Abstract
Objectives
To test the hypothesis that somatic PIK3CA mutations would be found in patients with more common disorders including isolated lymphatic malformation (LM) and Klippel-Trenaunay syndrome (KTS).
Study design
We used next generation sequencing, droplet digital PCR (ddPCR), and single molecule molecular inversion probes (smMIPs) to search for somatic PIK3CA mutations in affected tissue from patients seen at Boston Children’s Hospital who had an isolated LM (n=17), KTS (n=21), fibro-adipose vascular anomaly (FAVA; n=8), or congenital lipomatous overgrowth with vascular, epidermal, and skeletal anomalies syndrome (CLOVES; n = 33), the disorder for which we first identified somatic PIK3CA mutations. We also screened 5 of the more common PIK3CA mutations in a second cohort of patients with LM (n=31) from Seattle Children’s Hospital.
Results
Most individuals from Boston Children’s Hospital who had isolated LM (16/17) or LM as part of a syndrome, such as KTS (19/21), FAVA (4/8), and CLOVES (30/32) were somatic mosaic for PIK3CA mutations, with 5 specific PIK3CA mutations accounting for ~ 80% of cases. Seventy-four percent of patients with LM from Seattle Children’s Hospital also were somatic mosaic for 1 of 5 specific PIK3CA mutations. Many affected tissue specimens from both cohorts contained fewer than 10% mutant cells.
Conclusions
Somatic PIK3CA mutations are the most common cause of isolated lymphatic malformations and disorders in which lymphatic malformation is a component feature. Five PIK3CA mutations account for most cases. The search for causal mutations requires sampling of affected tissues and techniques that are capable of detecting low-level somatic mosaicism, because the abundance of mutant cells in a malformed tissue can be low.
Keywords: lymphatic malformation, PIK3CA, somatic mutation, Klippel-Trenaunay syndrome
Somatic mutations in PIK3CA, which encodes the catalytic subunit of the enzyme phosphatidylinositol 3-kinase (PI3K), occur frequently in human cancer 1. These activating mutations increase the enzyme’s baseline catalytic activity 2. The mutations may be insufficient to cause cancer on their own, but they enhance tumor growth in combination with other oncogenic mutations 3. Recently, several patients with malformative/overgrowth syndromes have been shown to contain the same somatic hypermorphic PIK3CA mutations found in tumors 4–9. The mechanism by which a PIK3CA mutation that arises during embryogenesis produces malformation and overgrowth is not understood, but is likely due to the key intermediate role that phosphatidylinositol 3-kinase plays in multiple signaling pathways, including the vascular endothelial growth factor, fibroblast growth factor, and insulin-like growth factor pathways 10.
Individuals with malformative syndromes resulting from somatic PIK3CA mutations have a spectrum of phenotypes, often non-overlapping (e.g., hemimegalencaphy versus macrodactyly) 4–9. Lymphatic malformations (LM), which arise most frequently as an isolated vascular anomaly, are a major component feature in patients that have CLOVES syndrome 11 and Klippel-Trenaunay syndrome (KTS) 12. Fatty-fibrous infiltration with adjacent venous-lymphatic malformation occurs in CLOVES syndrome and within skeletal muscles of patients with fibro-adipose vascular anomaly (FAVA) 13. Therefore, we hypothesized that patients with isolated LM and FAVA, in addition to patients with CLOVES and KTS, are somatic mosaic for PIK3CA mutations. Here, we report that most patients with these diseases have somatic PIK3CA mutations, and that 5 recurrent mutations account for the majority of cases.
METHODS
The Committee on Clinical Investigation at Boston Children’s Hospital approved this study. Participants were seen in the Vascular Anomalies Center and had a clinical diagnosis of LM, CLOVES, KTS, or FAVA. LM was diagnosed based on the presence of microcystic and/or macrocystic features by imaging and the absence of cutaneous or skeletal abnormalities 14. CLOVES syndrome was diagnosed based upon the presence of lipomatous overgrowth involving the torso, face, and/or extremity, cutaneous capillary-lymphatic malformation, and skeletal anomalies such as sandal gap toe, scoliosis, syndactyly, or polydactyly 11, 15. KTS was diagnosed based upon the presence of cutaneous capillary-lymphatic malformation, enlarged veins, and overgrowth of the affected extremity 12. Fibro-adipose vascular anomaly was diagnosed based upon a localized intramuscular fibrous and fatty infiltration with adjacent venous and lymphatic malformation 13.
Patients from an independent cohort with head and neck lymphatic malformations followed in the pediatric vascular malformation clinic at Seattle Children’s Hospital were also studied.
Prospectively, affected tissue was collected from participants during a clinically-indicated surgical procedure and stored frozen. Retrospectively, affected tissue was retrieved from archived formalin fixed paraffin embedded (FFPE) tissue blocks. DNA was extracted from fresh frozen or FFPE tissue using the QIAamp DNA Mini or QIAamp DNA FFPE Tissue Kit (Qiagen, Germantown, MD), respectively. We first performed whole-exome sequencing (WES) of DNA from affected tissue obtained from individuals with LM (n=7) and FAVA (n=8), and targeted capture sequencing (TCS) of DNA from affected tissue from individuals with CLOVES (n=22) and KTS (n=15). Next, we designed a low-cost, high throughput, droplet digital PCR (ddPCR) assay and used this assay to screen for 5 recurrent PIK3CA missense mutations in affected tissue samples from larger cohorts of individuals with LM, CLOVES, KTS, and FAVA. Finally, in samples for which a mutation was not identified, we used single molecule molecular inversion probes (smMIPs) to screen the entire PIK3CA coding sequence for novel mutations.
