown in Additional file 2: Table S1), PC1 and PC2 of 22 populations were given in Additional file 3: Table S2. Different geographic clusters of Asian and European were noted on the PCA plot. The most divergent were the clusters of African, Senegalese and Indian, as they did not cluster in a single group (Figure 2). Tibetans was plotted within the Asian group, the Uyghur was plotted within the European group, while the Kazakh was mapped between Asian and European groups. The neighbor-joining dendrogram was constructed, using the allelic frequency data of 12 KIR loci (KIR2DL1-3, 2DL5, 3DL1, 2DS1-5, 3DS1 and 2DP1) to show a relationship between the three groups and other previously reported populations [15] (Additional file 2: Table S1). In this dendrogram, there were four main clusters (Figure 3): European, African, Asian and Indian cluster. The Uyghur population was located in the European cluster, the Tibetan was located in the Asian cluster, while the Kazakh was located between European and Asian clusters. Genetic distance was shown in Figure 3.
Figure 2.
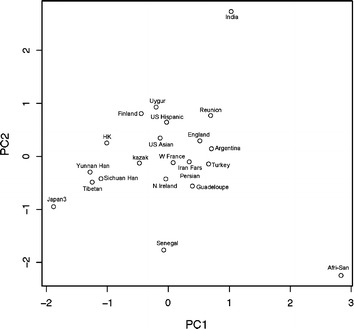
Principal component analysis (PCA) was conducted on the carrier frequencies of nine variable KIR genes. The PCA graph which was dependent on the frequencies of carriers with nine variably KIR genes (2DL1-3, 2DS1-4, 3DL1 and 3DS1) showed a worldwide perspective on the relationship between the three groups studied in this paper and other populations. PC1 and PC2 of 22 populations were given in Additional file 3: Table S2.
Figure 3.
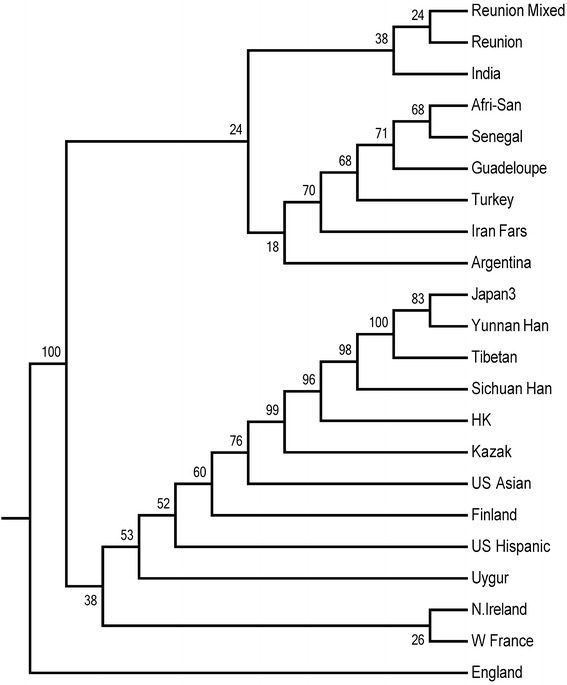
The neighbor-joining dendrogram represented the genetic distances and was created on the basis of the frequencies of 12 KIR gene (KIR2DL1-3, 2DL5, 3DL1, 2DS1-5, 3DS1 and 2DP1) in order to show the genetic relationship between the three groups and other previously reported populations. The KIR gene frequency of populations was given in Additional file 2: Table S1.
