Abstract
Prevention efforts for respiratory syncytial virus (RSV) have been advanced due to the recent isolation and characterization of antibodies that specifically recognize the prefusion conformation of the RSV fusion (F) glycoprotein. These potently neutralizing antibodies are in clinical development for passive prophylaxis and have also aided the design of vaccine antigens that display prefusion-specific epitopes. To date, prefusion-specific antibodies have been shown to target two antigenic sites on RSV F, but both of these sites are also present on monomeric forms of F. Here we present a structural and functional characterization of human antibody AM14, which potently neutralized laboratory strains and clinical isolates of RSV from both A and B subtypes. The crystal structure and location of escape mutations revealed that AM14 recognizes a quaternary epitope that spans two protomers and includes a region that undergoes extensive conformational changes in the pre- to postfusion F transition. Binding assays demonstrated that AM14 is unique in its specific recognition of trimeric furin-cleaved prefusion F, which is the mature form of F on infectious virions. These results demonstrate that the prefusion F trimer contains potent neutralizing epitopes not present on monomers and that AM14 should be particularly useful for characterizing the conformational state of RSV F-based vaccine antigens.
Author Summary
Respiratory syncytial virus (RSV) causes significant morbidity and mortality in children, yet an efficacious vaccine remains unavailable. Antibodies that preferentially recognize the prefusion conformation of the fusion (F) glycoprotein, particularly those that bind antigenic site Ø at the membrane-distal apex, potently neutralize infection and have aided vaccine design. Here we characterize AM14, a potent human antibody, which we show recognizes a novel epitope midway between the membrane-proximal region and the apex of the prefusion F trimer. The epitope is evenly distributed across two protomers, causing AM14 to be uniquely trimer-specific and, surprisingly, cleavage-dependent. These results indicate that the prefusion trimer is antigenically distinct from the monomer. Our findings also demonstrate that epitopes other than site Ø can be the target of extremely potent neutralizing antibodies and thus provide a new target for structure-based vaccine design. Recognition of this novel epitope could make AM14 an ideal candidate for strategies that combine passive prophylaxis with vaccination, since binding of AM14 would not block elicitation of antibodies against site Ø. Due to its unique specificity, AM14 will also be valuable for probing the conformation of RSV F-based vaccine antigens designed to be in the furin-cleaved trimeric prefusion conformation.
Introduction
Respiratory syncytial virus (RSV) is a ubiquitous paramyxovirus that infects nearly all children in the U.S. by two years of age [1]. In infants and young children RSV can cause acute lower respiratory tract infections, leading to bronchiolitis and pneumonia. In 2010, RSV was estimated to cause the deaths of more than 200,000 children, accounting for 2.3% of neonatal and 6.7% of infant deaths worldwide [2]. Although infant mortality in the U.S. due to RSV is low, the cost of hospital care for infected infants is estimated to be as high as $750 million per year [3, 4]. Prophylaxis with the humanized monoclonal antibody palivizumab (Synagis) is the only viable intervention for RSV but is limited to use in high-risk infants due to its cost and modest efficacy [5–7]. The development of very potent antibodies or an efficacious vaccine would bring protection to more children and reduce the financial burden of RSV.
Most RSV vaccine candidates contain at least one of the two viral surface glycoproteins: the fusion protein (F) and the attachment protein (G). Of these, only F is absolutely required for infection [8], and it is the target of palivizumab as well as the majority of neutralizing activity in human sera [9–12]. RSV F is a class I fusion protein that is initially synthesized as an inactive precursor. Proteolysis by a furin-like protease at two sites liberates a 27-amino-acid glycopeptide [13–15]. The N- and C-terminal polypeptides, F2 and F1, respectively, are connected by two disulfide bonds to form a single protomer [16–18]. Although three protomers eventually associate to form the mature trimeric F protein, the order and timing of F protein cleavage and trimerization are unknown.
To facilitate virus entry, the mature F protein is triggered to undergo a transition from a metastable prefusion conformation to a stable postfusion conformation, resulting in fusion of the viral and host-cell membranes. Crystal structures of both prefusion and postfusion F have recently been solved, providing molecular insight into this dramatic structural rearrangement [17–19]. Upon triggering, multiple secondary structure elements in the F1 N-terminus assemble into a long alpha helix that extends toward the target cell. This reorganization pulls the hydrophobic fusion peptide from the interior of the trimer and thrusts it into the cellular membrane, resulting in a prehairpin intermediate. The F1 C-terminus then migrates toward the N-terminus, irreversibly forming a six-helix bundle and driving fusion of the two membranes [20]. This rearrangement of F can also occur spontaneously, resulting in an abundance of postfusion F on the viral membrane [21].
RSV-neutralizing antibodies that bind both the pre- and postfusion conformations of F were the first to be isolated. These include antibodies 101F and the murine precursor of palivizumab, antibody 1129 [9, 10, 18, 22]. More recently, extremely potent antibodies that specifically target prefusion F have been characterized [19, 23, 24]. Three such antibodies—D25, AM22 and 5C4—all bind to the apex of the prefusion trimer at antigenic site Ø, which is dramatically reorganized during fusion [19]. Antibody MPE8, which cross-neutralizes four pneumoviruses, competes with palivizumab, yet preferentially recognizes the prefusion conformation [25]. In addition to their potential for passive prophylaxis, these antibodies were critical to the design of vaccine antigens stabilized in the prefusion conformation, which were shown to elicit much higher neutralizing activity than postfusion F in mice and rhesus macaques [26].
Although site Ø antibodies were originally thought to recognize quaternary epitopes on the prefusion trimer, they have subsequently been shown to react with monomers [27]. Here we characterize AM14, a potent RSV neutralizing antibody that was previously isolated from human PBMCs [23], and show that it recognizes a novel quaternary epitope on the native trimeric prefusion conformation of RSV F.
Results
AM14 potently neutralizes RSV
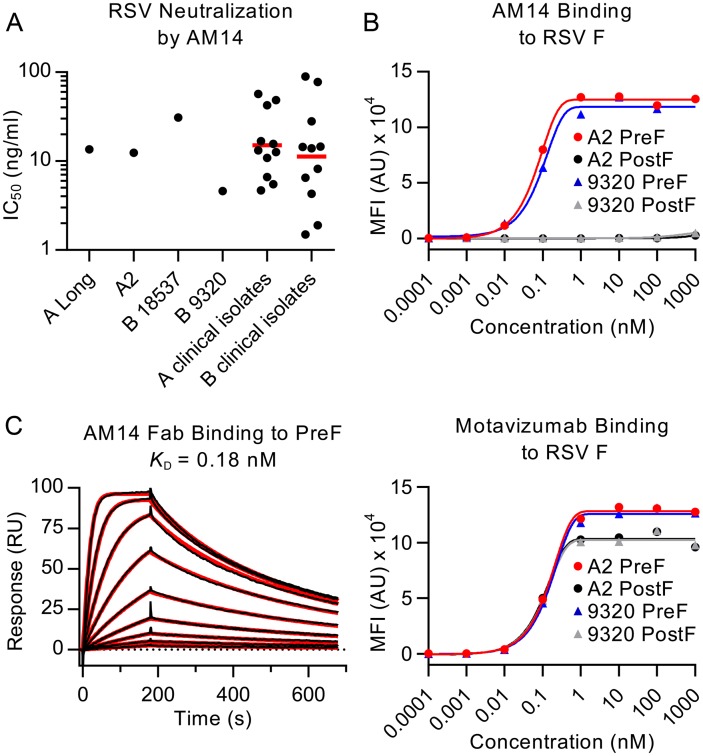
Microneutralization assays were performed to test the ability of AM14 to neutralize infection of HEp-2 cells by various RSV strains. AM14 potently neutralized all RSV strains tested, with IC50s of 13.6 ng/ml for strain A Long, 12.4 ng/ml for strain A2, 30.8 ng/ml for subtype B strain 18537 and 4.6 ng/ml for subtype B strain 9320 (Fig 1A). For comparison, palivizumab neutralized these strains with IC50s of 300 ng/ml, 320 ng/ml, 380 ng/ml and 120 ng/ml, respectively (S1A Fig). Clinical RSV isolates were also tested in the microneutralization assay (Fig 1A). AM14 neutralized subtype A clinical strains with a geometric mean IC50 of 15.1 ng/ml and a range of 4.7–56.9 ng/ml. Neutralization of subtype B clinical strains was similar, with a geometric mean IC50 of 11.3 ng/ml and a range of 1.5–89.2 ng/ml. The results obtained in this HEp-2 cell-based assay were similar to those previously reported for neutralization of strain A2 in a Vero cell-based assay (IC50 values of 2.1 ng/ml for AM14 and D25, and 209 ng/ml for palivizumab) [23, 28]. Similar to other prefusion-specific neutralizing antibodies tested previously, AM14 did not inhibit attachment of RSV to the surface of HEp-2 cells (S1B Fig) [19], suggesting that it prevents entry by blocking a step downstream of attachment. Collectively, these results demonstrate that AM14 is a potent neutralizer of RSV infection, capable of neutralizing both A and B subtypes equally well by a mechanism independent of viral attachment to the cell surface.