Whole Exome and Targeted Capture Sequencing
Genomic libraries, WES, and TCS were performed as previously described 4, using SureSelectXT Human All Exon V5 (Agilent Technology, Lexington, MA) for WES and a custom designed 1M SureSelect DNA Capture Array (Agilent Technology) containing the coding regions of 26 genes associated with the PI3K-AKT signaling pathway (AKT1, AKT2, AKT3, FKBP1A, IGF1, KCNH1, KCNH2, KCNK5, MDK, MTOR, NFATC1, NF1, NF2, NOS3, PIK3CA, PIK3CB, PIK3CG, PIK3R1, PIK3R2, PIK3R3, PPP3CA, PPP3CB, PPP3CC, PTEN, PTN, SLC12A7) for TCS. Samples were sequenced in 100-bp paired-end reads on the Illumina HiSeq 2000 sequencer (Illumina, Inc., San Diego, CA). Sequence was analyzed for mutations as previously described 4.
Droplet Digital PCR (ddPCR)
The ddPCR primers and probes used in this study (Table III; available at www.jpeds.com) were designed using Primer3 software 16 to target 5 PIK3CA mutations (p.C420R, p.E542K, p.E545K, p.H1047R, p.H1047L). Droplet digital PCR was performed as previously described 17. Briefly, the PCR mixture contained ddPCR Super Mix (Bio-Rad, Hercules, CA) (final concentration: 1X), 0.25 μM of mutant and reference probe, 0.9 μM of forward and reverse primer, and up to 30 ng of template DNA in 20 μL total volume. The 20 μL reaction mixture was emulsified into approximately 14,000 droplets using a QX100 Droplet Generator (Bio-Rad) according to the manufacturer’s instructions. PCR was performed with the following variables: 10 minues at 95°C, followed by 40 cycles of 30 seconds at 94°C, 60 seconds at 60°C, 30 sec at 72°C, and then kept at 4°C. Samples were analyzed within 24 hours using the QX100 Droplet Reader (Bio-Rad) and analyzed with QuantaSoft software (Bio-Rad).
In some cases, a p.H1047R, p.E542K, or p.E545K mutation found by WES, TCS, and/or ddPCR, was also confirmed using a hydrolysis probes (TaqMan) assay 18.
Single Molecule Molecular Inversion Probes (smMIPs)
smMIP capture and library construction were performed as previously described 19. In our experiments we used 500ng of affected tissue DNA and a 5000 to 1 molar excess of phosphorylated smMIPs during the capture process. Sequencing was performed on a MiSeq (Illumina, Inc., San Diego, CA) to obtain 150 bp paired-end reads, and sequence data were analyzed as previously described 4, with the exception that a different PCR duplicate removal strategy was employed because each smMIP has a unique bar-code 19.
Genotype-Phenotype Association
Medical records were reviewed for each participant from Boston Children’s Hospital to cull data regarding age, sex, location of malformation and overgrowth, and complications (Table V; available at www.jpeds.com). Patients were grouped by their individual PIK3CA mutation and univariate analyses using Fisher exact test and chi-square testing compared groups with a specific genotype to the 28 individual clinical and radiologic characteristics.
RESULTS
Individuals from Boston Children’s Hospital with LM (n=17), KTS (n=21), FAVA (n=8) and CLOVES (n=33) participated in our study. Figure 1 depicts features of LM, CLOVES, KTS, and FAVA that were determinate in assigning a clinical diagnosis. Table I summarizes the frequencies of features present in the Boston Children’s Hospital cohorts.
