Abstract
Zebrafish (Danio rerio) are a valuable model for studying immunity, infection, and hematopoietic disease and have recently been employed for transplantation assays. However, the lack of syngeneic zebrafish creates challenges with identifying immune-matched individuals. The MHC class I genes, which mediate allogeneic recognition in mammals, have been grouped into three broad lineages in zebrafish: the classical U genes on chromosome 19, the Z genes which have been reported to map to chromosome 1, and the L genes that map to multiple loci. Transplantations between individual zebrafish that are matched at the U locus fail to consistently engraft suggesting that additional loci contribute to allogeneic recognition. Although two full-length zebrafish Z transcripts have been described, the genomic organization and diversity of these genes have not been reported. Herein we define ten Z genes on chromosomes 1 and 3 and on an unplaced genomic scaffold. We report that neither of the Z transcripts previously described match the current genome assembly and classify these transcripts as additional gene loci. We characterize full-length transcripts for 9 of these 12 genes. We demonstrate a high level of expression variation of the Z genes between individual zebrafish suggestive of haplotypic variation. We report low level sequence variation for individual Z genes between individual zebrafish reflecting a possible nonclassical function, although these molecules may still contribute to allogeneic recognition. Finally, we present a gene nomenclature system for the Z genes consistent with MHC nomenclature in other species and with the zebrafish gene nomenclature guidelines.
Keywords: Teleost, MHCclassI, Evolution, Polymorphism, Haplotype
Introduction
Zebrafish (Danio rerio) recently have been shown to be a valuable model for studying human hematopoietic diseases through immune-matched hematopoietic transplantation assays (de Jong et al. 2011). Similar assays have historically been carried out in congenic mice as they are immunologically compatible. Attempts at generating congenic zebrafish lines have been largely unsuccessful, but the development of genotyping methods for identifying immune-matched donors and recipients is making it possible to take advantage of the many benefits of the zebrafish model (high fecundity, rapid maturation, small size, etc.) in transplantation assays. Genotyping methods have been and are being developed to differentiate between the multiple haplotypes of the zebrafish classical Major histocompatability complex (MHC) class I genes (de Jong and Zon 2012; Jill de Jong, personal communication). While the primary function of classical MHC class I molecules is presenting peptides to CD8+ T cells, they are also responsible for determining allograft compatibility. Although the MHC class I genes have been extensively characterized in humans and mice, the characterization of the zebrafish MHC Class I genes is incomplete and ongoing.
Multiple MHC class I multigene families have been identified in bony fish that form distinct phylogenetic lineages based on sequence alignments. The first MHC class I genes identified in the zebrafish are of the U lineage (Takeuchi et al. 1995; Michalova et al. 2000). The U lineage genes are considered classical MHC class I genes and have been identified in numerous teleost species. They have been mapped to chromosome 19 in zebrafish and six different haplotypes have been identified encoding multiple MHC class I U lineage genes (Sambrook et al. 2005; Jill de Jong, personal communication).
A second group of teleost MHC class I genes is designated the Z lineage. The first four Z lineage genes to be described, ZA, ZB, ZC, and ZD, were cloned from carp and based on their sequences, characterized as nonclassical MHC class I genes; however, orthologs of these genes have not been identified outside of carp species (Hashimoto et al. 1990; Okamura et al. 1993). In contrast, transcripts or genomic sequences representing a fifth Z lineage gene, ZE, have been identified in zebrafish, carp, salmon, flounder, medaka, barbus, and pufferfish (Kruiswijk et al. 2002; Srisapoome et al. 2004; Lukacs et al. 2010; Nonaka et al. 2011). Since the first description of the ZE genes in 2002 (Kruiswijk et al. 2002; Stet et al. 2003), they have been recognized for their contradictory characteristics: they have a historical association with the nonclassical ZA, ZB, ZC, and ZD genes in carp and relatively well-conserved extracellular α1 and α2 domains (which are normally polymorphic for a diverse peptide-binding repertoire), yet they exhibit conservation of the residues thought to be important for peptide binding and generally exhibit ubiquitous expression. In accordance with this unique display of sequence features of both classical and nonclassical MHC class I receptors, phylogenetic analyses place the ZE genes in a lineage related to, but distinct from, the carp ZA, ZB, ZC, and ZD genes. Functional data will be necessary to classify the ZE genes as classical or nonclassical as they may represent a separate or intermediate functional category yet to be defined (Stet et al. 2003). The third group of MHC class I genes to be identified in zebrafish is designated the L lineage, and based on their sequences, they have been described as nonclassical MHC class I genes (Dijkstra et al. 2007).
As the only MHC class I Z genes identified outside of carp species are the ZE genes, in this manuscript, we refer to the ZE genes as “Z” unless specifically discussing this MHC class I lineage in carp. The Z genes have been best characterized in Atlantic salmon in which four unique Z loci have been mapped to two linkage groups (Lukacs et al. 2010). In salmon, as well as in medaka, the Z genes are linked to U genes, suggesting a common origin for the U and Z genes (Lukacs et al. 2010; Nonaka et al. 2011). Thus far, the characterization of the Z genes lacks any description of the sequence or expression variability between individuals of the same species. The availability of individual zebrafish representing diverse genetic backgrounds including well-established laboratory lines, lines derived from wild-caught individuals, and clonal lines makes the zebrafish ideal for investigating the intraspecific diversification of this recently identified multigene family.
Here we present an analysis of the zebrafish MHC class I Z genes. We investigate the genomic organization and sequence diversity of these genes, demonstrate their variable expression between individual zebrafish, and propose a nomenclature system for them. From the reference genome (Zv9), we have identified ten unique zebrafish Z gene loci: four on chromosome 1, five on chromosome 3, and one on an unplaced genomic scaffold. Two additional genes map to a BAC clone that account for the initial full-length zebrafish Z gene transcripts (Kruiswijk et al. 2002) not present in the current genome assembly. Full-length transcripts have been identified for 9 of the 12 Z genes with transcript variants and variable expression patterns detected across individual zebrafish from different genetic backgrounds. The identification of loci not represented in the reference genome along with evidence obtained from both PCR-based and Southern blot strategies supports the hypothesis that the Z genes are subject to haplotypic transmission. It is plausible that they play a role in allogeneic recognition as the success of hematopoietic transplantation is not entirely dependent on matching MHC class I U genes on chromosome 19 (de Jong and Zon 2012). The data provided herein will lead to a better understanding of the evolution of the MHC class I Z lineage and may lead to improved success of transplantation assays in zebrafish.
Materials and methods
Gene nomenclature
The zebrafish MHC class I Z gene nomenclature employed herein (Dare-mhc1zxa) is based on the guidelines proposed for all species (Klein et al. 1990), reflects the nomenclature system in Atlantic salmon (Lukacs et al. 2010), and includes MHC nomenclature conventions previously established in zebrafish. The four letter abbreviation, Dare, refers to the first two letters of the genus and species, Danio rerio. The “mhc1” is included as it is for the nomenclature of the zebrafish MHC class I U lineage loci. The letter “z” corresponds to the MHC class I lineage. The variable second letter “x” designates the locus (named alphabetically). The third letter specifies “a” for class I. Accordingly, the previously described zebrafish Z transcripts (Kruiswijk et al. 2002) have been renamed Dare-mhc1zaa (previously Dare-ZE*0101) and Dare-mhc1zla (previously Dare-ZE*0201, see discussion for further clarification). Within this manuscript, we refer to individual Z genes as mhc1zxa with the corresponding proteins named Mhc1zxa or Zxa. Alleles are defined as having at least one amino acid difference from the corresponding protein predicted from the reference genome. Please note that Dare-mhc1zaa, Dare-mhc1zba, Dare-mhc1zca, and Dare-mhc1zda are not orthologous to the carp Cyca-ZA, Cyca-ZB, Cyca-ZC, and Caau-ZD genes (see “Phylogenetic analyses”).