Fig 1. AM14 is a prefusion-specific neutralizing antibody.
(A) Neutralization of laboratory strains and clinical isolates of RSV. Red bars are geometric means. (B) Binding of AM14 and motavizumab IgGs to immobilized RSV F proteins was measured using a Luminex system. (C) Binding of AM14 Fab to immobilized prefusion RSV F was measured by surface plasmon resonance. Best fit of the data to a 1:1 binding model is shown in red.
AM14 is specific for prefusion RSV F
Since the neutralization potency of AM14 was similar to that of the prefusion-specific antibody D25, we hypothesized that AM14 might also exclusively recognize the prefusion conformation. To test this possibility, we performed a Luminex-based binding assay using furin-cleaved RSV F ectodomains stabilized in the prefusion (DS-Cav1) or postfusion (F ΔFP) conformation [18, 26]. In this experiment, AM14 bound tightly to prefusion RSV F derived from A and B subtypes with EC50s of 0.63 nM and 0.18 nM, respectively (Fig 1B). In contrast, no binding to furin-cleaved postfusion F was detected. Motavizumab, which binds equally well to both RSV F conformations [18, 19], recognized all proteins tested in this assay, confirming the immobilization of RSV F proteins to the beads (Fig 1B).
To further characterize the binding of AM14 to prefusion RSV F, surface plasmon resonance experiments were performed (Fig 1C). AM14 Fab bound to immobilized prefusion RSV F with an equilibrium dissociation constant (K D) of 0.18 nM, with rapid association and dissociation rate constants of 1.87 x 107 M-1s-1 and 3.4 x 10-3 s-1, respectively. This is in contrast to D25, which bound prefusion RSV F with association and dissociation rate constants more than 10- and 30-fold slower, respectively (1.35 x 106 M-1s-1 and 9.65 x 10-5 s-1) (S2A Fig). Therefore, although AM14 binds specifically to prefusion RSV F with sub-nanomolar affinity, its kinetics are much faster than those of other potent antibodies such as D25.
Structure determination of AM14 in complex with prefusion F
To identify the epitope on prefusion RSV F recognized by AM14, the crystal structure of AM14 alone and in complex with prefusion F was determined. Crystals of AM14 Fab in space group P212121 diffracted X-rays to 2.0 Å, and after a molecular replacement solution was obtained, the structure was built and refined to an R work/R free of 18.6%/22.6% (S3 Fig). To obtain a structure of AM14 Fab bound to RSV F, a matrix of F proteins and Fabs was screened, including ten different protein complexes, eight of which formed crystals. Only four of those diffracted past 8 Å (S1 Table). Prefusion RSV F complexed with both AM14 and motavizumab Fabs formed rod-like crystals in spacegroup P21 that diffracted X-rays to 5.5 Å. The asymmetric unit was composed of one prefusion F trimer, three AM14 Fabs and three motavizumab Fabs (S4 Fig). The structure was refined in Phenix with NCS torsion restraints and reference-model restraints to an R work/R free of 21.1%/27.7%. Secondary structures and connecting loops fit the electron density well, particularly at the antibody–F protein interfaces (S5 Fig). Additionally, the electrostatic potential of the interface showed substantial charge complementarity, with a positively charged region on RSV F interacting with negatively charged residues in the complementarity-determining regions (CDRs) of the AM14 Fab (S6 Fig).
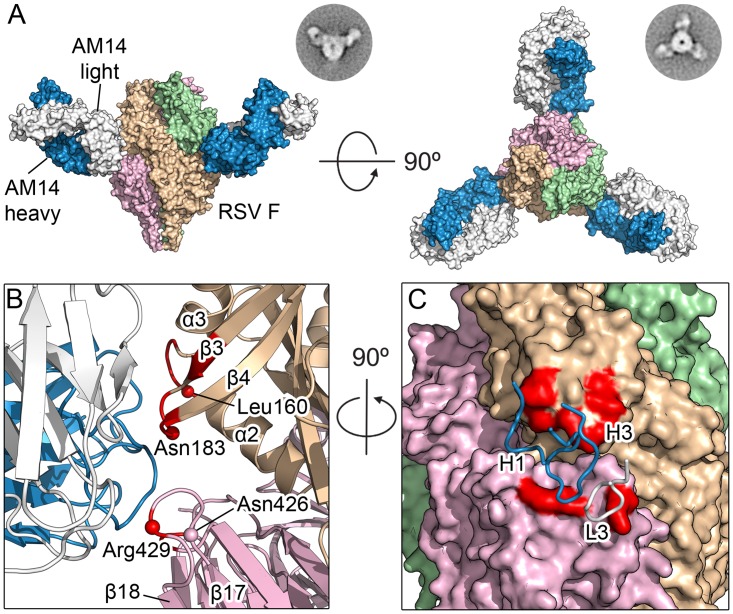
The orientation of the three AM14 Fabs was largely similar to that observed in negative-stain EM images of prefusion RSV F in complex with AM14, with the exception of the side-view, in which the apex of the trimer is not visible (Fig 2A). This difference is likely due to a combination of the trimer apex protruding from the stain and averaging of slightly different tilts of the complex. The crystal structure shows that AM14 binds at the junction of two protomers within the RSV F trimer (Fig 2B), with approximately 330 Å2 buried on the membrane-distal protomer and 520 Å2 buried on the membrane-proximal protomer. The heavy chain CDRs 1 and 3 make contact with RSV F, as does the light chain CDR 3 (Fig 2C). The AM14 contacts are localized to three regions of the RSV F primary sequence that fold together in the quaternary structure of the prefusion trimer. The first two regions map to the ends of two loops connecting α2 with α3 and β3 with β4, both of which undergo dramatic conformational changes in the pre- to postfusion transition, moving by nearly 100 Å [19]. These two regions together with α4 of antigenic site Ø form the continuous α5 helix of postfusion F, which forms the inner heptad repeat (HRA) of the six-helix bundle, explaining the prefusion-specificity observed for AM14 [19]. The third region of the epitope, located on an adjacent protomer, maps to the loop connecting β17 with β18, which partially overlaps with antigenic site IV and is in a similar conformation in pre- and postfusion structures [18, 19, 22]. Collectively, these data provide a structural basis for the prefusion specificity of AM14, and predict that AM14 is trimer-specific.
Fig 2. Structures of AM14 in complex with RSV F.
