Abstract
Scope
Grape seed polyphenol extract (GSPE) is receiving increasing attention for its potential preventative and therapeutic roles in Alzheimer’s disease (AD) and other age-related neurodegenerative disorders. The intestinal microbiota is known to actively convert many dietary polyphenols, including GSPE, to phenolic acids. There is limited information on the bioavailability and bioactivity of GSPE-derived phenolic acid in the brain.
Methods and Results
We orally administered GSPE to rats and investigated the bioavailability of 12 phenolic acids known to be generated by microbiota metabolism of anthocyanidins. GSPE treatment significantly increased the content of 2 of the phenolic acids in the brain: 3-hydroxybenzoic acid (3-HBA) and 3-(3′-hydroxyphenyl) propionic acid (3-HPP), resulting in the brain accumulations of the two phenolic acids at μM concentrations. We also provided evidence that 3-HBA and 3-HPP potently interfere with the assembly of β-amyloid (Aβ) peptides into neurotoxic Aβ aggregates that play key roles in AD pathogenesis.
Conclusion
Our observation suggests important contribution of the intestinal microbiota to the protective activities of GSPE (as well as other polyphenol preparations) in AD. Outcomes from our studies support future preclinical and clinical investigations exploring the potential contributions of the intestinal microbiota in protecting against the onset/progression of AD and other neurodegenerative conditions.
Keywords: Phenolic Metabolites, Grape seed polyphenol extract, bioavailability, Proanthocyanidins, β-amyloid
1. INTRODUCTION
Grape seed polyphenol extract (GSPE) has been investigated widely as a dietary supplement with widespread health benefits. We recently demonstrated the potential efficacy of GSPE in protecting against neuropathology and cognitive impairment in animal models of Alzheimer’s disease (AD) [1] and tau-mediated neurodegenerative disorder [2, 3]. GSPE has also been reported to be protective against cardiovascular disease [4–7], colonic cancer [7, 8], and breast cancer [9, 10]. GSPE is a rich source of simple and complex phenolic compounds including gallic acid (GA), catechin (C), epicatechin (EC) as well as complex proanthocyanidins (PAC) dimers (B1, B2, B3, and B4), trimers (C1, C2, etc.), tetramers, and polymers [1, 11, 12]. The growing body of experimental, preclinical, and clinical evidence supporting GSPE exerting beneficial biological activities in multiple medical conditions, including neurodegenerative disorders such as Alzheimer’s disease (AD) and tauopathies [1–3, 13, 14], cardiovascular disease [4–6, 15–17], and cancer [7–9], has led to increased interest in its bioavailability, metabolism, and distribution of the primary GSPE phenolic constituents, including GA, C, EC, as well as PAC dimers and larger oligomers. Prior studies have investigated the absorption and metabolism of GSPE [18–20]. Interestingly, evidence suggests that intestinal microbiota (gut microorganisms) significantly contribute to GSPE metabolism/absorption, as CT and EC, major components of GSPE, are both metabolized by colonic microbiota fermentation [21, 22], and colonic microbiota fermentation products of CT and EC have been identified in urine and plasma [23–25].
The potential clinical application of GSPE (or other polyphenol preparations) in AD and in tauopathies, such as fronto-temporal dementia and progressive supranuclear palsy, that are associated with pathological aggregation of the microtubule-associated protein, tau, in the brain, is largely hindered by our limited knowledge on the absorption, metabolism, and bioavailability of GSPE polyphenols, particularly in the central nervous system. We have recently described accumulation of multiple CT and EC polyphenol metabolites in the brain following GSPE consumption[18], implicating potential contributions of these polyphenol metabolites to the neuroprotective effects of GSPE. Moreover, we recently demonstrated that one of these brain-bioavailable GSPE metabolites, 3′-O-methyl-epicatechin-5-O-β-glucuronide (3′-O-Me-EC-Gluc), contributes to GSPE’s neuroprotective effects by improving synaptic plasticity in the brain [13], and ongoing investigations in our lab are continuing to explore the neuroprotective roles of the other brain-available GSPE metabolites that we have identified[18].
Major dietary polyphenolic components (e.g., CT and EC [26]as well as other polyphenols, including anthocyanins [21, 27], chlorogenic acid [28–30], quercetin [21], rutin, and naringin [21], can all serve as substrates for intestinal microbiota, resulting in the formation of small molecular phenolic acids. Intestinal microbiota metabolites of GSPE phenolic compounds, or PACs, have been investigated in vitro [31–33] and detected in human urine [34–38]. While GSPE is known to be metabolized by colonic microbiota fermentation [31, 32, 32, 33], there is currently no information on the potential contribution of GSPE colonic microbiota fermentation products to the neuroprotective effects of GSPE. The present study is designed to address this issue by characterizing the conversion of GSPE polyphenols by intestinal microbiota, their metabolic fate, and their tissue distribution, particularly in the brain, using Sprague-Dawley rats as an animal model.
The majority of dietary polyphenols consumed are not absorbed by the upper intestinal track, and are further broken down by gut microbiota in the colon into low-molecular-weight phenolic compounds, such as phenolic acids, that can be more efficiently absorbed by GI epithelial cells [39, 40]. In the case of orally consumed GSPE, it is anticipated that CT and EC components from the GSPE will be converted into multiple phenolic acids through ring fusion reactions facilitated by intestinal microbiota. In vitro studies have demonstrated that isolated human fecal microbiota is capable of metabolizing CT/EC into multiple phenolic acids, including 3-hydroxyphenolic acetic acid, 3,4-dihydroxyphenolactic acid, 3-(3′-hydroxyphenyl) propionic acid, and 3-(3′,4′-dihydroxyphenyl)propionic acid [41]. The formation of phenolic compounds from GSPE was anticipated based on previous observations from in vitro colonic microflora metabolism of flavonoids, including proanthocyanidins, and urinary output of phenolic acid following oral administration flavanoids [21, 30, 31, 35, 37].
A tentative rout by which gut microbiota metabolizes GSPE PAC into multiple phenolic acids is presented in Figure 1, based on previously published information on the metabolism of polyphenols and phenolic acids [42–44, 44, 45]. Based on this consideration, we surveyed 12 phenolic acids along this tentative metabolic pathway as potential phenolic acids that could be generated by gastrointestinal (GI) microbiota metabolism of GSPE: 1) ferulic acid (FA); 2) hippuric acid (HA); 3) 3-hydroxybenzoic acid (3-HBA); 4) 4-hydroxybenzoic acid (4-HBA); 5) 3-hydroxyhippuric acid (3-HHA); 6) 4-hydroxyhippuric acid (4-HHA); 7) 3-hydroxyphenyl acetic acid (3-HPA); 8) 3-(3′,4′-dihydroxyphenyl)propionic acid (3,4-diHPA); 9) 3-(3′-hydroxyphenyl) propionic acid (3-HPP); 10) 3-(3′,4′-dihydroxyphenyl)propionic acid (3,4-diHPP); 11) 5-(4′-hydroxyphenyl)valeric acid (5-HPV), and 12) phenylacetic acid (PA).
Figure 1. Tentative metabolic route of GSPE PAC and molecular formulas of PAC-derived phenolic acids.
Following cleavage of the interflavan bond, a monomeric PAC undergoes either C-ring fission [30]or A-ring fission [52] by the intestinal microbiota in the lower intestine. Three degradation routes of C-ring are shown as routes 1, 2, and 3. Routes 1 and 3 will form 3,4-diHBA or 3,4-diHPA, which further form 3-HBA and 4-HBA or 3-HPA and PA, respectively, by removal of a hydroxyl group. Route 2 will form 3,4-diHPP initially, followed by 3-HPP and FA, while 3-HPP is probably further degraded to 3-phenylpropionic acid (3-PP) by dehydroxylation, or 4-HHA and 3-HHA by β-oxidation. 3-PP can further form HA by β-oxidation reaction. On the other hand, there is also a possibility of A-ring cleavage to form 5-(3,4-dihydroxyphenyl)-γ-valerolactone (3,4-diHPVal) [52], which can further form 5-HPV. Dark arrows point to GSPE PAC-derived phenolic acids targeted for analysis in this study.
We now reported that two phenolic acid metabolites generated by intestinal microbiota fermentation of GSPE, namely 3-hydroxybenzoic acid (3-HBA) and 3-(3′-hydroxyphenyl) propionic acid (3-HPP), area found to reach and accumulate in the brain in a dose dependent manner. Most interestingly, in vitro studies revealed that both brain-accumulating phenolic acidscan potently interfere with the assembly of β-amyloid (Aβ) peptides into neurotoxic Aβ aggregates that play key roles in AD neuropathogenesis. Outcomes from our studies suggest that intestinal microbiota may help protect against the onset/progression of AD and other neurodegenerative conditions involving aberrant, pathological protein aggregations.
2. MATERIALS AND METHODS
2.1 General Experimental Procedures
We assessed phenolic acids contents in biological fluids (urine, plasma) and tissue specimens (cecum, colon, brain) in rats treated with GSPE or corresponding vehicles. We distinguished the large intestine into two separate compartments (i.e., the cecum and the colon) due to previous evidence that phenolic metabolism occurs separately in these two different compartments (Liu, et al., 2014). Twelve phenolic acids, listed in Figure 1A, were surveyed following previously published methods[46], with some modification. Analyses were conducted on an Agilent Technologies MSD-TOF mass spectrometer (G1969A) coupled to an Agilent Technologies 1100 High Performance Liquid Chromatography (HPLC) binary pump (Palo Alto, CA), using a Varian Polaris Amide C18 column (3 μm, 150×2.1mm i.d, Palo Alto, CA). Phenolic acids were resolved by gradient elution using mobile phases A (deionized water with 0.1% v/v formic acid) and B (acetonitrile with 0.1% v/v formic acid), from 10% B to 20% B in 12 min using a linear gradient, and were held from 12 to 15 min, followed by a linear gradient to 50 % B in 5 min, and a reset to 10% B from 21 to 25 min. Mobile phase flow rate was set at 0.3 mL/min, and the injection volume was 10 μL for all samples. Following the separation, flow was split 1:1 from the HPLC and introduced by negative mode electrospray ionization (ESI) into the MS-TOF. ESI capillary voltage was set at −3.5 kV, nebulizer gas (N2) pressure of 35 psi was applied to assist the spraying. Dry gas temperature was 350°C at a flow rate of 9.0 L/min. Fragmentor voltage was set to 145 V, skimmer 60 V, and octopole RF peak 250 V. Mass data was collected at m/z 60–1000 and analyzed using Agilent MassHunter Qualitative Analysis B.03.02 software (Agilent Technologies). Identification of FA, HA, 3-HBA, 4-HBA, 3-HPA, 3,4-diHPA, 3-HPP, 3,4-diHPP, and PA was accomplished by comparison with retention time and molecular ions of individual authentic standards, and quantitation was conducted using calibration curves with authentic standards. Identification of 3-HHA and 4-HHA were based on criteria from Gonthier et al. and Saura-Calixto et al [33, 47], and identification of 5-HPV was based on criteria from Déprez S. et al. [31, 47]. Liquid chromatography (LC)-Tandem mass spectrometry (MS/MS) parent/product ion pair, limit of detection (LOD), and limit of quantitation (LOQ) of the phenolic acids are shown in Figure 2A. Representative LC-MS/MS detections of authentic standard phenolic acids are shown in Figure 2B.
Figure 2. LC-MS/MS parent/product ion pair and Limit of Quantitation of phenolic acids.
A) Shown is information on LC-MS/MS parent/product ion pair, limit of detection (LOD), and limit of quantitation (LOQ) of the 12 phenolic acids we surveyed in this study. Identification of FA, HA, 3-HBA, 4-HBA, 3-HPA, 3,4-diHPA, 3-HPP, 3,4-diHPP, and PA was accomplished by comparison with retention time and molecular ions of individual authentic standards. Identification of 3-HHA and4-HHA were based on criteria from Gonthier et al. and Saura-Calixto et al [33, 47], and identification of5-HPV was based on criteria from Déprez S. et al. [31, 47]. B) Representative LC-MS/MS detection of authentic standard phenolic acids.
