Abstract
Stable isotope labeling with amino acids in cell culture (SILAC) was used to quantitatively study the host cell gene expression profile, in order to achieve an unbiased overview of the protein expression changes in BHK-21 cells infected with FMDV serotype Asia 1. The SILAC-based approach identified overall 2,141 proteins, 153 of which showed significant alteration in the expression level 6 h post FMDV infection (57 up-regulated and 96 down-regulated). Among these proteins, six cellular proteins, including three down-regulated (VPS28, PKR, EVI5) and three up-regulated (LYPLA1, SEC62 and DARs), were selected according to the significance of the changes and/or the relationship with PKR. The expression level and pattern of the selected proteins were validated by immunoblotting and confocal microscopy. Furthermore, the functions of these cellular proteins were assessed by small interfering RNA-mediated depletion, and their functional importance in the replication of FMDV was demonstrated by western blot, reverse transcript PCR (RT-PCR) and 50% Tissue Culture Infective Dose (TCID50). The results suggest that FMDV infection may have effects on the expression of specific cellular proteins to create more favorable conditions for FMDV infection. This study provides novel data that can be utilized to understand the interactions between FMDV and the host cell.
Introduction
Foot-and-mouth disease (FMD) is one of the most economically important diseases of cloven-hoofed animals because it severely compromises livestock production, resulting in high economic losses and international restrictions on the export of animals and animal products [1].The causative agent is the foot-and-mouth disease virus (FMDV) belonging to genus Aphthovirus of the family Picornaviridae, which is a non-enveloped virus possessing positive polarity and a single-stranded RNA genome around 8kb in length. The viral RNA is translated into one large polyprotein that is post-translationally cleaved by viral proteinase into individual structural and nonstructural proteins. Four structural proteins, VP1, VP2, VP3, and VP4, form the natural empty capsid with 60 copies of each protein. Nonstructural proteins, including Lpro, 2A, 2B, 2C, 3A, 3B, 3Cpro and 3Dpol, regulate viral replication, protein processing, and protein modification in the host [1, 2]. During infection, the virus affects the host cell in multiple ways through viral proteins. Accordingly, viral proteins can be grouped into different functional categories. P1 protein, especially VP1, has a role in virus attachment. Lpro, a papain-like proteinase, mediates autocatalytic cleavage from a polyprotein and is involved in modulating host response to infection by inhibiting the transcription of various immunological factors to antagonize the antiviral state of the host [3, 4]. 3Cpro can inhibit host cell transcription by cleavage of host proteins[5]. 2A is an autoproteinase involved in the cleavage of virus polyprotein[6, 7]. 2B, 2C and 3A proteins co-localize with the intracellular membrane system and may be related to viral RNA replication. 3B (VPg) can trigger virus genome replication by binding to the 5′ terminus of the genome [7]. 3Dpol interacts with 3A to form the viral replication complex and fulfill the function of a replicase[8].
FMDV infection results in multiple effects on the host cell, such as signaling pathways and morphology. During its whole life cycle, FMDV can interact with the host cell in several ways, ranging from activation of the innate immunity to establish an antiviral state in the cell, to utilizing cellular processes for efficient replication of FMDV[9–12]. Nevertheless, the pathogenesis of FMDV is not well understood at present. Novel insight into host-virus interactions gained from quantitative proteomic studies of the infected cells would drastically enhance our understanding of the FMDV infection processes, and may reveal novel cellular targets for the design of more effective antivirals and the development of new vaccine strategies. To this end, a global approach to investigate changes in the host cell proteome in response to FMDV infection, namely, stable isotope labeling with amino acids in cell culture (SILAC) was used in conjunction with LC–MS/MS in this study. The results showed that the abundance and expression patterns of 153 proteins were significantly changed upon FMDV infection. Among them, six proteins were selected and validated by using western blot, laser confocal microscopy and other functional assays. These studies confirmed there were significant alterations of cellular protein expression profile in FMDV-infected cells compared with mock-infected cells, and verified that variations in the expression of specific proteins would render differential effects on FMDV replication at different steps of the replication cycle, such as RNA replication, protein translation and virus titer.
Materials and Methods
Cells and virus
Baby hamster kidney 21 (BHK-21) from China Center for Type Culture Collection (CCTCC, Wuhan, China) were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS; HyClone), 100 U/mL penicillin and 100 mg/mL streptomycin, at 37°C with 5% CO2. FMDV strain Asia1/Jiangsu/China/2005 (GenBank Accession No. EF149009) was preserved at OIE/National Foot-and-Mouth Disease Reference Laboratory (Lanzhou, Gansu, P.R China) and propagated for 10 passages in BHK-21 cells.
Isotopic labeling in cell culture
DMEM was supplemented with 10% dialyzed fetal calf serum and all of the lacking amino acids (Invitrogen), except for L-lysine. The medium was then divided and supplemented with L-Lysine-2HCl or [U-13C6]-L-Lysine (Invitrogen) to produce light or heavy SILAC medium, respectively. BHK-21 cells were grown in parallel in both media, and passage was routinely performed every 2 to 3 days (as appropriate) at 80% to 90% confluence. After a minimum of seven population doublings, the cells were analyzed by mass spectrometry (MS) and achieved virtually complete incorporation of heavy L-lysine (S1 and S2 Figs).
Infection
Heavy-labeled cells were infected with FMDV, and BHK-21 cells cultured in medium with conventional amino acid source were used as mock-infected controls. Cells passaged in the two media were seeded in 75 cm2 cell culture flasks until they reached 75% confluence. The heavy-labeled cells were then infected with FMDV at a multiplicity of infection (MOI) of 1 and the light-labeled cells were mock-inoculated. Meanwhile, the dialyzed serum in the SILAC media was decreased to 2%. Cells were harvested at 6 hours post-infection (h.p.i.) using cell lysis buffer.
Gel electrophoresis and in-gel digestion
The total protein concentrations in the cell lysates were quantified and equivalent amounts of cell proteins from two independent biological replicates were diluted in SDS-PAGE loading buffer containing 5% β-mercaptoethanol prior to boiling, separated using one-dimensional SDS-PAGE (4% to 12% Novex Bis-Tris mini gel, Invitrogen), and then visualized by Coomassie blue stain (Pierce). The entire protein gel lane was excised in parallel and cut into 48 gel slices each. The gel slices were subsequently subjected to in-gel digestion with trypsin (20 ng/μL) at 37°C overnight, as described previously [13, 14]. The resulting tryptic peptides were extracted by 2.5% formic acid and 90% acetonitrile, lyophilized in a SpeedVac (Helena Biosciences), and resuspended in 0.1% formic acid and 2% acetonitrile before LC–MS/MS analysis.
LC–MS/MS
LC–MS/MS analysis was performed by Guangzhou Fitgene Biotechnology Co., Ltd., as described previously [14, 15]. In brief, the gel was cut into 48 slices from which proteins were digested and consequent peptides were extracted and then lyophilized before further analysis. The powder of peptides was resuspended in solvent A (2% acetonitrile, 0.1% formic acid in water) and loaded onto the C18 reverse phase column (100 μm in diameter, 15 cm long, 3 μm resin from Michrom Bioresources, Auburn, CA). Each mixture of peptide was separated with a linear gradient of solvent B from 5% to 15% within 15 minutes followed by a gradient from 15% to 35% over 85 minutes, end and sustain at 90% for the following 20 minutes. Eluted peptides were injected directly to LTQ-Obitrap XL (Thermo Fisher Scientific, Inc.) through a nanoelectrospray ion source (ProxeonBiosystems) with a voltage of 1.85kV and a temperature of transfer capillary of 200°C. Data-dependent mode was employed to acquire data using Xcalibur software (Thermo Electron). An accumulation of 106 ions was required to trigger a MS full scan, with maximal accumulation time of 500 ms and resolution of 60000 (m/z 400), ranging from 400–2000 Dalton. Six most intensive ions per MS scan were selected and fragmented by CID in LTQ to perform the MSMS scan with an accumulation of at least 5000 ions and the maximal accumulation time of 100 ms. The normalized collision energy was 35%, activation Q was 0.25, activation time was 30 ms, dynamic exclusion was enable with a maximum retention period of 90 s and a relative mass window of 10 ppm. The lock mass (PCM, MW445.12) was introduced to improve mass accuracy in survey scan.
Quantification, bioinformatics and protein analysis
Quantification was carried out using MaxQuant version [1.0.13.13], based on two-dimensional centroid of the isotope clusters within each SILAC pair, as previously described [14, 15]. Protein ratios were calculated as the median of all SILAC pair ratios belonging to peptides in the protein. The derived peak list was searched using the Mascot search engine (version 2.1.04; Matrix Science, London, UK) against the NCBI Cricetidae database under taxonomy ID 337677 (Date:2012-01-09,Total number of sequence:64093) and the decoy database (reverse database). The normalized H/L ratios, significance, and variability (%) were automatically produced by the MaxQuant 1.0.13.13 program. The resulting proteingroups.txt output file contained the peptide identification was imported into Microsoft Excel. The threshold of acceptance was set at false discovery rate (FDR) 1% at both peptide and protein level. Both unique and razor peptides included in the protein group were used for quantification with minimal ratio count of 1 (the ratio of protein group was calculated once a spectra with quantitative ratio was available). Ingenuity Pathways Analysis (Ingenuity Systems, www.ingenuity.com) was used to establish probable interaction networks of altered proteins, based on published reports and protein interaction databases. Data sets containing gene identifiers and corresponding expression values were used to generate the networks. Each gene in Ingenuity Pathways Knowledge Base was matched to the corresponding gene identifier.
Western blot analysis
Total protein concentrations in cell lysates prepared from FMDV-infected and uninfected cells harvested at 6 h p.i. were measured, and equivalent amounts of cell lysates from two independent biological replicates were denatured by heating at 100°C for 10 min in 5×sample loading buffer and separated using 10% SDS-PAGE. Proteins were electrotransferred to 0.45 μm nitrocellulose membranes (Bio-Rad), blocked with 5% nonfat milk in TBS containing 0.05% Tween-20 (TBST) overnight at 4°C. The membranes were then incubated for 1 h at room temperature with specific antibodies against DARS (Santa Cruz Biotechnology, Inc., USA), LYPLA1 (Abcam, UK), SEC62 (Sigma-Aldrich, USA), EIF2AK2 (Abcam, UK), EVI5 (Sigma-Aldrich, USA),VPS28 (Sigma-Aldrich, USA), Tublin (Santa Cruz Biotechnology, Inc., USA)or β-actin (Santa Cruz Biotechnology, Inc., USA). Incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies (Sigma) was performed at room temperature for 1 h, followed by washing with TBST three times. The protein bands were visualized using Amersham ECL Plus Western Blot Detection Reagents (GE Healthcare).