Zebrafish lines
AB and TU (Tübingen) fish were obtained through the Zebrafish International Resource Center (ZIRC; http://zebrafish.org). Additional TU individuals were provided by John Rawls (University of North Carolina, Chapel Hill). EKW zebrafish were purchased from EkkWill Waterlife Resources (Ruskin, FL). Individual zebrafish were also employed from a line with low stationary behavior (LSB) which is three generations away from true wild-type fish from the village of Gaighata in West Bengal, India (gift of John Godwin, North Carolina State University; Wong et al. 2012) and a double haploid, clonal golden zebrafish line (CG2; gift of Sergei Revskoy, Northwestern University; Mizgirev and Revskoy 2010). All experiments involving live zebrafish were performed in accordance with relevant institutional and national guidelines and regulations and were approved by the North Carolina State University Institutional Animal Care and Use Committee.
Genomic annotation and data mining
The mhc1zaa transcript mentioned above (previously named mhc1ze or Dare-ZE*0101 ; GenBank: NM_194425.1; Kruiswijk et al. 2002) was amplified from TU whole animal cDNA by RT-PCR, cloned, and sequenced by the methods described below. The predicted Mhc1zaa leader, α1–α3, and transmembrane domains were identified using SMART software (Letunic et al. 2012) in conjunction with other sources (Barclay 1997). The amino acid sequence for the leader and each alpha domain was then used as a query (tBLASTn) to search the zebrafish reference genome (Zv9). One chromosome 1 scaffold (Zv9_scaffold68; GenBank: NW_003334061.1), two chromosome 3 scaffolds (Zv9_scaffold307; GenBank: NW_003334229.1 and Zv9_scaffold312; GenBank: NW_003334232.1), and an unplaced scaffold (Zv9_NA257; GenBank: NW_003336703.1) were identified as encoding the Z genes. None of the ten genes identified from the reference genome exactly match the previously described mhc1zaa or mhc1zla transcripts (Kruiswijk et al. 2002).
The zebrafish Z gene alpha domains were also compared (BLASTn) to the zebrafish nonredundant nucleotide database as well as to the zebrafish EST database to aid in identifying predicted leader and transmembrane domains and in identifying genomic evidence of additional haplotypes. Additional comparisons (tBLASTn) were made to the de novo genomic assemblies from double haploid homozygous AB and Tübingen individuals. The assemblies were generated using Illumina GA sequencing technology by the Wellcome Trust Sanger Institute and are available on-line (www.sanger.ac.uk).
Phylogenetic analyses
Protein domains were identified with SMART software (Letunic et al. 2012). Sequences were aligned by ClustalW (Larkin et al. 2007). Phylogenetic trees were constructed from pairwise Poisson correction distances with 2,000 bootstrap replications by MEGA4 software (Tamura et al. 2007). Phylogenetic analyses included the following sequences: Dare-Zaa through Dare-Zla predicted from genomic scaffold and BAC sequences depicted in Fig. 1, Dare-Uba (GenBank: NP_571546.1), Dare-Uca (GenBank: CAK10898.1), Dare-Uda (GenBank: NP_571779.1), Dare-Uea (GenBank: NP_571780.1), Dare-Ufa (GenBank: CAD58763.1), Dare-Uga (GenBank: NP_956879.1; formerly Dare-Uxa2, Jill de Jong, personal communication), Dare-Uha (GenBank: NP_001070109.1), Dare-Uia (GenBank: AGL92229.1), Dare-Uja (GenBank: NP_956700.1), Dare-Uka (GenBank: NP_001038925.1; formerly LOC751750, Jill de Jong, personal communication), Dare-L (GenBank: NP_001017904.1), Cyprinus carpio Cyca-ZE*0101 (GenBank: CAD12793.1), Cyca-ZA1 (GenBank: AAA49212.1), Cyca-ZB1 (GenBank: L10420.1), Cyca-ZC1 (GenBank: L10421.1), Carassius auratus Caau-ZD1 (Okamura et al. 1993), and Oncorhynchus mykiss Onmy-LAA*0101 (GenBank: ABI21842.1).
Fig. 1.
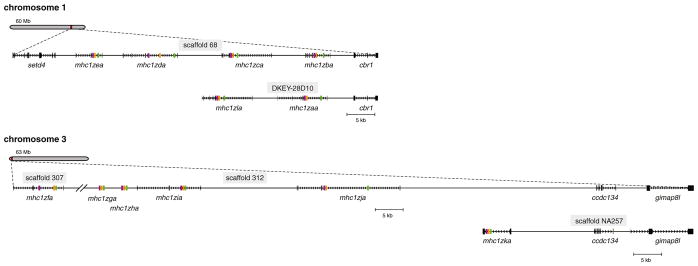
Genomic organization of zebrafish Z genes. The relative chromosomal positions of the Z genes are shown. The entire coding sequence is indicated for all genes except for mhc1zga and mhc1zha (alpha domains only) which lack identifiable start and stop codons. Exons are represented as rectangles while introns are represented as arrows pointing in the direction of transcription. Exons encoding α1, α2, and α3 domains are shown in green, orange, and purple, respectively. Predicted alternative haplotypes encoded on BAC clone DKEY-28D10 and unplaced genomic scaffold NA257 are shown below each chromosome along with their common flanking genes. Note that the Z genes on chromosome 3 are encoded by nonconsecutive scaffolds
Amplification of zebrafish cDNAs
Adult zebrafish were euthanized, frozen in liquid nitrogen, and pulverized by mortar and pestle. Total RNA was purified from either all or half of the individual (TRIzol® Reagent, Invitrogen) and reverse-transcribed into cDNA (SuperScript® III Reverse Transcriptase, Invitrogen). When available, genomic DNA was extracted from the other half of the individual (below). Forward and reverse primers were designed to amplify entire coding sequences based on the predicted Z gene sequences and used with a high fidelity DNA polymerase (KAPA HiFi HotStart ReadyMix, Kapa Biosystems) to amplify transcripts from a panel of single animal cDNAs (see Table 1 for primer sequences and PCR conditions). One primer set was used to amplify both mhc1zca and mhc1zla transcripts. Primers were designed to amplify mhc1zia, but failed to detect transcripts in all individuals evaluated. Primers were not designed to amplify mhc1zga and mhc1zha as exons 5′ and 3′ of the α1–α3 domains were not identifiable.
Table 1.
Primer sequences and PCR cycling parameters for cloning full-length cDNAs
| Primer sets | Primers | Annealing temp (°C) | Extension time (s) | No. of cycles (Fig. 4a) | No. of cycles (Fig. 6) | Amplicon length (bp) |
|---|---|---|---|---|---|---|
| mhc1zaa | Forwarda: ATGGCTGTTTTTGCCGTTTTG Reverseb: GGATGGTTCAATGGTTCCCGTTAG |
65 | 60 | 35 | – | 1,171 |
| mhc1zba | Forward: AGGCTTCAGAATGGGTAGTTTTGCCG Reverse: GTCGGATTTGGACTCCCTTACACA |
65 | 60 | 35 | 25 | 1,224 |
| mhc1zcac | Forward: GGCTTCAGAATGGCTGTTTTGGC Reverse: TTTCCTGGGAGCGAGGACTGCTTT |
66 | 60 | 35 | 35 | 1,135 |
| mhc1zda | Forward: GACACACACCGCGGCTTTAAAATGG Reverse: CTGTCAACAGATTTAACTGTCATTCTGCC |
65 | 60 | 40 | 35 | 1,280 |
| mhc1zea | Forward: CGCGACCTCAAAAGGCTGCAAAATG Reverse: TCAAAGAGCATTTCTCCATCCCATTC |
65 | 60 | 35 | 35 | 1,075 |
| mhc1zfa | Forward: CAACACAAGGCTTTTCTGATGATTATGG Reverse: CAATGTTGGTTTGATACTGACGGGTACC |
64 | 60 | 35 | 35 | 1,170 |
| mhc1zja | Forward: GGATTGACAATTATGACGAGCTTCG Reverse: GGTCCAGAAATGAAAGTTTGCAGAG |
64 | 60 | 35 | 40 | 1,189 |
| mhc1zka | Forward: TCAGTCATGATGTCAGGAGGAGTC Reverse: CCGACTGACACGCATGGTAAACTT |
65 | 60 | 35 | 35 | 1,190 |
| β actin | Forward: GGTATGGAATCTTGCGGTATCCAC Reverse: ATGGGCCAGACTCATCGTACTCCT |
65 | 60 | 25 | 25 | 301 |
Sequences corresponding to translational start codons are underlined
Sequences corresponding stop codons are underlined
The mhc1zca primers also amplified mhc1zla transcripts
The same PCR protocol was employed with a panel of cDNAs derived from spleen, kidney, intestine, liver, and gill tissues from different individual zebrafish. When sequencing was desired, PCR products were purified by phenol–chloroform extraction and ethanol precipitation, incubated with Taq DNA polymerase to introduce dA-overhangs (10 min reaction at 72 °C with GoTaq® Green Master Mix, Promega), ligated into pGEM®-T Easy (Promega), and sequenced (Electronic Supplementary File, Fig. S1). Sequence data have been deposited with GenBank under accession numbers KC607829–KC607872.