Analyses of the GSPE metabolites in cecum, colon, and brain tissues were completed using an Agilent 1200 HPLC coupled to a triple quadrupole MS/MS system (6460A) operated in Multiple Reaction Monitoring mode. Identical separation conditions were used as described above. MS/MS data were analyzed by Agilent Quantitative Analysis B.03.02 (Agilent Technologies) software. Identification was based on retention time and the respective m/z values of the parent and product ions, as shown in Figure 2A. A dwell time of 80 ms was used. Signal-to-noise ratios of 3 and 10 were used as the limit of detection (LOD) and limit of quantitation (LOQ). All tissue accumulation data were expressed as nmoles per gram wet tissue weight (nmol/g TW), while the phenolic concentration in urine and plasma were expressed as μmoles per day (μmoles/d) or μmoles per liter (μmol/L).
2.2 GSPE and Chemicals
The GSPE used in these studies was a gift from Polyphenolics, Inc. (Madera, CA). This GSPE is a highly purified, 100% water-soluble experimental polyphenolic extract from Vitis vinifera grape seeds (Polyphenolics, Inc., CA, USA). The polyphenolic component in this GSPE is comprised entirely of CT and EC, derivatives of CT and EC (e.g., eipcatechin gallate, in which epicatechin is modified with the addition of a gallic acid), and PACs, which are polymeric compounds derived from C, EC, and their derivatives[14, 18]. High-performance liquid chromatography analysis demonstrates that this GSPE is comprised primarily of CT and EC in the form of monomeric, oligomers, and polymeric PAC forms, with approximately 8% as monomers, 75% as oligomers, and 17% as polymers [14, 18]. β-glucuronidase (with sulfatase contamination) and the phenolic standards (+)-C, (−)-EC, 3-HPA, 3,4-diHPA, and HA, were obtained from Sigma Chemical Co. (St. Louis, MO). 3-3HPP, 3,4-diHPP, 3-HBA, 4-HBA, and PA were obtained from Lancaster Synthesis (Alfa Aesar, Ward Hill, MA). All extraction and HPLC solvents were certified HPLC and ACS grade and were purchased from J.T. Baker (Phillipsburg, NJ).
2.3 Animals, Diets, and Sampling
The animal experimental procedures were conducted under protocols approved by the Purdue University Animal Care and Use Committee, protocol #1111000165. Male Sprague-Dawley rats were purchased at 12 weeks of age from Harlan (Indianapolis, IN, USA) and ranged in body weight from 250–260 g. Upon arrival, rats were placed in individual stainless steel metabolic cages in a temperature controlled environment (23 °C±1) with a 12 hour light-dark cycle, and allowed to acclimatize for 5 days. Rats had access to an AIN 93M based polyphenol free diet (Dyet#101591, Dyets Inc, PA) and deionized water ad lib. Animals were gavaged once daily for 11 days with vehicle (control) or GSPE (at either 25 or 250 mg/Kg BW), n=4–6 per group.
Twenty-four hour urine specimens were collected prior to and on day 10 of the GSPE (or vehicle) treatment study. Urine samples were centrifuged at 5°C, 2,500 rpm for 10 min, total volumes were recorded, and the supernatant was kept in 1.5mL aliquots. All samples were purged with N2 and stored at −80°C until analysis. Animals were sacrificed six hours after the last GSPE dose at day 11 by an overdose of carbon dioxide. Plasma was collected from the dorsal aorta, and aliquots of 400 μL combined with 100 μL acidified saline (4:1) were prepared. All plasma samples were purged with N2 and stored at −80°C until analysis. After sacrifice, the vascular system was perfused with ice-cold physiological saline, and tissues including brain and large intestine (cecum and colon)were harvested and stored in a 10 ml solution of 0.2% ascorbic acid in saline.
2.4 Phenolic Acid Extraction
Urine, plasma, and tissue samples were extracted based on previously published methods [29, 46], with minor modification. Briefly, urine (1.5 mL) and plasma (140 μL) samples were incubated, respectively, with 4 mL or 1 mL enzyme solution (250 U β-glucuronidase + sulfatase contamination in 0.4 M NaH2PO4 pH 4.5) for 45 min at 37°C after purging with nitrogen. Perfused brain and intestinal tissue specimens (~600 mg) were diced finely and homogenized with 2 mL buffer (0.4 M NaH2PO4), and then homogenized with polytron to a uniform opaque liquid, followed by incubation with 1 mL enzyme solution (574 U β-glucuronidase + sulfatase contamination in 0.4 M NaH2PO4 pH 4.5). After incubation, all samples were acidified to pH <3 by hydrochloric acid, followed by extraction three times with ethyl acetate/0.01% 2,6-Di-tert-butyl-4-methylphenol. Combined ethyl acetate fractions were dried under vacuum, re-solubilized in 200 μL mobile phase (90% mobile phase A and 10% mobile phase B), and filtrated through a 0.45μm Nylon SYR filter (National Scientific, Rockwood, TN) prior to analysis.
2.5 Preparation of Synthetic β-amyloid 1-42 (Aβ1-42) for in vitro Aβ Aggregation Studies
Aβ1-42 peptides were synthesized, purified, and characterized as described previously [48]. Briefly, synthesis was performed on an automated peptide synthesizer (model 433A, Applied Biosystems, Foster City, CA) using 9-fluorenylmethoxycarbonyl-based methods. Peptides were purified using reverse-phase HPLC. Quantitative amino acid analysis and mass spectrometry yielded the expected compositions and molecular weights for each peptide. Purified peptides were stored as lyophilizates at −20 °C. A stock solution of glutathione S-transferase (GST; Sigma-Aldrich) was prepared by dissolving the lyophilizate to a concentration of 250 μm in 60 mm NaOH. Prior to use, aliquots were diluted 10-fold into 10 mm sodium phosphate, pH 7.4. Aggregate-free solutions of Aβ were prepared using size exclusion chromatography as previously described by Teplow [49]. In brief, 200 μl of a 2 mg/ml Aβ1-42 peptide solution in dimethyl sulfoxide was sonicated for 1 min using a bath sonicator (Branson Ultrasonics, Danbury, CT), and then centrifuged for 10 min at 16,000 × g. The resulting supernatant was fractionated on a Superdex 75 HR column using 10 mm phosphate buffer, pH 7.4, at a flow rate of 0.5 ml/min. The low-molecular-weight fraction consisting almost entirely of monomeric Aβ1-42 was collected. A 10-μl aliquot was taken for quantitative amino acid analysis, and the remaining low-molecular-weight Aβ1-42 was used immediately for aggregation studies using thioflavin-T (Th-T) fluorescence assay, photo-induced cross-linking of unmodified proteins (PICUP) assay, circular dichroism (CD) assays, or electron microscopy (EM).
2.6 Thioflavin-T (Th-T) Fluorescence Assay
Aβ1-42 peptide (25 μM), phenolic acid (3-HBA or 3-HPP at 4:1, 1:1 and 1:4 molar ration with Aβ)or vehicle, and Th-T(25 μM) were mixed together in 10 mm phosphate buffer, pH 7.4 in a final 200 μl volume. Fluorescence was determined at 0, 4, 8, 2, 16, 20 and 24 h using a Hitachi F-4500 fluorometer, as we have previously described [50]. Excitation and emission wavelengths were 445 and 490 nm, respectively.
2.7 Photo-Induced Cross-Linking of Unmodified Proteins (PI CUP) Assay
PICUP assays were conducted as previously described in Bitan et al.[51]. In brief, freshly isolated low-molecular-weight Aβ1-42 (25 μM) peptide (18 μl) was mixed with 1 μl of 1 mm tris-(2,2′-bipyridyl)dichlororuthenium(II) [Ru(Bpy)] and 1 μl of 20 mM ammonium persulfate in the absence or presence of individual phenolic acids (3-HBA or 3-HPP, at 1:1 or 1:4 molar ratio with Aβ) in 10 mM phosphate (pH 7.4). The mixture was irradiated for 1 s, quenched immediately with 10 μl of Tricine sample buffer (Invitrogen) containing 5% β-mercaptoethanol, and subjected to SDS-PAGE. Monomeric and multimeric Aβ peptide species were visualized by silver staining (SilverXpress; Invitrogen).
2.8 Circular Dichroism Spectroscopy
Aβ1-42 (25 μM) was incubated according to the aggregation protocol above in the presence or absence of phenolic acid (100 μM). CD spectra of the mixtures were acquired immediately after sample preparation (time 0) or following 3, 4, 6, 12, 24 hours of incubation at 37 °C, as we have previously described [50], using a J-810 spectropolarimeter (JASCO, Tokyo, Japan). Spectra were recorded at 22 °C from ~190–260 nm at 0.2 nm resolution with a scan rate of 100 nm/min. Ten scans were acquired and averaged for each sample. Raw data were manipulated by smoothing and subtraction of buffer spectra according to the manufacturer’s instructions.
2.9 Electron Microscopy
Aβ1-42 (25 μM) was incubated according to the aggregation protocol above in the presence or absence of phenolic acid (100 μM). Formation of Aβfibril was assessed by electron microscopy following 10 days of incubation at 37°C, as we have previously described [50]. In brief, a 10-μl aliquot of each sample was spotted onto a glow-discharged, carbon-coated Formvar grid (Electron Microscopy Sciences, Hatfield, PA) and incubated for 20 min. The droplet was then displaced with an equal volume of 2.5% (v/v) glutaraldehyde in water and incubated for an additional 5 min. Finally, the peptide was stained with 8 μl of 1% (v/v) filtered (0.2 μm) uranyl acetate in water (Electron Microscopy Sciences). This solution was wicked off, and then the grid was air-dried. Samples were examined using a JEOL CX100 transmission electron microscopy.
2.10 Statistics
GSPE bioavailability studies were analysed by SAS9.1 (Cary, NC) and presented as mean ± standard error of the mean (SEM). Raw data were transformed by Power Transformation using BoxCox method before statistical analysis. For single-variable analysis a t-test comparison was used, while analysis for multivariable studies used two-way factorial ANOVA with repetitive measures followed by Tukey-Kramer post hoc comparisons and significance was set at P<0.05to define group differences. All tissue accumulation data were calculated and expressed as μmoles per gram wet tissue weight (μmol/g WT), while the phenolic concentration in urine and plasma was expressed as μmoles per liter (μmol/L). For Aβ Th-T assay, one-way factorial ANOVA followed by Tukey-Kramer post hoc comparisons was used, and significance was set at P<0.05 to define group differences.
3. RESULTS
3.1 Regional distribution of dietary GSPE derived phenolic acid in rats
There is evidence that certain phenolic acids derived from GI microbiome metabolism of dietary polyphenol preparations are absorbed by GI epithelial cells, followed by distribution into the circulating blood [23, 39, 40, 52, 53]; phenolic acids that are not absorbed are cleared from the GI track through the feces [40]. Once in the circulatory system, phenolic acids may be distributed to targeted tissues and/or cleared through urinary excretion; some of the phenolic acids may remain in the blood [40]. Based on the consideration in this study, rats were treated with a daily dose of GSPE (250 mg/Kg BW) or vehicle for 11 consecutive days and initially monitored for accumulation of GSPE phenolic acid metabolites in multiple physiological compartments, including the GI track, e.g., colon and cecum, and biological fluids, e.g., urine and blood, to provide a more comprehensive assessment of targeted phenolic acid generations from orally administered GSPE. Cecum and colon specimens were collected 6 hours after the last dose of GSPE at day 11, followed by assessment of tissue phenolic acid contents.
Generation and accumulation of dietary GSPE derived phenolic acids in the large intestine
Among the 12 phenolic acids metabolites of GSPE surveyed in this study, 8 phenolic acids (HA, 3-HBA, 4-HBA, 3-HPA, 3,4-diHPA, 3-HPP, 3,4-diHPP and PA) were detectable in cecal tissues (Table IA). We found significantly higher contents of 3-HBA, 3-HPA, 3,4-diHPA, 3-HPP and 3,4-HPP in cecal samples of the GSPE-treated group compared to the vehicle-treated control group (Table IA). In contrast, we observed significantly lower levels of HA 4-HBA, and PA in cecal specimens from the GSPE-treated compared to the vehicle-treated control group (Table IA). No detectable accumulation of FA, 3-HHA, 4-HHA, or 5-HPV was observed in cecal samples from either the GSPE-treated or the vehicle-treated control group (Table 1A).
Table 1. Phenolic acid accumulation in the cecum, colon, urine, and plasma in response to GSPE treatment.
Rats were treated with GSPE (250 mg/Kg BW/day) or vehicle (control) for 11 days (n=per group). 24-hour urine specimens were collected at day 10. Animals were then sacrificed at day 11, 6 hours after the last dose of GSPE/vehicle treatment, followed by immediate collection of cecum, colon, and plasma specimens. Presented are accumulations of phenolic acids in cecum (1A), colon (1B), urine (1C), and plasma (1D) specimens collected. Data are displayed as mean ± SEM;
| A cecum (μmol/kg wet tissue weight) | B colon (μmol/kg wet tissue weight) | |||||
|---|---|---|---|---|---|---|
| Compounds | control | GSE | fold change (GSE/Ctl) | control | GSE | fold change (GSE/Ctl) |
| FA | — | — | — | — | — | — |
| HA | 37.11 ± 14.62 | 15.26 ± 1.26 * | 2.4