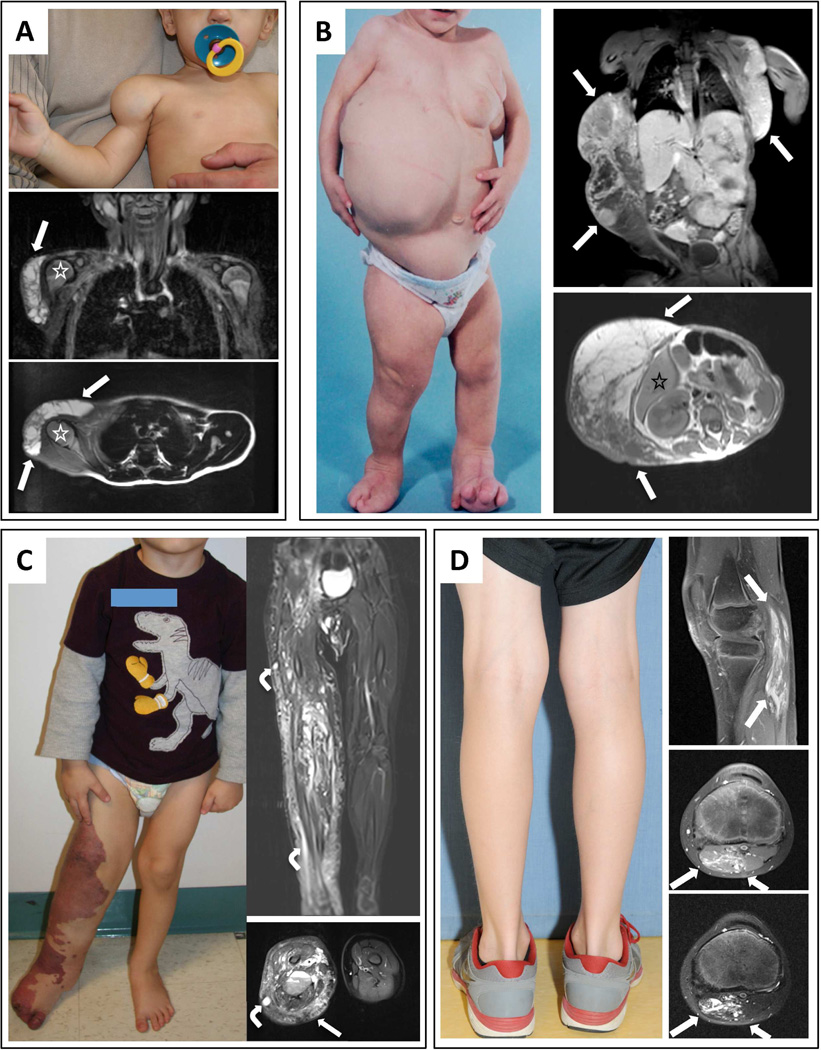
Figure 1.
Photographs and MRIs of participants with isolated LM, CLOVES, KTS, and FAVA. (A) 8-month-old boy (LM1) with isolated LM. Note swelling in deltoid region without cutaneous vascular signs. Coronal and sagittal fat-saturated T2-weighted MRI demonstrates macrocystic LM (a multilocular cystic mass) involving the anterolateral aspects of the right shoulder without muscular infiltration (arrows); humeral head (asterisk). (B) 19-month-old female (CL12) with CLOVES syndrome. Note asymmetric distribution of truncal lipomatous masses and bilateral lower extremity involvement. Coronal fat-saturated T1-weighted MRI following contrast administration demonstrates moderate heterogeneous enhancement of the bilateral truncal masses (arrows). Axial T1-weighted MRI without contrast depicts truncal lipomatous overgrowth (arrows); segment VI of the liver (asterisk). (C) 3-year-old boy (KT4) with KTS. Note capillary-lymphatic malformation and overgrowth involving right lower extremity. Coronal and axial fat-saturated T2-weighted MRI shows the persistent marginal vein system (bent arrows) and marked enlargement of the subcutaneous tissues due to a combination of lymphatic fluid and fat (straight arrow). There are also intramuscular venous malformations. (D) 9 - year-old boy (F8) with FAVA of the left gastrocnemius muscle; note absence of overgrowth and cutaneous vascular anomalies. Sagittal fat-saturated T1-weighted MRI following contrast administration demonstrates the longitudinal distribution of the diffuse, fibro-adipose vascular anomaly (arrows). Axial fat-saturated T2-weighted MRI with (upper) and without (lower) contrast. Note right head of the gastrocnemius muscle is diffusely replaced by a contrast enhancing heterogeneous soft tissue lesion (arrows).
Table 1.
Summary characteristics for the Boston Children’s Hospital cohorts. Truncal involvement indicates presence of one or more lipomatous masses on inspection, palpation, or MRI. Limb involvement indicates lipomatous overgrowth by inspection or MRI. Number of participants with limb length discrepancy (LLD), macrodactyly (macro), and syndactyly (syn) is indicated. CM – capillary malformation, LM – lymphatic malformation, VM – venous malformation. Number of participants with LM affecting a specific location (head or neck, trunk, and limb) is indicated, as is number of LMs that by MRI or ultrasonography were microcystic, macrocystic, or a combination.
| LM (n=17) |
CLOVES (n=33) |
KTS (n=21) |
FAVA (n=8) |
|
|---|---|---|---|---|
| Sex (M/F) | (11/6) | (18/15) | (13/18) | (3/5) |
| Truncal involvement | 0 | 26 | 2 | 0 |
| Limb involvement (LLD/macro/syn) |
4 (0,0,0) |
30 (17/22/6) |
21 (20/14/4) |
8 (0/0/0) |
| Cutaneous CM and/or LM | 0 | 28 | 21 | 0 |
| LM on imaging (head-neck/trunk/limb) [micro/macro/combination] |
17 (10/3/4) [1/8/8] |
33 (5/13/15) [4/19/10] |
21 (0/0/21) [1/11/9] |
4 |
| VM on imaging | 0 | 30 | 20 | 8 |
| Complications (INF/GIB/DVT/PVT/Wilms/death) |
(13/0/0/0/0/0) |
(12/7/6/4/1/2) |
(13/8/3/0/0/0) |
none |
Note, LM adjacent to fibro-adipose infiltrated muscle was a frequent histopathologic finding in FAVA. Number of participants with a VM detected by ultrasonography or MRI is indicated. VMs involved the marginal veins or large draining veins in CLOVES and KTS, and were adjacent to the fibro-adipose lesion in FAVA. Complications include infection at site of or originating from a vascular malformation (INF), gastro-intestinal bleed (GIB), deep vein thrombosis ± pulmonary embolism (DVT), portal vein thrombosis (PVT), Wilms tumor (Wilms), and death.