Genotyping PCR
Genomic DNA (gDNA) was purified from the same EKW, LSB, and CG2 individuals from which RNA was purified above. Additional TU individuals were employed for isolating matching RNA and gDNA samples. Forward and reverse primers were designed to amplify sequences spanning a single intron for mhc1zba, mhc1zja, and mhc1zka. PCRs were conducted with these primers on matching cDNA and gDNA samples to determine if the absence of a detectable transcript corresponded to the absence of the gene from that individual (see Table 2 for primer sequences and PCR conditions).
Table 2.
Primer sequences and PCR cycling parameters for genotyping PCR
| Primer sets | Primers | Annealing temp (°C) | Extension time (s) | No. of cycles | Amplicon length (bp) cDNA | Amplicon length (bp) gDNA |
|---|---|---|---|---|---|---|
| mhc1zbaa | Forward: ATGATGTCCGGTTTTCCAG Reverse: GCTGAAAGGTCCCATCTTC |
65 | 30 | 35 | 426 | 496 |
| mhc1zjaa | Forward: AGTTGAGAAACAGGGAAATGAACAT Reverse: TGTGGGACACAGAACAATCATAATT |
60 | 30 | 35 | 506 | 585 |
| mhc1zkaa | Forward: GAAAACCTCCTGTCTTTTGATGAGA Reverse: GAGTTTCAGCTTAGTTTCATCTTTA |
65 | 30 | 40 (cDNA) 30 (gDNA) |
252 | 331 |
| β actinb | Forward: GGTATGGAATCTTGCGGTATCCAC Reverse: ATGGGCCAGACTCATCGTACTCCT |
65 | 30 | 25 | 301 | 386 |
Primers span exon 3 (α2) to exon 4 (α3)
Primers span exon 4 to exon 5
Southern blot
A Southern blot was performed using genomic DNA from eight of the nine individuals of different genetic backgrounds used in genotyping PCR. DNA was digested with HindIII (New England BioLabs) according to the recommendations of Green and Sambrook (2012). Digested DNA (5 μg) was separated on a 0.8 % agarose gel at 30 V overnight. Following denaturation, DNA was transferred to a Zeta-Probe GT membrane (Bio-Rad) using the TurboBlotter rapid downward transfer system (Whatman) and alkaline transfer buffer. Probes were generated by PCR amplification of the well-conserved α1 domain from the most common alleles of mhc1zba (chromosome 1) and mhc1zfa (chromosome 3). A 265-bp mhc1zba probe was amplified from transcript 3651 with forward (AAACACTCCCTCTACTACATATACAC) and reverse (TATTGTGTCTCATTCGTTCCATCA) primers. A 260-bp mhc1zfa probe was amplified from transcript 3942 with forward (GAAGCACTCTTTGTATTACATCTACAC) and reverse (GTCTCATGCGGTCCATCAGTA) primers. Probes were independently labeled with an equivalent efficiency using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche) and mixed (1:1) for hybridization. Hybridization and detection were carried out using the same kit with a hybridization temperature of 42 °C and an exposure time of 25 min using the ChemiDoc MP imager (Bio-Rad).
Results and discussion
Identification of 12 MHC class I Z loci in zebrafish
In 2002, Kruiswijk et al. identified 2 full-length and 12 partial MHC class I Z cDNAs in zebrafish (Kruiswijk et al. 2002). At that time, it was unclear if these different sequences resulted from multiple loci or from allelic variation of a single locus. In 2005, a genome-wide survey of zebrafish MHC loci placed one of these full-length coding sequences on an unplaced scaffold as well as four similar genes on chromosome 1 (Zv4) but did not provide sequence data (Sambrook et al. 2005). BLAST searches of the most recent assembly of the zebrafish genome (Zv9) with the zebrafish Z transcript mhc1zaa (see “Materials and methods” for a description of Z gene nomenclature) as a query identified ten loci on genomic scaffolds that map to chromosome 1 or chromosome 3, or are currently unplaced in the reference genome (Fig. 1). Four unique loci were identified on Zv9_scaffold68 mapping to chromosome 1. These loci are designated mhc1zba through mhc1zea and correspond to the four genes identified on chromosome 1 by Sambrook et al. (2005). Five unique loci were identified on chromosome 3, one on Zv9_scaffold307 (mhc1zfa) and four on Zv9_scaffold312 (mhc1zga through mhc1zja). One unique locus, mhc1zka, was identified on the unplaced scaffold Zv9_NA257. It is likely that scaffold NA257 represents an alternative haplotype for chromosome 3 as both mhc1zja and mhc1zka are flanked by the ccdc134 (LOC100536187 ; GenBank: XM_003201673.1) and gimap8l (LOC100149190; GenBank: XM_001920324.3) genes in the same orientation but with different intergenic spacing. Similarly, a BLAST search of the zebrafish nonredundant nucleotide database identified a BAC clone, DKEY-28D10 (Genbank: BX005353.8), which encodes mhc1zaa and mhc1zla. These loci correspond to the original zebrafish Z transcripts Dare-ZE*0101 and Dare-ZE*0201 (Kruiswijk et al. 2002) which are not represented in the current reference genome (Zv9). This BAC clone likely represents an alternative haplotype for chromosome 1 as both mhc1zaa and mhc1zba are flanked by the cbr1 gene (NM_194406.1).
BLASTp searches of the zebrafish reference protein database identified 11 Z protein sequences. Phylogenetic analyses were employed to determine the relationships between the ten Z genes from the reference genome, the previously described Z transcripts now mapped to BAC DKEY-28D10 (Kruiswijk et al. 2002) and these 11 Z protein sequences (Electronic Supplementary File, Fig. S2). Based on these results and the new Z gene nomenclature: mhc1zba replaces the gene previously known as zgc:64115; zgc:136614 and zgc:111893 are predicted to be polymorphic variants of mhc1zca; zgc:162351 and si:ch211-287j19.6 are predicted to be polymorphic variants of mhc1zea; mhc1zfa replaces the predicted sequence LOC556263 ; mhc1zia replaces the predicted sequence LOC100150150; mhc1zja replaces the predicted sequence LOC571647; and mhc1zka replaces the predicted sequence LOC100536229 (Table 3 and Electronic Supplementary File, Fig. S2). The predicted transcript LOC563036 appears to encode two individual Z sequences described here as mhc1zga and mhc1zha. Transcripts have not been identified for either of these sequences and they may represent pseudogenes. To the extent of our knowledge, our mhc1zda transcript sequences are the first to be reported. The mhc1zga, mhc1zia, and mhc1zka sequences also can be equated with previously reported partial Z sequences Dare-ZE*0501, Dare-ZE*0701, and Dare-ZE*0601, respectively (Kruiswijk et al. 2002).