|
35.33 ± 17.69 | 71.23 ± 11.01 * |
2.0

|
| 3-HBA | 26.20 ± 5.09 | 72.06 ± 33.75 * |
2.8

|
12.13 ± 2.45 | 33.09 ± 5.45 * |
2.7

|
| 4-HBA | 113.72 ± 47.03 | 37.61 ± 14.80 * | 3.0

|
18.93 ± 3.72 | 43.94 ± 14.41 * |
2.3

|
| 3-HHA | — | — | — | — | — | — |
| 4-HHA | — | — | — | — | — | — |
| 3-HPA | 33.16 ± 14.33 | 375.69 ± 259.61 * |
11.3

|
20.07 ± 8.17 | 589.11 ± 46.98 * |
29.4

|
| 3,4-diHPA | 59.83 ± 8.85 | 323.60 ± 149.40 * |
5.4

|
19.83 ± 6.83 | 753.90 ± 302.74 * |
38.0

|
| 3-HPP | 190.51 ± 158.82 | 940.20 ± 52.56 * |
4.9

|
70.56 ± 59.42 | 2358.83 ± 865.71 * |
33.4

|
| 3,4-diHPP | 74.36 ± 15.07 | 496.22 ± 189.66 * |
6.7

|
42.41 ± 7.49 | 552.07 ± 169.46 * |
13.0

|
| 5-HPV | — | — | — | — | — | — |
| PA | 1167.88 ± 363.94 | 276.58 ± 65.50 * | 4.2

|
293.68 ± 93.38 | 705.93 ± 278.35 * |
2.4

|
| C urine (nmol/day) | D plasma (μM) | |||||
|---|---|---|---|---|---|---|
| Compounds | control | GSE | fold change (GSE/Ctl) | control | GSE | fold change (GSE/Ctl) |
| FA | 23.2 + 2.6 | 128.2 ± 56.6 * |
5.5

|
0.49 ± 0.09 | 0.47 ± 0.04 | 0.0 |
| HA | 7.3 + 1.0 | 22.2 ± 7.1 * |
3.0

|
3.14 ± 0.42 | 4.72 ± 0.48 * |
1.5

|
| 3-HBA | 0.0 + 0.0 | 92.0 ± 19.5 * |

|
— | — | — |
| 4-HBA | 448.0 + 70.0 | 366.2 ± 103.8 | 0.0 | — | — | — |
| 3-HHA | 21.3 + 0.9 | 54.8 ± 12.0 * |
2.6

|
— | — | — |
| 4-HHA | 0.0 + 0.0 | 241.5 ± 51.5 * |

|
— | — | — |
| 3-HPA | 1.5 + 0.1 | 1.0 ± 0.3 * | −1.5

|
0.32 ± 0.03 | 0.28 ± 0.01 | 0.0 |
| 3,4-diHPA | 4.7 + 0.3 | 2.5 ± 0.7 * | −1.9

|
— | — | |
| 3-HPP | 249.5 + 34.7 | 627.3 ± 127.5 * |
2.5

|
0.36 ± 0.03 | 0.92 ± 0.12 * |
2.6

|
| 3,4-diHPP | 455.3 + 85.0 | 590.6 ± 319.2 | 0.0 | 1.0 ± 0.05 | 1.25 ± 0.06 * |
1.3