Immunofluorescence assay
Coverslip-adhered BHK-21 cell monolayers were infected with FMDV at an MOI of 1. The plates were inoculated at 37°C over 1 h followed by replacement with fresh growth media, and were incubated at 37°C for another 5 h. The cells were fixed with 4% PFA and treated with 0.01% Triton X-100, with the following antibodies applied: DARS (Santa Cruz Biotechnology, Inc., USA), LYPLA1 (Abcam, UK), SEC62 (Sigma-Aldrich, USA), EIF2AK2 (Abcam, UK), EVI5 (Sigma-Aldrich, USA),VPS28 (Sigma-Aldrich, USA), or β-actin (Santa Cruz Biotechnology, Inc., USA). FMDV proteins were detected by pig anti-FMDV Asia 1 VLPs serum (primary antibody, 1:100) recognized by a TRITC-conjugated secondary antibody (Sigma). DAPI was used as counterstain to allow visualization of cell nuclei. Immunofluorescence confocal microscopy images were captured on an LSM510 META microscope (Carl Zeiss, Ltd.).
siRNA experiment
Three sets of siRNA per gene targeting three different sites within the coding region were purchased from Santa Cruz Biotechnology, Inc., USA. BHK-21 cells were seeded into six-well plates and transfected with Oligofectamine (Invitrogen) in a final concentration of 30 nM siRNA. In each experiment, siRNA universal negative controls (Sigma) were used as controls. The cells were then inoculated with FMDV at an MOI of 1 at 30 h after siRNA transfection. The cells were then washed and fresh culture medium was added 1 h after virus inoculation. Afterward, cells were obtained to assess the presence of infectious FMDV particles using Western blot, RT–PCR (reverse transcription–PCR), and 50% Tissue Culture Infective Dose (TCID50) at 6 h p.i. Each siRNA experiment was performed in triplicate.
TCID50 detection of FMDV after RNA interference
The TCID50 in the collected supernatants was determined by virus titration assay, as described previously [16]. Briefly, a BHK-21 cell suspension in DMEM with 5% FBS at a concentration of 1.5×106 cells/mL was dispensed at 50 μL per well into 96-well flat-bottomed tissue culture plates. The plates were rocked to achieve uniform suspension thickness and incubated at 37°C for 24 h to 36 h under 5% CO2 tension to attain 90% confluence. Serial 10-fold dilutions of virus stock prepared in FBS-less DMEM were added in 50 μL volumes to all wells. Plates were incubated at 37°C, 5% CO2 for 72 h, and the presence or absence of cytopathic effect (CPE) was then monitored. TCID50 was calculated by Reed–Muench method.
RT–PCR analyses of FMDV gene
Reverse transcription PCR analyses of FMDV gene post-RNA interference were performed to quantify FMDV RNA in BHK-21 cells. Total RNA was extracted using Trizol reagent (Ambion) according to the manufacturer’s protocol. Reverse primers for FMDV 3D or β-actin were used to synthesize cDNA fragments by using reverse transcriptase M-MLV (TaKaRa). Subsequently, PCR was performed using rTaq polymerase (TaKaRa) and specific primers for either FMDV 3D or β-actin (FMDV 3D primers, forward: 5-TTCGGCCTTTGATGCTAACCACTG-3, reverse: 5-GCATCCCGCCCTCAACAACAAT-3; β-actin primers, forward: 5-CGGCATCCACGAAACTAC-3, reverse: 5-ATCTTCATCGTGCTGGGCG-3). For replication of FMDV 3D gene or β-actin gene, the amplification program was set at 94°C for 25 s, 56°C for 25 s, 72°C for 20 sec for 20 cycles. The sizes and uniqueness of PCR products were verified by agarose gel electrophoresis.
Statistical analysis
The data of relative quantity in RT-PCR, western blot and TCID50 are presented as mean ± SD after analysis by Image J software. The statistical analysis of variance between groups was performed by SPSS Statistics 19.0 software. One-way ANOVA Comparison between groups using Least Significance Difference (LSD) was applied. Significant difference of all statistical tests was set at 0.05 (p < 0.05).
Results
Optimal time-point for collection of BHK-21 cells infected with FMDV serotype Asia 1
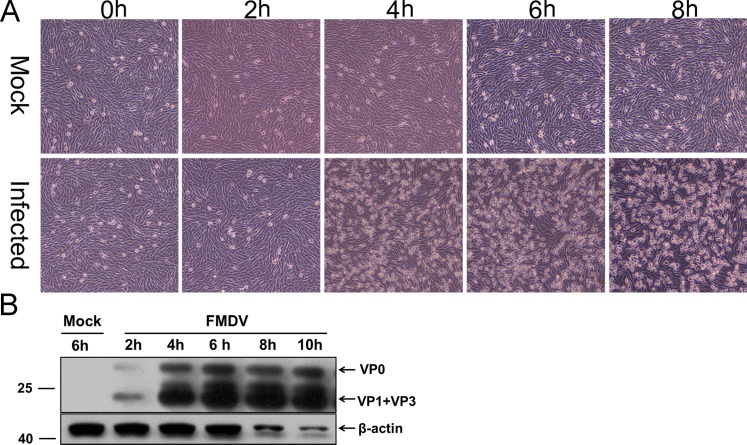
A distinctive feature in FMDV-infected BHK-21 cells is the formation of CPE, which denotes a virus-induced alteration of cells including general stress responses, especially cell death. Dead lytic cells plus FMDV-induced suppression of host cell protein synthesis cause a drastic decrease in the quantity of many cellular proteins, at times reaching undetectable. Thus, to ascertain a time-point for maximal effect with minimal negative effect of CPE after infection of FMDV, BHK-21 cells were infected with FMDV serotype Asia 1 at an MOI of 1 and microscopically monitored for CPE. Afterward, the FMDV protein was detected by Western blot against pig anti-FMDV Asia 1 serum over time. As shown in Fig 1A, CPE appeared at approximately 4 h p.i. and was readily observed at later time points. Consistently, the capsid protein of FMDV increased over time (Fig 1B). Although the FMDV protein expression reached the peak at 8h p.i., the expression of β-actin as a control was markedly decreased at this time point, suggesting virus-induced suppression of host cell protein synthesis and lysis or death of a majority of the cells at 8h p.i. Therefore, combining the results of CPE and the expression of FMDV and cellular proteins, we chose to examine the composition of cells at 6 h p.i. in the subsequent studies, at this time point the expression of virus structural proteins was just initialized and cellular proteins were kept stable.
Fig 1. FMDV infection in BHK-21 cells.
(A) Photomicrographs of BHK-21 cells infected with FMDV serotype Asia1 at MOI = 1 PFU/cell or mock-infected for the indicated hours post infection (h.p.i.). Images were taken at an original magnification of 100×. (B) Confirmation of FMDV capsid proteins in the cell lysate at the indicated h.p.i. by western blot using the pig anti-FMDV Asia 1 serum as primary antibody. Actin was used as loading control.
SILAC coupled with LC–MS/MS and bioinformatics analyses of FMDV-infected BHK-21 cells
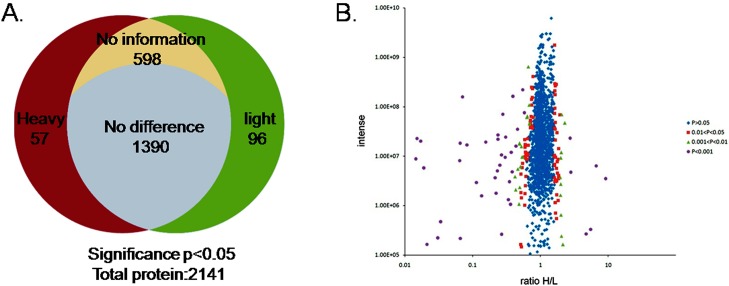
Although a previous paper used SILAC coupled with LC–MS/MS to identify and quantify proteome changes in IB-RS-2 cells infected with serotype O FMDV was published [15], no research related to quantitative proteomics of BHK-21 cells infected with FMDV serotype Asia 1 was available before this study. In this study, we obtained cellular proteomes from BHK-21 cells, known as the universal cell line for FMDV proliferation and production of FMD vaccine, and compared the differences in the levels of the isolated proteins by quantitative proteomics for the first time. Mock-infected cells were grown in media labeled with lightlysine medium, and cells infected with FMDV serotype Asia 1 at an MOI of 1 were grown in media containing heavy 13C6 HCl-Lys. Cells were collected at 6 h.p.i. Equal amounts of proteins were subsequently mixed, identified and quantified. For quantification, by setting the threshold at 0.05 in significance B, which was calculated from log protein ratios as a measurement of outlier significance score, proteins with values less than 0.05 were defined as significantly varied members and added to the candidate pool for investigating potential proteome changes between data sets through IPA (S1 Table). LC–MS/MS analysis identified 2,141 proteins in whole cellular fractions, which were identified by two or more peptides. Among these, 57 and 96 proteins were identified and quantified to be up-regulated and down-regulated at a significance B value≤0.05, as calculated by MaxQuant, respectively (Tables 1 and 2). Meanwhile, no significant change in abundance was observed for 1,390 proteins (64.92%), and 598 proteins (27.93%) could not be quantified (Fig 2).
Table 1. Proteins present in increased abundance in FMDV-infected cells.