(A) Crystal structure of three AM14 Fabs bound to the prefusion RSV F trimer, viewed from the side and the top of RSV F. Negatively stained EM class averages that may correspond to each view are shown in the upper right. AM14 heavy chain is blue, light chain is white, and RSV F protomers are tan, light green and pink (B) Close-up of the side view, colored as in (A). Prefusion RSV F residues with Cα atoms within 8 Å of AM14 Fab Cα atoms are colored red, and Cα atoms of resistance mutations are shown as spheres. (C) Ninety-degree rotation of the view in (B), showing a molecular surface representation of RSV F and the location of the three AM14 CDR loops that contact F. The binding surface spans the two neighboring protomers (tan and pink).
AM14 MARMs map to the three loops identified in the crystal structure
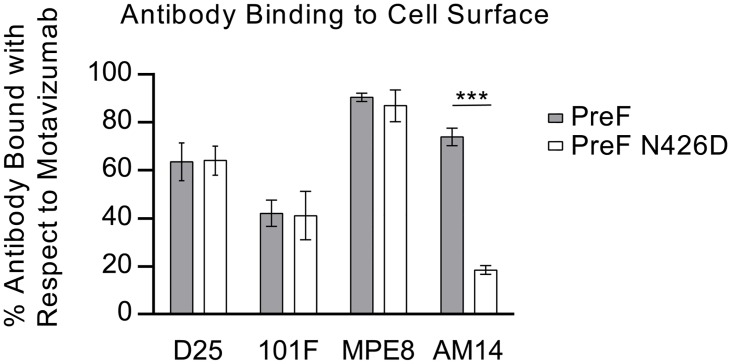
Due to the low resolution of the X-ray crystal structure, we sought to verify the AM14 epitope and identify critical interactions by generating monoclonal antibody-resistant mutants (MARMs). After three rounds of selection in HEp-2 cells, four unique AM14-escape viruses were isolated and sequenced. Three of the viruses each contained a single mutation in RSV F (L160S, N183K, or N426D), whereas the fourth virus contained three mutations in RSV F (I79M/R429S/H515N) (Table 1). Resistance of viruses to neutralization by AM14 was confirmed by microneutralization assays (S7 Fig). For one MARM, N426D, a full-length prefusion F variant was generated and expressed on the surface of HEK293 cells. Binding of AM14 IgG to these cells was reduced approximately four-fold compared to cells expressing wild-type prefusion F (Fig 3). Consistent with this result, binding of AM14 IgG to purified prefusion F N426D was reduced by approximately 100-fold in an ELISA (S8 Fig). Binding of antibody 101F to prefusion F N426D was also slightly reduced when measured by ELISA, which is not surprising given that the 101F epitope (residues 427–437) is close to this region [22]. The binding of motavizumab, MPE8 and D25 was not affected by the N426D mutation, consistent with the known locations of their epitopes.
Table 1. Amino acid changes in F protein of AM14 MARMs.
| Amino acid sequence (position number) | |||||||
|---|---|---|---|---|---|---|---|
| 79 | 160 | 183 | 426 | 429 | 514 | 515 | |
| RSV A Long | I | L | N | N | R | H | H |
| MARM 14 p3 | M | - | - | - | S | - | N |
| MARM 20 p3 | - | S | - | - | - | - | - |
| MARM 21 p3 | - | - | - | D | - | - | - |
| MARM 57 p3 | - | - | K | - | - | - | - |
Fig 3. N426D disrupts binding of AM14 to prefusion F.
Relative binding of D25, 101F, MPE8 and AM14 to cell surface-expressed prefusion F (grey) or prefusion F containing the AM14 escape mutation, N426D (white) was measured by flow cytometry. Data were normalized to motavizumab binding. Binding of D25, 101F and MPE8 to N426D was comparable to wild-type prefusion F, whereas AM14 binding to N426D was reduced four-fold (n = 3, P < 0.001; Tukey’s HSD, error bars show standard deviation).
Mapping of the MARMs on the prefusion F structure revealed that L160S and N183K are located on the loops connecting α2 with α3 and β3 with β4, respectively, in agreement with the crystal structure (Fig 2B). This verifies that AM14 makes a substantial interaction with the prefusion-specific region of the membrane-distal protomer. Two other MARMS, N426D and R429S, are located on the loop connecting β17 with β18 in the adjacent protomer, near antigenic site IV. The isolation of these MARMs further supports the low-resolution crystal structure and indicates that the membrane-proximal protomer is a critical component of the AM14 epitope. The final two MARMS, I79M and H515N, were part of the same RSV F variant harboring R429S, and are likely compensatory mutations since they are located more than 30 and 80 Å away from the AM14 epitope, respectively.
AM14 specifically recognizes cleaved, trimeric, prefusion F
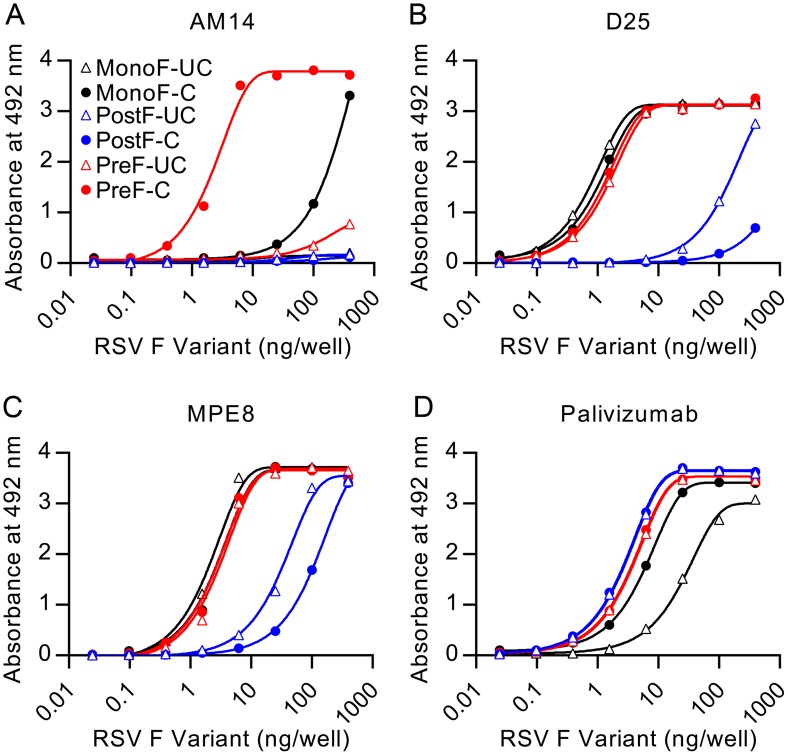
The crystal structure and MARMs suggested that AM14 is a quaternary-specific antibody with an epitope spanning two protomers. To test the quaternary specificity, we measured binding of immobilized AM14 to monomeric F and trimeric prefusion F by ELISA (Fig 4). The monomeric F was composed of RSV F residues 1–524 and lacked the foldon trimerization motif. Consistent with the hypothesized quaternary epitope, AM14 binding to monomeric F was reduced by nearly 100-fold compared to the prefusion F trimer (Fig 4A). The residual monomer-binding activity may be due to transient trimerization at high concentrations, which would be indistinguishable from monomeric F binding in this assay.
Fig 4. AM14 is specific for cleaved, trimeric RSV F.
Binding of antibodies (A) AM14, (B) D25, (C) MPE8, or (D) palivizumab to uncleaved monomeric RSV F (open black triangles), cleaved monomeric RSV F (black circles), uncleaved postfusion RSV F (open blue triangles), cleaved postfusion RSV F (blue circles), uncleaved prefusion RSV F (open red triangles) and cleaved prefusion RSV F (red circles) was measured by ELISA.