|
| 5-HPV | 0.0 + 0.0 | 78.7 ± 6.6 * |

|
0.0 ± 0.0 | 0.48 ± 0.07 * |

|
| PA | — | — | — | — | — | — |
P<0.05, GSPE-treated compared to vehicle-treated control.
Data in bold highlight phenolic acids whose contents are significantly elevated in biologic fluids and/or tissues in response to GSPE treatment. Dark and light arrows indicate, respectively, increase or decrease fold change in GSPE-treated vs. vehicle-treated control specimens.
The same8 phenolic acids detected in the cecum were also identified in the colon section of the large intestine (Table IB). We observed that GSPE dietary regimen significantly increased the accumulation of all of the 8 phenolic acids in colon specimens from the GSPE group vs. the vehicle-treated control group (Table IB). No detectable accumulation of FA, 3-HHA, 4-HHA, or 5-HPV was observed in colon samples from the vehicle-treated control or GSPE-treated groups (Table IB).
Our observation, that significantly higher levels of HA, 3-HBA, 4-HBA, 3-HPA, 3,4-diHPA, 3-HPP, 3,4-diHPP, and PA are accumulated in cecal and/or colon samples in the GSPE-treated group vs. vehicle-treated control group is consistent with generation of these phenolic acids from the orally administered GSPE by the GI microbiota.
Urinary secretion of dietary GSPE derived phenolic acid
In parallel studies, we continued to survey the excretion of the GSPE phenolic acids in urine samples collected over 24 hours on treatment day 10. We found that 11 of the 12 microbiota fermentation phenolic acids products(FA, HA, 3-HBA, 4-HBA, 3-HHA, 4-HHA, 3-HPA, 3,4-diHPA, 3-HPP, 3,4-diHPP, and 5-HPV) were detectable in the 24-hour urine specimens(Table IC). Compared to the vehicle-treated control group, we found that the GSPE-treated group had significantly elevated levels of 7 phenolic acid contents in 24-hour urine specimens: FA, HA,3-HBA, 3-HHA, 4-HHA, 3-HPPand 5-HPV(Table IC). This observation tentatively implicates generation of the 7 phenolic acids from orally administered GSPE, followed by GI absorption and clearance by urinary excretion. In contrast, lower levels of 3-HPA and 3,4-diHPA were observed in 24-hour urinary specimens from GSPE-treated, compared to vehicle-treated controls (Table IC). No detectable change in the 24-hour urinary contents for 4-HBA or 3,4-diHPP was observed in urinary specimens from the GSPE-treated group vs. the vehicle-treated control group. No detectable accumulation of PA was observed in urinary specimens from either the vehicle-treated control group or the GSPE-treated group (Table IC).
Circulatory levels of dietary GSPE-derived phenolic acids
Surveying contents of phenolic acids in plasma 6 hours after the last treatment of rats with GSPE at treatment day 11, we found that 6 of the GSPE microbiota fermentation phenolic acid metabolites (FA, HA, 3-HPA, 3-HPP, 3,4-diHPP and 5-HPV) were detectable in μM to sub-μM concentrations (Table ID). In comparison to vehicle-treated controls, 11 days of a repeated GSPE dietary regimen resulted insignificantly increased plasma contents of 4 phenolic acids: HA, 3-HPP,3,4-diHPP, and 5-HPV (Table ID). No detectable change in plasma contents for FA, HA, 3-HPA, 3-HPP, or 3,4-diHPP was observed in response to GSPE treatment (Table ID). No detectable plasma content of 3-HBA, 4-HBA, 3-HHA, 4-HHA, 3,4-diHPA, or PA were observed in plasma specimens from either the GSPE-treated group or vehicle-treated controls (Table ID).
These observations suggest that dietary GSPE treatment significantly increases the accumulation of all of the surveyed phenolic acids in the cecum, colon, urine, and/or circulation, and are consistent with the hypothesis that the phenolic acids are generated by GI microbiota fermentation of orally administered GSPE, possibly through metabolic pathways outlined in Figure 1.
3.2 Dose-dependent accumulation of GI microbial derived phenolic acids in the brain
We explored the dose-dependent accumulation of GI derived phenolic acid following 11 days treatment with a daily dose of GSPE (25 mg/Kg BW/day or 250 mg/Kg BW/day, followed by assessments of phenolic acids in perfused brain specimens); parallel studies using vehicle-treated rats served as controls.
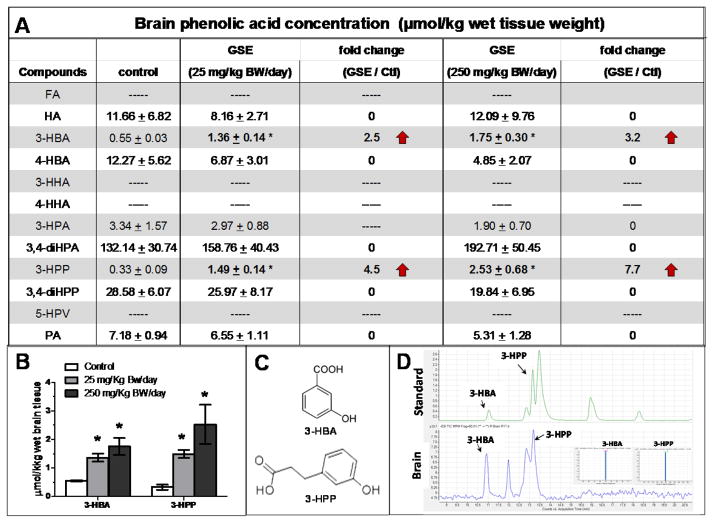
We found 8 phenolic acids (HA, 3-HBA, 4-HBA, 3-HPA, 3,4-diHPA, 3-HPP, 3,4-diHPP, and PA) that were detectable in perfused brain tissues at μM concentrations (Figure 3A). In contrast, FA, 3-HHA, 4-HHA, and 5-HPV were not detectable from perfused brain tissues (Figure 3A). We observed that 11 days of repeated GSPE treatment significantly increased brain accumulations for 2 of the GSPE-derived phenolic acids, 3-HBA and 3-HPP. Treatment with 250 mg/Kg BW/day GSPE significantly increased brain contents of 3-HBA and 3-HPPby 3.2-fold (p<0.05 compared to vehicle-treated controls) and 7.7-fold (p<0.05), respectively (Figure 3A, B). Treatment with the lower dose of 25 mg/Kg BW/day GSPE also significantly increased brain contents of 3-HBA and 3-HPPby 2.5-fold (p<0.05) and 5.5-fold (p<0.05), respectively, relative to vehicle-treated controls (Figure 3A, B). Moreover, we observed that both 3-HBA and 3-HPP accumulated in the brain in a dose-dependent manner, and that exposure to the higher GSPE dose (250 mg/Kg BW/day) was associated with accumulation of higher contents of 3-HBA and 3-HPP in the brain, compared to exposure to the lower GSPE dose (25 mg/Kg BW/day) (Figure 3B). No detectable changes in brain contents of HA, 4-HBA, 3-HPA, 3,4-diHPA, or 3,4-diHPP were observed following GSPE treatment at 250 or 25 mg/Kg BW/day (Figure 3A). Collectively, our observation suggests that two phenolic acids, 3-HBA and 3-HPP, derived from GSPE, are capable of penetrating and accumulating in the brain.
Figure 3. Detection of GSPE-derived phenolic acids in the brain.
Rats were treated with GSPE at 250 mg/Kg BW/day, 25 mg/Kg BW/day, or vehicle (control) for 11 days, and animals (n=4 per group) were sacrificed 6 hours after the last dose of GSPE/vehicle treatment. A) Detection of phenolic acids in perfused brain specimens from vehicle-treated control and GSPE treated groups. Data are displayed as mean ± SEM; * p<0.05, GSPE-treated compared to vehicle-treated control animals. 3-HBA and 3-HPP data are presented in bold to highlight significantly elevated contents for these two phenolic acids in brain specimen in response to GSPE treatment. Dark arrows indicate increased fold-change in GSPE-treated vs. vehicle-treated control brain specimens. B) Dose-responsive accumulation of 3-HBA and 3-HPP in brain specimens from animals treated with a daily dose of 250 or 25 mg/Kg BW/day GSPE. Bar graphs present mean ± SEM; * p<0.05, GSPE-treated compared to vehicle-treated control animals. C) Molecular structure of 3-HBA and 3-HPP. D) Representative HPLC resolution/detection of 3-HBA and 3-HPP from molecular standards (top spectrogram) and a brain specimen (bottom spectrogram). (Inset: MS-MS analysis of 3-HBA and 3-HPP from brain specimens following HPLC resolution).
Our observations suggest that select phenolic acids, generated from GI microbiota conversion of dietary polyphenols, are capable of penetrating through the blood brain barrier and accumulating in pharmacologically relevant, μM concentrations in the brain. This implicates the possibility that brain-accumulating phenolic acids (e.g., 3-HBA, 3-HBB, and perhaps additional phenolic acids not yet identified) derived from GI microbiota metabolism of GSPE may contribute to the efficacy of the GSPE in modulating AD neuropathogenic mechanisms.
3.3 Brain-accumulating phenolic acid metabolites interfere with the assembly of Aβ1-42 peptide in to neurotoxic Aβ aggregates, in vitro
We previously demonstrated that oligomeric Aβ is neurotoxic [50] and that treatment with GSPE prevents oligomerization of Aβ peptides in the brain and attenuates cognitive deterioration in a mouse model of Alzheimer’s disease [1]. Based on our new evidence that metabolism of GSPE by GI microbiota leads to generation and delivery of 3-HBA and 3-HPP to brain tissues, we tested whether the two brain-accumulating phenolic acid metabolites (3-HBA and 3-HPP purchased from Sigma Chemical Company) may contribute to anti-Aβ oligomerization bioactivity in the brain, using multiple in vitro assays.
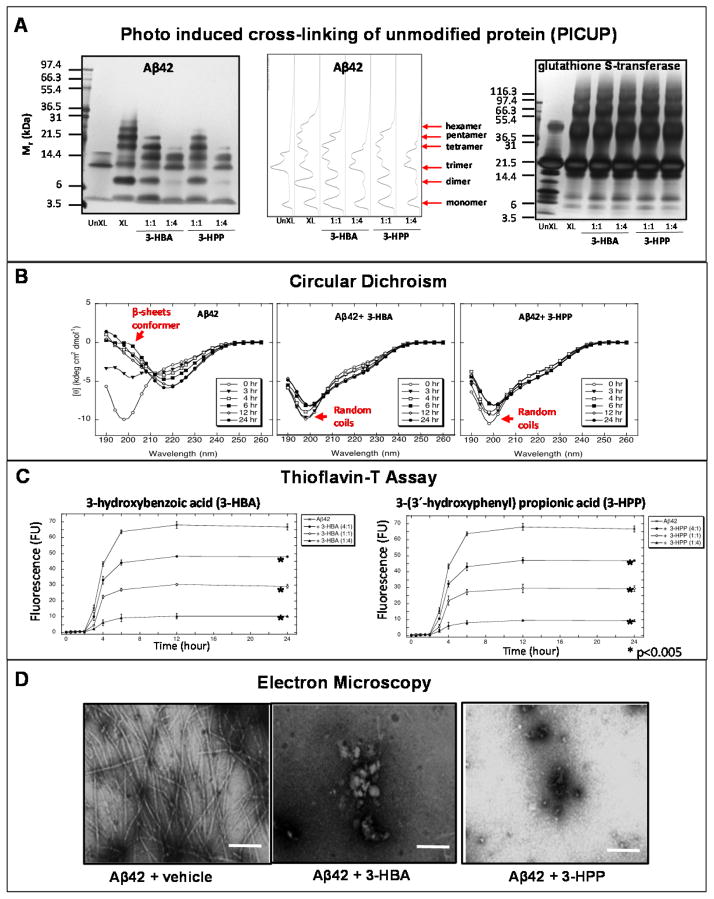
We used an in vitro PICUP assay to explore the potential impacts of 3-HPP and 3-HBA on modulating initial protein-protein interactions necessary for the assembly of synthetic monomeric Aβ1-42 peptides into neurotoxic oligomeric Aβ species. In the absence of cross-linking, only Aβ1-42monomers, trimmers, and tetramers (Figure 4A) were observed. The Aβ1–42 trimer and tetramer bands under non-crosslinking conditions are known SDS-PAGE-induced artifacts [54, 55]. As expected, a mixture of oligomer order from dimer to octamer was observed for Aβ1–42 following cross-linking (Figure 4A). We found that both 3-HBA and 3-HPP were effective in blocking formation of aggregated Aβ1–42 (Figure 4A) species. Almost complete inhibition of dimeric, pentameric, hexameric, heptameric, and octameric Aβ1–42 (Figure 4A) species were achieved with presence of 3-HPP or 3-HBA at 1:4 molar ratio of Aβ1-42 peptide: phenolic acid. At 1:1 molar ratio with the Aβ peptides, 3-HPP and, to a lesser extent, 3-HBA, also reduced the formation of higher order Aβ1–42 (Figure 4A) species. In control studies, we demonstrated that 3-HBA and 3-HPP do not interfere with the PICUP cross-link mechanism, as presence of either phenolic acid, at 1:1 or at 1:4 molar ratio with glutathione S-transferase (GST), does not interfere with assembly of GST into higher order aggregates (Figure 4A).
Figure 4. Brain-accumulating phenolic acids potently interfere with aggregation of Aβ peptides, in vitro.