Table 3.
Zebrafish Z gene information and accession numbers
| Gene | Genomic location | Aliases | Alias accession nos. | This report’s acc. nos. |
|---|---|---|---|---|
| mhc1zaa | DKEY-28D10 | Dare-ZE*0101a mhc1ze |
AJ420953 NM_194425.1 |
KC607829–KC607830 |
| mhc1zba | Chr1, scaffold 68 | zgc:64115 | NM_001110118.1 | KC607831–KC607834 |
| mhc1zca | Chr1, scaffold 68 | zgc:136614 zgc:111893 |
NM_001083545.1 NM_001039986.1 |
KC607835–KC607842 |
| mhc1zda | Chr1, scaffold 68 | – | – | KC607843–KC607849 |
| mhc1zea | Chr1, scaffold 68 | si:ch211-287j19.6 zgc:162351 |
NM_001045563.1 NM_001089550.1 |
KC607850–KC607854 |
| mhc1zfa | Chr3, scaffold 307 | LOC556263 | XM_003197994.1 | KC607855–KC607865 |
| mhc1zga | Chr3, scaffold 312 | Dare-ZE*0501a mhc1zel-002b LOC563036 |
AJ420975 XM_001919256.3 |
|
| mhc1zha | Chr3, scaffold 312 | LOC563036 | XM_001919256.3 | |
| mhc1zia | Chr3, scaffold 312 | Dare-ZE*0701a mhc1zel-001b LOC100150150 |
AJ420977 XM_001919268.3 |
|
| mhc1zja | Chr3, scaffold 312 | LOC571647 mhc1ze-001 (tv1)b mhc1ze-002 (tv2)b |
NM_001109718.1 | KC607866–KC607869 |
| mhc1zka | Scaffold NA257 | Dare-ZE*0601a LOC100536229 |
AJ420976 XM_003201674.1 |
KC607870 |
| mhc1zla | DKEY-28D10 | Dare-ZE*0201a | AJ420954 | KC607871–KC607872 |
Sequences from Kruiswijk et al. (2002)
From the Zebrafish Model Organism Database (ZFIN.org)
Loci encode highly similar MHC class I Z proteins
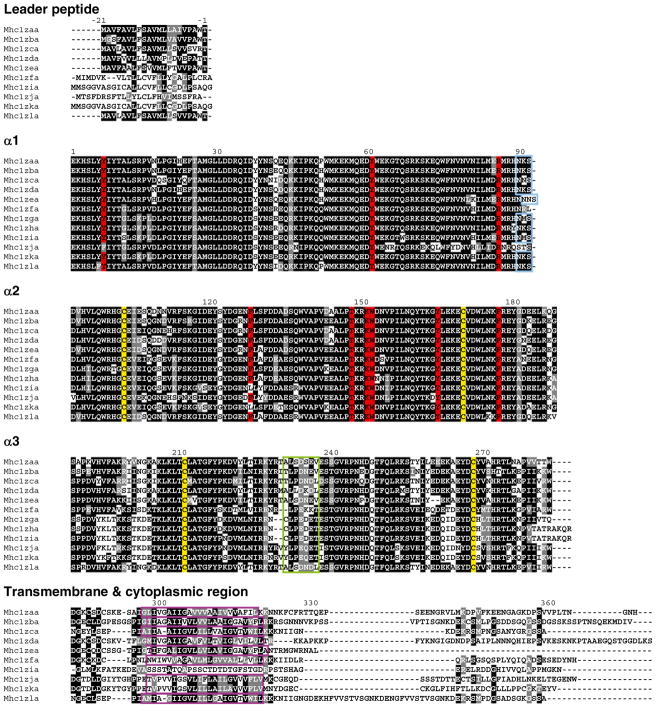
Aligning the amino acid sequences encoded by the Z genomic loci reported in Fig. 1 revealed that these 12 sequences encode highly similar proteins (Fig. 2). The α1 and α2 domains, which together are responsible for forming the peptide binding groove, are more similar than the α3 domains, suggesting that these genes may have a nonclassical function, i.e., a function other than antigen presentation. This trend has been reported before for the ZE lineage cDNAs identified in barbus and carp (Dixon and Stet 2001). Further inspection of the alignment, however, revealed that many important features of classical MHC class I genes have been conserved in the zebrafish Z proteins. Four cysteine residues that are important for forming the disulfide bond in an Ig fold, two in the α2 domain and two in the α3 domain, have been conserved in all 12 sequences. Nine residues that have been shown to function as anchors for the end of bound peptide (Kruiswijk et al. 2002) are also conserved in all sequences with just a few exceptions. A N-linked glycosylation site is present at the end of the α1 domain in all proteins except Mhc1zfa and Mhc1zja. The presence of acidic residues within a seven amino acid stretch of the α3 domains suggests that the Z proteins potentially associate with CD8 (Dijkstra et al. 2007). However, this interaction remains to be verified as the residues in this region are slightly less acidic relative to those in the proteins of the MHC class I U lineage. All full-length Z sequences possess a predicted transmembrane segment except for Mhc1zia.
Fig. 2.
Z protein sequences. The Z proteins predicted from the genomic loci described in Fig. 1 were organized into domains and aligned. The leader and alpha domains derive from single exons while the transmembrane and cytoplasmic domains derive from one to five exons (Electronic Supplementary File, Fig. S3). Numbering of amino acids is based on the Mhc1zaa sequence starting with the first residue of α1. Positions that are at least 50 % identical are shaded in black and similar residues are shaded in gray. Conserved cysteine residues likely involved in the Ig-fold are shaded in yellow. Nine residues implicated in peptide binding are shaded in red. A conserved N-linked glycosylation site at the end of the α1 domain is boxed in blue. A green box indicates a stretch of seven acidic residues that binds CD8 in mammals. The putative transmembrane segments are boxed in purple
The sequences comprising the transmembrane and cytoplasmic domains were investigated in further detail, exon by exon, in an attempt at identifying the evolutionary history of this multigene family within zebrafish (Electronic Supplementary File, Fig. S3). In general, this region is highly variable across the zebrafish Z genes, although a phylogenetic analysis of the transmembrane domains was able to differentiate the genes based on the chromosome on which they are located (data not shown). However, a phylogenetic comparison of transmembrane domains encoded by Z genes from different teleost species failed to provide evidence for definitive orthologous Z genes (data not shown). Additionally, three observations can be made with regard to the four exons that encode the cytoplasmic tail of Mhc1zla: (1) exons 6 and 7 encode nearly identical sequences, (2) exon 8 encodes a peptide sequence nearly identical to the sequence encoded by exon 7 of mhc1zba, and (3) exon 9 encodes a peptide sequence nearly identical to the sequence encoded by exon 7 of mhc1zca. These observations suggest that exon duplication and/or swapping has contributed to the sequence diversity of this multigene family.
Phylogenetic analyses
Phylogenetic analysis of the MHC class I proteins (α1 through α3 domains) of different lineages from zebrafish as well as other fish species (Fig. 3a) supports the inclusion of the Z genes encoded on zebrafish chromosome 1 and chromosome 3 within the ZE subgroup of the Z lineage. Representative genes of the MHC class I L lineage from zebrafish and trout group together and are more similar to the classical zebrafish U genes than to the Z genes. Analysis of the individual alpha domains from both the zebrafish U and Z genes (Fig. 3b) verifies that the predicted domains of the Z genes identified as α1, α2, or α3 indeed group with the correct alpha subgroup. Both panels a and b of Fig. 3 demonstrate that the mhc1zaa and mhc1zla genes encoded by BAC DKEY-28D10 group more closely with the Z genes on chromosome 1 while the mhc1zka gene encoded by unplaced genomic scaffold NA257 groups more closely with the Z genes on chromosome 3. These observations support the classification of these three genes as belonging to alternative haplotypes of either chromosome 1 or chromosome 3 (Fig. 1).