| Accession number | Protein description | Protein name | Ratio (infection /control) | Peptides | Functions |
|---|---|---|---|---|---|
| gi|354485463 | membrane-associated progesterone receptor component 2-like | PGRMC2 | 9.20 | 2 | Receptor for steroids |
| gi|344254339 | Aspartyl-tRNA synthetase | DARS | 6.70 | 1 | aminoacyl-tRNA ligase activity |
| gi|344249005 | Translocation protein SEC62 | SEC62 | 5.52 | 2 | transporter activity |
| gi|354488911 | acyl-protein thioesterase 1-like | LYPLA1 | 4.75 | 3 | translation elongation factor activity |
| gi|354475980 | DNA ligase 1-like | LIG1 | 2.82 | 1 | ATP binding/DNA binding |
| gi|354494845 | lamina-associated polypeptide 2 | TMPO | 2.75 | 17 | DNA binding |
| gi|344257723 | Serine/arginine repetitive matrix protein 2 | SRRM2 | 2.35 | 1 | RNA splicing |
| gi|354481274 | dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase comple | DLST | 2.30 | 2 | transferase activity, transferring acyl groups |
| gi|344244946 | Lipase maturation factor 2 | LMF2 | 2.16 | 4 | endoplasmic reticulum membrane |
| gi|344246379 | Cancer-associated gene 1 protein-like | SSR1 | 2.06 | 4 | Endoplasmic reticulum |
| gi|354481751 | glypican-4 | GPC4 | 2.04 | 1 | glycoprotein binding |
| gi|344253619 | Legumain | LGMN | 2.04 | 4 | endopeptidase activity |
| gi|354465940 | disks large homolog 1 isoform 1 | DLG1 | 2.02 | 2 | iron ion binding |
| gi|125204 | cAMP-dependent protein kinase catalytic subunit alpha | PRKACA | 2.00 | 1 | protein kinase activity |
| gi|344245500 | Muscle-related coiled-coil protein | MURC | 2.00 | 1 | Activator/Developmental protein |
| gi|354475251 | neutral cholesterol ester hydrolase 1-like | NCEH1 | 1.99 | 1 | carboxylesterase activity |
| gi|344251417 | Putative deoxyribose-phosphate aldolase | DERA | 1.97 | 5 | Catalyzes a reversible aldol reaction between acetaldehyde and D-glyceraldehyde 3-phosphate to generate 2-deoxy-D-ribose 5-phosphate |
| gi|344258664 | Metalloproteinase inhibitor 1 | TIMP1 | 1.93 | 5 | enzyme inhibitor activity |
| gi|354485259 | synaptobrevin homolog YKT6-like | YKT6 | 1.86 | 2 | endoplasmic reticulum membrane |
| gi|354488481 | dnaJ homolog subfamily A member 3 | DNAJA3 | 1.86 | 2 | purine nucleotide binding |
| gi|354485509 | mitochondrial import receptor subunit TOM70-like | TIMM70A | 1.86 | 1 | receptor activity |
| gi|354489198 | eukaryotic initiation factor 4A-III-like | EIF4A3 | 1.83 | 3 | translation initiation factor activity |
| gi|354467882 | ATP synthase mitochondrial F1 complex assembly factor 2-like | ATPAF2 | 1.82 | 15 | May play a role in the assembly of the F1 component of the mitochondrial ATP synthase |
| gi|354478986 | metaxin-1-like | MTX1 | 1.80 | 3 | Involved in transport of proteins into the mitochondrion |
| gi|4126964 | peroxisomal membrane anchor protein | PEX14 | 1.76 | 1 | protein N-terminus binding |
| gi|153581939 | glycerolphosphate dehydrogenase | GPD2 | 1.75 | 12 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity |
| gi|354506683 | sorting and assembly machinery component 50 homolog | SAMM50 | 1.75 | 1 | May be required for the assembly pathway of mitochondrial outer membrane proteins |
| gi|354482314 | 5'-AMP-activated protein kinase catalytic subunit alpha-1-like | PRKAA1 | 1.74 | 6 | protein kinase activity |
| gi|344255846 | BAG family molecular chaperone regulator 3 | BAG3 | 1.73 | 1 | chaperone binding |
| gi|354497226 | integrin alpha-5 | ITGA5 | 1.73 | 1 | receptor activity |
| gi|1730177 | Glucose-6-phosphate isomerase | GPI | 1.72 | 5 | glucose-6-phosphate isomerase activity |
| gi|354498914 | delta-1-pyrroline-5-carboxylate dehydrogenase | ALDH4A1 | 1.68 | 1 | 1-pyrroline-5-carboxylate dehydrogenase activity |
| gi|344251256 | Annexin A11 | ANXA11 | 1.67 | 1 | calcium ion binding |
| gi|344249997 | Mitochondrial import inner membrane translocase subunit Tim17-B | TIMM17B | 1.67 | 2 | transporter activity |
| gi|2499845 | CD44 antigen | CD44 | 1.67 | 1 | Receptor |
| gi|344237837 | Mitochondrial 2-oxoglutarate/malate carrier protein | SLC25A11 | 1.67 | 1 | signal transducer activity |
| gi|354485616 | hydroxymethylglutaryl-CoA lyase | HMGCL | 1.67 | 7 | lyase activity |
| gi|354474977 | SRA stem-loop-interacting RNA-binding protein | SLRIP | 1.66 | 8 | RNA binding |
| gi|124426 | Inosine-5'-monophosphate dehydrogenase 2 | IMPDH2 | 1.66 | 1 | DNA binding |
| gi|354488213 | citrate synthase, mitochondrial-like | CS | 1.66 | 1 | transferase activity, transferring acyl groups |
| gi|354494828 | protein NipSnap homolog 2-like | GBAS | 1.65 | 7 | mitochondrion/Inferred from sequence or structural similarity |
| gi|344245669 | Prohibitin | PHB | 1.65 | 16 | sequence-specific DNA binding RNA polymerase II transcription factor activity |
| gi|354498001 | endoplasmic reticulum metallopeptidase 1-like | ERMP1 | 1.65 | 9 | peptidase activity |
| gi|327248646 | ATP synthase alpha subunit 1 | ATP5A1 | 1.64 | 4 | receptor binding |
| gi|344248533 | Electron transfer flavoprotein subunit beta | ETFB | 1.64 | 2 | electron carrier activity |
| gi|344259127 | GPI transamidase component PIG-S | PIGS | 1.63 | 1 | Component of the GPI transamidase complex |
| gi|354469583 | complement component 1 Q subcomponent-binding protein | C1QBP | 1.63 | 4 | Binds to the globular "heads" of C1Q thus inhibiting C1 activation |
| gi|344249404 | Prostacyclin synthase | PTGIS | 1.63 | 2 | monooxygenase activity |
| gi|344257987 | Isocitrate dehydrogenase | IDH2 | 1.62 | 1 | isocitrate dehydrogenase activity |
| gi|344245224 | Annexin A5 | ANXA5 | 1.62 | 2 | calcium ion binding |
| gi|344243386 | ATP synthase subunit delta, mitochondrial | ATP2D | 1.62 | 2 | transporter activity |
| gi|344236740 | Succinyl-CoA:3-ketoacid-coenzyme A transferase 1 | OXCT1 | 1.62 | 3 | 3-oxoacid CoA-transferase activity |
| gi|354483129 | ras-related protein Rab-12-like | RAB12 | 1.61 | 5 | GTP binding |
| gi|344243359 | Basigin | BSG | 1.61 | 2 | mannose binding |
| gi|15721885 | calnexin | CANX | 1.61 | 1 | calcium ion binding |
| gi|344245614 | Cytochrome b-c1 complex subunit 2 | UQCRC2 | 1.60 | 9 | metalloendopeptidase activity |
Table 2. Proteins present in decreased abundance in FMDV-infected cells.
| Accession number | Protein description | Protein name | Ratio (infection /control) | Peptides | Functions |
|---|---|---|---|---|---|
| gi|334303194 | eukaryotic translation initiation factor 2 alpha kinase 2 variant 2kinase 2 | EIF2AK2 | 0.01 | 4 | translation factor activity, nucleic acid binding |
| gi|354488450 | hypothetical protein LOC100769587 | hypothetical protein | 0.01 | 7 | No report |
| gi|354488340 | peroxisomal carnitine O-octanoyltransferase isoform 2 | CROT | 0.02 | 2 | transferase activity, transferring acyl groups |
| gi|344240087 | Protein KRI1-like | KRI1 | 0.02 | 15 | No report |
| gi|344251862 | EVI5-like protein | EVI5 | 0.02 | 6 | small GTPase regulator activity |
| gi|354474640 | MAP7 domain-containing protein 2 | MAP7D2 | 0.02 | 1 | No report |
| gi|344236597 | Vacuolar protein sorting-associated protein 28-like | VPS28 | 0.03 | 3 | Component of the ESCRT-I complex, a regulator of vesicular trafficking process |
| gi|354500438 | potassium voltage-gated channel subfamily D member 3-like | KCND3 | 0.03 | 1 | transporter activity |
| gi|344242595 | Dolichyl-diphosphooligosaccharide—protein glycosyltransferase subunit DAD1 | DAD1 | 0.06 | 7 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity |
| gi|344237874 | Anionic trypsin-2 | PRSS2 | 0.07 | 12 | peptidase activity |
| gi|354486039 | DDB1- and CUL4-associated factor 4-like | DCAF4L1 | 0.07 | 6 | No report |
| gi|354493412 | cullin-5-like isoform 1 | CUL5 | 0.07 | 2 | enzyme binding |
| gi|354478266 | NEDD8 ultimate buster 1 isoform 2 | NUB1 | 0.08 | 1 | Specific down-regulator of the NEDD8 conjugation system |
| gi|354475105 | KN motif and ankyrin repeat domain-containing protein 2 | KANK2 | 0.11 | 1 | guanyl-nucleotide exchange factor activity |
| gi|354466146 | plastin-1 | PLS1 | 0.14 | 10 | No report |
| gi|344254912 | Cleavage stimulation factor 50 kDa subunit | CSTF1 | 0.16 | 5 | One of the multiple factors required for polyadenylation and 3'-end cleavage of mammalian pre-mRNAs |
| gi|17376715 | Golgi apparatus protein 1 | GLG1 | 0.22 | 20 | receptor binding |
| gi|354473428 | immunoglobulin-like and fibronectin type III domain-containing protein 1-like | lrfn1l | 0.22 | 1 | May be involved in the regulation of excitatory synapses |
| gi|344236075 | Integrin alpha-1 | ITGA1 | 0.23 | 3 | receptor activity |
| gi|344237623 | Scaffold attachment factor B1 | SAFB | 0.24 | 1 | small molecule binding |
| gi|354477755 | ectopic P granules protein 5 homolog | EPG5 | 0.24 | 1 | Involved in autophagy. May play a role in the degradation step of the autophagy pathway |
| gi|354473176 | inverted formin-2-like | INF2 | 0.25 | 6 | cytoskeletal protein binding |
| gi|344243254 | U4/U6.U5 tri-snRNP-associated protein 1 | SART1 | 0.27 | 1 | May play a role in mRNA splicing. May also bind to DNA. |
| gi|344249797 | Phosphatidylinositol-binding clathrin assembly protein | PICALM | 0.27 | 2 | phospholipid binding |
| gi|354485371 | small acidic protein-like | - | 0.28 | 42 | No report |
| gi|344238413 | Phosphoacetylglucosamine mutase | PGM3 | 0.30 | 24 | magnesium ion binding |
| gi|344251937 | Signal transducer and activator of transcription 3 | STAT3 | 0.31 | 3 | signal transducer activity |
| gi|354507834 | calcium-binding and coiled-coil domain-containing protein 1-like | CALCOCO1 | 0.34 | 9 | chromatin binding |
| gi|154243355 | stearoyl-CoA desaturase 1 | SCD1 | 0.35 | 8 | stearoyl-CoA 9-desaturase activity |
| gi|344257067 | Coiled-coil domain-containing protein 6 | CUL3 | 0.36 | 7 | Core component of multiple cullin-RING-based BCR (BTB-CUL3-RBX1) E3 ubiquitin-protein ligase complexes which mediate the ubiquitination and subsequent proteasomal degradation of target proteins |