The potential efficacy of 3-HBA and 3-HPP in interfering with aggregation of a synthetic Aβ1-42 peptide was assessed by multiple independent in vitro assays. A) PICUP assay. Aβ1–42 (25 μM) was cross-linked in the presence or absence of 3-HBA or 3-HPP at 1:1 or 1:4 molar ratio of Aβ1-42 peptide: phenolic acid. PICUP products were resolved by SDS-PAGE and visualized using silver staining. (Left panel) Cross linking in the presence of phenolic acids. Lane 1, control non-cross-linked Aβ peptides. Aβ1–42 trimers and tetramer bands under non-cross-linking conditions are known SDS-PAGE-induced artifacts [54, 55]. Lanes 2, cross-linked in the absence of phenolic acid resulting in formations of dimer, trimer, tetramer, pentamer, and hexamer bands. Lanes 3–6, cross-linked Aβ peptide in the presence phenolic resulted in dose-responsive reduction of multimeric Aβ bands. (Middle panel) The density of native Aβ1-42 and cross-linked Aβ1-42 peptide bands were quantified by densitometry scanning. (Right panel) Control studies exploring the impacts of 3-HBA and 3-HPP on photo-induced cross-linking of glutathione S-transferase (GST) confirmed that the addition of phenolic acids does not interfere with GST aggregation at 1:1 or 4:1 molar ratio of phenolic acid:GST peptide. B) Circular dichroism assay. Aβ1-42 peptide (25 μM) was incubated in the absence of phenolic acid (left panel) or in the presence of 3-HBA (100 μM, middle panel) or 3-HPP (100 μM, right panel) at 37°C for 7 days in 10 mm phosphate, pH7.4. Spectra were acquired immediately at the start of the incubation period (zero hour, ○), and after 3 (▼), 4 (□), 6 (■), 12 (◇) and 24 (●) hours. The spectra presented at each time are representative of those obtained during each of three independent experiments. Spectra characteristics of β-sheet conformer and random coils are noted. C) Thioflavin-T assay. Aβ1-42 was incubated for 7 days at 37 °C in 10 nm phosphate, pH7.4, in the presence of 3-HBA (left panel) or 3-HPP (right panel) at 4:1 (○), 1:1 (●), or 1:4 (▲) molar ratio of Aβ1-42 peptide: phenolic acid. Periodically, aliquots were removed, and thioflavin-T binding levels were determined. Binding is expressed as mean fluorescence (in arbitrary fluorescence units (FU)) ± S.E (error bars). *p < 0.005 (Tukey-Kramer post hoc test after ANOVA). Each figure comprises data obtained from three independent experiments. D) Electron microscopy. Aβ1-42 peptide (Aβ42 25 μM) was incubated at 37 °C for 7 days in 10 mm sodium phosphate, pH 7.4 in buffer alone (A and B) or in the presence of 3-HBA (100 μM, middle panel) or 3-HPP (100 μM, right panel) for 10 days. Fibril formation was assessed by electron microscopy. Scale bars indicate 100 nm.
Using a CD assay, we monitored the effects of 3-HBA and 3-HPP on conformational properties of the Aβ1-42 peptide (Figure 4B). Consistent with our previous observations [50], Aβ1-42 peptide, when incubated alone, presented a CD spectrum characteristic of random coil, and the secondary structures of Aβ1-42 progressively shifted to β-sheet conformer with increasing incubation time, with a large magnitude minimum centered at ~198 nm (Figure 4B, left panel). Co-incubation with 3-HBA (Figure 4B, middle panel) or 3-HPP (Figure 4C, right panel) completely interfered with transition of incubated Aβ1-42 peptide to β-sheet conformers.
We continued to explore the impacts of the two brain-accumulating GSPE-derived phenolic acids on modulating temporal changes in the transition of Aβ1-42 peptide from random to β-sheet conformers using a Th-T binding assay. In this assay, β-sheet formation over time was measured by monitoring Th-T fluorescence emission following intercalation of Th-T into β-sheet protein conformers. As we have previously reported [50], incubation of Aβ-1-42 over time led to progressively increasing Th-T fluorescence emission that reached maximal fluorescence emission by 12 hrs in this assay, reflecting an increasing shift to β-sheet conformers(Figure 4C, left and right panel). We found that co-incubation with 3-HBA (Figure 4C, left panel) or 3-HPP (Figure 4C, right panel) interfered with transition of Aβ1-42 from random coil to β-sheet in a dose-dependent. Co-incubation with increasingly higher contents of 3-HBA or 3-HPP at molar ratios of Aβ1-42 to phenolic acid from 4:1 to 1:4 showed increasing efficacy in reducing formation of Aβ1-42 β-sheet conformers (Figure 4C, left and right panels). When Aβ1-42 was co-incubated with 3-HBA at the lowest molar ratio of 4:1, the final fluorescence of 24 hours was significantly smaller than that without compounds (mean±S.D: 48.07±0.25 vs 66.81±1.97, p< 0.005) (Figure 4C, left panel). Similarly, in the co-incubation with 3-HPPat a 4:1 molar ratio, the final fluorescence of 24 hours was significantly smaller than that without compounds (mean±S.D: 46.98±0.67 vs 66.81±1.97, p< 0.005) (Figure 4C, right panel).
Lastly, we explored the effects of 3-HBA and 3-HPP on the morphologies of Aβ1-42 assemblies (Figure 4D). As we have previously reported [50], self -assembly of Aβ1-42 peptide during incubation in the absence of phenolic acids ultimately led to formation of Aβ1-42 peptide into classical amyloid fibrils that are readily visualized by EM as non-branched, helical filaments (Figure 4C, left panel. We observed that co-incubation with 3-HBA (Figure 4D, middle panel) or 3-HPP (Figure 4D, right panel) strongly inhibited formation of Aβ1-42 amyloid fibrils.
Collectively, our in vitro evidence demonstrates that 3-HBA and 3-HPP potently interfere with self-assembly of Aβ peptides into neurotoxic high-molecular-weight Aβ aggregates and AD-type amyloid fibrils.
DISUCUSSION
Increasing evidence from preclinical and in vitro experimental studies has shown that GSPE is effective in modulating onset/progression of AD dementia by interfering with Aβ- and tau-related neuropathogenic mechanisms [1, 3, 56]. Based on evidence demonstrating the capability of GI microbiota to metabolically convert dietary polyphenols to more readily absorbable phenolic acids, the present study was designed to explore potential GI microbiota conversion of orally administered GSPE into phenolic acids, bioavailability of these GSPE-derive phenolic acids in brain tissues, and potential bioactivities leading to modulation of underlying mechanisms in the brain.
We identified 12 phenolic acids that might be generated from GI microbiota metabolism of GSPE for investigation. We treated rats with repeated oral administration of GSPE for 11 days to simulate longer-term GSPE treatments used in biological assessments of GSPE in animal models of AD [1, 3], as well as long-term treatment paradigms that most likely will be necessary for development of GSPE for AD prevention and/or treatment in humans. Exploring accumulation of phenolic acids in multiple physiological compartments, we found that repeated oral administration with GSPE significantly increased the accumulation of all 12 phenolic acids we surveyed in the cecum, colon, urine, and/or circulating blood, implicating that the 12 phenolic acids are generated by GI microbiota metabolism of orally administered GSPE. We note that in our study, all biological samples were treated with β-glucuronidase before HPLC-MS analysis, which is a common approach to facilitate quantification of phenolics in tissue specimens. Thus, our analysis of the 12 phenolic acids did not distinguish the parent compounds or potential glucuronide derivatives. Ongoing studies are examining the conversion of parent and glucuronidated-phenolic acid forms from GSPE and distribution of these phenolic forms in urine, plasma, and tissues.
Our observation of elevated plasma accumulation of multiple intestinal microbiota-derived GSPE phenolic acid metabolites, e.g., HA, 3-HPP, 3,4-diHPP, and 5-HPV, in concentrations ranging from 0.5 to 4.7 μM (Table I), is consistent with the possibility that certain intestinal microbiota phenolic acid metabolites from GSPE may contribute to the protective actions of GSPE in disorders such as cardiovascular disease [4–6, 15–17], colonic cancer [7, 8], and breast cancer [9], where affected tissues are located peripherally. However, we note that there are appreciable discrepancies between the levels of these phenolic acids we detected in plasma and doses reported for bioactivities relevant for disease treatments. For example, following GSPE treatment, we detected plasma concentrations of FA and 3,4-diHPP at sub-μM to μM range concentrations. In contest, anti-proliferative activities of FA and 3,4-diHPP in cultured cancer cells may require doses of 50 μM or higher [57, 58]. More information will be necessary, including information on the dose-response bioactivities of HA, 3-HPP, 3,4-diHPP, and 5-HPV, and concentrations of these phenolic acids in peripheral tissues after GSPE treatment, before we can evaluate the potential contribution of intestinal microbiota GSPE fermentation products to the protective benefits of GSPE in cardiovascular disease and cancer.
We note that some phenolic acids can be generated from host bioconversion of amino acids. For example, L-tyrosine can be transformed to caffeic acid by the action of the enzyme tyrosine ammonia lyase [59]. Phenylalanine can be converted to cinnamic acid by the action of the enzyme phenylananine ammonia lyase [60] and, thereafter, a series of enzymatic hydroxylations and methylations may lead to generation of multiple phenolic acids, such as coumaric acid, ferulic acid, among others. Thus, background levels of the phenolic acids that we detected could represent that that are formed over time from normal digestion and absorption of protein.