Fig. 3.
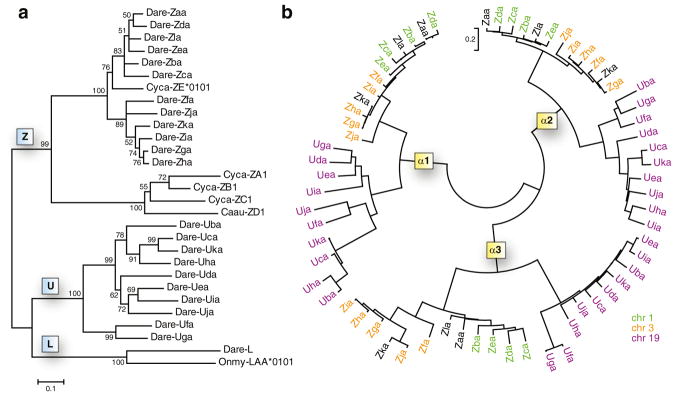
Phylogenetic analyses of MHC class I Z genes. a A neighbor-joining tree was constructed from the alignment of the α1 through α3 protein sequences from the different MHC class I lineages of zebrafish and other fish species. The consensus tree was based on 2,000 bootstrap replications. Bootstrap values below 50 % are not shown. Species indicators include Dare (Danio rerio, zebrafish), Cyca (Cyprinus carpio, common carp), Caau (Carassius auratus, goldfish), and Onmy (Oncorhynchus mykiss, rainbow trout). b A neighbor-joining tree was constructed from the amino acid alignment of individual alpha domains from all identified zebrafish Z and U genes. MHC genes from chromosomes 1, 3, and 19 are shown in green, orange, and purple, respectively
Variable expression of MHC class I Z genes
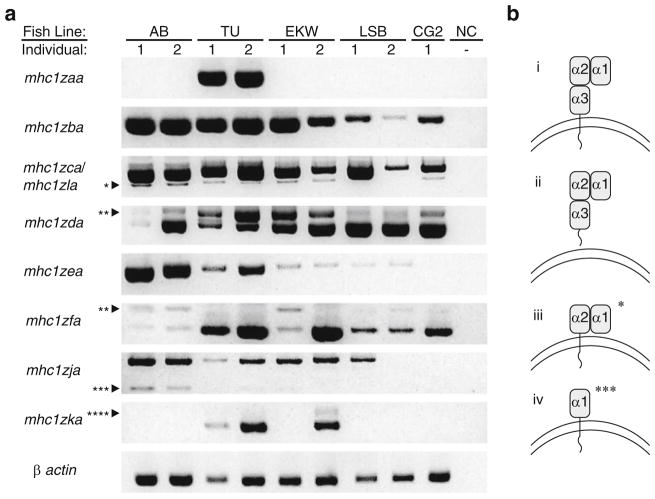
Individual zebrafish from different genetic backgrounds display variable Z gene expression patterns (Fig. 4a) similar to the haplotypic variation reported for the MHC class I U lineage genes (de Jong et al. 2011). Individual animals were selected from three standard (but polymorphic) laboratory lines: TU (the zebrafish reference genome is based on individuals from this line), AB, and EKW. Individuals from the LSB line that are three generations from true wild-type fish were also included with the expectation that they would show greater sequence variability. The CG2 zebrafish was included as it represents a homozygous diploid clonal line (Mizgirev and Revskoy 2010). RT-PCR was employed to amplify the coding sequences of nine Z genes. While transcripts for all nine of the genes could be amplified from the TU zebrafish, only transcripts for four Z genes were amplified from the CG2 individual, three encoded on chromosome 1 (scaffold 68 in Fig. 1) and one encoded on chromosome 3 (scaffold 307 in Fig. 1). Full-length mhc1zaa and mhc1zla transcripts were only detected in the TU individuals while transcripts from all other genes were detected across multiple fish lines. Expression was consistent across individuals of the same genetic background with only two exceptions: mhc1zja in LSB fish and mhc1zka in EKW fish.
Fig. 4.
Variable expression of Z genes across individual zebrafish. a RT-PCR was performed to detect expression of individual Z genes from individual zebrafish representing five different genetic backgrounds: AB, Tübingen (TU), EkkWill (EKW), wild type with low stationary behavior (LSB), and clonal golden (CG2). A negative control with no cDNA (NC) was included for each primer set. Full-length sequences were successfully amplified for 9 of the 12 Z genes (one primer set amplifies both mhc1zca and mhc1zla). β actin serves as a positive control for the quality of the cDNA. Bands of expected and unexpected sizes were cloned and sequenced revealing that some genes are subject to alternative splicing: one asterisk represents mhc1zca transcripts without an α3 domain (reading frame maintained), two asterisks represent mhc1zda or mhc1zfa transcripts with intronic sequence between the leader and α1 domains (reading frame not maintained), three asterisks represent mhc1zja transcripts without an α2 or α3 domain (reading frame maintained), and four asterisks represent mhc1zka transcripts with intronic sequence between the α2 and α3 domains (reading frame not maintained). b The predicted protein structures for a traditional MHC class I transcript (i) as well as for the nontraditional MHC class I transcripts resulting from alternative splicing (ii through iv) are shown. A secreted isoform of mhc1zca (ii) was identified but is not distinguishable from the regular isoform by standard gel electrophoresis shown in a. The structures shown in (iii) and (iv) correspond to the bands in a marked by one asterisk and three asterisks, respectively
In addition to sequencing the nine full-length cDNAs from each genetic background in which they were detected, bands of unexpected sizes were cloned and sequenced as well (Electronic Supplementary File, Fig. S1). This revealed that many of the Z genes are subject to alternative splicing. For example, mhc1zca was found as three different transcript variants: the expected transcript resulting in three alpha domains followed by a transmembrane domain (Fig. 4b(i)), a secreted variant lacking the transmembrane domain but maintaining the reading frame (Fig. 4b(ii)), and a variant lacking the α3 domain but again maintaining the reading frame (Fig. 4b(iii), bands marked by an asterisk in Fig. 4a). Transcript variants of mhc1zja lacking both the α2 and α3 domains also were identified (Fig. 4b(iv), bands marked by three asterisks in Fig. 4a) with the reading frame maintained. Several transcripts were identified that were incompletely spliced with introns present between the leader and α1 domains (bands marked by two asterisks in Fig. 4a) or between the α2 and α3 domains (bands marked by four asterisks in Fig. 4a). All identified transcripts with intronic sequence had a frameshift resulting in premature stop codons. It is unclear whether transcript variants with intact reading frames represent functional isoforms.