| gi|354472580 | sarcoplasmic/endoplasmic reticulum calcium ATPase 2-like | ATP2A2 | 0.37 | 1 | transporter activity |
| gi|28194647 | cathepsin L | CTSL1 | 0.38 | 23 | peptidase activity |
| gi|354470621 | myosin-3-like | MYH3 | 0.39 | 3 | cytoskeletal protein binding |
| gi|354468695 | calcium/calmodulin-dependent protein kinase type II subunit gamma-like | CAMK2G | 0.42 | 1 | protein kinase activity |
| gi|354504107 | endothelial differentiation-related factor 1-like | EDF1 | 0.43 | 2 | sequence-specific DNA binding |
| gi|354507394 | myosin regulatory light chain 2 | MYH2 | 0.44 | 1 | calcium ion binding |
| gi|344238999 | Isopentenyl-diphosphate Delta-isomerase 1 | IDI1 | 0.46 | 2 | hydrolase activity |
| gi|344247197 | putative E3 ubiquitin-protein ligase TRIP12 | TRIP12 | 0.47 | 5 | ligase activity |
| gi|354468967 | regulatory-associated protein of mTOR | PRTOR | 0.48 | 8 | Involved in the control of the mammalian target of rapamycin complex 1 (mTORC1) activity which regulates cell growth and survival |
| gi|354496956 | TAR DNA-binding protein 43-like isoform 2 | TARDBP | 0.49 | 7 | small molecule binding |
| gi|354465282 | E3 ubiquitin-protein ligase NEDD4-like | NEDD4 | 0.49 | 1 | ligase activity |
| gi|354482078 | exocyst complex component 4 | EXOC4 | 0.50 | 7 | Component of the exocyst complex involved in the docking of exocytic vesicles with fusion sites on the plasma membrane |
| gi|344237164 | RNA-binding protein 7 | RBM7 | 0.51 | 1 | small molecule binding |
| gi|344240655 | Integrin beta-3 | ITGB3 | 0.51 | 5 | receptor activity |
| gi|344242040 | Uncharacterized protein C1orf77-like | C11orf58 | 0.51 | 8 | No report |
| gi|344258772 | 40S ribosomal protein S6 | Rps6 | 0.52 | 1 | structural molecule activity |
| gi|344249426 | GTPase HRas | HRAS | 0.52 | 2 | GTP binding |
| gi|354471711 | microtubule-associated protein 1A-like | MAP1A | 0.52 | 2 | Structural protein involved in the filamentous cross-bridging between microtubules and other skeletal elements. |
| gi|354481682 | C-type mannose receptor 2 | MRC2 | 0.53 | 3 | receptor activity |
| gi|354478904 | ubiquitin-associated protein 2 isoform 1 | UBAP2L | 0.54 | 2 | binding of sperm to zona pellucida |
| gi|344250619 | Protein FAM49A | FAM49A | 0.54 | 1 | Inferred from electronic annotation |
| gi|344239224 | Vigilin | HDLBP | 0.55 | 1 | RNA binding |
| gi|344257977 | putative ATP-dependent RNA helicase DDX17 | DDX17 | 0.55 | 1 | helicase activity |
| gi|354494465 | developmentally-regulated GTP-binding protein 1 | DRG1 | 0.56 | 6 | GTP binding |
| gi|354498518 | suppressor of G2 allele of SKP1 homolog | SUGT1 | 0.58 | 6 | No report |
| gi|354483672 | spectrin beta chain | SPTB | 0.58 | 4 | cytoskeletal protein binding |
| gi|354476738 | lysosomal protective protein isoform 1 | CTSA | 0.59 | 3 | carboxypeptidase activity |
| gi|344256627 | DNA-directed RNA polymerase II subunit RPB3 | POLR2C | 0.59 | 2 | DNA-directed RNA polymerase activity |
| gi|354495341 | stathmin-like | STMN1 | 0.59 | 23 | Involved in the regulation of the microtubule (MT) filament system by destabilizing microtubules |
| gi|354502497 | cleavage and polyadenylation specificity factor subunit 7 isoform 1 | CPSF7 | 0.59 | 1 | small molecule binding |
| gi|344254882 | Maspardin | SPG21 | 0.60 | 4 | May play a role as a negative regulatory factor in CD4-dependent T-cell activation |
| gi|354479041 | cell division cycle 5-related protein-like | CDC5L | 0.60 | 1 | DNA binding |
| gi|344243055 | putative ATP-dependent RNA helicase DDX5 | DDX5 | 0.60 | 8 | helicase activity |
| gi|354488191 | SAP domain-containing ribonucleoprotein-like | Syncrip | 0.60 | 16 | No report |
| gi|354470481 | caprin-1-like | CAPRIN1 | 0.60 | 4 | RNA binding |
| gi|354504568 | DNA-directed RNA polymerase II subunit RPB7-like | RPB7 | 0.61 | 3 | DNA-directed RNA polymerase activity |
| gi|354473311 | 40S ribosomal protein S3-like | RPS3 | 0.61 | 1 | structural molecule activity |
| gi|354498985 | EF-hand domain-containing protein D2-like | EFHD2 | 0.61 | 1 | calcium ion binding |
| gi|354499471 | spectrin alpha chain, brain-like isoform 2 | SPTAN1 | 0.63 | 7 | Actin capping |
| gi|344237085 | U4/U6.U5 tri-snRNP-associated protein 2 | USP39 | 0.63 | 4 | ubiquitin thiolesterase activity |
| gi|354468104 | protein transport protein Sec23B isoform 1 | SEC23B | 0.64 | 1 | DNA-directed RNA polymerase activity |
| gi|344247943 | 40S ribosomal protein S3a | RPS3 | 0.64 | 1 | structural molecule activity |
| gi|344254132 | Dynamin-like 120 kDa protein | OPA1 | 0.65 | 10 | GTPase activity |
| gi|344250281 | Claudin-3 | CLDN3 | 0.65 | 1 | structural molecule activity |
| gi|344236900 | Pyridoxal-dependent decarboxylase domain-containing protein 1 | PDXDC1 | 0.65 | 5 | lyase activity |
| gi|354483820 | guanine nucleotide-binding protein subunit beta-2-like 1-like | GNB2L1 | 0.66 | 1 | ion channel inhibitor activity |
| gi|354492355 | eukaryotic translation initiation factor 4 gamma 2-like | EIF4G2 | 0.66 | 2 | translation initiation factor activity |
| gi|344255980 | Cullin-3 | Cul3 | 0.69 | 12 | enzyme binding |
| gi|354473860 | regulator of nonsense transcripts 1-like | GL50803_13452 | 0.69 | 2 | ATP binding |
| gi|344253625 | Actin-related protein 2/3 complex subunit 2 | ARPC2 | 0.69 | 2 | as actin-binding component of the Arp2/3 complex which is involved in regulation of actin polymerization and together with an activating nucleation-promoting factor (NPF) mediates the formation of branched actin networks. |
| gi|354486221 | long-chain-fatty-acid—CoA ligase 4-like | ACSL4 | 0.71 | 5 | ligase activity |
| gi|354506341 | protein transport protein Sec31A | SEC31A | 0.71 | 2 | protein transport |
| gi|344239124 | Protein transport protein Sec23A | Sec23A | 0.71 | 1 | zinc ion binding |
| gi|344251913 | Eukaryotic translation initiation factor 1 | EIF1 | 0.72 | 1 | translation initiation factor activity |
| gi|20373098 | RNA helicase | RNA-H | 0.72 | 3 | sequence-specific DNA binding transcription factor activity |
| gi|354493469 | transcription factor BTF3-like isoform 2 | BTF3 | 0.73 | 9 | transcription coactivator activity |
| gi|49652 | 40S ribosomal protein S24 | RPS24 | 0.74 | 1 | structural constituent of ribosome |
| gi|354476231 | coatomer subunit alpha isoform 2 | COPA | 0.74 | 3 | structural molecule activity |
| gi|344255737 | 40S ribosomal protein S26 | RPS26 | 0.75 | 23 | structural constituent of ribosome |
| gi|344248597 | Programmed cell death 6-interacting protein | PDCD61P | 0.75 | 6 | microtubule organizing center |
| gi|354497705 | 40S ribosomal protein S12-like | RPS12 | 0.76 | 3 | structural constituent of ribosome |
| gi|344240144 | Eukaryotic translation initiation factor 2 subunit 3 | EIF2 | 0.77 | 4 | translation initiation factor activity |
| gi|354488129 | nascent polypeptide-associated complex subunit alpha-like | Naca | 0.77 | 2 | DNA binding |
| gi|1083173 | carbohydrate-binding protein CBP30 | LGALS3 | 0.78 | 6 | sugar binding |
| gi|344258446 | 60S ribosomal protein L23a | RPL23A | 0.78 | 1 | Structural constituent of ribosome |
Fig 2. Distributions of proteins identified in BHK-21 cells infected with FMDV and proteome-wide accurate quantitation and significance.
(A) The left (black red color) represents up-regulated host proteins in virus-infected cells. The right (green color) represents down-regulated host proteins based on significance B ≤ 0.05. The other parts represent unchanged proteins in FMDV-infected cells. (B) Signal intensities (log10) of all quantified proteins in the FMDV-infected cells are shown as functions of their fold change (log2). The spread of the cloud is lower at high abundance, indicating that quantification is more precise. The criterion of being identified as a significantly regulated protein can be evaluated by the significance B level indicated in blue, red, purple, and green.
Based on the underlying biology evidence from the UniProtKB/Swiss-Prot, TrEMBL protein databases and the Gene Ontology database, 163 identified proteins were assigned to different molecular functional classes and subcellular annotations, in order to gain functional insights into the cellular proteome. The hamster genome database had poor annotation compared with the mouse genome, so the number of identified proteins was limited. To avoid missing information, gene identifications of proteins were converted to mouse protein GI numbers. Based on their underlying biological evidence from the literature database, the interacting pathways were constructed by importing the protein GI numbers and levels of regulation into the Ingenuity Pathways Analysis (IPA) tool. The underlying biological implication of the varied proteins was deciphered by enriching the annotations of Gene Ontology (GO) possessed by these proteins. The enrichment was conducted with GO miner in three main aspects comprising Biological Process (BP), Cellular Component (CC), and Molecular Function (MF) (Fig 3). In FMDV-infected cells, the up-regulated proteins are found to be mainly associated with cellular responses to stimuli, protein binding, localization and transport; whereas the down-regulated proteins are mainly related to the nucleotide and nucleoside activities, catabolic and hydrolase functions, alternative splicing, and cell adhesion. Fig 3 shows the relative proportion of proteins in each separate functional class (also see S3 Fig).
Fig 3. Classification of proteins in BHK-21 cells infected with FMDV according to their assigned fraction and biological function.
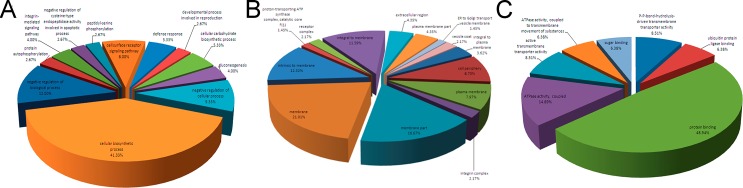
(A) Biological process; (B) Cellular component; (C) Molecular functions. More information is available in Supplemental material (S1 Table and S3 Fig).