Since previously described quaternary-specific antibodies against other class I fusion proteins have in some cases shown preference for the mature protease-cleaved fusion protein over the uncleaved protein [29], we sought to determine if cleavage of RSV F was required for AM14 binding. In these assays, AM14 failed to bind prefusion F with mutated furin sites (Fig 4A). In contrast, both D25 and MPE8 (Fig 4B and 4C, respectively) bound to cleaved and uncleaved monomeric and trimeric F proteins with profiles similar to palivizumab, an antibody not expected to have preference for the conformation, cleavage or trimerization of RSV F (Fig 4D). Although D25 was originally described as a quaternary-specific antibody, 90% of its epitope on prefusion F is located on a single protomer, explaining its ability to bind monomeric F (Fig 4B and [19]) and a peptide spanning RSV F residues 153–211 (S2B Fig). The approximately 30-fold decrease in affinity observed for D25 binding to peptide is likely due to the absence of contact residues in the F2 subunit and the neighboring protomer, as well as differences in peptide structure compared to the complete prefusion F. Collectively, these results demonstrate that AM14 is unique in its ability to discriminate cleaved, trimeric, prefusion F from the other forms tested.
Binding of AM14 is sufficient to stabilize RSV F ectodomain in the trimeric prefusion conformation
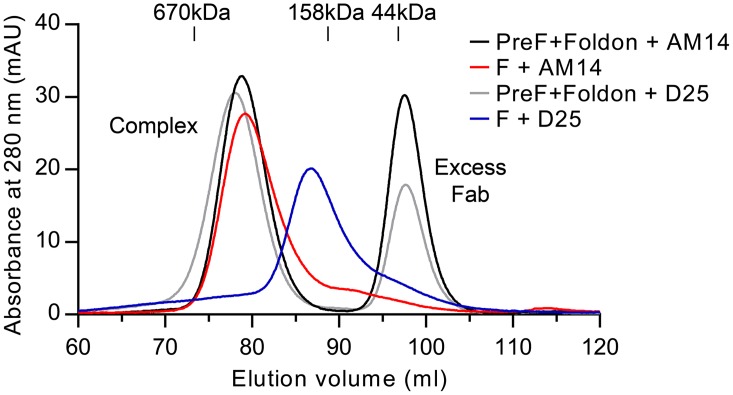
Production of soluble prefusion RSV F has thus far relied on the presence of the foldon motif at the C-terminus of F1 to stabilize the weak interprotomer interactions that are normally formed when RSV F is localized in the membrane [19, 26]. Having demonstrated that AM14 is a trimer-specific antibody, we sought to determine if the binding of AM14 was sufficient to stabilize trimeric prefusion F in the absence of a trimerization motif or stabilizing mutations. Since D25 exhibits some degree of interprotomer binding, we also tested this antibody. For this experiment, we first determined the gel filtration elution volumes for purified prefusion, trimeric RSV F (DS-Cav1) incubated with excess Fab. These were then compared to complexes formed by co-expression of each Fab with RSV F ectodomain lacking foldon and stabilizing mutations. Co-expression of AM14 Fab and the F ectodomain resulted in a complex with an elution profile very similar to that of AM14 complexed with DS-Cav1 (Fig 5, compare red and black traces). In contrast, co-expression of D25 and F ectodomain resulted in a complex that eluted later (i.e., smaller) than the corresponding complex of D25 with DS-Cav1 (Fig 5, compare blue and grey traces). Thus, D25 co-expression was not sufficient to stabilize the formation of prefusion F trimer in the absence of a trimerization motif. This property appears to be unique to AM14 due to its epitope being split evenly between two protomers.
Fig 5. AM14 stabilizes RSV F trimer in the absence of the foldon trimerization motif.
Size-exclusion chromatography profiles from a Superose 6 column are shown for AM14 Fab or D25 Fab complexed with prefusion RSV F containing the foldon trimerization motif (black and grey, respectively) and for AM14 Fab or D25 Fab co-expressed with RSV F ectodomain without foldon (red and blue, respectively).
Discussion
AM14 neutralization of RSV is similar in potency to that of the antigenic site Ø antibody D25 [19, 23]. It was originally hypothesized that site Ø-directed antibodies would be the most potent neutralizers due to the location of the epitope on the accessible apex of the prefusion F trimer [19]. Consistent with this hypothesis, the prefusion-specific antibody MPE8, which binds to a lower region on prefusion F, has decreased neutralization potency compared to D25 [25]. In contrast, the potency of AM14 was similar to D25, despite the equatorial binding observed in both the crystal structure and negative stain EM. This demonstrates that antibodies targeting other regions of prefusion F can be as potent as those binding to the apex, which may be an important consideration when optimizing RSV vaccine antigens. In addition, the location of this epitope could make AM14 a candidate for passive prophylaxis. AM14 would have the advantage of high potency and would not block site Ø, leaving this antigenic supersite accessible for inducing protective antibody responses induced by vaccination or infection of the upper airway.
In our ELISAs, prefusion-specific antibodies D25 and MPE8 both bound to uncleaved postfusion F, albeit with low affinity. One possible explanation is that the presence of pep27 in uncleaved postfusion F leads to a more flexible state in which the association of HRA and HRB is weaker and portions of these epitopes that are normally occluded in the cleaved postfusion state are accessible. This hypothesis is supported by the finding that the six-helix bundle-directed antibody 114F bound tighter to cleaved postfusion F (S9 Fig). It is further supported by the reduced SDS-stability of uncleaved compared to cleaved postfusion F trimers. Although uncleaved postfusion F is not biologically relevant, there are a number of subunit vaccines in development that are based upon uncleaved F. Our data suggest that for these proteins in particular, D25 and MPE8 may not be good indicators of prefusion F.
Besides AM14, other antibodies have been identified that recognize higher-order protein structures in both class I and class II viral fusion proteins. The specificity of these antibodies can be grouped into at least three categories. In the first, antibodies contact only one protomer, but recognize a conformation that exists only in the assembled fusion protein, as is the case for the HIV antibody 35O22, which binds to both gp120 and gp41 subunits [30]. This type of antibody has also been proposed for influenza HA antibodies [31]. In the second category, antibodies make direct contact with more than one protomer. This quaternary-specific binding is observed for AM14 and has previously been demonstrated for the influenza antibody HC63 and the HIV antibody PGT151 [29, 32–34]. Additionally, dengue virus antibodies have been identified that recognize both protomers within one dimer of the class II fusion protein E [35]. Interestingly, these antibodies have been reported to shift the monomer–dimer equilibrium, similar to the stabilization of trimeric RSV F that we observed here for AM14 [35]. A third category of higher-order specificity can exist due to the ordered array of glycoproteins on the surface of a virus, as has been described for class II fusion proteins of West Nile virus [36]. Antibody CR4354 binds across two E protein dimers, preventing the rearrangement of E into trimers after exposure to low pH [36]. Thus, the mode of AM14 binding is but one way in which antibodies are able to recognize higher-order structures in class I and II viral fusion proteins.
In addition to quaternary-specific binding, AM14 also exhibited a unique dependence on furin cleavage. RSV F is distinct from the F proteins of other paramyxoviruses due to the presence of two furin sites separated by a glycosylated 27 amino acid spacer, pep27, which is released from the protein after cleavage [15]. There are two likely explanations for the cleavage-dependence of AM14 binding. The first is that pep27 sterically inhibits binding of AM14 to the prefusion trimer. An alternative is that uncleaved F does not adopt the native trimeric state. This would be in contrast to the related paramyxovirus PIV5 F protein, which contains only one cleavage site, since the structures of the cleaved and uncleaved PIV5 prefusion F proteins are nearly identical [37, 38]. Future work on the structure of uncleaved RSV F will be needed to resolve these two possibilities.
The specificity of the potent RSV neutralizing antibody AM14 makes it a useful reagent for probing or isolating the cleaved trimeric state of prefusion F. AM14 may also allow an unparalleled view of prefusion F in its native state, as this antibody can be used to capture trimeric prefusion F without the use of a trimerization motif or stabilizing mutations, as was done for HIV Env using the recently identified trimer-specific antibody PGT151 [29]. AM14 can also be used to help unravel the role of furin cleavage in prefusion F trimerization and to track the order of these events in the secretory pathway, similar to what has been done using antibodies specific for trimeric influenza HA [39, 40]. Further, the high potency and properties of AM14 described here could make it well suited for passive prophylaxis.