WES yielded > 85% enrichment for target sequence and an average of 77-fold (77x) coverage (ranging from 42x to 133x) across the exome for the LM and FAVA samples. To minimize identifying false positive variants caused by library preparation or by sequencing error, we considered only variants observed in at least 2 independent reads at a locus with > 40x sequencing coverage. Using these criteria, we detected likely disease-causing PIK3CA mutations in 1/8 LM and 2/7 FAVA samples (Table II). Thus, although WES revealed mutations in several samples it did not identify mutations in most samples, potentially because the sequencing coverage and/or the mutant allele frequency were too low to distinguish a true, low-frequency mutation from a chance sequencing error. For example, WES from participants LM1, LM8, and FAVA5 each contained 1 variant read in a PIK3CA hotspot among 31, 58, and 31 total reads, respectively (Table II), and therefore were excluded by our filtering criteria because such observations often arise by chance in NGS datasets.
Table 2.
Summary of next generation sequencing (NGS), droplet digital PCR (ddPCR), and single molecule molecular inversion probe (smMIP) results for tissue samples from the Boston Children’s Hospital CLOVES, KTS, isolated LM, and FAVA cohorts. NGS, employing either whole exome sequencing (WES) or targeted capture sequencing of 26 genes (TCS), results are reported as mutant alleles/total alleles sequenced. ddPCR was performed for 5 specific mutations and a minimum of 2000 amplimer-containing droplets was analyzed for each ddPCR assay. The percent mutant allele is reported for the ddPCR assay; for example LM12, which had 0.8% mutant alleles, generated 41 droplets that fluoresced positive for the mutant probe among 4005 total amplimer-containing droplets. smMIP results are reported as mutant alleles/total alleles sequenced. Samples for which no mutant alleles were identified by NGS, ddPCR, or smMIP are indicated with a “-,” whereas samples not studied with a technique are left blank. Occasional large differences in mutant allele frequencies from the same study participant may be the consequence of DNA being extracted from different tissues sections for the NGS, ddPCR, and smMIP experiments. Participant for which DNA was extracted from fresh frozen tissue are identified in bold, while participants for which DNA was extracted from paraffin embedded tissue are listed using standard font. Some individuals with CLOVES syndrome (asterisked) had WES and TCS data published previously 4 and are included in this table as positive controls.
| CLOVES | KTS | LM | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pt. | Var. | NGS | ddPCR | smMIP | Pt. | Var. | NGS | ddPCR | smMIP | Pt. | Var. | NGS | ddPCR | smMIP |
| CL1* | H1047R | 5/24(TCS) | 17 | KT1 | - | - | - | - | LM1 | E542K | 1/31(WES)¶ | 8.5 | ||
| CL2* | H1047R | 13/64(TCS) | 20 | KT2 | E545K | - | 6.2 | LM2 | H1047R | 3/35(WES)¶ | 2.5 | |||
| CL3* | E542K | 1/11(WES)¶ | 8.4 | KT3 | E545G | - | - | 28/825 | LM3 | C420R | 9/58(WES) | 7.9 | ||
| CL4* | E542K | 2/16(WES)¶ | KT4 | H1047L | 4/84(TCS) | 4.7 | LM4 | E545K | 2/36(WES)¶ | 9.8 | ||||
| CL5* | C420R | 12/40(WES) | 15 | KT5 | E545K | - | 12 | LM5 | H1047R | - | 6.2 | |||
| CL6* | C420R | 8/44(WES) | 25 | KT6 | E545K | - | 7.4 | LM6 | H1047L | - | 5.4 | |||
| CL7 | H1047R | - | 3.3 | KT7 | E545K | - | 14 | LM7 | E542K | 1/58(WES)¶ | 2.8 | |||
| CL8 | - | - | - | - | KT8 | H1047R | - | 8.0 | LM8 | H1047R | 6.6 | |||
| CL9 | E545K | - | 8.0 | KT9 | E545K | - | 7.4 | LM9 | E542K | 4.9 | ||||
| CL10 | E545K | 10/180(TCS) | 9.4 | KT10 | H1047R | 2/58(TCS) | 8.0 | LM10 | - | - | ||||
| CL11 | H1047R | - | 14 | KT11 | H1047R | 2/44(TCS) | 7.4 | LM11 | H1047L | 6.0 | ||||
| CL12 | H1047R | 19/73(TCS) | 32 | KT12 | H1047R | - | 2.7 | LM12 | E545K | 0.8 | ||||
| CL13 | H1047R | - | ? | KT13 | E542K | 2/48(TCS) | 5.0 | LM13 | H1047L | 10 | ||||