Network pathway analysis
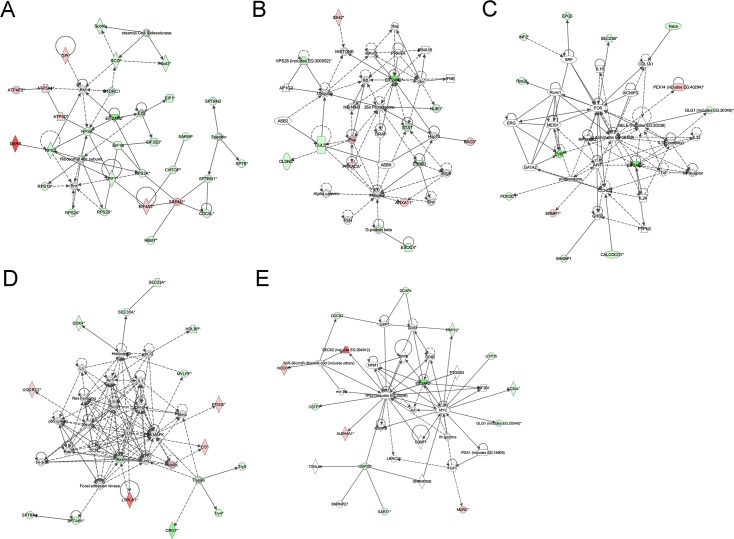
By importing the protein GI numbers and regulation levels into the Ingenuity Pathway Analysis (IPA) tool, the interacting pathways were constructed. A total of 14 pathways were identified at 95% or greater confidence level. Among them, 11 pathways have involved 11 or more significantly up- or down-regulated proteins (the so called “focus” members) (S4 Fig, S2 and S3 Tables). As suggested by network analysis, it appears that most proteins with altered abundance are involved in cellular assembly, organization, morphology, signaling and growth in FMDV-infected cells. For example, many of the proteins with significant regulation were linked to EIF2AK2 (also named PKR, double-stranded RNA-dependent protein kinase R) signaling network. Thus, combined with the significance of regulation, the five networks of interest are as follows: (1) Protein Synthesis, Gene Expression, Nucleic Acid Metabolism (Fig 4A); (2) Cell Death, Inflammatory Response, Infectious Disease (Fig 4B); (3) Hematological System Development and Function, Hematopoiesis, Cellular Development (Fig 4C); (4) Gene Expression, Cell Death, Cell Morphology (Fig 4D); (5) DNA Replication, Recombination and Repair, Cancer, Skeletal and Muscular Disorders (Fig 4E). Among these networks, up-regulated DARS and SEC62 proteins and down-regulated VPS28 and EVI5 significantly were involved in the PKR pathway. Only LYPLA1 protein bore no relation to PKR, but also was up-regulated significantly. As shown in Fig 4, identified up-regulated proteins present in the pathways are depicted in shades of red; proteins present in the pathways and identified as down-regulated are shown in green; and proteins present in the pathways and identified with no change in regulation are depicted in white. The intensity of color indicates the level of regulation.
Fig 4. Ingenuity pathway analysis of proteins that were significantly altered in BHK-21 cells infected with FMDV Asia 1.
The proteins indentified were involved in Protein Synthesis, Gene Expression, Nucleic Acid Metabolism (A); Cell Death, Inflammatory Response, Infectious Disease (B); Hematological System Development and Function, Hematopoiesis, Cellular Development (C); Gene Expression, Cell Death, Cell Morphology (D); DNA Replication, Recombination and Repair, Cancer, Skeletal and Muscular Disorders (E). Proteins shaded in green indicate decrease in abundance and proteins shaded in red indicate increase in abundance in the FMDV-infected cells compared with mock-infected cells. The color intensity corresponds to the degree of abundance. Proteins in white are those known to be in the network but were not identified in our study through the Ingenuity Pathways Knowledge Base. The shapes denote the molecular class of the protein. The solid lines indicate the promotional interactions while the dashed lines indicate the suppressive interaction.
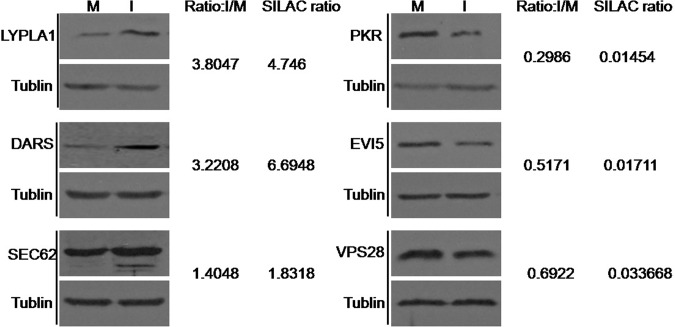
Validation of SILAC ratios by western blot
Combined with the results of IPA and calculation of MaxQuant, SEC62 and DARS in FMDV-infected cells are up-regulated significantly, whereasVPS28 and EVI5are down-regulated significantly. Although the LYPLA1 protein is not in the network related to EIF2AK2, its expression was also up-regulated significantly (ratio = 4.746). Thus, to rigorously validate the identification and the SILAC-determined protein ratios, we analyzed six selected proteins, including the up-regulated (SEC62, DARS, and LYPLA1) and the down-regulated (EIF2AK2, VPS28, and EVI5) proteins in infected and mock-infected cells by immunoblotting. As shown in Fig 5, the Western blot analysis results showed the consistency of the ratios of the six representative proteins in infected and uninfected cells with those obtained from SILAC.
Fig 5. Confirmation of six up-regulated or down-regulated proteins by immunoblot compared to the SILAC analysis.
Relative expression of six representative proteins (normalized to actin band) was determined using image densitometry by software Image J. The ratio of I/M is reflected by the band densitometry of infection compared to that of mock infection. M, Mock; I, infected.
Potential relocalization of cellular proteins in FMDV-infected cells
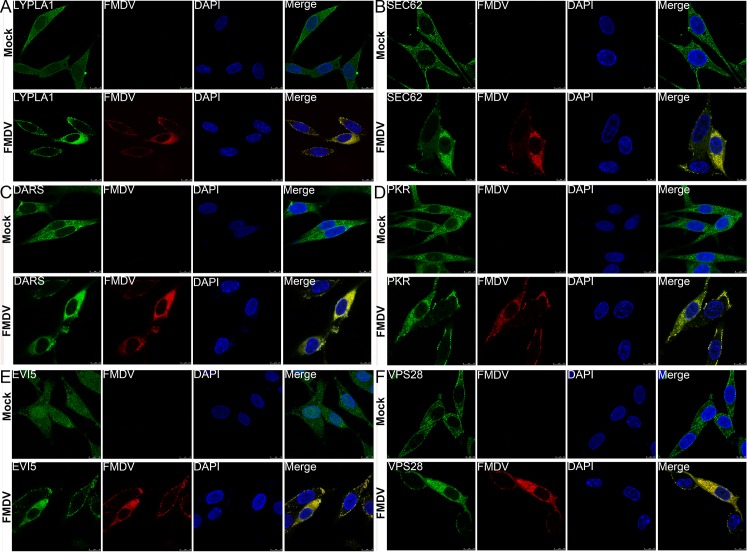
To investigate whether the six cellular proteins were redistributed in FMDV-infected cells or co-localized with FMDV capsid protein, BHK-21 cells were infected with FMDV, and their subcellular localization was determined at6 h.p.i by using specific antibody against six cellular proteins, respectively. As shown in Fig 6, except for the EVI5 protein which is localized to both the cytoplasm and the nucleus in mock-infected cells, other proteins including SEC62, DARS, PKR, LYPLA1 and VPS28 were localized in the cytoplasm. Upon FMDV infection, all six proteins were co-localized with the FMDV capsid proteins either in the cytoplasm or in the peripheral cytoplasm (Fig 6). EVI5 mainly re-localized with the FMDV capsid in the cytoplasm, with no remaining detection in the nucleus (Fig 6E). The translocation and colocalization of the six host proteins with the major FMDV proteins suggest that these host cell proteins may play a functional role in FMDV replication.
Fig 6. Subcellular localization of target proteins in mock- and FMDV-infected BHK-21 cells by confocal microscopy analysis.
Target proteins are stained green, FMDV proteins are shown in red, and the nuclei are stained blue with DAPI.
Potential importance of cellular proteins in the FMDV life cycle
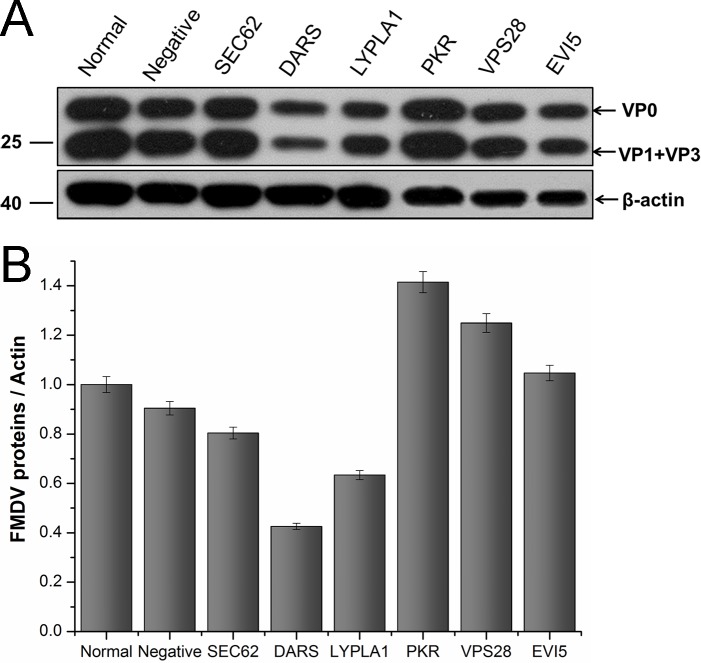
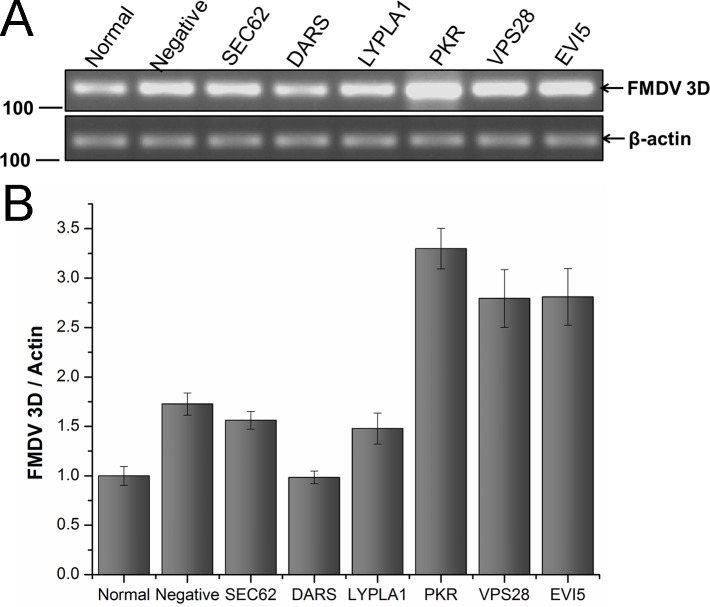
To investigate the importance of these cellular proteins in the FMDV life cycle, BHK-21 cells were transfected with specific siRNAs for each mRNA coding for the selected cellular target proteins. Cell pellets were collected at 30 h after siRNA transfection and analyzed by western blotting. The results indicated that the expression of all six proteins was decreased in the presence of specific siRNA, but not with the non-targeting siRNA (S5 Fig) at 30 h after transfection when compared to cells untreated with siRNA. In the subsequent experiments, the time point of 30 h post-siRNA transfection was chosen. At this time point, the transfected cells were infected with FMDV at an MOI of 1.To identify the functions of the six target proteins in FMDV infection, cells and cell culture medium were collected for western blot, RT–PCR, and TCID50 assay at 6 h after FMDV infection. Western blot analysis indicated that the abundance of FMDV capsid protein in SEC62-, DARS-, or LYPLA1-knockdown cells decreased compared with that of the negative control (nontargeting siRNA-treated cells) or normal cell control (untreated cells) (Fig 7). Among these cells, DARS-knockdown cells induced a significant decrease of FMDV capsid (Fig 7). However, in PKR-, VPS28-, or EVI5-knockdown cells, the abundance of FMDV capsid proteins increased, compared with that of the negative control (nontargeting siRNA-treated cells) or normal cell control (untreated cells) (Fig 7), which suggested the target proteins could affect the translation or expression of FMDV protein in cells. To identify whether the target proteins impact the replication of FMDV at the genome level, RT–PCR was used to investigate the abundance of FMDV 3D gene under different conditions using a specific primer pair (Fig 8). The data indicated that the amount of genomic RNA decreased in infected cells reduced of SEC62, DARS, or LYPLA1, but the amount of genomic RNA increased in infected cells depleted of PKR, VPS28, or EVI5 (Fig 8). It demonstrated that these host proteins could play a role during the replication of FMDV. To further study whether the target proteins may affect the assembly or release of FMDV, the FMDV titer in the cell culture medium was evaluated after the siRNA-mediated knockdown of these proteins (Fig 9). Variations in the FMDV titer were consistent with the viral protein and RNA levels, confirming that these host proteins may affect FMDV replication.