Materials and Methods
Production of RSV F proteins
Plasmids encoding RSV F prefusion (DS-Cav1) and postfusion (F ΔFP) proteins based on strain A2 [18, 26] were transfected into FreeStyle 293-F cells (Invitrogen). Uncleaved versions of prefusion and postfusion F proteins were produced by changing the basic residues of the two furin-cleavage sites to asparagine residues using site-directed mutagenesis. Proteins were purified from the media using Ni-NTA Superflow resin (Qiagen) and Strep-Tactin resin (IBA). Tags were removed by digestion with thrombin or HRV3C protease, followed by gel filtration using a Superose 6 column (GE Healthcare Biosciences). For crystallization, RSV F proteins were expressed in the presence of kifunensine (5 μM), digested with Endo H (10% w/w), mixed with a 1.5-fold molar excess of Fab, and purified using the Superose 6 column. AviTagged F proteins were biotinylated with biotin ligase BirA (Avidity) and separated from excess biotin by gel filtration with a Superdex 200 column (GE).
Production of IgGs and Fabs
Plasmids encoding antibody heavy and light chains were transfected into Expi293 cells (Invitrogen). IgGs and Fabs were purified using Protein A agarose (Fisher) or CaptureSelect IgG-CH1 affinity matrix (Life Technologies), respectively.
Luminex binding assay
Biotinylated proteins were coupled to avidin-coated MagPlex beads at 1 μg per 50,000 beads (Radix). Approximately 1000 beads per well were incubated with 10-fold serial dilutions (1 μM to 0.1 pM) of each antibody in a 384-well plate, washed with PBS plus 0.1% BSA with 0.05% Tween 20 and incubated with 0.33 μg/ml phycoerythrin (PE)-conjugated mouse anti-human IgG Fc secondary antibody (Southern Biotech). Beads were washed and emission at 575 nm was measured using the FLEXMAP 3D flow cytometer (Luminex).
Surface plasmon resonance
Biotinylated DS-Cav1 was immobilized on an SA sensor chip to a total of 292 response units using a Biacore X100 (GE). A buffer-only sample was injected over the DS-Cav1 and reference flow cells, followed by AM14 Fab 2-fold serially diluted from 5 nM to 19.5 pM in HBS-EP+, with a duplication of the 156 pM concentration. The data were double-reference subtracted and fit to a 1:1 binding model using the Biacore X100 analysis software.
Negative-stain electron microscopy
Samples were diluted to approximately 0.03 mg/ml, adsorbed to a freshly glow-discharged carbon-film grid for 15 sec, and stained with 0.7% uranyl formate. Images were collected semi-automatically using SerialEM [41] on a FEI Tecnai T20 with a 2k x 2k Eagle CCD camera at a pixel size of 0.22 nm/px. Particles were picked automatically and reference-free 2D classification was performed in EMAN2 [42].
Crystallization and data collection
AM14 Fab crystals were produced by hanging-drop vapor diffusion by mixing 1 μl of AM14 Fab (8.7 mg/ml) with 1 μl of reservoir solution containing 0.1 M sodium acetate pH 4.5, 0.2 M ammonium sulfate and 25% (w/v) PEG 4000. Crystals were soaked in reservoir solution supplemented with 30% (v/v) ethylene glycol and frozen in liquid nitrogen. Data to 2.0 Å were collected at the MacCHESS beamline (Cornell High Energy Synchrotron Source, Cornell University).
The ternary complex was produced by mixing Endo H-treated DS-Cav1 with a 1.5-fold molar excess each of AM14 Fab and motavizumab Fab before separation of the complex from excess Fab by gel filtration. Crystals were produced by hanging-drop vapor diffusion by mixing 0.67 μl of protein (5.6 mg/ml) with 1.33 μl of reservoir solution containing 11.4% (w/v) PEG 8000, 1.7% (v/v) 2-methyl-2,4-pentanediol and 0.1 M imidazole pH 6.5. Many cryopreservation solutions were tested, but diffraction was highest when the crystal was directly plunged into liquid nitrogen after removal of the cold gas layer [43]. Diffraction data were collected to 5.5 Å at the SBC beamline 19-ID (Advanced Photon Source, Argonne National Laboratory).
Structure determination, model building and refinement
Diffraction data were processed using the CCP4 software suite: data were indexed and integrated in iMOSFLM [44] and scaled and merged with AIMLESS [45]. A molecular replacement solution for the 2.0 Å AM14 Fab dataset was found by PHASER [46] using the heavy and light chains of PDB ID: 4ERS [47] and PDB ID: 4JHA [19], respectively, as search models. The structure was built manually in COOT [48] and refined using PHENIX [49].
A molecular replacement solution for the 5.5 Å ternary complex was obtained using PHASER with prefusion RSV F (PDB ID: 4JHW [19]), motavizumab Fab (PDB ID: 3IXT [11]) and the 2.0 Å AM14 Fab structures as search models. The asymmetric unit contained the prefusion trimer bound by three motavizumab Fabs and three AM14 Fabs. Rigid-body refinement was then performed in PHENIX, and several of the Fab constant domains were manually placed into the electron density using COOT, followed by another round of rigid-body refinement in PHENIX. Group B-factors and coordinates were refined in PHENIX with NCS torsion restraints and reference-model restraints. The reference models were the 2.4 Å prefusion F structure (PDB ID: 4MMS [26]), the 2.75 Å motavizumab Fab structure (PDB ID: 3IXT [11]), and the 2.0 Å AM14 Fab structure determined here. Data collection and refinement statistics for both structures are presented in S2 Table.
Isolation of MARMs and neutralization assay
RSV strain Long was incubated at 5 x 106 pfu/ml with 3 μg/ml of AM14 for 1 hr prior to infection of confluent HEp2 cells at an MOI of 1.0. Following a 2–3 hour infection, viral inoculum was removed and medium containing 3 μg/ml of antibody was added and incubated at 37°C for 5–7 days. Virus was harvested from wells containing cytopathic effect during the first round of selection and subjected to an additional 2 rounds of selection at 10 μg/ml. Following each round of selection, RNA was isolated from virally infected cells and analyzed by sequencing to determine F protein sequences. After the third round of selection, viruses were plaque purified and microneutralization assays were performed in HEp-2 cells as previously described [50].
Flow cytometric analysis of prefusion F and N426D variant
A plasmid encoding RSV F residues 1–574 and harboring the DS-Cav1 stabilizing mutations was created, along with an N426D variant, for expression on the cell surface. Expi293 cells were transfected, harvested 48 hours post-transfection, and washed twice with PBS, followed by incubation with RSV F-specific antibodies (1 μg/ml) for 1 hour at 4°C and Alexa488-conjugated goat anti-human secondary antibody (Invitrogen) (5 μg/ml) for 30 minutes at 4°C. Cells were washed, fixed with 0.5% paraformaldehyde, and evaluated by flow cytometry (LSR II instrument, Becton Dickinson). Data were analyzed using FlowJo software (Tree Star) and GraphPad Prism.
RSV F ELISA
96-well plates were coated with purified monoclonal antibody (AM14, D25, MPE8, palivizumab or 114F) at 6 μg/ml in PBS. Plates were blocked with 2% pig serum in PBS with 0.05% Tween 20 and washed with water. Purified F proteins were serially diluted 4-fold (8 μg/ml to 0.49 ng/ml) and added to plates, which were then washed before incubation with biotinylated anti-HisTag antibody (0.3 μg/ml) (Bio-Rad) and streptavidin-HRP (1:2000) (GE). After addition of o-phenylenediamine dihydrochloride (Sigma), reactions were stopped with 2 N sulfuric acid and absorbance was read at 490 nm.