Evidence for polymorphic, haplotypic, and transcriptional variation
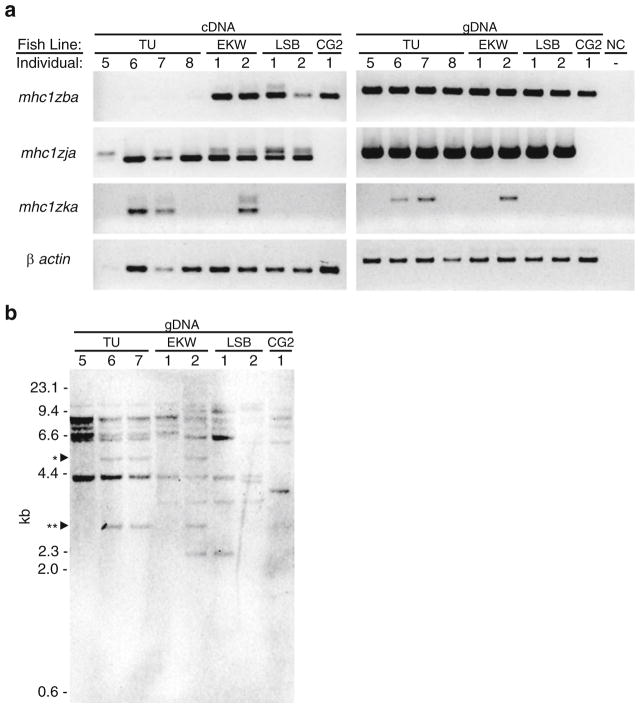
After observing variable expression of the Z genes across zebrafish of different genetic backgrounds, two hypotheses were considered. The first is that different zebrafish have different haplotypes at these loci encoding a variable number of Z genes. The second hypothesis is that transcripts were not detected in some individuals due to polymorphisms at the primer binding sites or due to transcriptional control mechanisms yet to be discovered. To test these hypotheses, we performed genotyping PCR using intron-spanning primers for PCR on gDNA and cDNA from the same individuals (Fig. 5a). The results were intriguing as they supported both hypotheses. In the case of mhc1zja and mhc1zka, the presence or absence of transcript in the cDNA correlated with the presence or absence of the gene in the gDNA. The finding that expression of mhc1zka is limited to the second EKW individual agreed with the PCR panel of full-length transcripts from Fig. 4a. However, the presence of mhc1zja in the second LSB individual detected in Fig. 5a suggests that the failure to detect full-length transcripts in this individual (Fig. 4a) is likely due to nucleotide polymorphisms at the primer binding sites. Genotyping PCR was performed for the mhc1zba gene with the expectation that it would serve as a positive control as full-length transcripts were detected in all individuals evaluated in Fig. 4a. Instead mhc1zba transcripts were not detected in new TU individuals despite the fact that the gene was detected by genomic PCR. These results suggest that the zebrafish Z genes are polymorphic and likely display haplotypic variation, and their transcription may be variably controlled.
Fig. 5.
Genotyping PCR and Southern blot analysis. a RT-PCR was performed on a panel of matching cDNA and gDNA samples from four additional TU individuals as well as the same EKW, LSB, and CG2 individuals as in Fig. 4a. Primers were designed to amplify a portion of each gene across a single intron. The faint, larger bands detected in the cDNA PCR panel all correspond to the size of the band from the gDNA PCR panel due to the presence of introns in the transcripts. b Southern blot analysis was performed on HindIII-digested genomic DNA from eight of the nine individuals used in a with a 1:1 mix of two probes designed to detect all Z genes (α1 domains of mhc1zba and mhc1zfa). The ~5.2-kb band marked by one asterisk correlates to the predicted size of the fragment containing mhc1zka and confirms the genotyping result observed in a. The ~3-kb band marked by two asterisks is likely associated with the mhc1zka haplotype
Data mining the de novo genomic assemblies from double haploid homozygous AB and TU individuals provided support for haplotypic variation of the Z loci on chromosome 3 (see Electronic Supplementary File, Table S1). All four chromosome 1 Z genes that are linked on scaffold 68 were identified in both assemblies, although not all alpha domains of mhc1zda could be identified in the AB individual. As for the chromosome 3 Z genes, mhc1zfa from scaffold 307 was found in both assemblies, but the AB individual seems to encode the scaffold 312 haplotype (mhc1zga, mhc1zha, mhc1zia, and mhc1zja) while the TU individual encodes the NA257 haplotype (mhc1zka). This suggests that the two scaffolds encoding Z genes from chromosome 3 are not linked and are instead transmitted as separate haplotypes. This is supported by the RT-PCR data in Fig. 4a in which the second LSB fish and the CG2 fish express mhc1zfa from scaffold 307 but not mhc1zja from scaffold 312.
In addition, a Southern blot was performed employing a mixture of probes designed to detect all zebrafish Z genes and genomic DNA from eight of the nine individuals used for PCR-based genotyping (see Electronic Supplementary File, Table S2 for predicted fragment sizes). The observed differences in hybridization patterns indicate gene copy number variation across different genetic backgrounds. While in general, it is difficult to resolve bands correlating to specific Z genes, the presence of ~5.2 kb fragment (marked by an asterisk in Fig. 5b) in TU individuals 6 and 7 and EKW individual 2 matches the predicted size of the HindIII genomic fragment containing mhc1zka and confirms the genotyping result reported in Fig. 5a. There are also bands around 3 kb (marked by two asterisks in Fig. 5b) found only in the same three individuals suggesting that there may be an additional Z gene associated with the mhc1zka haplotype.
In analyzing the Southern blot, special attention was given to the hybridization pattern of the CG2 individual as this individual is homozygous for its Z gene haplotype. While the RT-PCR analysis (Fig. 4) showed the CG2 individual to express four Z genes (mhc1zba, mhc1zca, mhc1zda, and mhc1zfa), the Southern blot shows five distinct bands in the CG2 lane. There is a ~4-kb band that is unique to the CG2 line. While this could represent mhc1zla, there is no other evidence of this transcript being encoded by CG2 fish. It is likely that the CG2 individual represents a unique haplotype not present in other laboratory lines. The founder fish for the golden line, and thus the CG2 line, came from a private dealer and the fish have been inbred to maintain the mutant allele causing the golden phenotype for over 30 years (Chakrabarti et al. 1983). This may explain why multiple bands observed in the CG2 lane of the Southern blot do not match the predicted fragment sizes and why the Z gene alleles identified in the CG2 line have anywhere from 5 to 33 amino acid changes compared to the reference genome (see below).
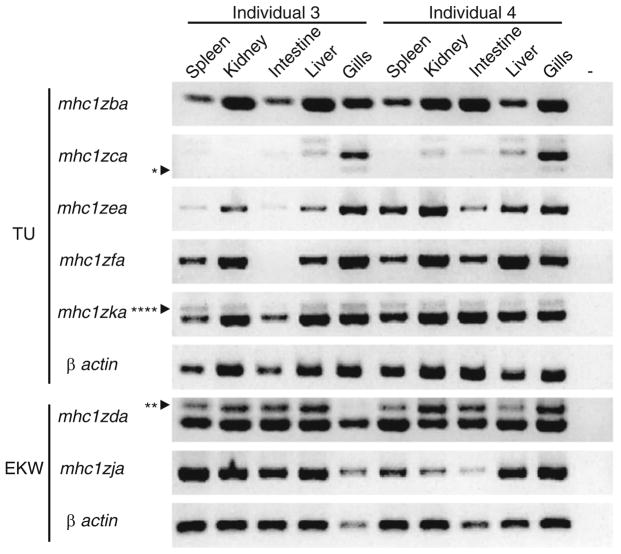
Zebrafish Z genes primarily show ubiquitous expression
As mentioned previously, a hallmark of classical MHC class I genes is that they are expressed ubiquitously on all nucleated cells while nonclassical genes typically exhibit tissue-specific expression. Seven of the zebrafish Z genes were successfully amplified from a panel of tissue-specific cDNA, and nearly all of them showed ubiquitous expression in the five tissue types tested (Fig. 6). The tissue-specific expression of mhc1zaa was unable to be assessed as transcripts of the gene were only ever identified in whole animal cDNA from two individuals of the TU background. Only two Z genes were identified that displayed tissue-specific expression. Our data demonstrate that mhc1zca is expressed strongly in the gills while only faintly expressed or absent in the other tissues. The same pattern was observed for mhc1zca in four other individuals of the AB and EKW backgrounds (data not shown). In contrast, mhc1zfa transcripts were detected in all tissues evaluated except for the intestine of one of the TU individuals. This result was not observed in additional zebrafish and no consistent pattern of mhc1zfa expression was identified. This data further supports the ambiguous classification of the zebrafish Z genes as classical or nonclassical as most but not all Z genes display ubiquitous expression.
Fig. 6.