Genetic relationships between three ethnic populations centromeric and telomeric clusters of KIR genes (CxTx)
We found that the three groups carried all four subsets of Bx genotypes (CxT4, C4Tx, C4T4, and CxTx, Figure 1). 55 with the expression of C4Tx genotypes carried three to five activating KIR genes, among them, 49.1% (27/55) carried three activating KIRs and 45.3% (25/55) carried five activating KIRs. 108 with the expression of CxT4 genotypes carried three to six activating KIR genes, among them, 73.2% (79/108) carried four activating KIRs. 1 with the expression of C4T4 genotypes carried six activating KIRs. 73 individuals displaying CxTx carried one to five activating KIRs, among them, 50.7% (37/73) carried two activating KIRs, while 37.0% (27/73) carried four activating KIRs (Figure 1). The Uyghurs (25.4%, 33/130) comprised higher frequencies of Bx genotypes with C4Tx than the Kazakhs (11.2%, 14/125) and Tibetans (3.6%, 8/224, Figure 4).
Figure 4.

Subsets of KIR genetic content profiles and their frequency in populations. The frequencies of unique KIR genotype subsets in Tibetan, Kazakh and Uyghur populations were elucidated.
Discussion
KIR genotypes show extensive variation in populations according to different geographical regions and different ethnic groups, due to the presence or absence of different KIRs in individual haplotypes and allelic polymorphism of KIR genes [17]. In this study, Tibetans were detected to display a lower level of genotypic diversity, with the predominant genotypes ID1 (AA genotypes) [15] and ID2. The cumulative frequency of the top five genotypes 1–4, 6 was 84.8% (190/224), which showed that KIR genotype distribution in Tibetan populations was relatively concentrated. Meantime, Uyghurs and Kazakhs displayed a higher genotypic diversity, with the predominant genotypes ID1. Intriguingly, Uyghur displayed consistency with Caucasians on the level of KIR allelic frequency [9], which comprised high frequencies of Bx genotypes (absence of KIR3DS1-2DL5-2DS5-2DS1), while Tibetans shared a lower frequencies consistent with Northeast Asian (Han Chinese [21], Korean [22] and Japanese [23]). Our data on Kazakhs showed that the KIR allelic frequency is between that of Caucasians and Northeast Asians. Many studies on KIR gene diversity have been reported on Han populations in China [18–21]. Expanding our understanding of ethnic intermarriage and possibly random demographic factors for ethnic minorities could help us to determine the variation of KIR gene frequencies, which might be useful for future research on ethnicity-based diseases.
Activating KIR genes were found to show greater variability in frequencies than inhibitory KIR genes, which was similar to those previously reported in other populations [1]. KIR2DS4 is the only activating gene in AA genotypes, with the other 5 activating genes (KIR2DS1-3, 2DS5 and 3DS1) being absent [5, 21]. In this study, KIR2DS4-v was found to have a higher frequency in Uyghurs and Kazakhs, which was consistent with the frequency in Caucasians [9]. Tibetans had a lower KIR2DS4-v frequency when compared with Kazakhs and Uyghurs, more similar to Asian populations (Han Chinese, Japanese and Koreans) [21–23].
The PCA and the generated dendrogram aligned with the origins of ancestry distributing across vast geographic regions. In our data, the Uyghur was located in the European cluster, showing a similarity to the origins of European Caucasian ancestry. This was consistent with previously report that Uyghur population, settled in Xinjiang, was a population presenting a typical admixture of Eastern and Western anthropometric traits [25]. The Kazakh was between Eastern Asian and European Caucasian groups, demonstrating an array of mixed anthropological features of Europeans and Asians. This could help to further understand the origins of ancestry, the pattern of human migration, the intermarriage between ethnicities and the genetic distances between different populations or ethnic groups.
Conclusions
The KIR gene frequencies of the Tibetan and Uyghur populations were consistent with the geographic and ethnic distribution, while the Kazakhs showed unique genetic content and allelic polymorphisms. Better understanding the ethnic intermarriage and possibly random demographic factors could help determine the factors in the variation of KIR gene frequencies, which could be useful for future research on ethnicity-based diseases.