Analysis of prefusion F complexes by size exclusion chromatography
RSV F ectodomain consisting of residues 1–513 with a C-terminal thrombin cleavage site, 6x His-tag and Strep-tag II was co-expressed with either AM14 or D25 Fab in FreeStyle 293-F cells. Complexes were purified using Ni-NTA and Strep-Tactin resins. Separately, DS-Cav1 was produced as described above and mixed with a 1.5-fold molar excess of either AM14 or D25 Fab prior to analysis. Tags were removed from both complexes by thrombin digestion before gel filtration using a Superose 6 XK 16/70 column (GE).
Supporting Information
(A) Neutralization of RSV strains A2, Long, 18537 and 9320 were measured using a microneutralization assay. Two-fold serial dilutions of AM14 (red) or palivizumab (black) were incubated with RSV before infection of HEp-2 cells and detection of RSV F on the surface of infected cells by ELISA. In all strains tested, AM14 was greater than one log more potent than palivizumab. (B) A flow cytometry-based attachment assay was modified from a previously described adherent cell assay [22, 51]. Antibodies and heparin were four-fold serially diluted and incubated with RSV A2 prior to attachment to HEp-2 cells and detection of RSV F on the surface of cells by flow cytometry. Although heparin (grey), a known attachment inhibitor, prevented binding of RSV to the cell surface, palivizumab (black) and AM14 (red) did not.
(TIF)
(A) Binding of D25 Fab to prefusion RSV F was measured by surface plasmon resonance. Prefusion RSV F was immobilized on an SA chip to a total of 300 RU and binding to D25 Fab was measured in 2-fold serial dilutions from 10 to 0.04 nM, with a duplicate of the 0.31 nM concentration. Best fit of the data to a 1:1 binding model is shown in red. D25 Fab bound to immobilized prefusion RSV F with an equilibrium dissociation constant (K D) of 0.07 nM, with association and dissociation rate constants of 1.35 x 106 M-1s-1 and 9.65 x 10-5 s-1, respectively. (B) Binding of D25 to HRA peptide was measured by biolayer interferometry. His-Tagged HRA peptide (RSV A2 F residues 153–211) was immobilized to anti-His sensors and two-fold serial dilutions of D25 IgG from 50 nM to 0.8 nM were assessed for binding on an Octet QK. Red lines are the fit of a global association and then dissociation algorithm in GraphPad Prism, from which the 0.8 nM concentration was excluded. This algorithm was used to calculate an equilibrium dissociation constant (K D) of 1.9 nM for D25 IgG binding to HRA peptide.
(TIF)
Structure of the AM14 Fab with heavy chain shown in blue and light chain shown in white. Complementarity-determining regions (CDRs) are labeled for both the heavy and light chains. The CDR H3 loops of the two Fabs in the asymmetric unit were in different conformations, the second of which is shown in purple. The dotted line indicates a region that was not modeled due to poor electron density. For the complex with RSV F, the Fab with the ordered CDRH3 was used as the search model.
(TIF)
The asymmetric unit of the ternary complex was composed of one RSV F trimer, three AM14 Fabs and three motavizumab Fabs. The side view and top view of the complex are shown, with RSV F protomers colored tan, light pink and light green. AM14 heavy chains are blue, motavizumab heavy chains are magenta and both AM14 and motavizumab light chains are white.
(TIF)
The main chains are shown as ribbons within the 2Fo—Fc electron density maps for the refined structure (blue), contoured at 1σ. Only one AM14 Fab and the RSV F trimer are shown for clarity. Chains are colored as in S4 Fig. The side view and top view are shown, with zoomed views for the interface between AM14 and the F trimer. The CDR H3 is labeled for both views and fits the electron density well.
(TIF)
Molecular surface representations of prefusion F and AM14 Fab colored according to electrostatic surface potential (red to blue, -8.4 to +8.4 kT/e, respectively). The surfaces are shown in an open-book representation of the binding interface with the approximate footprint for prefusion F and AM14 Fab outlined in yellow and black. A negatively charged region on the AM14 Fab complements a positively charged region on the lower protomer of prefusion F.
(TIF)
Two-fold serial dilutions of AM14 (red) or palivizumab (black) were incubated with (A) RSV A Long I79M/R429S/H515N, (B) L160S or (C) N183K before infection of HEp-2 cells and detection of RSV F on the surface of infected cells by ELISA. (D) In a separate assay, two-fold serial dilutions of AM14 were incubated with mKate RSV A2 (grey) or mKate RSV A2 N426D (blue) before infection of HEp-2 cells and measurement of fluorescence (excitation of 588 nm and emission at 635 nm) with a plate reader as previously described [52]. All MARMs were neutralized by palivizumab but not by AM14.
(TIF)
Plates were coated with (A) stabilized prefusion F or (B) the N426D mutant (1 μg/ml each) and were washed, blocked and incubated with four-fold serial dilutions of motavizumab (black), D25 (blue), 101F (grey), MPE8 (open blue) and AM14 (red) before detection with HRP-conjugated goat anti-human IgG (Santa Cruz Biotechnology) and Super AquaBlue substrate (eBiosciences Inc.). An ELISA plate reader (Molecular Devices, Inc.) was used to read signal at 538 nm. Binding of motavizumab, MPE8 and D25 was similar for both proteins. 101F binding to N426D was slightly reduced, whereas AM14 binding to N426D was reduced approximately 100-fold compared to wild-type prefusion F.
(TIF)
(A) Coomassie-stained reducing SDS-PAGE gel of the six proteins used in the ELISAs here and in Fig 4. (B) Binding of 114F, an antibody specific for the six-helix bundle of postfusion F, to the uncleaved monomeric RSV F (open black triangles), cleaved monomeric RSV F (black circles), uncleaved prefusion RSV F (open red triangles), cleaved prefusion RSV F (red circles), uncleaved postfusion RSV F (open blue triangles) and cleaved postfusion RSV F (blue circle) was measured by ELISA. 114F bound more tightly to cleaved postfusion F than to uncleaved postfusion F.
(TIF)
(PDF)
(PDF)
Acknowledgments
The authors would like to thank Peter D Kwong for providing plasmids, Emilie Shipman for assistance with protein expression and purification, members of the McLellan lab for critical reading of the manuscript, the A1 beamline staff at CHESS, Cornell High Energy Sychrontron Source and the 19-ID beamline staff at the Structural Biology Center at the Advanced Photon Source, Argonne National Laboratory.
Data Availability
Atomic coordinates and structure factors for the structures of AM14 unbound and in complex with RSV F have been deposited in the Protein Data Bank under accession codes 4ZYK and 4ZYP, respectively.