Fig 7. Effects of representative protein on FMDV replication in BHK-21 cells were confirmed by western blot analysis after RNA interference.
FMDV capsid protein was detected by the pig anti-FMDV Asia 1 serum. Relative expression of FMDV proteins was determined using image densitometry by software Image J. The relative quantity of FMDV protein was showed by normalization of ratio of FMDV capsid protein to actin with that ratio of normal cells. Bars represented the means±standard deviations (SD) of three independent experiments.
Fig 8. Quantification of FMDV gene in BHK-21 cells after knockdown genes of representative proteins.
The cells transfected with untargeting siRNA was set as negative control, the cells untransfected was blank control. FMDV RNA was indicated by the FMDV 3D gene which is conservative and stable in the FMDV genome. Relative quantity of gene was determined using image densitometry by software Image J. The relative quantity of FMDV gene was showed by normalization of ratio of FMDV 3D gene to actin gene with that ratio of normal cells. Bars represented the means±SD of three independent experiments.
Fig 9. FMDV titer was detected by the TCID50 after transfection with siRNA or untargeting siRNA.
The cells transfected with untargeting siRNA was set as negative control, the cells untransfected was blank control. Bars represent the means±SD of three independent experiments.
Discussion
A comprehensive knowledge of host cell protein expression profiles during FMDV infection would provide significant insights into the molecular mechanisms that regulate viral replication and pathogenesis. This may lead to the identification of cellular targets for the development of a new generation of antiviral drugs against essential host cell factors. This study aims to provide an unbiased global overview of the protein expression profiles in cells infected with FMDV through quantitative proteomics and subsequent bioinformatics analysis using IPA. The study highlights new interactions between FMDV and cellular proteome in BHK-21 cells in vitro. As we know, there are seven serotypes of FMDV which have different biological characteristics in infection of animals, suggesting that it would be necessary to comparatively study the interaction of each FMDV serotype with the host cell. Extending these types of analyses to other cell types and FMDV serotypes could lead to the identification of common and unique features in host interaction with individual serotypes and may provide an approach for ideal antiviral therapies. As far as we were aware, this study may represent the first quantitative proteomic analysis of cells infected with FMDV serotype Asia 1.The reason for selection of FMDV serotype Asia 1/Jiangsu/China/2005 as a model in this study is due to the fact that this strain is widely used as a vaccine strain in China and its biological property is well defined. The quantitative proteomic analysis on serotype Asia 1/Jiangsu/China/2005 would be the most representative for other FMDV serotype Asia 1 which is endemic in the Asia. In addition, BHK-21 cell line was selected as model because it is commonly used in investigating FMDV host–pathogen interactions, antiviral screening and vaccine production. Although the hamster genome database has poorer annotation compared with the mouse genome and many proteins are unassigned or uncharacterized, the origin of BHK-21 cell is hamster, which belongs to rodent species and has close relationship with mice. Hence, gene identifications of the identified proteins can be converted to mouse protein GI numbers, which can minimize the possibility of information missing error. It showed a majority of the cellular proteome remained relatively unchanged in virus-infected BHK-21 cells, suggesting that only specific changes occurred in response to virus infection. Proteins were identified and quantified at 6 h post-infection, prior to the appearance of major CPE. At this time point, the majority of the infected cells would remain viable for downstream processing. In this study, network pathway analysis indicated that the cellular proteins in the proteomes could be linked to signaling cascades, such as PKR. Interestingly, several significant up- and down-regulated proteins, including DARs, SEC62, VPS28 and EVI5, were related to PKR in different networks. Hence, in this study, DARs, SEC62, VPS28, EVI5 and PKR were selected to verify their functional importance in FMDV infection. Although, LYPLA1 seems to have no relationship with PKR, its expression increased significantly in BHK-21 cells infected with FMDV. Hence, LYPLA1 was also studied in this report.
PKR is a serine and threonine protein kinase that was expressed ubiquitously and identified as an antiviral and antitumor protein induced by interferon (IFN)[17]. This kinase contributes to inflammation and immune regulation through several signaling pathways and can be activated by multiple stimuli [17, 18]. Active PKR can mediate the activation of mitogen-activated protein kinases [19, 20], inhibitor of κB kinase [21, 22], and IFN-β-promoter simulator 1 (IPS-1) signaling [23]. Thus, PKR has multifaceted functions in the regulation of inflammatory and immune responses. As for the relationship of PKR and FMDV, previous research showed an important function of PKR in blocking FMDV replication at the protein translation level. In support of these findings, porcine and bovine cells treated with 2-aminopurine, an inhibitor of PKR, increased the virus yield for several folds compared with that of the untreated infected cells [24]. In addition, immediate-early induction of IFN-mRNA and mRNAs for some IFN-stimulated gene products, such as 2′5-oligoadenylate synthetase, Mx1 and PKR could be reduced by FMDV Lpro protein in swine cells. Knockdown of cellular PKR by RNA interference did not show obvious effect on wild-type virus yield, but resulted in a higher yield of A12-LLV2, a mutant virus lacking the Lpro protein. These results indicated a direct function of PKR to control FMDV replication in the natural host and the relationship of FMDV Lpro and PKR[25]. Thus, PKR was selected as a positive control in our study. In quantitative proteomics, the expression of PKR decreased in cells infected with FMDV, suggesting that there is an antagonism between PKR and FMDV. Consistent with this speculation, cellular PKR down-regulation by RNA interference resulted in a higher titer of FMDV, which further confirmed a direct function of PKR in controlling FMDV replication combined with previous research. Therefore, previously published data were also reflected by the bioinformatics analysis of the current quantitative proteomics data, and on the other hand indicated that the results of RNA interference assay in our study were reliable and positive.
The endoplasmic reticulum (ER) is the cellular site for production of secretory proteins, lipids, cell membranes and sterol. With the aid of molecular chaperons, newly synthesized proteins are folded and glycosylated in the ER lumen before budding and transport to the Golgi apparatus. A number of cellular stress conditions may interfere with the ER function and cause the accumulation of misfolded and unfolded proteins in the ER, leading to the initiation of a group of signal transduction pathways, known as the ER stress response or unfolded protein response [26, 27]. Interestingly, ER is also a cellular organelle exploited by many viruses for replication and maturation [28, 29]. In virus-infected cells, ER stress response can be induced, either by overwhelming the protein folding capability or by directly disrupting the ER structure and function, to facilitate viral replication and pathogenesis in some cases [30, 31]. Certain nonstructural proteins, such as 2B, 2C and 3A from some picornaviruses, are implicated in disrupting the ER integrity and functions [32–34]. On the other hand, some viruses in the picornavirus family may require ER membranes for the biogenesis of viral proteins and for anchoring the replication complexes [34–36]. In the present study, SEC62 was up-regulated after FMDV infection, suggesting that SEC62 proteins are beneficial to FMDV proliferation. This finding is consistent with the decrease of FMDV replication after knockdown of SEC62 protein expression.Sec62 has been found to interact with the ribosome, which suggests its function in co-translational translocation in mammalian cells [37]. Sec62 and Sec63 are important components that act in post-translational translocation at the mammalian ER for precursor chain of proteins [38]. Thus, SEC62–SEC63 balance is expected to contribute to the development of human tumorigenesis, possibly by relocation of the uncomplexed SEC62 to a yet undetermined cellular location along the secretory pathway. However, the association of the overexpression of SEC62 after FMDV infection with ER stress, as well as the mechanisms of ER stress that affect FMDV proliferation, is not clearly elucidated. The clarification of this mechanism, especially, the relationship between ER stress and FMDV replication, is beneficial in understanding the FMDV pathogenesis.
The basic function of aminoacyl-tRNA synthetases (ARSs) is to ensure the accurate transfer of information directed by the genetic code and ensure the correct expression of proteins[39]. During evolution, certain ARSs also acquire appended domains responsible for a range of noncanonical activities unrelated to aminoacylation[40]. These noncanonical activities include translation control, transcription regulation, cell migration, signal transduction, inflammation, angiogenesis, and tumorigenesis. Defects in both canonical and noncanonical ARS functions may cause or contribute to human disease [41].DARS belongs to class II ARSs and exists as a dimer. The pathophysiology of the disorder, including slowly progressive cerebellar ataxia, spasticity and dorsal column dysfunction, is elucidated with the discovery of mutations in the DARS-encoding gene [42, 43]. However, no reports are available on DARS in virus-infected cells. DARS is up-regulated after FMDV infection in BHK-21 cells by SILAC. This finding suggests that DARS could be useful for FMDV proliferation, especially, for FMDV protein translation, as confirmed by the RNA interference result of the DARS-encoding gene. Interestingly, DARS gene is the first known calcium-regulated tRNA synthetase in Dictyosteliumdiscoideum, suggesting a novel mechanism by calcium to regulate the translation machinery [44]. Combined with previous research, the gradual enhancement of membrane permeability because of the formation of membrane-embedded pores of picornovirus 2B protein, which disrupts intracellular Ca2+ homeostasis, is shown [45–48]. The increase in cellular Ca2+ after FMDV infection could promote the expression of DARS gene, which is consistent with the result of SILAC and RNA interference. This finding would warrant further studies of the relationship between DARS and the expression of 2B protein as well as the 2B-mediated regulation of Ca2+concentration during FMDV infection.