More studies will be necessary to explore whether mechanisms additional to GI microbiota metabolism of GSPE may contribute to the observed increases in the contents of phenolic acids we surveyed in multiple physiological compartments following repeated GSPE administration. Ongoing studies in our group are exploring the generation and tissue distribution of GSPE-derived phenolic acid in gnotobiotic mouse models to establish a cause-effect relationship between GI microbiota metabolism and tissue accumulation of select phenolic acids in response to repeated oral GSPE administration.
Most importantly for the goal of our investigation in exploring the protective effects of GSPE in neurodegenerative disorders such as AD, we identified two intestinal microbiota GSPE metabolites, 3-HBA and 3-HPP, that are capable of penetrating through the blood brain barrier and accumulating in the brain at μM concentrations (Figure 3). In our present study, we did not distinguish brain accumulations of 3-HBA and 3-HPP in either parent or glucuronidated phenolic forms. Nonetheless, we note that non-glucuronidated 3-HBA and 3-HPP are both observed in human plasma and/or urine under analytical conditions in the absence of β-glucuronidase treatment [33, 61], supporting the bioavailability of 3-HBA and 3-HPP in tissues, including the brain. Ongoing studies are evaluating how much of the 3-HBA and 3-HPP that we found in the brain are the parent phenolic acids and how much are the glucuronidated forms.
There is evidence that phenolic compounds at physiologically relevant μM to sub-μM concentrations may have an impact on brain physiological activities. For example, we recently reported that one of the GSPE polyphenol metabolites, 3′-O-Me-EC-Gluc, is accumulated in the brain at a concentration of ~0.4 μM, and that this concentration of 3′-O-Me-EC-Gluc is effective in promoting basal synaptic transmission and long-term potentiation in brain hippocampus slices through mechanisms associated with cAMP response element binding protein signaling [13]. Thus, it is likely that the concentrations of 3-HBA and 3-HPP we found in the perfused brain (1.75 and 2.53 μM, respectively) following GSPE treatment are sufficient to support physiological activities. There is little information on why 3-HBA and 3-HPP, but not other GSPE-derived phenolic acids, are capable of accumulation in the brain. Membrane carriers are required for transport of most phenolic compounds across the blood brain barrier [62]. More investigation will be required to clarify why only certain subset(s) of the absorbed phenolic acids derived from intestinal microbiota fermentation are able to target brain tissues.
In follow-up in vitro studies exploring potential bioactivities of 3-HBA and 3-HPP in the brain, we demonstrated that both brain-accumulating phenolic acids potently interfere with self-assembly of Aβ peptides into neurotoxic high-molecular-weight Aβ aggregates and AD-type amyloid fibrils. Oyr observation supports the notion that intestinal microbiota may contribute to the protective activities of GSPE in AD by converting PAC components from GSPE to brain-accumulating phenolic acids that might exert disease-modifying activities in the brain. Outcomes from our studies establish that select GSPE polyphenol metabolites derived from intestinal microbiota fermentation are capable of penetrating the blood brain barrier and of accumulating in the brain, and are able to interfere with Aβ pathogenic mechanisms. This implicates important contribution of intestinal microbiota in protection against AD.
Processes underlying assembly of Aβ peptides into misfolded, neurotoxic Aβ protein aggregates are also relevant for generation of additional misfolded proteins central to neuropathogenesis of many other neurodegenerative conditions. This includes misfolding and aberrant aggregation of the microtubule associated protein tau that is a key neuropathologic feature shared by multiple neurodegenerative disorders, such as frontotemporal dementia, corticobasal degeneration, Pick’s disease, argyrophilic grain disease, and Alzheimer’s disease, that are collectively referred to as tauopathies. Key pathogenic roles for misfolded proteins are also observed in Huntington’s disease and in Parkinson’s disease, with signature aberrant accumulations of, respectively, misfolded huntingtin [63] and α-synuclein [64] protein aggregates in the brain. A current hypothesis suggests that aggregations of misfolded proteins in these diverse neurodegenerative disorders follow a nucleation model [65]. The first step involves protein–protein interactions among monomeric misfolded proteins that lead to the formation of oligomers. Oligomeric conformers then serve as ordered nuclei to catalyze further growth of the polymers [65]. Our present novel evidence, that both 3-HBA and 3-HPP are able to interfere with initial protein-protein interactions necessary for the assembly of Aβ peptides into neurotoxic aggregated Aβ forms, suggests that these two brain-bioavailable GSPE metabolites generated from the intestinal microbiome might also be similarly effective in interfering with aberrant assembly of neurotoxic tau, huntingtin, and/or α-synuclein protein aggregates. Consistent with this, we recently demonstrated that dietary supplementation with GSPE is effective in attenuating development of mutant tau-mediated tau neuropathology and cognitive dysfunction in independent JNPL3 and TMHT mutant tau mouse models[2, 3]. While additional investigations will be needed to confirm the efficacy of 3-HBA and 3-HPP to interfere with the assembly of tau, huntingtin, and α-synuclein protein aggregates, our evidence supports the potential contribution of the intestinal microbiota in protection against AD and many other neurodegenerative disorders, and supports continual exploration of GSPE and other bioactive polyphenol preparations for treating these neurodegenerative conditions.
The doses of GSPE (250 and 25 mg/kg BW/day) that we used in rats in this study are equivalent to human doses of 2780 and 278 mg/day, calculated using the standard FDA conversion table. We observed significant elevation of both 3-HBA and 3-HPP in the brain following treatment with both GSPE doses (Figure 3A, B). Thus, human doses around 278 mg/day will likely suffice to deliver 3-HBA and 3-HPP to the brain. We note that daily intake of total polyphenols in human has been estimated to be 1 g/day [66] and that long-term treatments in mice using a dose of GSPE equivalent to a human dose of up to 1000 mg/day [1] and in humans with doses up to 1000 mg/day GSPE [4, 5]are not associated with detectable aversive effects. Thus, GSPE doses around 278 mg/day in human are highly tolerated for long-term administrations.
There is increasing interest in the role of intestinal microbiota in governing health and disease states [67, 68]. Outcomes from our studies links intestinal microbiota with neuroprotective activities of GSPE have extensive implications for the development of GSPE (or other bioactive polyphenol preparations) for novel clinical application for brain protection, and as therapeutic and preventative applications, particularly in early stages of neurodegenerative diseases.
Collectively, our evidence supports the notion that intestinal microbiota may contribute to the protective activities of GSPE in neurodegenerative disorders, and in other diseases, by converting PAC components from GSPE to phenolic acid metabolites capable of accumulating in target tissues, such as the brain, and of exerting disease-modifying activates. Thus, GSPE phenolic acid metabolites derived from intestinal microbial fermentation, along with previously reported PACs and their conjugated metabolites should be further investigated as potential contributors to the observed health benefits associated with GSPE consumption. Moreover, outcomes from our studies support future preclinical and clinical investigations of the mechanisms behind PACs absorption, bioavailability, and potential preventative and therapeutic application targeting neuropathological features of age related disorders and other conditions [69–75].
Acknowledgments
The GSPE sample used in this study was generously provided by Polyphenolics (Madera, CA). Funding was provided by NIH-NCCAM grant P01AT004511-01 as part of the Mount Sinai School of Medicine Center of Excellence in Complementary and Alternative Medicine for Alzheimer’s Disease (to GM Pasinetti) and by the Endeavour International Postgraduate Research Scholarship (IPRS), awarded by Australia government, and the Graduate School International Travel Award (GSITA) from The University of Queensland.
This material is the result of work supported in part with resources and the use of facilities at the James J. Peters Veterans Affairs Medical Center, Bronx, NY. In addition, Dr. Pasinetti holds a Career Scientist Award in the Research and Development unit and is the Director of the Basic and Biomedical Research and Training Program, GRECC, James J. Peters Veterans Affairs Medical Center. We also acknowledge that the contents of this manuscript do not represent the views of the U.S. Department of Veterans Affairs or the United States Government.
Abbreviations
- Aβ
β-amyloid
- AD
Alzheimer’s disease
- CT
catechin
- CD
circular dichroism
- EC
epicatechin
- EM
electron microscopy
- ESI
electrospray ionization
- FA
ferulic acid
- GA
gallic acid
- GI
gastrointestinal
- GSPE
Grape seed polyphenol extract
- GST
glutathione S-transferase
- HA
hippuric acid
- MS/MS
tandem mass spectrometry
- LC
liquid chromatography
- LOD
limit of detection
- LOQ
limit of quantitation
- PA
phenylacetic acid
- PAC
proanthocyanidin
- HPLC
high-performance liquid chromatography
- PICUP
photo-induced cross-linking of unmodified proteins
- Ru(Bpy)
tris-(2,2′-bipyridyl)dichlororuthenium(II)
- SEM
standard error of the mean
- Th-T
thioflavin-T
- 3-HBA
3-hydroxybenzoic acid
- 3-HHA
3-hydroxyhippuric acid
- 3-HPA
3-hydroxyphenyl acetic acid
- 3-HPP
3-(3′-hydroxyphenyl) propionic acid