Funding Statement
Data collection at CHESS is supported by the NSF & NIH/NIGMS via NSF award DMR-1332208, and the MacCHESS resource is supported by NIH/NIGMS award GM-103485. Results shown in this report were also derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. Work performed by UB was funded in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. NKP, KK, QZ and NDU were employed by MedImmune Inc, which funded the data collection and analysis of their experiments (neutralization assays, MARM isolation and affinity of D25 for peptide). JAM received funding from “Plan Nacional I+D+I” (Ministerio de Economía y Competitividad), grant SAF2012-31217. BSG was supported by funding from the intramural program of the National Institute of Allergy and Infectious Diseases. JSM received funding from the National Institutes of Health, grant 1R43AI112124. With the exception of MedImmune’s involvement stated above, the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Glezen WP, Taber LH, Frank AL, Kasel JA. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child. 1986;140(6): 543–546. [DOI] [PubMed] [Google Scholar]
- 2. Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859): 2095–2128. 10.1016/S0140-6736(12)61728-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Byington CL, Wilkes J, Korgenski K, Sheng X. Respiratory syncytial virus-associated mortality in hospitalized infants and young children. Pediatrics. 2015;135(1): e24–31. 10.1542/peds.2014-2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leader S, Kohlhase K. Recent trends in severe respiratory syncytial virus (RSV) among US infants, 1997 to 2000. J Pediatr. 2003;143(5 Suppl): S127–132. [DOI] [PubMed] [Google Scholar]
- 5. Homaira N, Rawlinson W, Snelling TL, Jaffe A. Effectiveness of Palivizumab in Preventing RSV Hospitalization in High Risk Children: A Real-World Perspective. Int J Pediatr. 2014;2014: 571609 10.1155/2014/571609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. The IMpact-RSV Study Group. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics. 1998;102(3 Pt 1): 531–537. [PubMed] [Google Scholar]
- 7. Kamal-Bahl S, Doshi J, Campbell J. Economic analyses of respiratory syncytial virus immunoprophylaxis in high-risk infants: a systematic review. Arch Pediatr Adolesc Med. 2002;156(10): 1034–1041. [DOI] [PubMed] [Google Scholar]
- 8. Karron RA, Buonagurio DA, Georgiu AF, Whitehead SS, Adamus JE, Clements-Mann ML, et al. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci U S A. 1997;94(25): 13961–13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beeler JA, van Wyke Coelingh K. Neutralization epitopes of the F glycoprotein of respiratory syncytial virus: effect of mutation upon fusion function. J Virol. 1989;63(7): 2941–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson S, Oliver C, Prince GA, Hemming VG, Pfarr DS, Wang SC, et al. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J Infect Dis. 1997;176(5): 1215–1224. [DOI] [PubMed] [Google Scholar]
- 11. McLellan JS, Chen M, Kim A, Yang Y, Graham BS, Kwong PD. Structural basis of respiratory syncytial virus neutralization by motavizumab. Nat Struct Mol Biol. 2010;17(2): 248–250. 10.1038/nsmb.1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sastre P, Melero JA, Garcia-Barreno B, Palomo C. Comparison of affinity chromatography and adsorption to vaccinia virus recombinant infected cells for depletion of antibodies directed against respiratory syncytial virus glycoproteins present in a human immunoglobulin preparation. J Med Virol. 2005;76(2): 248–255. [DOI] [PubMed] [Google Scholar]
- 13. Gonzalez-Reyes L, Ruiz-Arguello MB, Garcia-Barreno B, Calder L, Lopez JA, Albar JP, et al. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc Natl Acad Sci U S A. 2001;98(17): 9859–9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bolt G, Pedersen LO, Birkeslund HH. Cleavage of the respiratory syncytial virus fusion protein is required for its surface expression: role of furin. Virus Res. 2000;68(1): 25–33. [DOI] [PubMed] [Google Scholar]
- 15. Begona Ruiz-Arguello M, Gonzalez-Reyes L, Calder LJ, Palomo C, Martin D, Saiz MJ, et al. Effect of proteolytic processing at two distinct sites on shape and aggregation of an anchorless fusion protein of human respiratory syncytial virus and fate of the intervening segment. Virology. 2002;298(2): 317–326. [DOI] [PubMed] [Google Scholar]
- 16. Scheid A, Choppin PW. Two disulfide-linked polypeptide chains constitute the active F protein of paramyxoviruses. Virology. 1977;80(1): 54–66. [DOI] [PubMed] [Google Scholar]
- 17. Swanson KA, Settembre EC, Shaw CA, Dey AK, Rappuoli R, Mandl CW, et al. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc Natl Acad Sci U S A. 2011;108(23): 9619–9624. 10.1073/pnas.1106536108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McLellan JS, Yang Y, Graham BS, Kwong PD. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol. 2011;85(15): 7788–7796. 10.1128/JVI.00555-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340(6136): 1113–1117. 10.1126/science.1234914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McLellan JS, Ray WC, Peeples ME. Structure and function of respiratory syncytial virus surface glycoproteins. Curr Top Microbiol Immunol. 2013;372: 83–104. 10.1007/978-3-642-38919-1_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liljeroos L, Krzyzaniak MA, Helenius A, Butcher SJ. Architecture of respiratory syncytial virus revealed by electron cryotomography. Proc Natl Acad Sci U S A. 2013;110(27): 11133–11138. 10.1073/pnas.1309070110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McLellan JS, Chen M, Chang JS, Yang Y, Kim A, Graham BS, et al. Structure of a major antigenic site on the respiratory syncytial virus fusion glycoprotein in complex with neutralizing antibody 101F. J Virol. 2010;84(23): 12236–12244. 10.1128/JVI.01579-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kwakkenbos MJ, Diehl SA, Yasuda E, Bakker AQ, van Geelen CM, Lukens MV, et al. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat Med. 2010;16(1): 123–128. 10.1038/nm.2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Magro M, Mas V, Chappell K, Vazquez M, Cano O, Luque D, et al. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc Natl Acad Sci U S A. 2012;109(8): 3089–3094. 10.1073/pnas.1115941109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Corti D, Bianchi S, Vanzetta F, Minola A, Perez L, Agatic G, et al. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature. 2013;501(7467): 439–443. 10.1038/nature12442 [DOI] [PubMed] [Google Scholar]
- 26. McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GB, Yang Y, et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342(6158): 592–598. 10.1126/science.1243283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Swanson KA, Balabanis K, Xie Y, Aggarwal Y, Palomo C, Mas V, et al. A monomeric uncleaved respiratory syncytial virus F antigen retains prefusion-specific neutralizing epitopes. J Virol. 2014;88(20): 11802–11810. 10.1128/JVI.01225-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johnson S, Griego SD, Pfarr DS, Doyle ML, Woods R, Carlin D, et al. A direct comparison of the activities of two humanized respiratory syncytial virus monoclonal antibodies: MEDI-493 and RSHZl9. J Infect Dis. 1999;180(1): 35–40. [DOI] [PubMed] [Google Scholar]
- 29. Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Pena AT, et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40(5): 669–680. 10.1016/j.immuni.2014.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014;515(7525): 138–142. 10.1038/nature13601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Magadan JG, Khurana S, Das SR, Frank GM, Stevens J, Golding H, et al. Influenza A virus hemagglutinin trimerization completes monomer folding and antigenicity. J Virol. 2013;87(17): 9742–9753. 10.1128/JVI.00471-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, et al. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40(5): 657–668. 10.1016/j.immuni.2014.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gigant B, Barbey-Martin C, Bizebard T, Fleury D, Daniels R, Skehel JJ, et al. A neutralizing antibody Fab-influenza haemagglutinin complex with an unprecedented 2:1 stoichiometry: characterization and crystallization. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 8): 1067–1069. [DOI] [PubMed] [Google Scholar]
- 34. Barbey-Martin C, Gigant B, Bizebard T, Calder LJ, Wharton SA, Skehel JJ, et al. An antibody that prevents the hemagglutinin low pH fusogenic transition. Virology. 2002;294(1): 70–74. [DOI] [PubMed] [Google Scholar]
- 35. Rouvinski A, Guardado-Calvo P, Barba-Spaeth G, Duquerroy S, Vaney MC, Kikuti CM, et al. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. 2015;520(7545): 109–113. 10.1038/nature14130 [DOI] [PubMed] [Google Scholar]