Author’s contributions
QZ and JW designed and performed the experiments, analyzed and interpreted the data, and drafted the manuscript; ZH and XL collected and informed consents of all samples used in this study; SM, SH and GB participated in the experiments and helped analyzed data; FM designed the experiments and wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
We thank Dr. Bo Chen and Ms. Ka Yi Li at Institute of Blood Transfusion, CAMS and PUMC, for their generous help in proofreading the manuscript and providing constructive advice. This work was sponsored by National Natural Science Foundation of China (NSFC 81001341) to Jue Wang.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Additional files
Figure S1. Map of China showing the city of three study populations. DNA samples of the Kazakh and Uyghur ethnic minority populations were collected from the Xinjiang autonomous region (Urumqi) of Northwest China, the Tibetan ethnic minority populations were collected from the Tibet autonomous region (Lhasa) of Southwest China.
Table S1. KIR diversity in three ethnic minority populations in China with other populations.
Table S2. PC1 and PC2 of 22 populations.
Footnotes
Qiongxiu Zhou and Jue Wang are co-first authors
Contributor Information
Qiongxiu Zhou, Email: qiongxiuzhou@163.com.
Jue Wang, Email: Wjue007@qq.com.
Zhi He, Email: hezhixj@sina.cn.
Xiaojuan Li, Email: 1846836379@qq.com.
Song Mao, Email: maosong327@gmail.com.
Shu Huang, Email: seazer7@126.com.
Guohui Bian, Email: joyey_love@126.com.
Feng Ma, Email: mafeng@hotmail.co.jp.
References
- 1.Ashouri E, Farjadian S, Reed EF, Ghaderi A, Rajalingam R. KIR gene content diversity in four Iranian populations. Immunogenetics. 2009;61:483–492. doi: 10.1007/s00251-009-0378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schleinitz N, Hamidou M, Vély F. NK cells: new insights on physiology and clinical implication in diseases. Rev Med Interne. 2006;27:465–472. doi: 10.1016/j.revmed.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 3.Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature. 2011;476:96–100. doi: 10.1038/nature10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson MJ, Torkar M, Haude A, Milne S, Jones T. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci USA. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faure M, Long EO. KIR2DL4 (CD158d), an NK cell-activating receptor with inhibitory potential. J Immunol. 2002;168:6208–6214. doi: 10.4049/jimmunol.168.12.6208. [DOI] [PubMed] [Google Scholar]
- 6.Parham P. MHC class Imolecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 7.Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR, et al. Global diversity and evidence for coevolution of KIR and HLA. Nat Genet. 2007;39:1114–1119. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- 8.McQueen KL, Dorighi KM, Guethlein LA, Wong R, Sanjanwala B, Parham P. Donor–recipient combinations of group A and B KIR haplotypes and HLA class I ligand affect the outcome of HLA-matched, sibling donor hematopoietic cell transplantation. Hum Immunol. 2007;68:309–323. doi: 10.1016/j.humimm.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heather AH, Maria P, Glenda EG, Louise K, Caroline TT. KIR2DS4 allelic variants: differential effects on in utero and intrapartum HIV-1 mother-to-child transmission. Clin Immunol. 2013;149:498–508. doi: 10.1016/j.clim.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Middleton D, Gonzalez A, Gilmore PM. Studies on the expression of the deleted KIR2DS4*003 gene product and distribution of KIR2DS4 deleted and nondeleted versions in different populations. Hum Immunol. 2007;68:128–134. doi: 10.1016/j.humimm.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Maxwell LD, Williams F, Gilmore PA. Investigation of killer cell immunoglobulin-like receptor gene diversity: II. KIR2DS4. Hum Immunol. 2004;65:613–621. doi: 10.1016/j.humimm.2004.02.028. [DOI] [PubMed] [Google Scholar]