Z gene expression. The same primers used in Fig. 4a were used to detect full-length Z transcripts from cDNA derived from the tissues of two TU or EKW individuals (different individuals than those used in Fig. 4). A negative control with no cDNA was included for each gene (−). Splice variants are labeled as in Fig. 4: one asterisk represents mhc1zca without an α3 domain (reading frame maintained), two asterisks represent mhc1zda with intronic sequence between the leader and α1 domains (reading frame not maintained), and four asterisks represent mhc1zka with intronic sequence between the α2 and α3 domains (reading frame not maintained)
Z gene polymorphism and genetic background
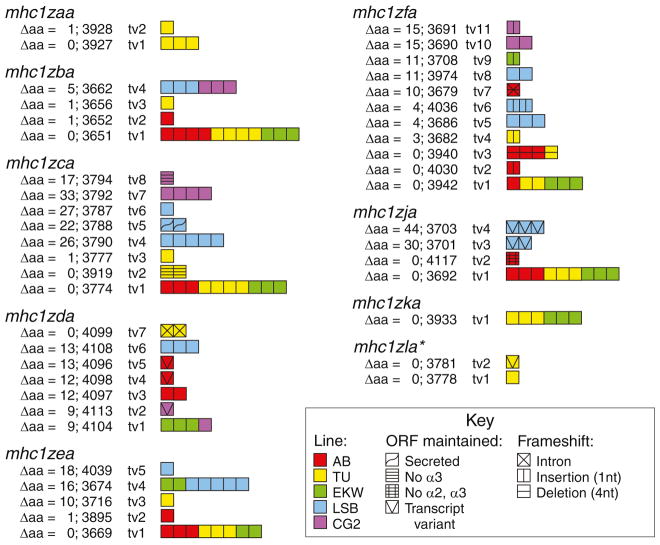
Sequencing transcripts of the nine Z genes amplified from five individual zebrafish from different genetic backgrounds revealed that the level of polymorphism ranges from 0 to 44 amino acid changes as compared to the genomic sequences reported in Fig. 1 (Fig. 7). The overall trend is that alleles with no or few amino acid changes were identified in the standard laboratory lines (AB, TU, EKW) while the more polymorphic alleles were predominately found in the LSB and CG2 zebrafish. The TU zebrafish express alleles of all nine Z genes that exactly match the reference sequence. This is as expected as the zebrafish reference genome (Zv9) and the BAC clone encoding the mhc1zaa and mhc1zla genes (DKEY-28D10) are derived from animals of the TU background. As diploid animals, the AB, TU, EKW, and LSB fish all express either one or two alleles for each gene while the clonal CG2 individual only expresses one allele per gene. Although the secreted isoform of mhc1zca was only identified in a LSB zebrafish and the isoform lacking the α3 domain was only detected in TU and CG2 zebrafish, it is likely that these splice variants are present in the fish of other genetic backgrounds, based on the bands observed in the PCR panel of Fig. 4a, and would be identified with sequencing of additional transcripts. In contrast, the band representing the mhc1zja isoform lacking both the α2 and α3 domains was only observed in AB zebrafish (Fig. 4a) which was also the only genetic background in which a transcript of the isoform was identified.
Fig. 7.
Allelic variation of Z genes. Full-length Z transcripts amplified from individual zebrafish from different genetic backgrounds were cloned and sequenced (Electronic Supplementary File, Fig. S1). Each sequenced transcript is represented by a square. Transcripts are grouped by gene and within each grouping they are classified into alleles based on the number of amino acid differences between its predicted protein sequence and the protein sequence predicted by the genomic loci described in Fig. 1 (Electronic Supplementary File, Fig. S4). Each allele is assigned a four digit number. The squares are color coded to indicate from which genetic background the transcript was identified. Different symbols are overlaid on squares representing transcript variants. The symbols are grouped by those that maintain the reading frame and those that cause an unproductive frameshift. *Note: mhc1zca and mhc1zla transcripts were amplified by the same primers
Transcripts of mhc1zfa with mutations causing unproductive frameshifts were recovered from all genetic backgrounds. In fact, nonfunctional mhc1zfa transcripts were more common in AB zebrafish than functional transcripts. Observed mutations included a consistent insertion of a single A nucleotide within a stretch of nine A’s, a consistent deletion of four nucleotides, and the inclusion of the intron between the leader and α1 domain.
The correlation between genetic background and polymorphism may help to explain the variable intensity of the PCR bands observed in Fig. 4a. For example, the bands for mhc1zba are much stronger in the AB, TU, and EKW fish than in LSB and CG2 fish. Accordingly, the sequencing results shown in Fig. 7 indicate that the mhc1zba transcripts from AB, TU, and EKW fish all had zero or one amino acid change while the transcripts from LSB and CG2 fish all had five amino acid changes. These more divergent transcripts from the LSB and CG2 fish suggest the possibility of polymorphism at the primer binding sites in these animals leading to less efficient amplification of the transcripts by PCR.
Transcripts 3701 and 3703 were isolated from a wild-derived LSB zebrafish using primers designed to amplify mhc1zja; however, based on the following observations, it is plausible that these transcripts represent an additional Z gene and a third chromosome 3 haplotype. First, when compared to all other characterized Z gene transcripts, the proteins encoded by transcripts 3701 and 3703 are two of the most divergent from the reference genome (Electronic Supplementary File, Fig. S2) differing by up to 11 % on the amino acid level (Fig. 7), suggesting that they may not be encoded by mhc1zja. Second, both of these transcripts include a cytoplasmic exon that is not present in mhc1zja (scaffold 312); however, a sequence highly similar to this novel exon can be identified within an intron of mhc1zka (scaffold NA257). These observations, along with the previous observation that exon swapping may have contributed to the sequence diversity of the Z gene family (Electronic Supplementary File, Fig. S3), suggest that transcripts 3701 and 3703 represent an additional Z gene that shares similarity to both mhc1zja and mhc1zka. It is predicted that these transcripts are encoded by a third chromosome 3 haplotype that is present in a subset of zebrafish in the wild, but not yet detected from laboratory lines.
Concluding remarks
Altered levels of Z gene expression have been reported in response to different immune stimuli and disease states. The gene zgc:64115 which has herein been reclassified as mhc1zba was reported to undergo a 12-fold decrease in expression in response to siRNA silencing of the zebrafish peptidoglycan recognition protein (zfPGRP-SC), a protein thought to function as a bacterial recognition molecule (Chang et al. 2009). In studying the metastatic properties of osteosarcoma, it was discovered that transformed mesenchymal stem cells (MSCs) capable of forming osteosarcoma in mice induced a host response when injected into zebrafish embryos that included decreased expression of a Z gene as compared to the injection of nontransformed MSCs (Mohseny et al. 2012). However, this data is confounding as the sequence of the probes used in the microarray analysis were specific to the locus defined here as mhc1zaa while the primers used in the qRT-PCR analysis were specific to mhc1zja. It has also been reported that immunization of zebrafish with Edwardsiella tarda live attenuated vaccine causes a 3-fold increase in levels of si:ch211-187j19.6, or mhc1zea, transcripts in the liver (Yang et al. 2012). While incomplete, these data suggest that the Z genes do play a role in zebrafish immune function.
The Z genes are an interesting gene family as they are not easily classified into either the classical or nonclassical MHC class I subgroups, but seem to fit into a separate or intermediate classification. This is based on their characterization as having low level polymorphism primarily concentrated in the α3 domain, their exhibition of primarily ubiquitous expression, and their maintenance of important peptide binding motifs. In order to better understand the function of this unique class of molecules, it remains to be determined if they bind and present peptide, associate with β2-microglobulin at the cell surface, and interact with CD8+ T cells. While polymorphism has proven to play a role in the ability to detect transcripts of Z genes, haplotypic variation is likely involved in diversifying the Z gene repertoire across different individual zebrafish. This is supported by genomic sequence data, genotyping PCR, and Southern blot analyses indicating variable Z gene copy number across different genetic backgrounds. Efforts to accurately identify the haplotypes that exist within this species will rely on genomic sequencing of numerous individual zebrafish from a variety of genetic backgrounds. This knowledge would aid in the ability to identify zebrafish that are immune-matched at multiple MHC class I loci for improved success in hematopoietic and other transplantation experiments.