| CL14 | E545K | 4/86(TCS) | 5.6 | KT14 | H1047R | - | 16 | LM14 | H1047R | 4.2 | ||||
| CL15 | E545K | 5/68(TCS) | 10 | KT15 | E545K | - | 5.5 | LM15 | H1047R | 8.4 | ||||
| CL16 | H1047L | 4/43(TCS) | 16 | KT16 | E545K | 8.2 | LM16 | E542K | 4.4 | |||||
| CL17 | C420R | - | 9.4 | KT17 | H1047R | 8.9 | LM17 | E545K | 2.8 | |||||
| CL18 | E545K | 62/228(TCS) | 27 | KT18 | E542K | 10 | ||||||||
| CL19 | E545K | - | 11 | KT19 | E545K | 8.7 | ||||||||
| CL20 | E542K | - | 8.9 | 88/903 | KT20 | E545K | 13 | FAVA | ||||||
| CL21 | T1025A | - | - | 8/275 | KT21 | E542K | 3.7 | Pt. | Var. | NGS | ddPCR | smMIP | ||
| CL22 | E545K | - | 4.3 | F1 | - | - | - | - | ||||||
| CL23 | H1047L | 48/149(TCS) | 24 | F2 | E545K | 2/30(WES)¶ | 8.9 | |||||||
| CL24 | C420R | - | 10 | F3 | Q546K | 4/53(WES) | 6.9# | |||||||
| CL25 | C420R | - | 25 | F4 | - | - | - | |||||||
| CL26 | C420R | 135/527(TCS) | 20 | F5 | E542K | 1/31(WES)¶ | 8.4 | |||||||
| CL27 | E542K | 1.2 | F6 | - | - | - | - | |||||||
| CL28 | E542K | 9.8 | F7 | H1047R | 26/238(WES) | 20 | ||||||||
| CL29 | N345K | - | 314/837 | F8 | E545K | - | 5.0 | |||||||
| CL30 | H450R! | - | 11/459 | |||||||||||
| CL31 | E542K | 5.0 | 110/1362 | |||||||||||
| CL32 | - | - | ||||||||||||
| CL33 | E545K | 9.2 | ||||||||||||
indicate samples that would have been missed by our NGS filtering strategy due to insufficient number of reads containing the mutation.
indicates sample for which the p.E545K ddPCR assay indicated an alternate mutation.
To increase sequencing throughput, we employed a targeted capture array containing 26 genes associated with the PI3K-AKT signaling pathway and applied this array to CLOVES (n=21) and KTS (n=15) samples. We obtained sequence enrichments > 45% with targeted capture sequencing (TCS). Still requiring 2 independent mutant reads at a locus, we detected PIK3CA mutations in 10/21 CLOVES and 4/15 KTS samples (Table II).
Because 5 different PIK3CA mutations (p.C420R, p.E542K, p.E545K, p.H1047R, p.H1047L) occurred commonly in patients with CLOVES, KTS, LM, and FAVA, and knowing that these same mutations are common in patients with hemimegalencephaly, fibroadipose hyperplasia, macrodactyly, and cancer 5, 6, 9, 20, we developed a ddPCR assay for these 5 mutations. ddPCR, is method that can detect low frequency mutant alleles and provide accurate measurements of wild-type and mutant allele abundances. Briefly, DNA was recovered from 1000 – 4000 cells in an affected tissue sample and partitioned into ~ 14,000 nanoliter-sized PCR droplets, after which PCR was performed. Droplets were then digitally counted using fluorescent probes that could differentiate wild-type and mutant amplimers. This approach allowed us to screen these 5 mutations at greater depth (sample > 2000 alleles) and at low cost (~ $15/sample). To determine the sensitivity of the assay, we performed ddPCR on genomic DNA from a cell line heterozygous for the p.H1047R mutation that was serially diluted with healthy control genomic DNA. We reliably detected a mutant allele frequency as low as 0.1% (1 mutant per 1000 total alleles) with this assay (data not shown). To determine the specificity of the assay, we performed ddPCR using water and healthy control DNA, instead of affected tissue DNA. In control experiments, where no template was added, generally no positive droplets were detected, although occasionally 1 to 10 droplets representing a wild-type amplimer observed. Rarely, 1 to 5 droplets representing mutant amplimer were detected in healthy, control DNA. We concluded these “false positive” droplets were the result of contamination occurring during set up of the reaction 21. Because affected tissue samples yielded > 2000 amplimer containing droplets, we set a minimum cut-off frequency of 0.5% (1 mutant per 200 total alleles) to call a DNA sample positive for a PIK3CA mutation by ddPCR. Representative ddPCR outputs are shown in Figure 2.
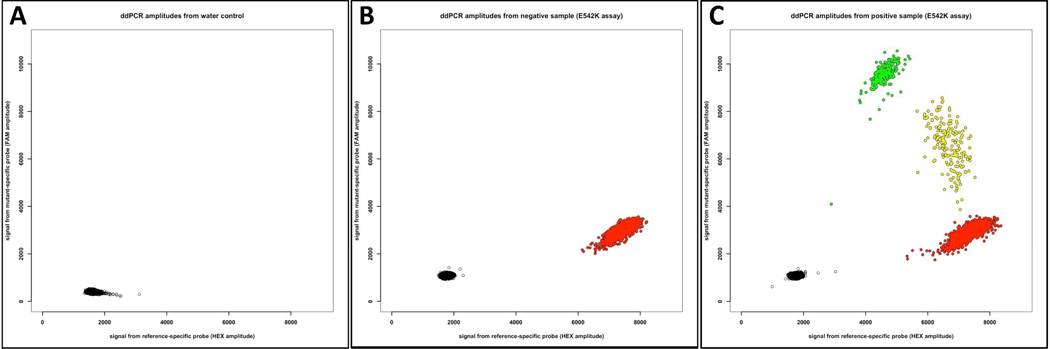
Figure 2.