- 12.Bao X, Hou L, Sun A. An allelic typing method for 2DS4 variant used in study of haplotypes of killer cell immunoglobulin-like receptor gene. Int J Lab Hematol. 2010;32:625–632. doi: 10.1111/j.1751-553X.2010.01234.x. [DOI] [PubMed] [Google Scholar]
- 13.Gllez-Hawkins GM, Franck AE, Li X. Expression of activating KIR2DS2 and KIR2DS4 genes after hematopoietic cell transplantation: relevance to cytomegalovirus infection. Biol Blood Marrow Transplant. 2011;17:1662–1672. doi: 10.1016/j.bbmt.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merino A, Malhotra R, Morton M, Mulenga J. Impact of a functional KIR2DS4 allele on heterosexual HIV-1 transmission among discordant Zambian couples. J Infect Dis. 2011;203:487–495. doi: 10.1093/infdis/jiq075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The allele frequency net database. Available at: http://www.allelefrequencies.net/
- 16.Badwe R, Joshi N, Carrington M. Comparison of the rapidly evolving KIR locus in Parsis and natives of India. Immunogenetics. 2008;60:121–129. doi: 10.1007/s00251-008-0279-1. [DOI] [PubMed] [Google Scholar]
- 17.Wang HD, Zhang FX, Shen CM. Genetic polymorphism analysis of killer cell immunoglobulin-like receptor genes in the Chinese Uygur population. Mol Biol. 2012;39:3017–3028. doi: 10.1007/s11033-011-1064-z. [DOI] [PubMed] [Google Scholar]
- 18.Zhan XT, Wang S, Wang J. Relationship between KIR and HLA ligands in Han population of Sichuan Marrow Donor Program. Chin J Blood Transfus. 2012;25:327–332. [Google Scholar]
- 19.Zhu FM, Jiang K. Zhejiang Han population of killer cell immunoglobulin-like receptor gene polymorphism. Chin J Exp Hematol. 2005;13:1109–1112. [PubMed] [Google Scholar]
- 20.Su PC, Yang TH, Zou H. Polymorph ism of killer cell immunoglobulin-like receptors gene in Yunnan Han population, China. Chin J Blood Transfus. 2008;21:268–272. [Google Scholar]
- 21.Bao X, Wang M, Zhou H, Wu X, Yang L. Characterization of killer cell immunoglobulin-like receptor (KIR) genotypes and haplotypes in Chinese Han population. Tissue Antigens. 2013;82:327–337. doi: 10.1111/tan.12211. [DOI] [PubMed] [Google Scholar]
- 22.Whang DH, Park H, Yoon JA. ParkMH: haplotype analysis of killer cell immunoglobulin-like receptor genes in 77 Korean families. Hum Immunol. 2005;66:146–154. doi: 10.1016/j.humimm.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 23.Yawata M, Yawata N, McQueen KL, Cheng NW, Guethlein LA, Rajalingam R, et al. Predominance of group A KIR haplotypes in Japanese associated with diverse NK cell repertoires of KIR expression. Immunogenetics. 2002;54:543–550. doi: 10.1007/s00251-002-0497-x. [DOI] [PubMed] [Google Scholar]
- 24.National Bureau of Statistics of China: Chinese Statistical Yearbook 2000. Beijing, China, Statistics Publishing Group 2001
- 25.Wang JX, Liu J, Yao FZ, Wen GX, Li JL, Huang Y, et al. Prevalence, incidence, and residual risks for transfusion transmitted HIV-1/2 infection among Chinese blood donors. Transfusion. 2013;53(6):1240–1249. doi: 10.1111/j.1537-2995.2012.03940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tabulation on the 2010 population census of the Peoples Republic of China. Available at: http://www.stats.gov.cn/tjsj/pcsj/rkpc/6rp/indexch.htm
- 27.Xu SH, Huang W, Qian J, Jin L. Analysis of genomic admixture in Uyghur and its implication in mapping strategy. Am J Hum Genet. 2008;82:883–894. doi: 10.1016/j.ajhg.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell KS, Martin MP, Carrington M. KIR genotyping by multiplex PCR-SSP//Campbell, SK. Natural killer cell protocols:methods in molecular biology. New York: Humana Press; 2010. pp. 365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Felsenstein J (1989) PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5:164–166
- 30.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]