Supplementary Material
Acknowledgments
The zebrafish Z gene nomenclature has been approved by the Zebrafish Nomenclature Committee. We thank Brian Dixon (University of Waterloo), Hans (JM) Dijkstra (Fujita Health University), and Amy Singer (Zebrafish Model Organism Database, University of Oregon) for very helpful discussions and advice on MHC gene nomenclature, and Jill de Jong and Sean McConnell (University of Chicago) for sharing their knowledge regarding the MHC class I U genes. We thank John Rawls (University of North Carolina), John Godwin (NC State University), and Sergei Revskoy (Northwestern University) for sharing their TU, LSB, and CG2 fish, respectively. We thank Ivan Rodriguez-Nunez (North Carolina State University) and Carlos Rivera (Millbrook High School) for assistance with experiments. H.D. is supported in part by a National Institutes of Health Biotechnology Traineeship (T32 GM008776) and by a Joseph E. Pogue Fellowship through the UNC Royster Society of Fellows.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00251-013-0748-z) contains supplementary material, which is available to authorized users.
Contributor Information
Hayley Dirscherl, Department of Molecular Biomedical Sciences, North Carolina State University, 1060 William Moore Drive, Raleigh, NC 27607, USA. The Joint Biomedical Engineering Graduate Program, University of North Carolina–North Carolina State University, 911 Oval Drive, Raleigh, NC 27695, USA.
Jeffrey A. Yoder, Email: Jeff_Yoder@ncsu.edu, Department of Molecular Biomedical Sciences, North Carolina State University, 1060 William Moore Drive, Raleigh, NC 27607, USA. Center for Comparative Medicine and Translational Research, North Carolina State University, 1060 William Moore Drive, Raleigh, NC 27607, USA
References
- Barclay AN. The leucocyte antigen factsbook. Academic; San Diego: 1997. [Google Scholar]
- Chakrabarti S, Streisinger G, Singer F, Walker C. Frequency of gamma-ray induced specific locus and recessive lethal mutations in mature germ cells of the zebrafish, Brachydanio rerio. Genetics. 1983;103:109–123. doi: 10.1093/genetics/103.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MX, Wang YP, Nie P. Zebrafish peptidoglycan recognition protein SC (zfPGRP-SC) mediates multiple intracellular signaling pathways. Fish Shellfish Immunol. 2009;26:264–274. doi: 10.1016/j.fsi.2008.11.007. [DOI] [PubMed] [Google Scholar]
- de Jong JL, Zon LI. Histocompatibility and hematopoietic transplantation in the zebrafish. Adv Hematol. 2012;2012:282318. doi: 10.1155/2012/282318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong JL, Burns CE, Chen AT, Pugach E, Mayhall EA, Smith AC, Feldman HA, Zhou Y, Zon LI. Characterization of immune-matched hematopoietic transplantation in zebrafish. Blood. 2011;117:4234–4242. doi: 10.1182/blood-2010-09-307488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra JM, Katagiri T, Hosomichi K, Yanagiya K, Inoko H, Ototake M, Aoki T, Hashimoto K, Shiina T. A third broad lineage of major histocompatibility complex (MHC) class I in teleost fish; MHC class II linkage and processed genes. Immunogenetics. 2007;59:305–321. doi: 10.1007/s00251-007-0198-6. [DOI] [PubMed] [Google Scholar]
- Dixon B, Stet RJ. The relationship between major histocompatibility receptors and innate immunity in teleost fish. Dev Comp Immunol. 2001;25:683–699. doi: 10.1016/s0145-305x(01)00030-1. [DOI] [PubMed] [Google Scholar]
- Green MR, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2012. [Google Scholar]
- Hashimoto K, Nakanishi T, Kurosawa Y. Isolation of carp genes encoding major histocompatibility complex antigens. Proc Natl Acad Sci U S A. 1990;87:6863–6867. doi: 10.1073/pnas.87.17.6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Bontrop RE, Dawkins RL, Erlich HA, Gyllensten UB, Heise ER, Jones PP, Parham P, Wakeland EK, Watkins DI. Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics. 1990;31:217–219. doi: 10.1007/BF00204890. [DOI] [PubMed] [Google Scholar]
- Kruiswijk CP, Hermsen TT, Westphal AH, Savelkoul HF, Stet RJ. A novel functional class I lineage in zebrafish (Danio rerio), carp (Cyprinus carpio), and large barbus (Barbus intermedius) showing an unusual conservation of the peptide binding domains. J Immunol. 2002;169:1936–1947. doi: 10.4049/jimmunol.169.4.1936. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs MF, Harstad H, Bakke HG, Beetz-Sargent M, McKinnel L, Lubieniecki KP, Koop BF, Grimholt U. Comprehensive analysis of MHC class I genes from the U-, S-, and Z-lineages in Atlantic salmon. BMC Genomics. 2010;11:154. doi: 10.1186/1471-2164-11-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalova V, Murray BW, Sultmann H, Klein J. A contig map of the Mhc class I genomic region in the zebrafish reveals ancient synteny. J Immunol. 2000;164:5296–5305. doi: 10.4049/jimmunol.164.10.5296. [DOI] [PubMed] [Google Scholar]
- Mizgirev IV, Revskoy S. A new zebrafish model for experimental leukemia therapy. Cancer Biol Ther. 2010;9:895–902. doi: 10.4161/cbt.9.11.11667. [DOI] [PubMed] [Google Scholar]
- Mohseny AB, Xiao W, Carvalho R, Spaink HP, Hogendoorn PC, Cleton-Jansen AM. An osteosarcoma zebrafish model implicates Mmp-19 and Ets-1 as well as reduced host immune response in angiogenesis and migration. J Pathol. 2012;227:245–253. doi: 10.1002/path.3998. [DOI] [PubMed] [Google Scholar]
- Nonaka MI, Aizawa K, Mitani H, Bannai HP, Nonaka M. Retained orthologous relationships of the MHC Class I genes during euteleost evolution. Mol Biol Evol. 2011;28:3099–3112. doi: 10.1093/molbev/msr139. [DOI] [PubMed] [Google Scholar]
- Okamura K, Nakanishi T, Kurosawa Y, Hashimoto K. Expansion of genes that encode MHC class I molecules in cyprinid fishes. J Immunol. 1993;151:188–200. [PubMed] [Google Scholar]
- Sambrook JG, Figueroa F, Beck S. A genome-wide survey of Major Histocompatibility Complex (MHC) genes and their paralogues in zebrafish. BMC Genomics. 2005;6:152. doi: 10.1186/1471-2164-6-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisapoome P, Ohira T, Hirono I, Aoki T. Genes of the constant regions of functional immunoglobulin heavy chain of Japanese flounder, Paralichthys olivaceus. Immunogenetics. 2004;56:292–300. doi: 10.1007/s00251-004-0689-7. [DOI] [PubMed] [Google Scholar]
- Stet RJ, Kruiswijk CP, Dixon B. Major histocompatibility lineages and immune gene function in teleost fishes: the road not taken. Crit Rev Immunol. 2003;23:441–471. doi: 10.1615/critrevimmunol.v23.i56.50. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Figueroa F, O’hUigin C, Klein J. Cloning and characterization of class I Mhc genes of the zebrafish, Brachydanio rerio. Immunogenetics. 1995;42:77–84. doi: 10.1007/BF00178581. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Wong RY, Perrin F, Oxendine SE, Kezios ZD, Sawyer S, Zhou L, Dereje S, Godwin J. Comparing behavioral responses across multiple assays of stress and anxiety in zebrafish (Danio rerio) Behaviour. 2012;149:1205–1240. [Google Scholar]
- Yang D, Liu Q, Yang M, Wu H, Wang Q, Xiao J, Zhang Y. RNA-seq liver transcriptome analysis reveals an activated MHC-I pathway and an inhibited MHC-II pathway at the early stage of vaccine immunization in zebrafish. BMC Genomics. 2012;13:319. doi: 10.1186/1471-2164-13-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.