- 3′-O-Me-EC-Gluc
3′-O-methyl-epicatechin-5-O-β-glucuronide
- 3,4-diHPA
3,4-dihydroxyphenylacetic acid
- 3,4-diHPP
3-(3′,4′-dihydroxyphenyl)propionic acid
- 3-PP
3-phenylpropionic acid
- 4-HBA
4-hydroxybenzoic acid
- 4-HHA
4-hydroxyhippuric acid
- 4-HPA
4-hydroxyphenylacetic acid
- 4-HPP
3-(4′-hydroxyphenyl) propionic acid
- 5-HPV
5-(4′-hydroxyphenyl) valeric acid
Reference List
- 1.Wang J, Ho L, Zhao W, Ono K, et al. Grape-derived polyphenolics prevent Abeta oligomerization and attenuate cognitive deterioration in a mouse model of Alzheimer’s disease. J Neurosci. 2008;28 (25):6388–6392. doi: 10.1523/JNEUROSCI.0364-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santa-Maria I, az-Ruiz C, Ksiezak-Reding H, Chen A, et al. GSPE interferes with tau aggregation in vivo: implication for treating tauopathy. Neurobiol Aging. 2012;33 (9):2072–2081. doi: 10.1016/j.neurobiolaging.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, Santa-Maria I, Ho L, Ksiezak-Reding H, et al. Grape derived polyphenols attenuate tau neuropathology in a mouse model of Alzheimer’s disease. J Alzheimers Dis. 2010;22 (2):653–661. doi: 10.3233/JAD-2010-101074. [DOI] [PubMed] [Google Scholar]
- 4.Sivaprakasapillai B, Edirisinghe I, Randolph J, Steinberg F, Kappagoda T. Effect of grape seed extract on blood pressure in subjects with the metabolic syndrome. Metabolism. 2009;58 (12):1743–1746. doi: 10.1016/j.metabol.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 5.Ward NC, Hodgson JM, Croft KD, Burke V, et al. The combination of vitamin C and grape-seed polyphenols increases blood pressure: a randomized, double-blind, placebo-controlled trial. J Hypertens. 2005;23 (2):427–434. doi: 10.1097/00004872-200502000-00026. [DOI] [PubMed] [Google Scholar]
- 6.Ras RT, Zock PL, Zebregs YE, Johnston NR, et al. Effect of polyphenol-rich grape seed extract on ambulatory blood pressure in subjects with pre- and stage I hypertension. Br J Nutr. 2013;110 (12):2234–2241. doi: 10.1017/S000711451300161X. [DOI] [PubMed] [Google Scholar]
- 7.Dinicola S, Cucina A, Pasqualato A, Proietti S, et al. Apoptosis-inducing factor and caspase-dependent apoptotic pathways triggered by different grape seed extracts on human colon cancer cell line Caco-2. Br J Nutr. 2010;104 (6):824–832. doi: 10.1017/S0007114510001522. [DOI] [PubMed] [Google Scholar]
- 8.Singletary KW, Meline B. Effect of grape seed proanthocyanidins on colon aberrant crypts and breast tumors in a rat dual-organ tumor model. Nutr Cancer. 2001;39 (2):252–258. doi: 10.1207/S15327914nc392_15. [DOI] [PubMed] [Google Scholar]
- 9.Agarwal C, Sharma Y, Zhao J, Agarwal R. A polyphenolic fraction from grape seeds causes irreversible growth inhibition of breast carcinoma MDA-MB468 cells by inhibiting mitogen-activated protein kinases activation and inducing G1 arrest and differentiation. Clin Cancer Res. 2000;6 (7):2921–2930. [PubMed] [Google Scholar]
- 10.Kim H, Hall P, Smith M, Kirk M, et al. Chemoprevention by grape seed extract and genistein in carcinogen-induced mammary cancer in rats is diet dependent. J Nutr. 2004;134 (12 Suppl):3445S–3452S. doi: 10.1093/jn/134.12.3445S. [DOI] [PubMed] [Google Scholar]
- 11.Manach C, Scalbert A, Morand C, Remesy C, Jimenez L. Polyphenols: food sources and bioavailability. Am J Clin Nutr. 2004;79 (5):727–747. doi: 10.1093/ajcn/79.5.727. [DOI] [PubMed] [Google Scholar]
- 12.Sano A, Yamakoshi J, Tokutake S, Tobe K, et al. Procyanidin B1 is detected in human serum after intake of proanthocyanidin-rich grape seed extract. Biosci Biotechnol Biochem. 2003;67 (5):1140–1143. doi: 10.1271/bbb.67.1140. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Ferruzzi MG, Ho L, Blount J, et al. Brain-targeted proanthocyanidin metabolites for Alzheimer’s disease treatment. J Neurosci. 2012;32 (15):5144–5150. doi: 10.1523/JNEUROSCI.6437-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho L, Pasinetti GM. Polyphenolic compounds for treating neurodegenerative disorders involving protein misfolding. Expert Rev Proteomics. 2010;7 (4):579–589. doi: 10.1586/epr.10.69. [DOI] [PubMed] [Google Scholar]
- 15.Du Y, Lou H. Catechin and proanthocyanidin B4 from grape seeds prevent doxorubicin-induced toxicity in cardiomyocytes. Eur J Pharmacol. 2008;591 (1–3):96–101. doi: 10.1016/j.ejphar.2008.06.068. [DOI] [PubMed] [Google Scholar]
- 16.Leifert WR, Abeywardena MY. Cardioprotective actions of grape polyphenols. Nutr Res. 2008;28 (11):729–737. doi: 10.1016/j.nutres.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Sato M, Bagchi D, Tosaki A, Das DK. Grape seed proanthocyanidin reduces cardiomyocyte apoptosis by inhibiting ischemia/reperfusion-induced activation of JNK-1 and C-JUN. Free Radic Biol Med. 2001;31 (6):729–737. doi: 10.1016/s0891-5849(01)00626-8. [DOI] [PubMed] [Google Scholar]
- 18.Ferruzzi MG, Lobo JK, Janle EM, Cooper B, et al. Bioavailability of gallic acid and catechins from grape seed polyphenol extract is improved by repeated dosing in rats: implications for treatment in Alzheimer’s disease. J Alzheimers Dis. 2009;18 (1):113–124. doi: 10.3233/JAD-2009-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura Y, Tonogai Y. Metabolism of grape seed polyphenol in the rat. J Agric Food Chem. 2003;51 (24):7215–7225. doi: 10.1021/jf030250+. [DOI] [PubMed] [Google Scholar]
- 20.Tsang C, Auger C, Mullen W, Bornet A, et al. The absorption, metabolism and excretion of flavan-3-ols and procyanidins following the ingestion of a grape seed extract by rats. Br J Nutr. 2005;94 (2):170–181. doi: 10.1079/bjn20051480. [DOI] [PubMed] [Google Scholar]
- 21.Aura AM, O’Leary KA, Williamson G, Ojala M, et al. Quercetin derivatives are deconjugated and converted to hydroxyphenylacetic acids but not methylated by human fecal flora in vitro. J Agric Food Chem. 2002;50 (6):1725–1730. doi: 10.1021/jf0108056. [DOI] [PubMed] [Google Scholar]
- 22.Cueva C, Sanchez-Patan F, Monagas M, Walton GE, et al. In vitro fermentation of grape seed flavan-3-ol fractions by human faecal microbiota: changes in microbial groups and phenolic metabolites. FEMS Microbiol Ecol. 2013;83 (3):792–805. doi: 10.1111/1574-6941.12037. [DOI] [PubMed] [Google Scholar]
- 23.Gonthier MP, Donovan JL, Texier O, Felgines C, et al. Metabolism of dietary procyanidins in rats. Free Radic Biol Med. 2003;35 (8):837–844. doi: 10.1016/s0891-5849(03)00394-0. [DOI] [PubMed] [Google Scholar]
- 24.Wang LQ, Meselhy MR, Li Y, Nakamura N, et al. The heterocyclic ring fission and dehydroxylation of catechins and related compounds by Eubacterium sp. strain SDG-2, a human intestinal bacterium. Chem Pharm Bull (Tokyo) 2001;49 (12):1640–1643. doi: 10.1248/cpb.49.1640. [DOI] [PubMed] [Google Scholar]
- 25.Unno T, Tamemoto K, Yayabe F, Kakuda T. Urinary excretion of 5-(3′,4′-dihydroxyphenyl)-gamma-valerolactone, a ring-fission metabolite of (−)-epicatechin, in rats and its in vitro antioxidant activity. J Agric Food Chem. 2003;51 (23):6893–6898. doi: 10.1021/jf034578e. [DOI] [PubMed] [Google Scholar]
- 26.Hasslauer I, Oehme A, Locher S, Valotis A, et al. Flavan-3-ol C-glycosides--preparation and model experiments mimicking their human intestinal transit. Mol Nutr Food Res. 2010;54 (11):1546–1555. doi: 10.1002/mnfr.201000003. [DOI] [PubMed] [Google Scholar]
- 27.Keppler K, Humpf HU. Metabolism of anthocyanins and their phenolic degradation products by the intestinal microflora. Bioorg Med Chem. 2005;13 (17):5195–5205. doi: 10.1016/j.bmc.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Gonthier MP, Verny MA, Besson C, Remesy C, Scalbert A. Chlorogenic acid bioavailability largely depends on its metabolism by the gut microflora in rats. J Nutr. 2003;133 (6):1853–1859. doi: 10.1093/jn/133.6.1853. [DOI] [PubMed] [Google Scholar]
- 29.Olthof MR, Hollman PC, Buijsman MN, van Amelsvoort JM, Katan MB. Chlorogenic acid, quercetin-3-rutinoside and black tea phenols are extensively metabolized in humans. J Nutr. 2003;133 (6):1806–1814. doi: 10.1093/jn/133.6.1806. [DOI] [PubMed] [Google Scholar]
- 30.Rechner AR, Smith MA, Kuhnle G, Gibson GR, et al. Colonic metabolism of dietary polyphenols: influence of structure on microbial fermentation products. Free Radic Biol Med. 2004;36 (2):212–225. doi: 10.1016/j.freeradbiomed.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 31.Deprez S, Brezillon C, Rabot S, Philippe C, et al. Polymeric proanthocyanidins are catabolized by human colonic microflora into low-molecular-weight phenolic acids. J Nutr. 2000;130 (11):2733–2738. doi: 10.1093/jn/130.11.2733. [DOI] [PubMed] [Google Scholar]
- 32.Wang D, Williams BA, Ferruzzi MG, D’Arcy BR. Different concentrations of grape seed extract affect in vitro starch fermentation by porcine small and large intestinal inocula. J Sci Food Agric. 2013;93 (2):276–283. doi: 10.1002/jsfa.5753. [DOI] [PubMed] [Google Scholar]
- 33.Gonthier MP, Cheynier V, Donovan JL, Manach C, et al. Microbial aromatic acid metabolites formed in the gut account for a major fraction of the polyphenols excreted in urine of rats fed red wine polyphenols. J Nutr. 2003;133 (2):461–467. doi: 10.1093/jn/133.2.461. [DOI] [PubMed] [Google Scholar]
- 34.Prior RL, Rogers TR, Khanal RC, Wilkes SE, et al. Urinary excretion of phenolic acids in rats fed cranberry. J Agric Food Chem. 2010;58 (7):3940–3949. doi: 10.1021/jf9028392. [DOI] [PubMed] [Google Scholar]
- 35.Ward NC, Croft KD, Puddey IB, Hodgson JM. Supplementation with grape seed polyphenols results in increased urinary excretion of 3-hydroxyphenylpropionic Acid, an important metabolite of proanthocyanidins in humans. J Agric Food Chem. 2004;52 (17):5545–5549. doi: 10.1021/jf049404r. [DOI] [PubMed] [Google Scholar]
- 36.Baba S, Osakabe N, Natsume M, Muto Y, et al. Absorption and urinary excretion of (−)-epicatechin after administration of different levels of cocoa powder or (−)-epicatechin in rats. J Agric Food Chem. 2001;49 (12):6050–6056. doi: 10.1021/jf010965h. [DOI] [PubMed] [Google Scholar]