Protein acylation, including isoprenylation (farnesylation and geranylgeranylation), glycosyl phosphatidylinositol anchoring, myristoylation and palmitoylation, refers to the covalent addition of a fatty acid prosthetic group onto an acceptor protein. Among these modifications, myristoylation and palmitoylation are the two major types of acylmodification. Protein acylation would provide hydrophobicity for proteins to be localized at the membrane and may facilitate protein–protein interactions [49, 50]. Several viral capsid or envelope proteins have been found to be modified bymyristoylation and palmitoylation. This may enhance the hydrophobic characteristics of viral proteins for their correct subcellular localization as well as to facilitate the assembly and disassembly of infectious virions[51]. A number of protein acyltransferase and acylproteinthioesterase (APT or LYPLA1) catalyze the palmitoylation and depalmitoylation processes. Among them, APT1 is the enzyme responsible for cleavage of the palmitoyl group[52]. As previously noted, most viral acyl proteins are capsid or envelope constituents involved in capsid assembly or in the entry of viruses into the cells. We speculated that palmitoylation and depalmitoylation processes are involved in virion assembly and production of infectious progeny. LYPLA1 expression was up-regulated after FMDV infection and the virus replication decreased after knockdown of LYPLA1 expression, which demonstrated that LYPLA1 could improve FMDV replication, especially for assembly and production of infectious progeny. However, the detailed mechanism in which LYPLA1 modifies the FMDV capsid proteins should be further studied.
Endosomal sorting complex required for transport (ESCRT) complexes and associated protein play a function in membrane fission events, such as multi vesicular body (MVB) formation and terminal stages of cytokinesis [53, 54]. The ESCRT machinery is also required for the budding of numerous enveloped viruses to cut the membranous neck that connects the virion to the plasma membrane [55]. This machinery includes ESCRT-I, ESCRT-II, and ESCRT-III, as well as other associated proteins, such as ATPase vacuolar protein sorting 4 (VPS4). ESCRT-I proteins (such as tumor susceptibility gene 101, Tsg101) and associated proteins (Alix) have function at the early stage in this pathway, but ESCRT-III proteins (charged MVB proteins, Chmp) have function in the late stage and are disassembled by VPS4 [56]. Previous research has also demonstrated that mammalian TSG101 binds VPS28 directly to form ESCRT-I and TSG101. Its interacting components are directly involved in endosomal sorting, which suggests that VPS28 could have function in virus infection [57]. Although limited information on non-enveloped viruses is available, previous studies have shown that α2β1 integrin clustering triggers α2-MVBs. This phenomenon depends on ESCRT complexes that seem to crucial for promoting non-enveloped picornavirus echovirus 1 infection [58]. Hepatitis A virus (HAV) released from cells is wrapped in host-derived membranes, which may facilitate virus escape from neutralizing antibodies and promote virus spread within the liver. These enveloped viruses (“eHAV”) resemble exosomes, which are small vesicles whose biogenesis is dependent on host proteins associated with ESCRT, VPS4B, and ALIX. Interestingly, in this study, we showed that FMDV infection down-regulated VPS28 protein expression and the virus titer increased after knockdown of the VPS28 gene. This finding suggests that the VPS28 protein may function as a suppressor in FMDV infection, which is inconsistent with a previous study [58]. Further studies would be required to clarify the relationship between FMDV and VPS28.
EVI5 is a Rab11-binding protein that acts as a GAP protein for Rab11. Rab11 regulates intracellular transport and has specific functions in endosome recycling and cytokinesis [59, 60].Many studies have shown that Rab11 involved in cellular recycling endosome pathway and has a function in enveloped viral budding and release, such as respiratory syncytial virus [61, 62], influenza A virus [63], Andes virus [64] and Sendai virus [65], suggesting a possible function for the Rab11 pathway akin to that of the ESCRT machinery. A previous study has demonstrated that FMDV infection occurs predominantly from within early endosome and does not require virus trafficking to the late endosomal compartments. Expression of dominant-negative Rab11 mutants inhibited infection by FMDV up to 35% [66], which indicates that Rab11 also functions during FMDV infection. However, the present study showed that EVI5, a rab11-binding protein, was down-regulated after FMDV infection. FMDV titer increased after knockdown of EVI5 expression. The results suggested that EVI5 protein could inhibit FMDV infection, which further implies the opposite function of Rab11 and EVI5 for FMDV infection. It would be interesting to elucidating the relationship of Rab11 and EVI5 in FMDV infection.
During viral replication cycle, the genome is rapidly translated into polyprotein upon FMDV entry into a cell. The functional capacity of the polyprotein involved in viral replication, protein translation and protein modification in the host cells, will increase when cellular changes occur during viral replication, and will further affect the cellular pathways, which are critical for viruses [67]. It suggests that the virus proteins could also affect cellular response during virus proliferation in cells. Thus, determining the relationship of certain virus proteins through cellular interactome is also necessary in the future.
Conclusions
In this study, we have applied SILAC to study the regulation of 2,141 cellular proteins in BHK-21 cells infected with FMDV serotype Asia 1. Most proteins measured by this nonbiased approach were not altered significantly with L/H ratios around 1.0, confirming that no attempt to enrich for any subpopulation of proteins or specific modifications was made experimentally. The results showed that the abundance and expression patterns of 153 proteins were significantly changed upon FMDV infection. Network pathway analysis indicated that cellular proteins with significant regulation could be linked to signaling cascades, such as PKR. Interestingly, several significant up- and down-regulated proteins, including DARs, SEC62, VPS28 and EVI5, were related to PKR in different signaling networks. Furthermore, DARs, SEC62, VPS28, EVI5, PKR and LYPLA1 were selected and validated. The results suggest that FMDV infection may have effects on the expression of specific cellular proteins to create more favorable conditions for FMDV infection. This study may lead to the identification of cellular targets for the development of a new generation of antiviral drugs against essential host cell factors.
Supporting Information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(XLSX)
(DOCX)
(DOCX)
Acknowledgments
We thank Miss. Yu Zhang for cell culture, Mr. Dan Yan for analysis of some results.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by grants from the National Science and Technology Support Program (2013BAD12B00), International Science &Technology Cooperation Program of China (2014DFA31890), the Fundamental Research Funds for the Chinese Academy of Agricultural Sciences (2015ZL062), and the "Twelfth Five-Year" National Science and technology program in rural areas (2011AA10A211). Guangzhou Fitgene biotechnology Co. Ltd. provided support in the form of salaries for author [XJL], but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of this author are articulated in the ‘author contributions’ section.
References
- 1. Grubman MJ, Baxt B. Foot-and-Mouth Disease. Clin Microbiol Rev. 2004;17(2):465–93. 10.1128/cmr.17.2.465-493.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grubman MJ, Morgan DO, Kendall J, Baxt B. Capsid intermediates assembled in a foot-and-mouth disease virus genome RNA-programmed cell-free translation system and in infected cells. J Virol. 1985;56(1):120–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de los Santos T, Diaz-San Segundo F, Zhu J, Koster M, Dias CCA, Grubman MJ. A conserved domain in the leader proteinase of foot-and-mouth disease virus is required for proper subcellular localization and function. J Virol. 2009;83(4):1800–10. 10.1128/jvi.02112-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang D, Fang LR, Li P, Sun L, Fan JX, Zhang QY, et al. The leader proteinase of foot-and-mouth disease virus negatively regulates the type i interferon pathway by acting as a viral deubiquitinase. J Virol. 2011;85(8):3758–66. 10.1128/jvi.02589-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belsham GJ, McInerney GM, Ross-Smith N. Foot-and-mouth disease virus 3C protease induces cleavage of translation initiation factors eIF4A and eIF4G within infected cells. J Virol. 2000;74(1):272–80. 10.1128/jvi.74.1.272-280.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Donnelly ML, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, et al. Analysis of the aphthovirus 2A/2B polyprotein 'cleavage' mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal 'skip'. J Gen Virol 2001;82(Pt 5):1013–25. . [DOI] [PubMed] [Google Scholar]
- 7. Lubroth J, Brown F. Identification of native foot-and-mouth disease virus non-structural protein 2C as a serological indicator to differentiate infected from vaccinated livestock. Res Vet Sci 1995;59(1):70–8. 10.1016/0034-5288(95)90034-9 [DOI] [PubMed] [Google Scholar]
- 8. O'Donnell VK, Pacheco JM, Henry TM, Mason PW. Subcellular distribution of the foot-and-mouth disease virus 3A protein in cells infected with viruses encoding wild-type and bovine-attenuated forms of 3A. Virology. 2001;287(1):151–62. 10.1006/viro.2001.1035 . [DOI] [PubMed] [Google Scholar]
- 9. Belov GA, Altan-Bonnet N, Kovtunovych G, Jackson CL, Lippincott-Schwartz J, Ehrenfeld E. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J Virol. 2007;81(2):558–67. 10.1128/JVI.01820-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beske O, Reichelt M, Taylor MP, Kirkegaard K, Andino R. Poliovirus infection blocks ERGIC-to-Golgi trafficking and induces microtubule-dependent disruption of the Golgi complex. J Cell Sci. 2007;120(Pt 18):3207–18. 10.1242/jcs.03483 . [DOI] [PubMed] [Google Scholar]
- 11. Choe SS, Dodd DA, Kirkegaard K. Inhibition of cellular protein secretion by picornaviral 3A proteins. Virology. 2005;337(1):18–29. 10.1016/j.virol.2005.03.036 . [DOI] [PubMed] [Google Scholar]
- 12. Moffat K, Knox C, Howell G, Clark SJ, Yang H, Belsham GJ, et al. Inhibition of the secretory pathway by foot-and-mouth disease virus 2BC protein is reproduced by coexpression of 2B with 2C, and the site of inhibition is determined by the subcellular location of 2C. J Virol. 2007;81(3):1129–39. 10.1128/jvi.00393-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal Chem. 1996;68(5):850–8. 10.1021/ac950914h . [DOI] [PubMed] [Google Scholar]
- 14. Fan H, Ye Y, Luo Y, Tong T, Yan G, Liao M. Quantitative proteomics using stable isotope labeling with amino acids in cell culture reveals protein and pathway regulation in porcine circovirus type 2 infected PK-15 cells. J Proteome Res. 2011;11(2):995–1008. 10.1021/pr200755d . [DOI] [PubMed] [Google Scholar]
- 15. Ye Y, Yan G, Luo Y, Tong T, Liu X, Xin C, et al. Quantitative proteomics by amino acid labeling in foot-and-mouth disease virus (FMDV)-infected cells. J Proteome Res. 2012;12(1):363–77. 10.1021/pr300611e . [DOI] [PubMed] [Google Scholar]