Scatterplots depicting ddPCR results specifically for the p.E542K mutation assay. The X-axis indicates fluorescence intensity in droplets for the probe targeting the wild-type amplimer and the Y-axis indicates fluorescence intensity in droplets for the probe targeting the p. E542K mutant amplimer. Individual droplets that contain no amplimer are pseudocolored black, wild-type amplimer red, p.E542K mutant amplimer green, and wild-type and p.E542K mutant amplimers yellow. (A) Scatterplot when water-alone was used as template. Neither wild-type nor mutant amplimer was present in any of the 15,633 droplets in this experiment. (B) Plot when affected tissue DNA from participant LM2, who has a different PIK3CA mutation (p.C420R), was used as template for the p.E542K assay. Of the 13,932 droplets analyzed, 11,670 contain no amplimer and 2,262 contain amplimer that is wild-type at the p.E542K locus. (C) Scatterplot when affected tissue DNA from participant LM1, who was previously found to have a p.E542K mutation by whole exome sequencing, was used as template. Of 13,146 droplets analyzed, 3,811 contain wild-type amplimer, 242 contain mutant amplimer, 193 contain wild-type and mutant amplimer, and 8,900 contain no amplimer.
We identified 1 of the 5 mutant PIK3CA alleles we tested by ddPCR in 27/33 CLOVES (82%), 19/21 KTS (90%), 16/17 LM (94%), and 4/8 FAVA (50%) samples (Table II). Droplet digital PCR always identified the same mutation as had been identified by WES or TCS. Additionally, ddPCR detected a low frequency PIK3CA mutation in many samples that yielded no detectable mutation with NGS. Another indication of the high specificity of the ddPCR assay is that affected tissue positive for one PIK3CA mutation was never positive for another mutation.
In an independent cohort from Seattle Children’s Hospital comprising affected tissue DNA from patients with isolated LM, ddPCR detected 1 of the 5 common PIK3CA mutations in 23/31 (74%) of individuals (data not shown).
Next we used smMIPs to study samples with sufficient remaining DNA and without somatic PIK3CA mutations detected by WES, TCS, or ddPCR (n=9). Two additional samples with previously identified PIK3CA mutations were included as positive controls. Except for samples CL8 and CL21, which were prepared from FFPE tissue and performed poorly in this assay, smMIP sequencing yielded >400x coverage for >78%, >200x coverage for >91%, and >100x coverage for > 98% of the PIK3CA coding sequence. We identified likely disease-causing PIK3CA mutations in 4 of the 9 affected tissue samples, with mutant allele frequencies ranging from 2.8% (8/275) to 11.4% (4/35), and in both positive controls (Table 2). Two samples without an identified PIK3CA mutation, F1 and F6, had >400x coverage for >98% of the PIK3CA coding sequence covered by smMIP sequencing.
We observed no significant correlation between a specific PIK3CA mutation, a disease phenotype, macrocystic versus microcystic LM, or clinical severity (data not shown); however, we were underpowered to detect modest genotype-phenotype correlations.
DISCUSSION
Most patients with isolated LM, KTS, FAVA, and CLOVES have somatic mosaic PIK3CA mutations. Five recurrent mutations, which can be quickly and inexpensively screened by ddPCR, account for most cases. Gene focused NGS using smMIPs identified additional PIK3CA mutations.
We did not identify a PIK3CA mutation in every patient. This may be because some patients have a mutation in a region of PIK3CA for which we had low WES, TCS, or smMIP sequence coverage. Alternatively, it is possible that a PIK3CA mutation is present, but at a frequency that falls below the sensitivity of the ddPCR assay or our NGS filtering algorithm. Lastly, we cannot preclude these disorders also being caused by mutations in other genes, as has been reported for individuals with hemimegalencephaly 6, 22. Our WES and TCS data did not suggest candidate mutations at alternative loci in samples that lacked a detectable PIK3CA mutation. Therefore, in contrast to patients with hemimegalencephaly for whom a mutation in 1 of several genes is sufficient to cause brain overgrowth, PIK3CA is responsible for most, if not all, of the LMs and other vascular malformations seen in patients with CLOVES and KTS.
Many reports characterize clinical features in patients with LM and KTS, and several pathogenic mechanisms have been proposed 14. KTS has been recognized for more than a century 23; we defined the condition as comprised of overgrowth, cutaneous capillary and lymphatic malformation, and persistence of embryonic vasculature in an extremity 12, 14. All of the KTS participants in this study fit this narrow definition with respect to appendicular involvement. Centers that use less rigid criteria to clinically diagnosis KTS, may inadvertently consider patients with other overgrowth disorders, such as Beckwith-Wiedemann 24, SOLAMEN 25, or Bockenheimer 26 syndromes, as having KTS. Thus, our finding that most patients with KTS have PIK3CA mutations is likely a consequence of the strict clinical definition we employ. CLOVES syndrome and FAVA are recently recognized clinical entities, although an 1867 report described a patient who likely had CLOVES syndrome 27, 28.