- 36. Kaufmann B, Vogt MR, Goudsmit J, Holdaway HA, Aksyuk AA, Chipman PR, et al. Neutralization of West Nile virus by cross-linking of its surface proteins with Fab fragments of the human monoclonal antibody CR4354. Proc Natl Acad Sci U S A. 2010;107(44): 18950–18955. 10.1073/pnas.1011036107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439(7072): 38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Welch BD, Liu Y, Kors CA, Leser GP, Jardetzky TS, Lamb RA. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc Natl Acad Sci U S A. 2012;109(41): 16672–16677. 10.1073/pnas.1213802109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Copeland CS, Doms RW, Bolzau EM, Webster RG, Helenius A. Assembly of influenza hemagglutinin trimers and its role in intracellular transport. J Cell Biol. 1986;103(4): 1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yewdell JW, Yellen A, Bachi T. Monoclonal antibodies localize events in the folding, assembly, and intracellular transport of the influenza virus hemagglutinin glycoprotein. Cell. 1988;52(6): 843–852. [DOI] [PubMed] [Google Scholar]
- 41. Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1): 36–51. [DOI] [PubMed] [Google Scholar]
- 42. Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1): 38–46. [DOI] [PubMed] [Google Scholar]
- 43. Warkentin M, Berejnov V, Husseini NS, Thorne RE. Hyperquenching for protein cryocrystallography. J Appl Crystallogr. 2006;39(6): 805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4): 271–281. 10.1107/S0907444910048675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 7): 1204–1214. 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4): 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Koth CM, Murray JM, Mukund S, Madjidi A, Minn A, Clarke HJ, et al. Molecular basis for negative regulation of the glucagon receptor. Proc Natl Acad Sci U S A. 2012;109(36): 14393–14398. 10.1073/pnas.1206734109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1): 2126–2132. [DOI] [PubMed] [Google Scholar]
- 49. Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11): 1948–1954. [DOI] [PubMed] [Google Scholar]
- 50. Wu H, Pfarr DS, Tang Y, An LL, Patel NK, Watkins JD, et al. Ultra-potent antibodies against respiratory syncytial virus: effects of binding kinetics and binding valence on viral neutralization. J Mol Biol. 2005;350(1): 126–144. [DOI] [PubMed] [Google Scholar]
- 51. Budge PJ, Li Y, Beeler JA, Graham BS. RhoA-derived peptide dimers share mechanistic properties with other polyanionic inhibitors of respiratory syncytial virus (RSV), including disruption of viral attachment and dependence on RSV G. J Virol. 2004;78(10): 5015–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen M, Chang JS, Nason M, Rangel D, Gall JG, Graham BS, et al. A flow cytometry-based assay to assess RSV-specific neutralizing antibody is reproducible, efficient and accurate. J Immunol Methods. 2010;362(1–2): 180–184. 10.1016/j.jim.2010.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Neutralization of RSV strains A2, Long, 18537 and 9320 were measured using a microneutralization assay. Two-fold serial dilutions of AM14 (red) or palivizumab (black) were incubated with RSV before infection of HEp-2 cells and detection of RSV F on the surface of infected cells by ELISA. In all strains tested, AM14 was greater than one log more potent than palivizumab. (B) A flow cytometry-based attachment assay was modified from a previously described adherent cell assay [22, 51]. Antibodies and heparin were four-fold serially diluted and incubated with RSV A2 prior to attachment to HEp-2 cells and detection of RSV F on the surface of cells by flow cytometry. Although heparin (grey), a known attachment inhibitor, prevented binding of RSV to the cell surface, palivizumab (black) and AM14 (red) did not.
(TIF)
(A) Binding of D25 Fab to prefusion RSV F was measured by surface plasmon resonance. Prefusion RSV F was immobilized on an SA chip to a total of 300 RU and binding to D25 Fab was measured in 2-fold serial dilutions from 10 to 0.04 nM, with a duplicate of the 0.31 nM concentration. Best fit of the data to a 1:1 binding model is shown in red. D25 Fab bound to immobilized prefusion RSV F with an equilibrium dissociation constant (K D) of 0.07 nM, with association and dissociation rate constants of 1.35 x 106 M-1s-1 and 9.65 x 10-5 s-1, respectively. (B) Binding of D25 to HRA peptide was measured by biolayer interferometry. His-Tagged HRA peptide (RSV A2 F residues 153–211) was immobilized to anti-His sensors and two-fold serial dilutions of D25 IgG from 50 nM to 0.8 nM were assessed for binding on an Octet QK. Red lines are the fit of a global association and then dissociation algorithm in GraphPad Prism, from which the 0.8 nM concentration was excluded. This algorithm was used to calculate an equilibrium dissociation constant (K D) of 1.9 nM for D25 IgG binding to HRA peptide.
(TIF)
Structure of the AM14 Fab with heavy chain shown in blue and light chain shown in white. Complementarity-determining regions (CDRs) are labeled for both the heavy and light chains. The CDR H3 loops of the two Fabs in the asymmetric unit were in different conformations, the second of which is shown in purple. The dotted line indicates a region that was not modeled due to poor electron density. For the complex with RSV F, the Fab with the ordered CDRH3 was used as the search model.
(TIF)
The asymmetric unit of the ternary complex was composed of one RSV F trimer, three AM14 Fabs and three motavizumab Fabs. The side view and top view of the complex are shown, with RSV F protomers colored tan, light pink and light green. AM14 heavy chains are blue, motavizumab heavy chains are magenta and both AM14 and motavizumab light chains are white.
(TIF)
The main chains are shown as ribbons within the 2Fo—Fc electron density maps for the refined structure (blue), contoured at 1σ. Only one AM14 Fab and the RSV F trimer are shown for clarity. Chains are colored as in S4 Fig. The side view and top view are shown, with zoomed views for the interface between AM14 and the F trimer. The CDR H3 is labeled for both views and fits the electron density well.
(TIF)
Molecular surface representations of prefusion F and AM14 Fab colored according to electrostatic surface potential (red to blue, -8.4 to +8.4 kT/e, respectively). The surfaces are shown in an open-book representation of the binding interface with the approximate footprint for prefusion F and AM14 Fab outlined in yellow and black. A negatively charged region on the AM14 Fab complements a positively charged region on the lower protomer of prefusion F.
(TIF)
Two-fold serial dilutions of AM14 (red) or palivizumab (black) were incubated with (A) RSV A Long I79M/R429S/H515N, (B) L160S or (C) N183K before infection of HEp-2 cells and detection of RSV F on the surface of infected cells by ELISA. (D) In a separate assay, two-fold serial dilutions of AM14 were incubated with mKate RSV A2 (grey) or mKate RSV A2 N426D (blue) before infection of HEp-2 cells and measurement of fluorescence (excitation of 588 nm and emission at 635 nm) with a plate reader as previously described [52]. All MARMs were neutralized by palivizumab but not by AM14.
(TIF)
Plates were coated with (A) stabilized prefusion F or (B) the N426D mutant (1 μg/ml each) and were washed, blocked and incubated with four-fold serial dilutions of motavizumab (black), D25 (blue), 101F (grey), MPE8 (open blue) and AM14 (red) before detection with HRP-conjugated goat anti-human IgG (Santa Cruz Biotechnology) and Super AquaBlue substrate (eBiosciences Inc.). An ELISA plate reader (Molecular Devices, Inc.) was used to read signal at 538 nm. Binding of motavizumab, MPE8 and D25 was similar for both proteins. 101F binding to N426D was slightly reduced, whereas AM14 binding to N426D was reduced approximately 100-fold compared to wild-type prefusion F.
(TIF)
(A) Coomassie-stained reducing SDS-PAGE gel of the six proteins used in the ELISAs here and in Fig 4. (B) Binding of 114F, an antibody specific for the six-helix bundle of postfusion F, to the uncleaved monomeric RSV F (open black triangles), cleaved monomeric RSV F (black circles), uncleaved prefusion RSV F (open red triangles), cleaved prefusion RSV F (red circles), uncleaved postfusion RSV F (open blue triangles) and cleaved postfusion RSV F (blue circle) was measured by ELISA. 114F bound more tightly to cleaved postfusion F than to uncleaved postfusion F.
(TIF)
(PDF)
(PDF)
Data Availability Statement
Atomic coordinates and structure factors for the structures of AM14 unbound and in complex with RSV F have been deposited in the Protein Data Bank under accession codes 4ZYK and 4ZYP, respectively.