- 16. Rweyemamu MM, Booth JC, Head M, Pay TWF. Microneutralization tests for serological typing and subtyping of foot-and-mouth disease virus strains. J Hyg (Lond). 1978;81(1):107–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Balachandran S, Barber GN. PKR in innate immunity, cancer, and viral oncolysis. Methods Mol Biol. 2007;383:277–301. 10.1007/978-1-59745-335-6_18 [DOI] [PubMed] [Google Scholar]
- 18. Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, et al. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70(4):1032–60. 10.1128/MMBR.00027-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goh KC, deVeer MJ, Williams BRG. The protein kinase PKR is required for p38 MAPK activation and the innate immune response to bacterial endotoxin. Embo J. 2000;19(16):4292–7. 10.1093/emboj/19.16.4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nakamura T, Furuhashi M, Li P, Cao H, Tuncman G, Sonenberg N, et al. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell. 2010;140(3):338–48. 10.1016/j.cell.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zamanian-Daryoush M, Mogensen TH, DiDonato JA, Williams BRG. NF-kappaB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-kappaB-inducing kinase and IkappaB kinase. Mol Cell Biol. 2000;20(4):1278–90. 10.1128/mcb.20.4.1278-1290.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bonnet MC, Weil R, Dam E, Hovanessian AG, Meurs EF. PKR stimulates NF-kappaB irrespective of its kinase function by interacting with the IkappaB kinase complex. Mol Cell Biol. 2000;20(13):4532–42. 10.1128/mcb.20.13.4532-4542.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang P, Samuel CE. Induction of protein kinase PKR-dependent activation of interferon regulatory factor 3 by vaccinia virus occurs through adapter IPS-1 signaling. J Biol Chem. 2008;283(50):34580–7. 10.1074/jbc.M807029200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chinsangaram J, Koster M, Grubman MJ. Inhibition of L-deleted foot-and-mouth disease virus replication by alpha/beta interferon involves double-stranded RNA-dependent protein kinase. J Virol. 2001;75(12):5498–503. 10.1128/jvi.75.12.5498-5503.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de los Santos T, de Avila Botton S, Weiblen R, Grubman MJ. The leader proteinase of foot-and-mouth disease virus inhibits the induction of beta interferon mRNA and blocks the host innate immune response. J Virol. 2006;80(4):1906–14. 10.1128/jvi.80.4.1906-1914.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin JH, Walter P, Yen TSB. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol. 2008;3(1):399–425. 10.1146/annurev.pathmechdis.3.121806.151434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yoshida H. ER stress and diseases. FEBS J. 2007;274(3):630–58. 10.1111/j.1742-4658.2007.05639.x . [DOI] [PubMed] [Google Scholar]
- 28. He B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006;13(3):393–403. 10.1038/sj.cdd.4401833 . [DOI] [PubMed] [Google Scholar]
- 29. Braakman I, Van Anken E. Folding of viral envelope glycoproteins in the endoplasmic reticulum. Traffic. 2000;1(7):533–9. 10.1034/j.1600-0854.2000.010702.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith JA, Schmechel SC, Raghavan A, Abelson M, Reilly C, Katze MG, et al. Reovirus induces and benefits from an integrated cellular stress response. J Virol. 2006;80(4):2019–33. 10.1128/jvi.80.4.2019-2033.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu C-Y, Hsu Y-W, Liao C-L, Lin Y-L. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J Virol. 2006;80(23):11868–80. 10.1128/jvi.00879-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Kuppeveld FJM, Hoenderop JGJ, Smeets RLL, Willems PHGM, Dijkman HBPM, Galama JMD, et al. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. Embo J. 1997;16(12):3519–32. 10.1093/emboj/16.12.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Doedens JR, Giddings TH, Kirkegaard K. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J Virol. 1997;71(12):9054–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Suhy DA, Giddings TH, Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J Virol. 2000;74(19):8953–65. 10.1128/jvi.74.19.8953-8965.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bienz K, Egger D, Troxler M, Pasamontes L. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J Virol. 1990;64(3):1156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Egger D, Bienz K. Intracellular location and translocation of silent and active poliovirus replication complexes. J Gen Virol. 2005;86(3):707–18. 10.1099/vir.0.80442-0 . [DOI] [PubMed] [Google Scholar]
- 37. Muller L, de Escauriaza MD, Lajoie P, Theis M, Jung M, Muller A, et al. Evolutionary gain offunction for the ER membrane protein sec62 from yeast to humans. Mol Biol Cell. 2010;21(5):691–703. 10.1091/mbc.E09-08-0730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Johnson N, Habdenteufel S, Theis M, Paton AW, Paton JC, Zimmermann R, et al. The signal sequence influences post-translational ER translocation at distinct stages. Plos One. 2013;8(10):e75394. doi: ARTN e75394. 10.1371/journal.pone.0075394 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Woese CR, Olsen GJ, Ibba M, SOll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev. 2000;64(1):202–36. 10.1128/mmbr.64.1.202-236.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guo M, Yang X-L, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat Rev Mol Cell Biol. 2010;11(9):668–74. 10.1038/nrm2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim S, You S, Hwang D. Aminoacyl-tRNA synthetases and tumorigenesis: more than housekeeping. Nat Rev Cancer. 2011;11(10):708–18. 10.1038/nrc3124 [DOI] [PubMed] [Google Scholar]
- 42. Lin J, Chiconelli Faria E, Da Rocha AJ, Rodrigues Masruha M, Pereira Vilanova LC, Scheper GC, et al. Leukoencephalopathy With Brainstem and Spinal Cord Involvement and Normal Lactate: A New Mutation in the DARS2 Gene. J Child Neurol. 2010;25(11):1425–8. 10.1177/0883073810370897 . [DOI] [PubMed] [Google Scholar]
- 43. Messmer M, Florentz C, Schwenzer H, Scheper GC, van der Knaap MS, Marechal-Drouard L, et al. A human pathology-related mutation prevents import of an aminoacyl-tRNA synthetase into mitochondria. Biochem J. 2011;433(3):441–6. 10.1042/bj20101902 . [DOI] [PubMed] [Google Scholar]
- 44. Jaiswal JK, Nanjundiah V. Calcium regulates the expression of a Dictyostelium discoideum asparaginyl tRNA synthetase gene. J Biosci 2003;28(6):697–707. 10.1007/bf02708430 . [DOI] [PubMed] [Google Scholar]
- 45. Aldabe R, Irurzun A, Carrasco L. Poliovirus protein 2BC increases cytosolic free calcium concentrations. J Virol. 1997;71(8):6214–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nieva JL, Agirre A, Nir S, Carrasco L. Mechanisms of membrane permeabilization by picornavirus 2B viroporin. FEBS Lett 2003;552(1):68–73. 10.1016/S0014-5793(03)00852-4 . [DOI] [PubMed] [Google Scholar]
- 47. Campanella M, de Jong AS, Lanke KWH, Melchers WJG, Willems P, Pinton P, et al. The coxsackievirus 2B protein suppresses apoptotic host cell responses by manipulating intracellular Ca2+ homeostasis. J Biol Chem. 2004;279(18):18440–50. 10.1074/jbc.M309494200 . [DOI] [PubMed] [Google Scholar]
- 48. van Kuppeveld FJM, de Jong AS, Melchers WJG, Willems P. Enterovirus protein 2B po(u)res out the calcium: a viral strategy to survive? Trends Microbiol 2005;13(2):41–4. 10.1016/j.tim.2004.12.005 . [DOI] [PubMed] [Google Scholar]
- 49. Greaves J, Chamberlain LH. Palmitoylation-dependent protein sorting. J Cell Biol. 2007;176(3):249–54. 10.1083/jcb.200610151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8(1):74–84. 10.1038/nrm2084 . [DOI] [PubMed] [Google Scholar]
- 51. Hruby DE, Franke CA. Viral acylproteins: Greasing the wheels of assembly. Trends Microbiol 1993;1(1):20–5. . [DOI] [PubMed] [Google Scholar]
- 52. Veit M, Schmidt MFG. Enzymatic depalmitoylation of viral glycoproteins with acyl-protein thioesterase 1 in vitro. Virology. 2001;288(1):89–95. 10.1006/viro.2001.1063 . [DOI] [PubMed] [Google Scholar]
- 53. McDonald B, Martin-Serrano J. No strings attached: the ESCRT machinery in viral budding and cytokinesis. J Cell Sci. 2009;122(13):2167–77. 10.1242/jcs.028308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wollert T, Yang D, Ren X, Lee HH, Im YJ, Hurley JH. The ESCRT machinery at a glance. J Cell Sci. 2009;122(13):2163–6. 10.1242/jcs.029884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344(1):55–63. 10.1016/j.virol.2005.09.044 . [DOI] [PubMed] [Google Scholar]
- 56. Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136(1):97–109. 10.1016/j.cell.2008.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bishop N, Woodman P. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J Biol Chem. 2001;276(15):11735–42. 10.1074/jbc.M009863200 . [DOI] [PubMed] [Google Scholar]
- 58. Karjalainen M, Rintanen N, Lehkonen M, Kallio K, Mäki A, Hellström K, et al. Echovirus 1 infection depends on biogenesis of novel multivesicular bodies. Cell Microbiol. 2011;13(12):1975–95. 10.1111/j.1462-5822.2011.01685.x . [DOI] [PubMed] [Google Scholar]
- 59. Westlake CJ, Junutula JR, Simon GC, Pilli M, Prekeris R, Scheller RH, et al. Identification of Rab11 as a small GTPase binding protein for the Evi5 oncogene. Proc Natl Acad Sci U S A 2007;104(4):1236–41. 10.1073/pnas.0610500104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Laflamme C, Assaker G, Ramel D, Dorn JF, She D, Maddox PS, et al. Evi5 promotes collective cell migration through its Rab-GAP activity. J Cell Biol. 2012;198(1):57–67. 10.1083/jcb.201112114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Brock SC, Goldenring JR, Crowe JE. Apical recycling systems regulate directional budding of respiratory syncytial virus from polarized epithelial cells. Proc Natl Acad Sci U S A 2003;100(25):15143–8. 10.1073/pnas.2434327100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Utley TJ, Ducharme NA, Varthakavi V, Shepherd BE, Santangelo PJ, Lindquist ME, et al. Respiratory syncytial virus uses a Vps4-independent budding mechanism controlled by Rab11-FIP2. Proc Natl Acad Sci U S A 2008;105(29):10209–14. 10.1073/pnas.0712144105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bruce EA, Digard P, Stuart AD. The Rab11 pathway is required for influenza A virus budding and filament formation. J Virol. 2010;84(12):5848–59. 10.1128/jvi.00307-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rowe RK, Suszko JW, Pekosz A. Roles for the recycling endosome, Rab8, and Rab11 in hantavirus release from epithelial cells. Virology. 2008;382(2):239–49. 10.1016/j.virol.2008.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chambers R, Takimoto T. Trafficking of Sendai virus nucleocapsids is mediated by intracellular vesicles. Plos One. 2010;5(6):e10994 10.1371/journal.pone.0010994 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Johns HL, Berryman S, Monaghan P, Belsham GJ, Jackson T. A dominant-negative mutant of rab5 inhibits infection of cells by foot-and-mouth disease virus: implications for virus entry. J Virol. 2009;83(12):6247–56. 10.1128/jvi.02460-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lin J-Y, Chen T-C, Weng K-F, Chang S-C, Chen L-L, Shih S-R. Viral and host proteins involved in picornavirus life cycle. J Biomed Sci. 2009;16(1):103 10.1186/1423-0127-16-103; PubMed Central PMCID: PMC2785775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(XLSX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.