Cells containing activating mutations were present at low frequencies (<10%) in many affected tissue samples (Table II). Biologically, this likely reflects the ability of mutant cells to recruit wild-type cells during the malformative/overgrowth process, although we cannot preclude difficulty in enriching solely for affected tissue from biopsies as an alternative explanation. We did not examine DNA obtained from normal skin or blood from most participants and therefore cannot preclude the possibility that mutant cells can reside in “normal” tissue. Regardless of mechanism, the low frequency of mutant alleles in many affected tissue specimens has important diagnostic implications, as detection of such mutations in patients requires approaches that can ascertain low-frequency alleles with high sensitivity and specificity. High sensitivity and specificity are essential in laboratories that offer clinical testing. Our results were obtained in the research setting and the ddPCR assay criteria we employed missed mutant allele frequencies less than 0.5% in order to avoid obtaining false positive results.
Although each of our genotyping strategies successfully identified PIK3CA mutations in affected tissue, we suggest initially performing ddPCR as an efficient and cost effective strategy for screening tissue samples from patients with isolated LM, CLOVES, KTS, and FAVA. Approximately 80% of affected tissue samples contained 1 of the 5 mutations screened by our ddPCR assay. For samples that did not have 1 of these 5 recurrent PIK3CA mutations, high-throughput NGS sequencing such as with smMIPs identified additional disease-causing PIK3CA mutations in another 8% of samples. PIK3CA inhibitors are currently in development as cancer therapeutics. These inhibitors may also benefit patients with LM, CLOVES, KTS, and FAVA.
Somatic mosaic mutations in PIK3CA cause LM as well as syndromes in which LM is a common feature. Currently, we do not know why the same somatic mutation in PIK3CA causes an isolated LM in one individual and a different disorder in another. Likely explanations include the stage during embryonic development when the mutation arises, the location of the first mutated cell in the embryo, and the stem and pluripotent progenitor cells that descend from the original mutant. Also worthy of further investigation is whether a PIK3CA mutation alone is sufficient to produce disease or whether additional genetic or environmental factors are required. Mouse models with a mutant Pik3ca allele that can be conditionally activated in a temporal- and tissue-specific manner 3, 29, 30 should help address these questions as well as facilitate the search for better therapies.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank their patients for participating in this study, Maayke van Ruler (Leiden University) for performing the TaqMan assays, and Joseph Hiatt, MD, and Jay Shendure, MD (both from University of Washington), for providing the smMIP probes.
Supported by the Manton Center for Orphan Disease Research at Boston Children’s Hospital, the Stuart and Jane Weitzman Fund for Vascular Anomalies Research at Boston Children’s Hospital, the Howard Hughes Medical Institute, and the National Institutes of Health (AR064231).
Abbreviations
- LM
lymphatic malformation
- KTS
Klippel-Trenaunay syndrome
- CLOVES
Congenital Lipomatous Overgrowth with Vascular, Epidermal, and Skeletal anomalies
- FAVA
fibroadipose vascular anomaly
- PIK3CA
Phosphatidylinositol-4,5-bisphospate 3-kinase, catalytic subunit alpha
- NGS
next generation sequencing
- WES
whole exome sequencing
- TCS
targeted capture sequencing
- ddPCR
droplet digital PCR
- smMIPs
single molecule molecular inversion probes
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
REFERENCES
- 1.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 2.Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kinross KM, Montgomery KG, Kleinschmidt M, Waring P, Ivetac I, Tikoo A, et al. An activating Pik3ca mutation coupled with Pten loss is sufficient to initiate ovarian tumorigenesis in mice. J Clin Invest. 2012;122:553–557. doi: 10.1172/JCI59309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurek KC, Luks VL, Ayturk UM, Alomari AI, Fishman SJ, Spencer SA, et al. Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am J Hum Genet. 2012;90:1108–1115. doi: 10.1016/j.ajhg.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JH, Huynh M, Silhavy JL, Kim S, Dixon-Salazar T, Heiberg A, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nature genetics. 2012;44:941–945. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riviere JB, Mirzaa GM, O'Roak BJ, Beddaoui M, Alcantara D, Conway RL, et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nature genetics. 2012;44:934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rios JJ, Paria N, Burns DK, Israel BA, Cornelia R, Wise CA, et al. Somatic gain-of-function mutations in PIK3CA in patients with macrodactyly. Human molecular genetics. 2013;22:444–451. doi: 10.1093/hmg/dds440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maclellan RA, Luks VL, Vivero MP, Mulliken JB, Zurakowski D, Padwa BL, et al. PIK3CA activating mutations in facial infiltrating lipomatosis. Plastic and reconstructive surgery. 2014;133:12e–19e. doi: 10.1097/01.prs.0000436822.26709.7c. [DOI] [PubMed] [Google Scholar]