- 37.Prasain JK, Peng N, Dai Y, Moore R, et al. Liquid chromatography tandem mass spectrometry identification of proanthocyanidins in rat plasma after oral administration of grape seed extract. Phytomedicine. 2009;16 (2–3):233–243. doi: 10.1016/j.phymed.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lotito SB, Frei B. Consumption of flavonoid-rich foods and increased plasma antioxidant capacity in humans: cause, consequence, or epiphenomenon? Free Radic Biol Med. 2006;41 (12):1727–1746. doi: 10.1016/j.freeradbiomed.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 39.Calani L, Dall’Asta M, Derlindati E, Scazzina F, et al. Colonic metabolism of polyphenols from coffee, green tea, and hazelnut skins. J Clin Gastroenterol. 2012;46 (Suppl):S95–S99. doi: 10.1097/MCG.0b013e318264e82b. [DOI] [PubMed] [Google Scholar]
- 40.Monagas M, Urpi-Sarda M, Sanchez-Patan F, Llorach R, et al. Insights into the metabolism and microbial biotransformation of dietary flavan-3-ols and the bioactivity of their metabolites. Food Funct. 2010;1 (3):233–253. doi: 10.1039/c0fo00132e. [DOI] [PubMed] [Google Scholar]
- 41.Aura AM, Mattila I, Seppänen-Laakso T, Miettinen J, et al. Microbial metabolism of catechin stereoisomers by human faecal microbiota: Comparison of targeted analysis and a non-targeted metabolomics method. Phytochemistry Letters. 2008;1:18–22. [Google Scholar]
- 42.Monagas M, Urpi-Sarda M, Sanchez-Patan F, Llorach R, et al. Insights into the metabolism and microbial biotransformation of dietary flavan-3-ols and the bioactivity of their metabolites. Food Funct. 2010;1 (3):233–253. doi: 10.1039/c0fo00132e. [DOI] [PubMed] [Google Scholar]
- 43.Rechner AR, Spencer JP, Kuhnle G, Hahn U, Rice-Evans CA. Novel biomarkers of the metabolism of caffeic acid derivatives in vivo. Free Radic Biol Med. 2001;30 (11):1213–1222. doi: 10.1016/s0891-5849(01)00506-8. [DOI] [PubMed] [Google Scholar]
- 44.Feng WY. Metabolism of green tea catechins: an overview. Curr Drug Metab. 2006;7 (7):755–809. doi: 10.2174/138920006778520552. [DOI] [PubMed] [Google Scholar]
- 45.Rechner AR, Smith MA, Kuhnle G, Gibson GR, et al. Colonic metabolism of dietary polyphenols: influence of structure on microbial fermentation products. Free Radic Biol Med. 2004;36 (2):212–225. doi: 10.1016/j.freeradbiomed.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 46.Hwang IK, Yoo KY, Kim DS, Jeong YK, et al. Neuroprotective effects of grape seed extract on neuronal injury by inhibiting DNA damage in the gerbil hippocampus after transient forebrain ischemia. Life Sci. 2004;75 (16):1989–2001. doi: 10.1016/j.lfs.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 47.Saura-Calixto F, Perez-Jimenez J, Tourino S, Serrano J, et al. Proanthocyanidin metabolites associated with dietary fibre from in vitro colonic fermentation and proanthocyanidin metabolites in human plasma. Mol Nutr Food Res. 2010;54 (7):939–946. doi: 10.1002/mnfr.200900276. [DOI] [PubMed] [Google Scholar]
- 48.Walsh DM, Lomakin A, Benedek GB, Condron MM, Teplow DB. Amyloid beta-protein fibrillogenesis. Detection of a protofibrillar intermediate. J Biol Chem. 1997;272 (35):22364–22372. doi: 10.1074/jbc.272.35.22364. [DOI] [PubMed] [Google Scholar]
- 49.Teplow DB. Preparation of amyloid beta-protein for structural and functional studies. Methods Enzymol. 2006;413:20–33. doi: 10.1016/S0076-6879(06)13002-5. [DOI] [PubMed] [Google Scholar]
- 50.Ono K, Condron MM, Ho L, Wang J, et al. Effects of grape seed-derived polyphenols on amyloid beta-protein self-assembly and cytotoxicity. J Biol Chem. 2008;283 (47):32176–32187. doi: 10.1074/jbc.M806154200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bitan G, Lomakin A, Teplow DB. Amyloid beta-protein oligomerization: prenucleation interactions revealed by photo-induced cross-linking of unmodified proteins. J Biol Chem. 2001;276 (37):35176–35184. doi: 10.1074/jbc.M102223200. [DOI] [PubMed] [Google Scholar]
- 52.Appeldoorn MM, Vincken JP, Aura AM, Hollman PC, Gruppen H. Procyanidin dimers are metabolized by human microbiota with 2-(3,4-dihydroxyphenyl)acetic acid and 5-(3,4-dihydroxyphenyl)-gamma-valerolactone as the major metabolites. J Agric Food Chem. 2009;57 (3):1084–1092. doi: 10.1021/jf803059z. [DOI] [PubMed] [Google Scholar]
- 53.Aura AM. Microbial metabolism of dietary phenolic compounds in the colon. Phytochem Rev. 2008;7:407–429. [Google Scholar]
- 54.Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, et al. Amyloid beta -protein (Abeta) assembly: Abeta 40 and Abeta 42 oligomerize through distinct pathways. Proc Natl Acad Sci U S A. 2003;100 (1):330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bitan G, Fradinger EA, Spring SM, Teplow DB. Neurotoxic protein oligomers--what you see is not always what you get. Amyloid. 2005;12 (2):88–95. doi: 10.1080/13506120500106958. [DOI] [PubMed] [Google Scholar]
- 56.Wang YJ, Thomas P, Zhong JH, Bi FF, et al. Consumption of grape seed extract prevents amyloid-beta deposition and attenuates inflammation in brain of an Alzheimer’s disease mouse. Neurotox Res. 2009;15 (1):3–14. doi: 10.1007/s12640-009-9000-x. [DOI] [PubMed] [Google Scholar]
- 57.Gao K, Xu A, Krul C, Venema K, et al. Of the major phenolic acids formed during human microbial fermentation of tea, citrus, and soy flavonoid supplements, only 3,4-dihydroxyphenylacetic acid has antiproliferative activity. J Nutr. 2006;136 (1):52–57. doi: 10.1093/jn/136.1.52. [DOI] [PubMed] [Google Scholar]
- 58.Serafim TL, Carvalho FS, Marques MP, Calheiros R, et al. Lipophilic caffeic and ferulic acid derivatives presenting cytotoxicity against human breast cancer cells. Chem Res Toxicol. 2011;24 (5):763–774. doi: 10.1021/tx200126r. [DOI] [PubMed] [Google Scholar]
- 59.Lin Y, Yan Y. Biosynthesis of caffeic acid in Escherichia coli using its endogenous hydroxylase complex. Microb Cell Fact. 2012;11:42. doi: 10.1186/1475-2859-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cui JD, Qiu JQ, Fan XW, Jia SR, Tan ZL. Biotechnological production and applications of microbial phenylalanine ammonia lyase: a recent review. Crit Rev Biotechnol. 2014;34 (3):258–268. doi: 10.3109/07388551.2013.791660. [DOI] [PubMed] [Google Scholar]
- 61.Urpi-Sarda M, Monagas M, Khan N, Llorach R, et al. Targeted metabolic profiling of phenolics in urine and plasma after regular consumption of cocoa by liquid chromatography-tandem mass spectrometry. J Chromatogr A. 2009;1216 (43):7258–7267. doi: 10.1016/j.chroma.2009.07.058. [DOI] [PubMed] [Google Scholar]
- 62.Faria A, Pestana D, Teixeira D, Couraud PO, et al. Insights into the putative catechin and epicatechin transport across blood-brain barrier. Food Funct. 2011;2 (1):39–44. doi: 10.1039/c0fo00100g. [DOI] [PubMed] [Google Scholar]
- 63.Clabough EB. Huntington’s disease: the past, present, and future search for disease modifiers. Yale J Biol Med. 2013;86 (2):217–233. [PMC free article] [PubMed] [Google Scholar]
- 64.Duyckaerts C. Neurodegenerative lesions: seeding and spreading. Rev Neurol (Paris) 2013;169 (10):825–833. doi: 10.1016/j.neurol.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 65.Dimakopoulos AC. Protein aggregation in Alzheimer’s disease and other neoropathological disorders. Curr Alzheimer Res. 2005;2 (1):19–28. doi: 10.2174/1567205052772795. [DOI] [PubMed] [Google Scholar]
- 66.Scalbert A, Williamson G. Dietary intake and bioavailability of polyphenols. J Nutr. 2000;130 (8S Suppl):2073S–2085S. doi: 10.1093/jn/130.8.2073S. [DOI] [PubMed] [Google Scholar]
- 67.Kim BS, Jeon YS, Chun J. Current status and future promise of the human microbiome. Pediatr Gastroenterol Hepatol Nutr. 2013;16 (2):71–79. doi: 10.5223/pghn.2013.16.2.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Power SE, O’Toole PW, Stanton C, Ross RP, Fitzgerald GF. Intestinal microbiota, diet and health. Br J Nutr. 2014;111 (3):387–402. doi: 10.1017/S0007114513002560. [DOI] [PubMed] [Google Scholar]
- 69.Basli A, Soulet S, Chaher N, Merillon JM, et al. Wine polyphenols: potential agents in neuroprotection. Oxid Med Cell Longev. 2012;2012:805762. doi: 10.1155/2012/805762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cimini A, Gentile R, D’Angelo B, Benedetti E, et al. Cocoa powder triggers neuroprotective and preventive effects in a human Alzheimer’s disease model by modulating BDNF signaling pathway. J Cell Biochem. 2013;114 (10):2209–2220. doi: 10.1002/jcb.24548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Magrone T, Marzulli G, Jirillo E. Immunopathogenesis of neurodegenerative diseases: current therapeutic models of neuroprotection with special reference to natural products. Curr Pharm Des. 2012;18 (1):34–42. doi: 10.2174/138161212798919057. [DOI] [PubMed] [Google Scholar]
- 72.Smid SD, Maag JL, Musgrave IF. Dietary polyphenol-derived protection against neurotoxic beta-amyloid protein: from molecular to clinical. Food Funct. 2012;3 (12):1242–1250. doi: 10.1039/c2fo30075c. [DOI] [PubMed] [Google Scholar]
- 73.Tavares L, Figueira I, McDougall GJ, Vieira HL, et al. Neuroprotective effects of digested polyphenols from wild blackberry species. Eur J Nutr. 2013;52 (1):225–236. doi: 10.1007/s00394-012-0307-7. [DOI] [PubMed] [Google Scholar]
- 74.Virmani A, Pinto L, Binienda Z, Ali S. Food, nutrigenomics, and neurodegeneration--neuroprotection by what you eat! Mol Neurobiol. 2013;48 (2):353–362. doi: 10.1007/s12035-013-8498-3. [DOI] [PubMed] [Google Scholar]
- 75.Wang J, Tang C, Ferruzzi MG, Gong B, et al. Role of standardized grape polyphenol preparation as a novel treatment to improve synaptic plasticity through attenuation of features of metabolic syndrome in a mouse model. Mol Nutr Food Res. 2013;57 (12):2091–2102. doi: 10.1002/mnfr.201300230. [DOI] [PMC free article] [PubMed] [Google Scholar]