- 9.Lindhurst MJ, Parker VE, Payne F, Sapp JC, Rudge S, Harris J, et al. Mosaic overgrowth with fibroadipose hyperplasia is caused by somatic activating mutations in PIK3CA. Nature genetics. 2012;44:928–933. doi: 10.1038/ng.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanhaesebroeck B, Stephens L, Hawkins P. PI3K signalling: the path to discovery and understanding. Nature reviews Molecular cell biology. 2012;13:195–203. doi: 10.1038/nrm3290. [DOI] [PubMed] [Google Scholar]
- 11.Alomari AI. Characterization of a distinct syndrome that associates complex truncal overgrowth, vascular, and acral anomalies: a descriptive study of 18 cases of CLOVES syndrome. Clinical Dysmorphology. 2009;18:1–7. doi: 10.1097/MCD.0b013e328317a716. [DOI] [PubMed] [Google Scholar]
- 12.Cohen MM. Klippel-Trenaunay syndrome. American journal of medical genetics. 2000;93:171–175. doi: 10.1002/1096-8628(20000731)93:3<171::aid-ajmg1>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 13.Alomari AI, Spencer SA, Arnold RW, Chaudry G, Kasser JR, Burrows PE, et al. Fibro-adipose vascular anomaly: clinical-radiologic-pathologic features of a newly delineated disorder of the extremity. Journal of pediatric orthopedics. 2014;34:109–117. doi: 10.1097/BPO.0b013e3182a1f0b8. [DOI] [PubMed] [Google Scholar]
- 14.Mulliken JB, Burrows PE, Fishman SJ, Mulliken JB. Mulliken and Young's vascular anomalies : hemangiomas and malformations. 2nd ed. Oxford: Oxford University Press; 2013. [Google Scholar]
- 15.Sapp JC, Turner JT, van de Kamp JM, van Dijk FS, Lowry RB, Biesecker LG. Newly delineated syndrome of congenital lipomatous overgrowth, vascular malformations, and epidermal nevi (CLOVE syndrome) in seven patients. American journal of medical genetics Part A. 2007;143A:2944–2958. doi: 10.1002/ajmg.a.32023. [DOI] [PubMed] [Google Scholar]
- 16.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3--new capabilities and interfaces. Nucleic acids research. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Analytical chemistry. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Eijk R, Licht J, Schrumpf M, Talebian Yazdi M, Ruano D, Forte GI, et al. Rapid KRAS, EGFR, BRAF and PIK3CA mutation analysis of fine needle aspirates from non-small-cell lung cancer using allelespecific qPCR. PloS one. 2011;6:e17791. doi: 10.1371/journal.pone.0017791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiatt JB, Pritchard CC, Salipante SJ, O'Roak BJ, Shendure J. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 2013;23:843–854. doi: 10.1101/gr.147686.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic acids research. 2014 doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urban C, Gruber F, Kundi M, Falkner FG, Dorner F, Hammerle T. A systematic and quantitative analysis of PCR template contamination. Journal of forensic sciences. 2000;45:1307–1311. [PubMed] [Google Scholar]
- 22.Poduri A, Evrony GD, Cai X, Elhosary PC, Beroukhim R, Lehtinen MK, et al. Somatic Activation of AKT3 Causes Hemispheric Developmental Brain Malformations. Neuron. 2012;74:41–48. doi: 10.1016/j.neuron.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klippel M, Trenaunay P. Du naevus variqueux osteo-hypertrophique. Arch Gen Med. 1900;185:641–672. [Google Scholar]
- 24.Weksberg R, Shuman C, Beckwith JB. Beckwith-Wiedemann syndrome. European journal of human genetics. 2010;18:8–14. doi: 10.1038/ejhg.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caux F, Plauchu H, Chibon F, Faivre L, Fain O, Vabres P, et al. Segmental overgrowth, lipomatosis, arteriovenous malformation and epidermal nevus (SOLAMEN) syndrome is related to mosaic PTEN nullizygosity. European journal of human genetics. 2007;15:767–773. doi: 10.1038/sj.ejhg.5201823. [DOI] [PubMed] [Google Scholar]
- 26.Kubiena HF, Liang MG, Mulliken JB. Genuine diffuse phlebectasia of Bockenheimer: dissection of an eponym. Pediatr Dermatol. 2006;23:294–297. doi: 10.1111/j.1525-1470.2006.00239.x. [DOI] [PubMed] [Google Scholar]
- 27.Friedberg H. Riesenwuchs des rechten Beines. Virchow Arch. 1867;40:353–360. [Google Scholar]
- 28.Alomari AI, Thiex R, Mulliken JB. Hermann Friedberg's case report: an early description of CLOVES syndrome. Clinical genetics. 2010;78:342–347. doi: 10.1111/j.1399-0004.2010.01479.x. [DOI] [PubMed] [Google Scholar]
- 29.Yuan W, Stawiski E, Janakiraman V, Chan E, Durinck S, Edgar KA, et al. Conditional activation of Pik3ca(H1047R) in a knock-in mouse model promotes mammary tumorigenesis and emergence of mutations. Oncogene. 2013;32:318–326. doi: 10.1038/onc.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu P, Cheng H, Santiago S, Raeder M, Zhang F, Isabella A, et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med. 2011;17:1116–1120. doi: 10.1038/nm.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.